Abstract
Increasing availability of high-dimensional clinical data, which improves the ability to define more specific phenotypes, as well as molecular data, which can elucidate disease mechanisms, is a driving force and at the same time a major challenge for translational and personalized medicine. Successful research in this field requires an approach that ties together specific disease and health expertise with understanding of molecular data through statistical methods. We present PEAX (Phenotype-Expression Association eXplorer), built upon open-source software, which integrates visual phenotype model definition with statistical testing of expression data presented concurrently in a web-browser. The integration of data and analysis tasks in a single tool allows clinical domain experts to obtain new insights directly through exploration of relationships between multivariate phenotype models and gene expression data, showing the effects of model definition and modification while also exploiting potential meaningful associations between phenotype and miRNA-mRNA regulatory relationships. We combine the web visualization capabilities of Shiny and D3 with the power and speed of R for backend statistical analysis, in order to abstract the scripting required for repetitive analysis of sub-phenotype association. We describe the motivation for PEAX, demonstrate its utility through a use case involving heart failure research, and discuss computational challenges and observations. We show that our visual web-based representations are well-suited for rapid exploration of phenotype and gene expression association, facilitating insight and discovery by domain experts.
Keywords: personalized medicine, hypothesis testing, visual analytics, gene expression, multidimensional data exploration
1. Introduction
The diversity of modern multidimensional clinical data enables researchers to define and study subtle, complex sub-phenotypes of patients. For example, rather than defining mere presence or absence of disease, certain types of cancer can be more precisely graded or staged when specific gene sequences and expression characteristics are known [1], and patients with different genotypes may exhibit differential response to drug treatment [2]. Phenotype characterization is a broad challenge in the field of phenomics [3], with much recent effort (e.g, [4]) towards associating phenotype with single nucleotide polymorphisms (SNPs) in phenome-wide association studies (PheWAS). But the art of meaningful classification using all available clinical and genetic data still requires significant domain expertise, especially when novel, complex sub-groups are defined. Consequently, clinical domain experts often define phenotypes and generate hypotheses using a certain set of tools and manually curated knowledge sources, whereas statisticians separately perform statistical analyses using an appropriate set of tools, most of which are poorly suited for phenotype definition. This cyclical process involves different users and tools, and therefore tends to be too slow and tedious for efficient collaborative work, presenting a major challenge in translational medical research.
From our collaboration with clinical experts and statisticians we have derived three primary observations regarding this inefficient workflow. First, datasets rich in both clinical and gene expression data may lead to novel insights when a specific, interesting phenotype is defined, and that phenotype is associated with the expression data in a biologically plausible manner. Second, data can and should be treated differently based on the domain knowledge and understanding of the audience as well as natural relationships and modeling techniques that are most appropriate for a particular class of data. Finally, an integrative analysis that is both visual and dynamically responsive is more likely to facilitate an iterative and collaborative analytical process that can generate useful insight than text-based scripts and comparisons of static data representations.
Given these observations, we have developed PEAX (Phenotype-Expression Association eXplorer), which allows domain experts to define and explore novel sub-phenotype correlation with gene expression using visual analytics. By integrating phenotype definition with statistical processing in a single tool on a single screen, we seek to inspire novel insight from exploratory analysis, in a more intuitive and collaborative approach than existing tools. We describe the iterative, agile development process driven specifically by use case requirements of cardiology experts; however, we also address general, fundamental challenges identified in personalized medicine involved in clinical phenotype definition, multiple testing, and integrated analysis of heterogenous and missing data [5]. Although issues such as “data dredging” and sparse data cannot be completely avoided, providing clear, comprehensive, and responsive metrics to a domain expert formulating a hypothesis may mitigate some of these effects. With PEAX, we contributed a tool that integrates web visualization with statistical analysis, and its application to a specific biomedical task shows that interactivity and responsiveness can improve existing methods and workflows for data analysis and exploration.
2. Domain Background
The motivating clinical research for PEAX is the analysis of drug efficacy in the clinical trial on the “Effect of β-blockers on Structural Remodeling and Gene Expression in the Failing Human Heart (BORG)” [6]. Heart failure has a devastating and costly impact within the United States, and is responsible for one million hospital visits and 280,000 deaths annually [7]. Patients enrolled in BORG were diagnosed with idiopathic dilated cardiomyopathy (IDCM), a form of heart failure primarily affecting the left ventricle of the heart, and were randomized to one of three different β-blocker treatments. The patients were monitored for up to a year from initial treatment; they exhibited a variable improvement in left ventricular ejection fraction (LVEF) with β-blocker treatment as observed previously [8]. Biopsies from ventricular tissue for each patient were performed prior to β-blocker treatment and at 3 and 12 months; myocardial gene expression of ~ 34,000 human mRNAs and ~ 7800 miRNAs was measured, producing longitudinal in vivo whole-transcriptome gene expression data in human IDCM patients. The clinical outcome used to measure drug response was improvement in LVEF; the primary aim of the study was to understand molecular mechanisms of LVEF improvement with β-blockers and to identify predictive clinical and/or molecular biomarkers to predict LVEF improvement.
Although the patient size is relatively small, the depth of the data represents a typical clinical trial scenario involving a primary research question of a single outcome tested against thousands of potential biomarkers. Previous analysis of BORG data included standard t-testing of associations between a subset of mRNA and miRNA probes with differential expression changes between responders and non-responders to accomplish the primary aim of the study. A previous collaborative analysis [9] used the machine-learning software package Weka [10] to discover predictive C4.5 trees for miRNA expression that may be associated as a biomarker for β-blocker drug response. In this prior work, decision trees were seen to be a simpler and accurate predictive model compared to machine learning methods such as support vector machines and random forest, yielding a model of drug responsiveness that was better accepted by cardiologists for description of phenotype. Additional analysis of learning was limited in analyzing the vast potential search space of clinical phenotype and molecular interaction, so that only a fraction of the data were used that could potentially provide knowledge regarding heart failure, as well as the mechanisms of disease and repair (remodeling and reverse remodeling, respectively).
Primary results of the BORG data analysis and new research in the field prompted additional research questions. However, there was a desire to shorten the loop between hypothesis generation and testing, and to move cardiology experts closer to the data analysis phase. In some cases, merely understanding the cohort size and distribution of patients that met specific criteria was important in deciding whether to proceed with further analysis. These research questions, and anticipation of further data exploration, motivated the development of a new workflow.
3. Related Work
Several toolkits can present statistical analysis with visualization. Rattle [11] uses a GUI to provide support for data exploration and output of statistical testing. It is useful for general statistical analysis methods but is not particularly enhanced for biomedical data analysis. Tools like JavaStat [12] provide a Java/R interface to support the combined development in Java and R and potentially harness the strengths of both languages. However, development requires an integrated mix of Java and R code; as a result the functional code is not as easily portable and leveragable from existing tools, new tools, and legacy code from user groups that are written natively in either of the individual languages. Several tools address the desire for a GUI by running R as a web server, and providing a web interfacing API and functionality. These include server tools such as RApache [13], and R packages like RServe [14]. RStudio’s Shiny [15], on the other hand, provides a framework that supports an R analytical backend that can be tied to browser-based visual displays as well, but also handles the interactivity between visual inputs and outputs, so that extensive coding is not required for this process, while analytical R scripts can generally be leveraged from native R-based projects.
Clinical hypothesis testing uses established statistical methodologies, by separating patients into differentiated groups, defined by distinct, measurable features a priori; for example, drug responders vs. non-responders. Candidate features, such as mRNA expression are tested for significant differences in expression between patient classes. The popular analysis pipeline Bioconductor [16] contains functions, such as edgeR [17] for differential gene expression. The Gene Expression Omnibus (GEO), a public repository for gene expression experiments, includes online tools such as heatmaps for analysis and display of differentially expressed genes [18]. The actual statistical testing of differential expression in these cases is used to answer specific hypotheses about phenotype-biomarker association.
Testing for associations between defined classes and potential predictors is also generally supported by machine learning techniques. Machine learning can be used to build models based on a subset of features, such as gene expression data, that are used to classify or predict membership of an instance, such as a patient, in a defined class group. Weka [10] is a popular tool that wraps support of several supervised learning techniques, such as C4.5 trees [19], random forests, and support vector machines. Predictive models can be tested and cross-validated. These techniques are based on settled definition of phenotype, so that each model must be individually and iteratively tested for association, requiring an additional step or solution for exploration.
Phenotype discovery can be considered generally as sub-group of latent class analysis, which is a technique applied not just in biological sciences, but also in social and behavioral sciences [20]. Such grouping can be aided by cluster analysis, which is a type of unsupervised learning method that takes all of the known features, and groups patients into distinct clusters based on aggregate similarity using selected features and defined metrics. Clustering has been used for identifying novel disease phenotypes [21]. R contains functions such as hclust and knn, and Weka contains clustering support as well. But unsupervised clustering may result in groupings that are driven by features which are less important or relevant to the discovery process, so feature selection, interpretation and refinement by an expert is still critical, especially in large datasets. Topological data analysis is also a powerful way to explore high-dimensional data, but tools like the Ayasdi Platform [22] are not open source, and such models are not used as widely in clinical medicine as decision trees.
Because of the regulatory relationship between miRNA and gene expression, simultaneous profiling and integrated analysis of expression data is useful in further understanding regulatory networks. Experiments that include such profiling look for anti-correlated expression of miRNA and respective mRNA targets, which may be searched in aggregated databases in packages like multiMiR [23] that contain information about observed and predicted miRNA-mRNA interactions. Tools such as mirConnx [24] use this prior knowledge to construct regulatory networks, which can be augmented by expression data from a particular experiment. In general, once a candidate list of potential interacting miRNA/genes is obtained, they can serve as inputs to other methods to look for enrichment that suggests biological interaction. The cBioPortal for Cancer Genomics [25] provides visualization and analysis tools, but this support is tied directly to cancer datasets only.
4. Analysis Task Definition
To define features for our system, as well as prototype, test, and refine, we began with two research questions that supported aims of the BORG study. Even though we use BORG as a driving dataset, these tasks are likely to be supportive of similar research that is rich in clinical, mRNA, and miRNA expression data.
Since BORG utilized three different types of the same class of drug, we want to analyze whether patients with specific drug treatments exhibit different fold changes in gene and/or miRNA expression. Different drugs in the same class may function differently in different patients, potentially exhibiting different side effects and, at a basic level, affect changes in gene expression. First, we sought the ability to compare molecular expression change and association after drug treatment when all patients on β-blocker treatments were analyzed in a pooled fashion, versus a separate analysis of patients grouped by one of three specific β-blocker drugs. Second, we want to test whether drug receptor polymorphisms exhibited different associations with molecular expression data. Even a single nucleotide difference can have drastic effects on individual response to drugs. Therefore, comprehensive understanding of differential drug response should ideally include genotype information of important drug-related SNPs. The BORG dataset includes SNP data of several adrenergic receptors (e.g. rs2234888, rs1801252, and rs1801253), known or suspected to affect β-blocker response [26, 27]. SNP variants are represented as a categorical variable representing whether a given SNP locus is heterozygous, or either of the homozygous combinations. For the second analysis task, we desired to stratify patients by genotype information for at least one of the SNPs, and compare resulting gene associations.
5. Visual Analytics Approach
The overall goal of our design is to enable informed clinicians who are experts in a certain disease domain to generate and test hypotheses regarding user-specified phenotypes and their associations with gene and miRNA expression data; it was motivated by our previous collaborative work with researchers in the Division of Cardiology in the CU Medical School. In order to bridge the statistical, scientific, and clinical analysis of the BORG dataset, we used a visual analytics approach. The visual analytics principles [28] in PEAX are used to integrate broad and heterogeneous data, and present the analysis results at a variable level of detail in order to facilitate insight from clinical experts. An improved analysis process also allows a group of users to obtain more rapid feedback regarding disease biomarkers and pathological processes, reducing the need for independent work by a dedicated statistician during hypothesis formation and refinement.
We now discuss our design methodology and infrastructure, including some of the visual analytics design choices used in PEAX. (The tool, demo, and code are available at http://compbio.ucdenver.edu/PEAX). Based on our initial observations, several major design goals and constraints were immediately apparent for our analysis engine:
GUI for responsive phenotype definition (Input)
Support for powerful statistical analysis (Processing)
Visual display of processed data (Output)
Additional considerations included use of open-source software to provide maximum capability of dissemination and re-use in the research community, and use of web-based applications to reduce installation and platform-dependence overhead. Given these considerations, R is a compelling choice for statistical analysis software, due to popularity and speed of statistical analysis. Using R would also ensure the ability to adapt and integrate ever-increasing library of analysis packages, and R-based analysis results were familiar to our domain experts. However, GUI support is not as well-supported in native use of R.
In order to wrap the power of R statistical analysis in a web-friendly GUI, we employed RStudio’s Shiny software as a visual front-end. It provides the ability to design webpages easily with responsive controls and full access and use of Javascript user-interface elements, as well as HTML capability, tied to an R-backend that could be used for data processing. Shiny also provides the capability of “reactive” inputs and outputs, in which changes in inputs can automatically trigger processing events, while newly updated data and UI input elements can be used to trigger updates of data outputs. Finally, we expanded our visual output display by using Data Driven Documents (D3), which is a powerful Javascript-based system for data visualization [29].
Taken together, Shiny allowed us to integrate GUI and data inputs on a webpage. These inputs are visible by R and used to construct a model. The model is processed and tested using statistical algorithms in R, and then displayed as webpage outputs using HTML, Javascript, and D3. Using a GUI removes the requirements of being able to script and interpret R code directly, increasing the size of the audience of researchers capable of participating in the analysis process, as well as moving the clinical expert closer to the data analysis. An overview of the PEAX GUI is shown in Figure 1.
Fig. 1.
Sample Screenshot of PEAX. On the left are various controls for defining sub-phenotype, including the depth of the decision tree (A), the decision variable for each node (B), and the decision variable threshold. A histogram of the sub-population of patients is shown below each decision node drop-down box. Together, these controls define the decision tree (C). A boxplot (D) shows distribution of a selected gene from a candidate list of genes (E), while a separate adjacent table (F) shows the top correlating miRNA expression levels with the selected gene. Selectable thresholds (G) allow for highlighting of significant associations in either table. Displayed data are random for illustrative purposes, due to confidentiality of clinical research work described in this paper. The entire set of controls, and resulting tree, boxplot, and associative tables, are replicated on the right side to allow for definition, viewing, and comparing of juxtaposed models.
We used several visual analytics concepts and design elements to represent data and analysis results. We chose to use binary decision trees as visual representations of complex phenotypes. In previous collaborative work, we used C4.5 trees [19] to discover a biomarker associated with drug response [9]. Decision trees are already familiar to clinicians, who work with decision-support systems in diagnosing and defining phenotype, as well as bioinformaticians, who are familiar with common machine-learning techniques that use decision tree analysis. They are even used for crowdsourced cancer gene expression analysis by users with a variety of levels of research experience and education [30]. A decision tree can cleanly and quickly present the data and relevant cutoffs for decisions in a way that representations through simple tables cannot achieve.
In PEAX, trees are implemented as D3 collapsible trees defined in JSON format. Trees are displayed with the vertical axis representing decision variables, and the horizontal axis representing patient/sample subgrouping. We augmented the decision tree with visual cues to show information about the data, primarily by varying line thickness between nodes, so that thicker lines represent a higher percentage of a sub-population of patients that meet a given decision-tree criterion. This becomes more evident when the phenotype definition is changed slightly, providing immediate feedback on distribution of phenotype. The leaves of the tree show the size of each group meeting the phenotype definition. Only samples which have their phenotype fully defined by all previous nodes are included in final analysis, so that patients with missing data are automatically excluded from analysis. By noting the number of patients in the final groupings compared to the total population, a researcher can discern how much data are missing.
PEAX also supports the comparison of two decision trees, presented side-by-side. This juxtaposition enables several potential use cases and scenarios, in which patients were stratified by drug treatment and/or genotype in different ways. More generally, an analyst can more easily compare two different phenotype definitions, perhaps with slight adjustments, and not have to rely on brain memory to examine differences in phenotype distribution and association. The ability to use and compare multiple, juxtaposed trees provides additional flexibility by providing an extra dimension of exploration and comparison. Since our decision trees are relatively small (they are rarely deeper than three levels) a simple juxtaposition is sufficient for a visual comparison and more advanced approaches for comparing larger trees are not required.
Phenotype variables, shown as decision-tree selection nodes, can be selected with drop-down boxes and adjusted via sliders; a histogram positioned directly above each slider shows the distribution of patients hierarchically and dynamically split by all higher nodes. The histogram quickly shows the data distribution based on a particular node and can provide insight into the distribution of the population, thus being useful to determine interesting cutoff values. Augmenting sliders with histograms is an example implementation of a “scented widget,” in which a GUI element is integrated with an embedded visualization, shown to help increase the number of discoveries in data [31].
After a phenotype is defined, a list of mRNAs is displayed, sorted by significance of association with the defined phenotype. Several selectable methods, currently including ANOVA and Kruskal-Wallis, provide statistical testing of association. Individual mRNAs can be selected from the list, upon which a boxplot is displayed that shows the difference in distribution of the selected mRNA. A red line indicates a separation between gene expression values that have increased or decreased. The boxplot can provide insight into directionality of upregulation/downregulation of specific genes, or possible dose-response or comparative relationships between more than two phenotypes. If a pattern of expression is found to be interesting based on a selectable threshold for upregulation/downregulation, a researcher can search for other genes that exhibit the same pattern. Once an mRNA is selected, a separate ordered list of the top-correlating miRNA expression values is shown. Each mRNA and miRNA is hyperlinked to online databases, and associations meeting a selectable threshold are highlighted in a different color. The combination of sorted lists, boxplots, and database links provide the capability for the user to achieve “details on demand” [28] when exploring a hypothesis.
Although investigating individual gene associations is rapid, we identified the performance bottleneck as the sheer number of analysis-of-variance (aov) tests run when the phenotype definition is modified. In order to improve performance to provide a responsive interfacewe distributed the aov calculations across the number of available cores. We achieved a noticeable improvement in speed, but the system still lagged by 5–10 minutes for each input adjustment. We further optimized the code by parallelizing several independent steps of the calculation instead of using the built-in aov function; namely, independent, parallelized computation of column means and sums. Finally, we filtered out low-variant expression data (a common pre-processing step for other forms of analysis [32]), leaving us with a set of 3893 fold-change gene expression values (reduced from the original ~34,000 values). Using the filtered gene-fold values, PEAX responded to input changes in approximately 30 seconds or less, so as to provide our clinical expert with a system that could meet research needs as well as be sufficiently responsive for data exploration.
6. Case Study
Based on our previous collaborative work with a group of cardiologists, we designed a prototype of PEAX to support the primary research tasks. We then met several times over the course of several weeks with a cardiology expert and refined and tested our system based on initial feedback. Being a researcher as well, this expert was also familiar with script-based analysis in R, and was able to provide comparative feedback with respect to the visual and interactive capabilities of the tool.
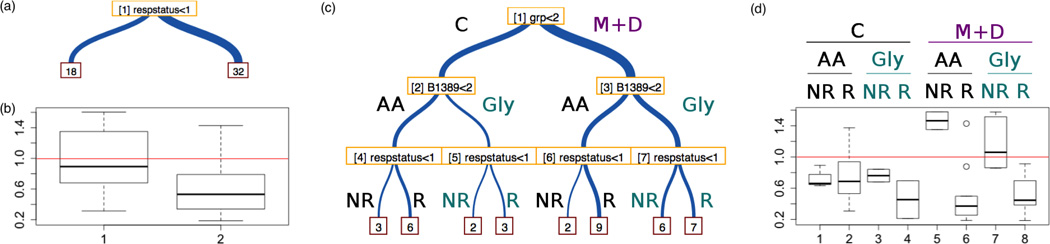
To gain trust and experience with PEAX, the domain expert first recreated previous research from his group. He looked into LVEF improvement, the clinical biomarker used for drug response, and its association with gene expression, and was able to verify that the tool was able to provide the same results through a visual interface rather than previous scripting. The domain expert then investigated two additional research aims of the BORG study: examining differences between drug treatment groups (Task I), and examining the effects of β-adrenergic SNPs on drug responsiveness (Task II). Now that the domain expert was more familiar with the tool, he combined these two tasks into a single exploratory analysis. These tasks were completed with a combination of unsupervised, self-directed usage of the tool, succeeded by a follow-up discussion of the results. The researcher stated that he was able to complete the task of initial investigation within half an hour, whereas it would have taken most of a day or more to set up, refine, test, and verify script-based results; he was also able to examine several genes of interest that he may otherwise not have investigated. For these tasks, he designed two trees (Figure 2a and Figure 2c), which are nominally shown side-by-side in PEAX. The first tree simply involves drug responsiveness, whereas the second tree combined drug treatment group with the β1-adrenergic receptor Arg389Gly SNP, and drug responsiveness. He investigated several potential genes of interest, and quickly identified a gene association (p < 0.0005) that had a more pronounced difference when the SNP and drug treatment group interaction was considered. The gene has a moderate differential response between responders and non-responders (Figure 2b), but this difference is mostly due to the drug treatment group (Figure 2d). He commented that the side-by-side comparison gave him additional insight regarding potential interactions when he was able to stratify patients by additional variables and compare to the more general LVEF response vs. non-response gene expression associations to explore possible differences in relative gene expression according to drug treatment and SNP. He suggested that the patterns of differentiation which change when additional variables are added are evident due to the visual side-by-side presentation.
Fig. 2.
Sample output from Tasks I and II. The analyst created a 1-node tree for drug responsiveness (a), and examined boxplot distribution for a particular gene of interest (b). He then created a 3-level tree that used drug treatment group grp, as well as SNP B1389 (corresponding to dbSNP rs1801253), in addition to drug responsiveness respstatus (c). He noticed that the same gene selected previously exhibited a differential response between responders and non-responders in group 3 (d). Tree branch group labels in (c) match those in (d), and are not generated by PEAX, but were added post-analysis by our analyst when showing to a collaborator.
After applying PEAX to Tasks I and II, he then annotated a screen shot (Figure 2c and 2d), which he sent to another member of the clinical research team. This showed us that combined phenotype and association output from the tool could be used quickly for transmitting and discussing results for collaboration within a team of colleagues, and suggested future feature enhancements.
7. Discussion and Lessons Learned
The development of PEAX, including the design phase and preliminary testing, highlighted several insights, challenges, and functional considerations that we used to refine the tool. The use of Shiny to create a rapid, functional, GUI-based prototype with statistical analysis is quite helpful in designing and refining development in an agile fashion. The continuous use of R for analysis allows researchers and developers that already trust R to retain their confidence in underlying methods and support for statistical computing. To our knowledge, PEAX is the first open-source based tool to combine powerful statistical analysis with interactive decision-tree models to enable clinicians to analyze heterogenous clinical and expression data. Below, we discuss lessons learned during the development and usage of PEAX.
Interactive statistical analysis requires a reactive graphical interface
In order to direct the user in exploratory analysis, an interface must be reactive so as to show the effect of changing aspects of a hypothesis. Implementing our tool in Shiny abstracts the reactive handling of asynchronous user input changes. Comparing different phenotype trees side-by-side, or seeing the difference in data distribution (including the amount of missing data) when making changes to a phenotype definition, for example, provides immediate visual feedback not readily apparent in text-based R-scripting, while still allowing for the power and support of statistical analysis in R.
Interactivity is a driver for optimization of statistical algorithms
The largest performance bottleneck with our test dataset was the number of ANOVA calculations necessary for thousands of candidate genes. While the GUI allowed for easier definition for phenotypes for subsequent testing, a long lag of several minutes caused the GUI to appear unresponsive and slow, and optimization of this step was crucial to achieve acceptable usability. Instead of using R’s built-in linear model (lm) or analysis of variance functions (aov), which provide model results and calculations unnecessary for our interactive display, we decomposed the ANOVA function to optimize the calculation of the F-value. We achieved a considerable speedup by using col Means and col Sums for intermediate calculations, as well as parallelization using mclapply from the parallel library to distribute the calculation of column means and the explained group variance for each of the defined phenotype groups. This algorithmic refactoring allowed us to experiment with a reactive and responsive system, as we were able to demonstrate our prototype on an 8-core system. Parallelization should allow for scalability on larger systems. We also observed that our framework took less than 1GB of memory while running, suggesting that use cases similar to ours are likely to be computationally intensive more than data intensive and therefore potentially more parallelizable.
True “real-time” responsiveness is not always necessary for useful data exploration
Even with performance optimizations, refreshes of the visual display of our data and analysis can take tens of seconds and the tool reactivity cannot be considered “real time.” However, we were surprised to find that our collaborator considered this a vast improvement over more familiar script-based techniques, which were more likely to require tedious and careful setup and checking of data setup before testing an individual hypothesis. In combination with other visual cues that suggested possible interesting patterns, the speed of visual updating encouraged our analyst to continue data exploration and experimentation.
A GUI-based tool allows the domain expert to drive analysis and collaboration
In our iterative design process and case analysis, we found a small, targeted audience of cardiology experts to be responsive and enthusiastic in suggesting features and desire to experiment with the tool, moreso than when similarly presented with an abstract, static list of potential features. With a working prototype, the clinical researcher was able to move closer to the data analysis, and the group was enthusiastic about using the tool to begin analysis tasks that supported prior research grants. Additionally, our collaborator explored data relationships in ways that were surprising to us, by exploration of both previously understood as well as novel gene associations, and by using screenshots of the tool to communicate with colleagues.
Discovery through exploration is based on a comprehensive analysis of all available data
A natural criticism of explorative approaches is the potential for false discovery rates (FDR), especially through “data dredging,” or drawing erroneous conclusions based on excessive analysis of numerous possible relationships. Like any tool, PEAX can and should be used appropriately for the appropriate task. We provide features for adjusting for multiple testing, such as displaying the cumulative number of statistical tests during a session of exploration. The analyst is responsible for appropriate adjustment of resulting significant associations. Another possible use case, not initially envisioned but very manageable with our system, would be to split the dataset into a training and test (holdout) set, whereupon a hypothesis is generated on the training set, and then tested on the holdout set [33]. This approach is especially amenable to our system of allowing for two different decision trees, where the holdout set could be verified side-by-side with the training set.
The evidence for biological plausibility is a comprehensive picture that depends on phenotype input as well as mRNA and miRNA correlation. Phenotype classification is in itself a subjective task performed by expertise, and a motivating factor of the interactivity of phenotype definition and data exploration was to provide expert feedback into the system using a supervised method. The problem of identifying potentially interesting, unknown phenotype definitions has not yet been adequately solved. Therefore, we focused on facilitating domain expert expression of phenotypes and integration of associated molecular data. Correlative mRNA and miRNA biomarkers are more compelling when, taken together, they relate to a plausible biological scenario, which will be determined by the domain expert. PEAX can provide information regarding the data, but interpretation, presentation of results, and experimental follow-up must be done with scientific care.
8. Conclusion
This paper presents a new tool, PEAX, for integrated analysis of complex phenotype definition and association with molecular expression data. This analytic scenario is targeted for clinical experts with mixed datasets involving deep clinical, mRNA, and miRNA expression data, and is designed to abstract statistical analysis scripts while providing useful feedback for exploration of data. We developed a prototype tool, demonstrated its utility through a case study with a domain expert on a cardiology dataset, and report initial observations and lessons learned. We chose decision trees as a simplified model familiar to clinical researchers, and present analysis results on a single interactive screen in order to streamline analysis.
We found the combination of visual interactivity in a web browser, with the statistical analysis capabilities of R, to be a compelling combination to make this type of analysis more rapid, exploratory, and collaborative. We found the generalized functions of defining and testing sub-classes visually to be faster, less error-prone, and more efficient than a process that uses “one-off,” proprietary scripts for very specifically subdividing patient groups based on specific features.
The web-based nature PEAX provides opportunities for scalability in performance and distribution, although clinical data sensitivity issues may require internal application usage. By supporting Javascript and D3 for GUI support, PEAX can continue to leverage new web-based visualizations; and by using R for statistical computation, we can add analysis algorithms based on the extensive and growing library of R functions. In addition to further exploration on BORG and other datasets, planned additional features include highlighting known miRNA-mRNA associations to direct further exploration of plausible miRNA regulation of mRNA related to a hypothesized complex phenotype.
By necessity, PEAX sped up association calculations using analysis of variance by parallelizing several key steps. There is still a lag between user input and redisplay of output, so that the interaction was not considered real-time in our scenario, but to our surprise, our clinical expert was pleased in being able to see new results presented within tens of seconds, as opposed to his previous techniques which involved tedious, text-based scripting. This observation is important for large, multidimensional datasets: interaction may not need to be real-time, if it is tolerably responsive and represents an improvement in the amount of time required to analyze and check through existing techniques, with a reduced cognitive demand for formulating and checking correct syntax of scripts.
Improper use of data-mining techniques to analyze high-dimensional data can lead to spurious, false associations. Therefore, an investigator must draw conclusions based on comprehensive consideration of all of the evidence, and particularly important observations should be validated independently. Nevertheless, a growing number of large datasets are available in which important biological questions have gone unexplored and undiscovered, perhaps because of computational complexity, or proprietary scripting. To address the big data challenges of personalized medicine, integrated statistical and visual analysis tools such as PEAX are needed for rapid data exploration, collaboration, and communication to drive hypothesis generation and testing by clinical experts.
Acknowledgments
This work was supported in part by NIH grants R01 LM008111, 2R01 HL48013, 1R01 HL71118, P20 HL101435-01, and T32 HL007822-12, and by grants from GlaxoSmithKline and AstraZeneca.
Contributor Information
Michael A. Hinterberg, Email: michael.hinterberg@ucdenver.edu.
David P. Kao, Email: david.kao@ucdenver.edu.
Michael R. Bristow, Email: michael.bristow@ucdenver.edu.
Lawrence E. Hunter, Email: larry.hunter@ucdenver.edu.
J. David Port, Email: david.port@ucdenver.edu.
Carsten Görg, Email: carsten.goerg@ucdenver.edu.
References
- 1.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, et al. Proceedings of the National Academy of Sciences of the United States of America. 2005 Sep;102:13550. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, et al. Clinical pharmacology and therapeutics. 2007 Mar;81:328. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Houle D, Govindaraju DR, Omholt S. Nature reviews. Genetics. 2010 Dec;11:855. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- 4.Shameer K, Denny JC, Ding K, et al. Human genetics. 2014 Jan;133:95. doi: 10.1007/s00439-013-1355-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Listgarten J, Stegle O, Morris Q, et al. Pacific Symposium on Biocomputing. 2014;19:247. doi: 10.1142/9789814583220_0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ClinicalTrials.gov. Effect of Beta-blockers on Structural Remodeling and Gene Expression in the Failing Human Heart (BORG, NCT01798992) 2013 http://www.clinicaltrials.gov/ct2/show/NCT01798992.
- 7.Roger VL, Go AS, Lloyd-Jones DM, Adams RJ, Berry JD, Brown TM, Carnethon MR, Dai S, de Simone G, Ford ES, Fox CS, Fullerton HJ, et al. Circulation. 2011 Feb;123:e18. doi: 10.1161/CIR.0b013e3182009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Investigators, The Beta-Blocker Evaluation of Survival Trial. The New England journal of medicine. 2001 May;344:1659. [Google Scholar]
- 9.Hinterberg MA, Kao D, Karimpour-Fard A, Sucharov K, Hunter LE, Port D, Bristow M. Journal of the American College of Cardiology. 2013;61 [Google Scholar]
- 10.Hall M, National H, Frank E, et al. ACM SIGKDD explorations newsletter. 2009;11:10. [Google Scholar]
- 11.Williams GJ. The R Journal. 2009;1:45. [Google Scholar]
- 12.Harner EJ, Luo D, Tan J. Computational Statistics. 2008 Sep;24:295. [Google Scholar]
- 13.Horner J. RApache: Web application development with R and Apache. 2013 http://www.rapache.net. [Google Scholar]
- 14.Urbanek S. Rserve: binary R server. 2013 https://rforge.net/Rserve. [Google Scholar]
- 15.RStudio. 2013 http://www.rstudio.com/shiny/
- 16.Gentleman RC, Carey VJ, Bates DM, et al. Genome biology. 2004 Jan;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson MD, McCarthy DJ, Smyth GK. Bioinformatics (Oxford, England) 2010 Jan;26:139. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R. Nucleic Acids Research. 2002 Jan;30:207. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Quinlan R. C4.5: Programs for Machine Learning. Morgan Kaufmann; 1992. [Google Scholar]
- 20.Collins LM, Lanza ST. Latent Class and Latent Transition Analysis: With Applications in the Social, Behavioral, and Health Sciences(Google eBook) John Wiley & Sons; 2010. [Google Scholar]
- 21.Haldar P, Pavord ID, Shaw DE, et al. American journal of respiratory and critical care medicine. 2008 Aug;178:218. doi: 10.1164/rccm.200711-1754OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ayasdi. 2013. http://www.ayasdi.com/ [Google Scholar]
- 23.Ru Y, Kechris KJ, Tabakoff B, Hoffman P, et al. Nucleic Acids Research. 2014 Jul;:1. doi: 10.1093/nar/gku631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang GT, Athanassiou C, Benos PV. Nucleic acids research. 2011 Jul;39:W416. doi: 10.1093/nar/gkr276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cerami E, Gao J, Dogrusoz U, Gross B, et al. Cancer discovery. 2012;2:401. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bristow MR, Murphy GA, Krause-Steinrauf H, Anderson JL, Carlquist JF, Thaneemit-Chen S, Krishnan V, Abraham WT, Lowes BD, et al. Circulation. Heart failure. 2010 Jan;3:21. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 27.O’Connor CM, Fiuzat M, Carson PE, et al. PloS one. 2012 Jan;7:e44324. doi: 10.1371/journal.pone.0044324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keim D, Andrienko G, Fekete J-D, Görg C, Kohlhammer J, Melançon G. Information Visualization: Human-Centered Issues and Perspectives. 2008:154. [Google Scholar]
- 29.Bostock M, Ogievetsky V, Heer J. IEEE Transactions on Visualization and Computer Graphics. 2011 Dec;17:2301. doi: 10.1109/TVCG.2011.185. [DOI] [PubMed] [Google Scholar]
- 30.Good BM, Loguercio S, Griffith OL, et al. arXiv preprint arXiv. 2013:1. [Google Scholar]
- 31.Willett W, Heer J, Agrawala M. IEEE Transactions on Visualization and Computer Graphics. 2007;13:1129. doi: 10.1109/TVCG.2007.70589. [DOI] [PubMed] [Google Scholar]
- 32.Bandyopadhyay S, Mallik S, Mukhopadhyay A. Computational Biology and Bioinformatics, IEEE/ACM Transactions on. 2014 Jan;11:95. doi: 10.1109/TCBB.2013.147. [DOI] [PubMed] [Google Scholar]
- 33.Young SS, Karr A. Significance. 2011;8:116. [Google Scholar]