Abstract
Directed evolution, the laboratory process by which biological entities with desired traits are created through iterative rounds of genetic diversification and library screening or selection, has become one of the most useful and widespread tools in basic and applied biology. From its roots in classical strain engineering and adaptive evolution, modern directed evolution came of age twenty years ago with the demonstration of repeated rounds of PCR-driven random mutagenesis and activity screening to improve protein properties. Since then, numerous techniques have been developed that have enabled the evolution of virtually any protein, pathway, network or entire organism of interest. Here we recount some of the major milestones in the history of directed evolution, highlight the most promising recent developments in the field, and discuss the future challenges and opportunities that lie ahead.
Introduction
Faced with the size, scope, and limitless complexity of the natural world, humankind has always sought the means by which to harness its resources for the improvement of life. For early societies, this was manifested simply in the hunting and gathering of wild fauna and flora as a mode of subsistence. A key paradigm shift occurred, however, when the mutability of nature was recognized as a feature to be exploited. An implicit awareness of the evolvability of the natural world, though it would perhaps not be formalized until Darwin’s On the Origin of Species in 1859, inspired the development of the millennia-old practices of selective breeding and domestication. Of course, early practitioners of these techniques could exhibit control only on the screening of target organisms for desired traits; the mechanisms by which variation could be introduced to a population and the means by which these variations could be controlled (or even influenced) were completely unknown. Nevertheless, great results were achieved that revolutionized all of human civilization.
In the mid-twentieth century, evolution was brought into the laboratory, primarily as a means of recreating and exploring natural evolutionary processes. With the discovery of chemical mutagens came the first methods of purposely introducing mutations to a host organism at an increased frequency, albeit with no control over the targeting of such mutations. Lerner and coworkers provided an early example when, in 1964, they utilized chemical mutagenesis to induce a xylitol utilization phenotype in the bacterium Aerobacter aerogenes in an effort to better elucidate the mechanisms by which new functions arise in nature.1 Although their importance to the field of directed evolution would only be noted in retrospect, pioneering studies in in vitro selection were carried out in the laboratory of Sol Spiegelman.2–4 In these studies, purified RNA replicases were reconstituted in vitro with their homologous RNA templates, and the fate of the resulting RNA molecules was monitored through several generations under different selective pressures. Again, this work was devised largely as an exploit in scientific curiosity – attempts to emulate the precellular world to witness firsthand the fundamental principles of the development of life. The authors went so far as to state their interest in answering the question, “What will happen to the RNA molecules if the only demand made on them is the Biblical injunction, multiply, with the biological proviso that they do so as rapidly as possible?”2 In the 1980s, in vitro selections would become more applications-driven, as exemplified by phage display.5 This technique enables the enrichment of a particular peptide that exhibits desired binding properties from a phage-expressed library, with clear relevance to fields such as antibody engineering.
Though the term had been occasionally applied for decades to describe adaptive evolution experiments, directed evolution in the modern sense began to take root in earnest in the 1990s. In broad terms, directed evolution can be defined as an iterative two-step process involving first the generation of a library of variants of a biological entity of interest, and second the screening of this library in a high-throughput fashion to identify those mutants that exhibit better properties, such as higher activity or selectivity. The best mutants from each round then serve as the templates for the subsequent rounds of diversification and selection, and the process is repeated until the desired level of improvement is attained. As compared to rational protein design, which had been pioneered several years prior,6 directed evolution provided the distinct advantage of requiring no knowledge of the protein structure or of the effects of specific amino acid substitutions, which were then (and still are now) very difficult to predict a priori.7
Early on, the target of a directed evolution experiment was most often an individual protein of interest, with genetic variation introduced either in a random or position-specific manner. Such proteins were often evolved with the goal of designing a more stable or more active biocatalyst for increased compatibility with industrial processing conditions.8–10 More recent trends, however, have moved toward the evolution of proteins in the context of pathways, or even the evolution of entire pathways or genomes themselves, to create novel whole cell biocatalysts for the synthesis of value-added chemicals, biofuels, and pharmaceuticals.11,12 Additionally, while improving the functionality of an existing protein or pathway was a common goal of early directed evolution experiments, more recent efforts have set the ambitious goal of designing completely novel functionalities not found in nature.13 Finally, while industrial applications often serve as a primary motivator, directed evolution also remains an invaluable tool in the elucidation of natural evolutionary principles and the testing of evolutionary hypotheses.
In this perspective, we will highlight some of the most significant technical developments and advancements in directed evolution at the level of individual proteins, metabolic pathways, circuits, and genomes (Figure 1). In addition, we will describe some insights into natural evolution that have been gained through directed evolution experiments. Finally, we will discuss the future of directed evolution and the challenges and opportunities likely to be faced.
Figure 1.
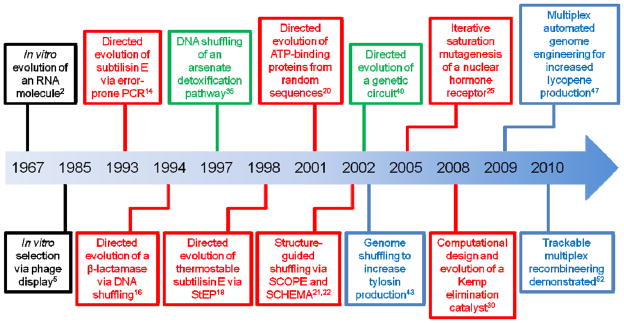
Timeline of selected directed evolution milestones at the protein (red), pathway (green), and genome (blue) level.
Directed Evolution of Proteins
To facilitate the myriad of chemical transformations that constitute a living cell, nature’s primary catalyst is the protein. As modular biopolymer chains of twenty canonical amino acid monomers, proteins are highly evolvable through mutations in their amino acid sequences, which are directly encoded via their cognate genes. Because of this, proteins in nature have continued to evolve over countless generations, gaining and losing functionalities to best suit their host organism. In the laboratory, this inherent evolvability has made the protein the most common target for directed evolution experiments, where a host of techniques have been developed to manipulate individual gene sequences for the creation of diverse protein libraries (Figure 2). From the reductionist perspective that has dominated much of the biological sciences in the modern era, proteins provide defined, discrete elements that can readily be isolated and studied under highly controlled conditions, making them ideal experimental subjects. The attractiveness of protein targets is further manifested in the industrial setting by their ability to catalyze reactions with extremely high regio- and stereo-selectivity as well as specificity, albeit only under benign conditions that may be incompatible with upstream or downstream processing steps.
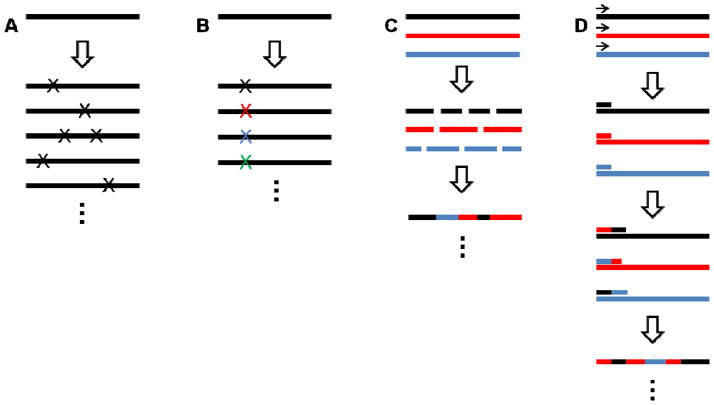
Figure 2.
(a) In error-prone PCR, random mutations are introduced in a gene sequence. (b) In site-saturation mutagenesis, codons for all 20 amino acids are substituted at a chosen position. (c) In DNA shuffling, multiple parent sequences are broken into smaller pieces and randomly reassembled. (d) In StEP, chimeric progenies are synthesized via sequential annealing, extension, and dissociation. For detailed descriptions of these and other techniques, refer to the references cited.
Random Evolution Strategies
Given the aforementioned properties of proteins, it is no wonder that the earliest modern directed evolution experiments focused almost exclusively on protein targets. As the a priori prediction of the effect of mutations on a given protein is often difficult, the earliest techniques focused simply on random mutagenesis. A landmark example in this field is the evolution of subtilisin E, a serine protease useful in several industrial applications, for increased activity in dimethylformamide.14 In this pioneering study, random mutations were introduced to the subtilisin E gene using an error-prone PCR amplification strategy. After three sequential rounds of mutagenesis and screening, a mutant was identified with six additional point mutations that exhibited 256-fold higher activity in 60 % dimethylformamide. This effort clearly demonstrated the power of a sequential, evolutionary protein engineering strategy to identify multiple cooperative mutations for vast protein improvement. In contrast, previous protein engineering efforts typically employed parallel single rounds of selection, identifying individual mutations that were by no means guaranteed to combine beneficially.
While error-prone mutagenesis is an effective means to introduce gradual changes to a protein, in natural evolution this is supplemented by an additional mechanism: recombination. Recombination allows larger pieces of similar genes to be exchanged, significantly diversifying the resultant pool of variants. Thus, to harness this power for directed evolution experiments, techniques were developed to mimic natural recombination in vitro. A key example of this is the DNA shuffling method developed in 1994 by Willem Stemmer of the Affymax Research Institute. In this method, a library of similar genes (e.g., homologs from different organisms or mutants identified from previous engineering efforts) is PCR amplified and digested into smaller fragments with DNase I. Following subsequent isolation of these fragments, a primer-free PCR-like assembly step recombines the fragments from diverse parent genes into new chimeric hybrids, which can then be cloned into an expression vector and screened.15 As an example of the power of this approach, a β-lactamase was evolved to improve the resistance of its host Escherichia coli strain to the antibiotic cefotaxime.16 After three cycles of shuffling and two cycles of backcrossing (to remove non-essential mutations), a mutant was identified that increased the minimum inhibitory concentration (MIC) of the host by 32,000-fold, compared to the 16-fold increase observed when non-recombinogenic methods were employed.
In subsequent years, numerous refined recombination-based directed evolution strategies were developed, which have been reviewed elsewhere.17 One such example is the staggered extension process (StEP) for in vitro recombination.18 Similar to DNA shuffling, this approach generates chimeric progenies from a set of parent genes. However, in StEP, the full-length recombined genes are synthesized in the presence of the parent genes without the intermediate step of generating and purifying short fragments. This is accomplished by sequential annealing of the nascent polynucleotide to different templates with abbreviated extension times, allowing only a small portion of the gene to be filled in before dissociating and annealing to a new template. Using a StEP-based approach, subtilisin E was evolved to exhibit thermostability equal to that of thermitase, a thermophilic homolog from the extremophile Thermoactinomyces vulgaris.19
While error-prone mutagenesis and recombination approaches both seek to develop improved protein functionalities via random variation of existing scaffolds, a more extreme approach to random evolution of proteins is to start from completely random sequences. This tactic has been employed by Keefe and Szostak to identify novel ATP-binding proteins from a library of completely random 80-mer polypeptides.20 From a library of 6 × 1012 proteins, each covalently bound to its cognate mRNA, they employed 18 rounds of selection on ATP-agarose to identify four with ATP-binding functionality. Of course, such a system is dependent upon both the ability to synthesize very large libraries and, most importantly, screen them in a high-throughput manner, which may be difficult for certain target properties. Nevertheless, this work serves as an impressive example of the power of a random evolutionary approach.
Targeted Evolution Strategies
While random evolution strategies have a proven record of success, the solution space they explore for any given protein is large – too large, in fact, for many screening methods to feasibly allow sufficient library coverage. However, it is apparent that with any random mutagenesis strategy, the vast majority of the mutants generated will exhibit no improvement or even inferior performance relative to the parent protein. As a result, a number of techniques have been developed to leverage the substantial amount of protein structural data available for the design of smaller, targeted libraries enriched in variants most likely to exhibit improved properties. Two examples of this are structure-based combinatorial protein engineering (SCOPE) and the SCHEMA algorithm, both presented in 2002.21,22 These strategies employ protein structural data to first identify discrete units of protein secondary structure. By biasing a recombination-based evolution strategy such that recombination occurs only between these units and not within them, the chances of the resulting chimeric proteins maintaining a correct folding pattern, and thus functionality, increase. Additionally, SCHEMA and SCOPE carry the benefit of enabling shuffling of parent proteins with low sequence identity, provided that they share similar folds. Besides these techniques, a number of others have been developed, which have been reviewed elsewhere.23,24
While SCHEMA and SCOPE focus on structural elements of a target protein, other approaches focus instead on the functional elements. To evolve a nuclear hormone receptor to bind a new ligand, for example, Chockalingam and coworkers employed stepwise saturation mutagenesis of active site residues to target only those that are expected to play a role in contacting the ligand.25 Similarly, Reetz and coworkers utilized an iterative saturation mutagenesis approach to increase the thermostability of a lipase from Bacillus subtilis by targeting those residues that showcased the highest degrees of thermal motion based on X-ray data.26 In multi-domain proteins, each domain can be independently evolved in the context of the holoenzyme. As an example of this approach, each domain of cytochrome P450BM3 from Bacillus megaterium was separately evolved using a combination of random, saturation, and site-directed mutagenesis. When the beneficial mutations in each domain were combined, the resulting protein was able to hydroxylate propane, a nonnative substrate, with nativelike coupling efficiency of cofactor utilization.27
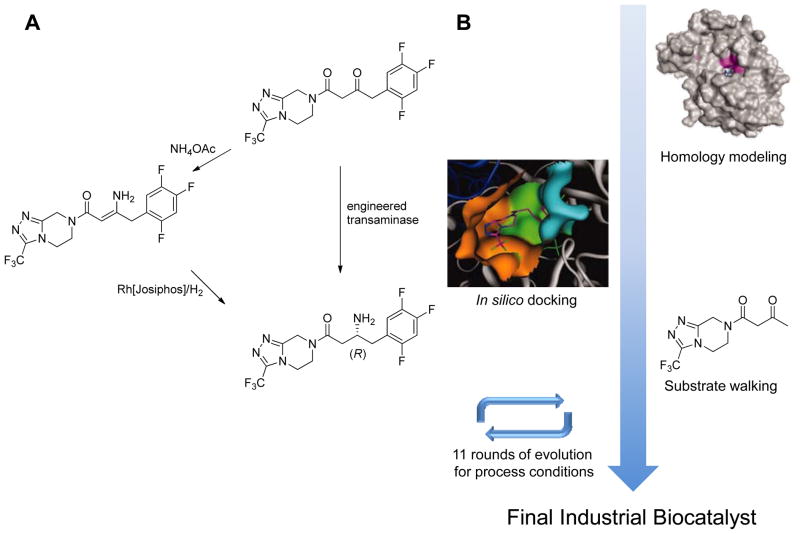
When structural information for a particular target protein is unavailable, computational modeling can be employed to guide directed evolution experiments. A particularly impressive example of this is the evolution of a transaminase for the industrial synthesis of the antidiabetic drug sitagliptin by researchers at Codexis and Merck.28 Motivated by the desire to replace a rhodium-catalyzed enamine hydrogenation (necessitating high pressure and extra purification steps) with a more efficient enzymatic process (Figure 3), they first sought a transaminase with activity towards the prositagliptin ketone. While a particular enzyme with the desired stereospecificity was identified, it had no activity toward the desired substrate. As a result, a homology model of the transaminase was built in silico to guide reconstruction of the active site. Applying multiple rounds of targeted mutagenesis, detectable activity toward the prositagliptin ketone was detected. Subsequently, the enzyme was evolved to function under process conditions, yielding a practical biocatalyst for the industrial process.
Figure 3.
(a) To replace a less efficient chemocatalytic process (left path), a transaminase enzyme was engineered (right path.) (b) The enzyme was engineered via subsequent rounds of homology modeling, docking, substrate walking, and directed evolution.28
Beyond evolving a given protein scaffold for activity with a new substrate, multiple directed evolution efforts in recent years have set the ambitious goal of evolving the scaffold for completely novel activities. In 2006, Park and coworkers presented a method to do so called simultaneous incorporation and adjustment of functional elements (SIAFE).29 In this approach, functional elements (including active site loops involved in catalysis and in substrate binding) are systematically and combinatorially incorporated into a chosen template sequence. As proof of concept, they evolved β-lactamase activity from an αβ/βα metallohydrolase scaffold through deletion, insertion and remodeling of active site functional elements. At the end of the process, although the kinetic properties of the new β-lactamase protein were inferior to those of natural β-lactamases, it no longer exhibited its former metallohydrolase activity. For this experiment, the choice of functional elements was guided by the structural and mechanistic data available for known β-lactamases of the αβ/βα superfamily; it would likely be significantly more difficult, therefore, to evolve a completely novel function using this approach. Nevertheless, this has been achieved by Röthlisberger and coworkers to create an enzyme capable of catalyzing the Kemp elimination reaction, for which no known natural enzymes exist.30 To accomplish this goal, a computational design strategy was employed, coupled with directed evolution. First, an ideal active site for the desired Kemp elimination enzyme was designed in silico based on quantum mechanical transition state calculations. Next, further computational analysis was used to identify the protein scaffolds that could best support the designed active site. Eventually, 59 different designs were experimentally characterized, of which eight exhibited the desired function at a detectable level. After seven rounds of directed evolution, comprising both random mutagenesis and shuffling, an increase of greater than 200-fold was observed in kcat/Km for the best mutant, yielding an overall rate enhancement of 1.18 × 106 relative to the uncatalyzed reaction. Another example of coupled in silico design and directed evolution was recently provided by Karanicolas and coworkers, who engineered a heterodimerization interface between two unrelated proteins.31 While the in silico design alone yielded a pair with a measured Kd of 130 nM, subsequent directed evolution decreased this value almost 1000-fold to 180 pM. These two examples clearly illustrate the synergistic relationship between computational tools and directed evolution, a trend that is likely to continue to develop in the years to come.
Directed Evolution of Pathways
Beyond its success in protein engineering, directed evolution has also been extended to metabolic pathway engineering. In industry, microorganisms have been increasingly engineered to produce value-added products. However, to enable cost-effective microbial synthesis, the associated endogenous or heterologous metabolic pathways often need to be optimized in the production host.32,33 While a few simple pathways are understood explicitly, rational design approaches are usually impeded by limited information of the genetic regulation mechanisms and metabolic networks with which the target pathways are associated.34 Conversely, directed evolution circumvents the limitations of rational design by harnessing targeted random mutagenesis and selection, which has led to a few notable successes in pathway engineering.
General strategies for directed evolution at this level are often analogous to those employed at the protein level, but in the context of an entire pathway. For example, Crameri and coworkers engineered an arsenate resistance operon from Staphylococcus aureus by recursive rounds of DNA shuffling and selection.35 The operon contains the genes arsR, arsB, and arsC, encoding a repressor regulatory protein, arsenite membrane pump, and arsenate reductase, respectively. Using E. coli as a host, after three rounds of DNA shuffling, non-silent mutations were identified in arsB and arsC. The resulting strain survived in up to 0.5 M arsenate, which was 40 times greater than the wild type resistance, and has a potential application in bioremediation of arsenate from gold metallurgy. Accelerating rate limiting steps to increase flux through a desired pathway, also called debottlenecking, is an intuitive strategy often applied in pathway optimization. Doramectin (commercially named Dectomax) is a veterinary pharmaceutical used for parasite treatment. Researchers at Codexis and Pfizer engineered aveC, a key gene in the Streptomyces avermitilis doramectin pathway, by iterative semi-synthetic DNA shuffling for minimized byproduct yield. In the end, the ratio between the desired product and the undesired product reached 14.3:1.36
Carotenoid biosynthesis pathways have often been used as model systems to demonstrate the concept of pathway evolution because libraries can be easily screened visually according to the color of the colonies. Additionally, many carotenoid natural products have relevance in the pharmaceutical and food industries.12 With the same idea of debottlenecking, Wang and coworkers evolved the rate-controlling enzyme geranylgeranyl diphosphate synthase, originally from Archaeoglobus fulgidus, and increased lycopene production by 100%.37 Besides targeting a higher productivity, directed evolution is also used to generate product variants of interest. Schmidt-Dannert and coworkers first shuffled phytoene desaturases from two different organisms and identified a new pathway that produced a fully conjugated carotenoid, 3,4,3′,4′-tetradehydrolycopene. Then, a library of shuffled lycopene cyclases was introduced to extend the new pathway. A variety of colored products was produced, in which the cyclic carotenoid torulene was generated for the first time in E. coli.38
For pathways in which the desired product is not as readily detectable, the Keasling group has recently demonstrated a transcription factor-based method applicable to both screening and selection for production of a desired small molecule product.39 This method employs a transcription factor that binds the desired product as well as its cognate promoter, which controls an antibiotic resistance gene. Thus, host fitness is coupled to the small molecule production phenotype. As proof of concept, this method was used to optimize a 1-butanol biosynthetic pathway in E. coli, yielding a 35% improvement in specific productivity. Such a technique could have broad applicability in the evolution of diverse small molecule biosynthetic pathways, including those for biofuel production.
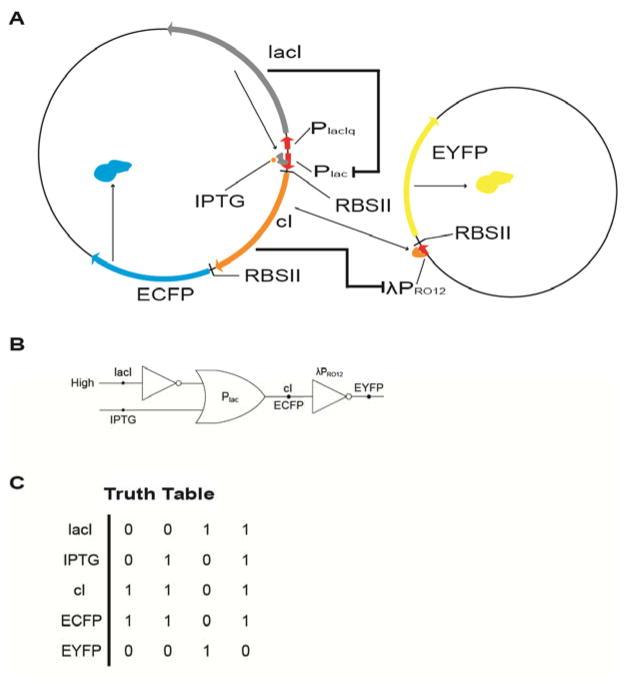
Directed evolution has also been applied to create novel regulatory machineries. Leveraging the current knowledge of transcriptional and translational regulatory mechanisms, researchers have attempted to construct biological circuits simulating electronic logical circuits. However, due to the complexity of intracellular chemistry, biochemical regulatory machineries usually do not correspond with each other and behave as expected. Yokobayashi and coworkers constructed a logical circuit with two regulatory machineries in E. coli to control fluorescence signals with the addition of the small-molecule inducer isopropyl-β-D-1-thiogalactopyranoside (IPTG) (Figure 4). The circuit was not functional at the very beginning since leaky expression of the repressor CI already exceeded the threshold of the λPR operator and shut off the expression of enhanced yellow fluorescent protein (EYFP). After the CI repressor and its ribosome binding site were engineered by directed evolution, the fluorescence signal was able to change according to the level of IPTG.40 Of note, although a number of advances in pathway evolution have been reported, most of them have focused on individual enzymes in the context of metabolic pathways, often in simple organisms like prokaryotes. With newly developed tools for efficient pathway construction,41,42 it can be predicted that more directed evolution studies on the whole pathways and in more complicated organisms will be seen in the near future.
Figure 4.
(a) Design of the gene circuit. The expression of the CI repressor and ECFP was controlled by IPTG and the lac repressor. EYFP expression is repressed by CI. (b) The gene circuit represented by elementary logic gates. Plac acted as an “IMPLIES” gate while λPPRO12 acted as an inverter. (c) Truth table of the logic circuit.40
Directed Evolution of Genomes
On an even larger scale, directed evolution has been applied to improve functions of interest on the level of the entire genome for the generation of novel whole cell biocatalysts. In fact, this approach can be traced back to the early years of human civilization, perhaps best exemplified by the evolutionary adaptation of brewer’s yeast for the fermentation of beverages. Modern studies in directed evolution of genomes incorporate accelerated mutagenesis and the ability to link phenotype with genotype. For example, Maxygen applied genome shuffling by recursive protoplast fusion between four different Streptomyces fradiae strains to improve the production of the antibiotic tylosin.43 After two rounds of genome shuffling, an eight-fold increase in tylosin titer was achieved. Comparatively, 20 rounds of classic strain improvement were required to reach a similar titer. The same technique was used by Codexis to improve acid tolerance of an industrial Lactobacillus strain.44 Due to increasing concerns about energy security, sustainability, and global warming, much effort has been made to efficiently convert biomass to biofuels. Directed evolution is often applied for improving microorganisms for biofuel production. For example, Kuyper and coworkers significantly increased the xylose consumption rate of an engineered Saccharomyces cerevisiae strain through adaptive evolution in lab-scale bioreactors.45 Similarly, Liu and coworkers used adaptive evolution to circumvent this by improving the tolerance of S. cerevisiae and Scheffersomyces stipitis to fermentation inhibitors from lignocellulosic biomass hydrolysate including furfural and hydroxymethylfurfural via conversion to less toxic products.46
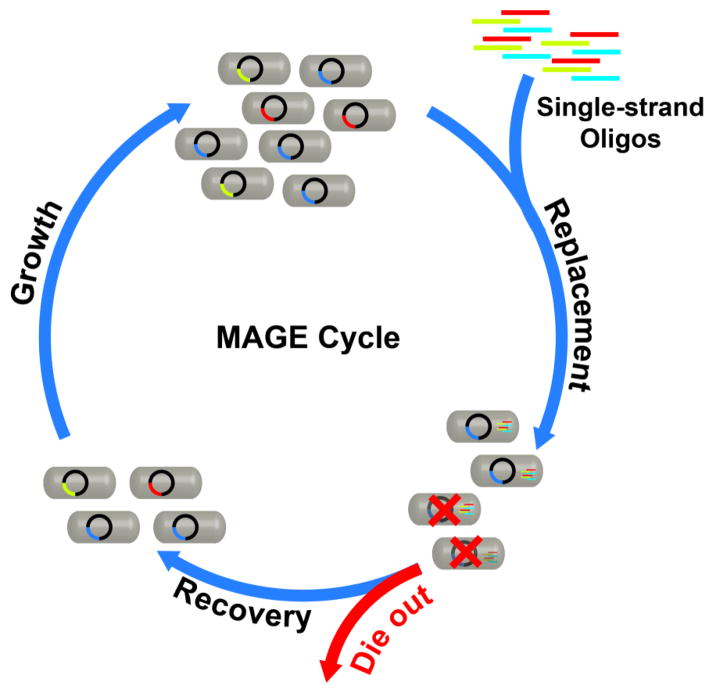
In recent years, more systematic mutagenic approaches have been reported. Wang and coworkers developed a multiplex automated genome engineering (MAGE) technique by iterative single stranded oligo-mediated allelic replacement in E. coli (Figure 5). The oligos contained degenerate bases, which introduced insertions and deletions to the genetic targets and created 4.3 billion combinatorial variants in a day. Within three days, lycopene productivity was increased by five-fold.47 Since then, the efficiency of this technique has been improved significantly,48,49 and it has been applied to such tasks as codon replacement50 and in vivo His6-tagging.51 Similarly, Warner and coworkers have also generated genomic libraries with oligo insertions. In their method, a “barcode” sequence was added to each oligo in order to simultaneously map traits to genetic modifications by DNA microarray. With this trackable multiplex recombineering (TRMR) method, the expression levels of 95% of the genes in E. coli were modified in a day and mapped with traits in a week.52 Genome-scale directed evolution has already made an impact on industrial bioprocessing, and recent advances like MAGE and TRMR are likely to further facilitate this powerful approach.
Figure 5.
A schematic of a MAGE cycle. Mutations were introduced iteratively by transforming E. coli cells with single-stranded oligos containing degenerate bases.47
Insights from Directed Evolution
As has been shown in the various examples presented here, directed evolution is a powerful engineering tool with demonstrated utility in the development of protein and whole cell biocatalysts for industrial processes. However, to consider only the applied aspects of directed evolution would be to ignore its utility as a tool for answering fundamental scientific questions about evolutionary processes, both in the laboratory and in nature. In a practical sense, lessons learned from previous directed evolution experiments can inform future studies, chiefly by guiding library design strategies. For example, in 1999 Miyazaki and Arnold noted that by applying a random mutation strategy to evolve a protein, the sequence space is limited to only those amino acid substitutions that can be achieved by swapping one nucleotide in a given codon.53 To evaluate the consequences of these limitations, they first identified two thermostable mutants of the protease subtilisin S41 using a random mutagenesis strategy. Next, they performed site-saturation mutagenesis at both of the identified sites to determine if mutations at the target positions requiring two or three sequential nucleotide substitutions could increase thermostability even further. This was in fact the case, illustrating the complementarity of random and targeted mutagenesis approaches. A natural follow-up question to this is, given multiple beneficial point mutations in a given protein scaffold (i.e., multiple hits from a round of screening), which position should be chosen first for saturation mutagenesis? This question was recently addressed by Gumulya and coworkers, who systematically explored all 24 of the possible evolutionary trajectories for iterative saturation mutagenesis of four identified positions in the Aspergillus niger epoxide hydrolase protein.54 Notably, they found that along some of the trajectories, local minima in the fitness landscape were reached such that saturation mutagenesis at a particular site yielded no improvement. Nevertheless, by choosing non-improved or even inferior mutants from such libraries and proceeding to the next position, they found that all 24 trajectories led to the identification of significantly improved mutants when screened for enhanced stereoselectivity. This result underscores the context dependence of individual mutations, which may appear neutral in one background but beneficial in another.
In the field of evolutionary biology, a significant limitation on the study of natural evolution is the fact that, by and large, we only have access to proteins and organisms in their current state. As Charles Darwin pointed out, this is like having “a history of the world, imperfectly kept and written in changing dialect. Of this history, we possess the last volume alone. Of this volume, only here and there a short chapter has been preserved; and of each page only here and there few lines.”55 As a result, evolutionary relationships and histories must be inferred from current states, which can be a difficult prospect. In this respect, directed evolution experiments provide the distinct advantage of enabling the visualization of a full evolutionary trajectory on a reasonable timescale. For example, the Tawfik group used an evolutionary approach to study the formation of protein folds.55 By first generating a library of diverse ~100 amino acid polypeptides based on the 236 amino acid Tachylectin-2 scaffold (a 5-bladed β-propeller with five sugar-binding sites), they were able to identify several that could individually assemble into homo-pentamers with sugar-binding activities comparable to the wild type Tachylectin-2 (Figure 6). Such a result substantiates the claim that protein folds with high internal symmetry, such as β-propellers, evolved via the duplication, fusion, and rearrangement of smaller units.
Figure 6.
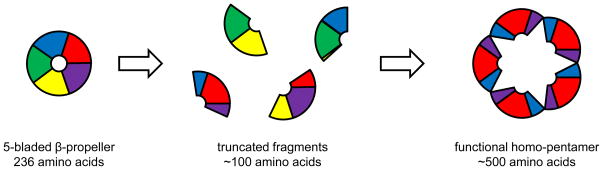
Exploring the evolution of protein folds. From the monomeric Tachylectin-2 protein, a library of truncated fragments was generated. Using an ELISA-based screening method, functional homo-pentameric lectins were identified.55
Another facet of the evolution of proteins that has been studied through directed evolution is the nature of promiscuity and the divergence of function.56 To better understand the process by which extant proteins evolve novel functions, Aharoni and coworkers evolved three proteins: serum paraoxonase, a bacterial phosphotriesterase, and carbonic anhydrase II.57 Selective pressure was applied with the objective of increasing a promiscuous activity of each enzyme, and early evolutionary progenies were identified and characterized. Overall, the goal of this study was to determine how these promiscuity-enhancing mutations affect the native function of each enzyme. Interestingly, it was found for all three enzymes that the mutants with enhanced activity toward the promiscuous substrate showed comparatively little change in their native activities. Such an observation lends credence to the theory that proteins with novel functions evolve first through the acquisition of promiscuous activity, followed by gene duplication and subsequent divergent evolution. On the other hand, directed evolution has also been used to study neutral drift – the accumulation of mutations when a protein is subjected to pressure to maintain its original function.55 Remarkably, following rounds of neutral drift selection, many members of the resulting libraries exhibited substantial changes in promiscuous activities, specificities, and inhibition, corroborating theoretical predictions for such ‘neutral networks.’
It should be noted, of course, that one must always be cautious in the extrapolation of results obtained from directed evolution experiments to the natural world. In the laboratory, the evolving protein is subjected to a well-defined, isolated selection pressure and screened in a very specific manner. In nature, however, the protein evolves under a complicated mix of biological constraints, and as a result mutations that may seem neutral or even deleterious in a laboratory setting can actually have significant improvements in a natural environment, or vice versa.58 Still, this can also be viewed as an advantage in that directed evolution allows the researcher to explore protein space limited only by what is physically possible, rather than what is biologically relevant.59 An additional caveat with directed evolution studies is that some techniques such as saturation mutagenesis explore evolutionary pathways that are generally inaccessible by natural means, and thus are not reflective of natural evolutionary trajectories. Nevertheless, directed evolution studies allow a rare glimpse at evolution in real time, affording insights otherwise unattainable through observation of the natural world alone.
Perspectives and Future Trends
From its conceptual origin in the earliest selective breeding and domestication endeavors, directed evolution itself has evolved into a powerful, versatile tool for the engineering of protein and whole cell biocatalysts for a variety of industrially relevant processes.8,60,61 The products of directed evolution have made contributions to such diverse fields as human health, energy, and the environment, and are poised to continue to do so for years to come. Additionally, directed evolution experiments have granted evolutionary biologists a rare glimpse at evolution in real time, elucidating principles that characterize the inherent evolvability of natural biological systems. In most directed evolution experiments, the biggest obstacle is the development of a suitable screening or selection method. While this is inherently dependent upon the nature of both the evolving entity and the desired improvement, the future will likely see the continued development of versatile techniques that facilitate screening or selection in a high-throughput manner. A promising example of this is the application of microfluidic platforms. While their use in directed evolution is currently limited, the immense capability of such platforms for high-throughput screening (~108 samples per day) and demonstrated versatility with different biological assays bodes well for future directed evolution endeavors.62,63 Alternatively, the need for such high-throughput capabilities can be obviated through the synergistic application of rational design with directed evolution in the creation of smaller libraries enriched in active variants.24
While the bulk of the directed evolution experiments carried out to date focus specifically on individual proteins, a likely trend for the near future will be continued expansion to the pathway and genome levels. Already, a number of techniques have been developed with this in mind. For example, the Cornish group and our own group have each demonstrated methods for the in vivo assembly of libraries of pathways in S. cerevisiae.33,64 Similarly, Pfleger and coworkers have demonstrated combinatorial assembly of intergenic regions in bacterial operons.65 Such techniques could readily be adapted to an iterative screening or selection approach for pathway evolution. Beyond even the genome level, one can also envision evolutionary strategies being applied at the level of microbial consortia to concurrently evolve multiple target organisms to achieve a desired concerted function.66
Another high-profile research area in which directed evolution will likely continue to be invaluable is synthetic biology. While a number of techniques have already been developed for the sophisticated manipulation of genetic elements, the fact remains that biological parts assembled from disparate sources typically are ill-suited to function together in the manner intended.67,68 Similarly, directed evolution is also expected to continue to flourish in the computational design of enzymes. While impressive progress has been made in the de novo design of protein catalysts, the fact remains that designed enzymes are typically significantly inferior to those found in nature.69 By applying directed evolution, we can mitigate the limitations of our ability to perfectly predict protein functionality and fine-tune designer enzymes for their desired purposes.
Acknowledgments
We thank the National Institutes of Health (GM077596) and the National Science Foundation as part of the Center for Enabling New Technologies through Catalysis (CENTC), CHE-0650456, for financial support in our directed evolution projects. R. Cobb also acknowledges support from the US National Institutes of Health under Ruth L. Kirschstein National Research Award 5 T32 GM070421 from the National Institute of General Medical Sciences and the Henry Drickamer Fellowship from the Department of Chemical and Biomolecular Engineering at the University of Illinois at Urbana-Champaign.
References
- 1.Lerner SA, Wu TT, Lin ECC. Evolution of a catabolic pathway in bacteria. Science. 1964;146:1313–1315. doi: 10.1126/science.146.3649.1313. [DOI] [PubMed] [Google Scholar]
- 2.Mills DR, Peterson RL, Spiegelman S. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci U S A. 1967;58:217–224. doi: 10.1073/pnas.58.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Levisohn R, Spiegelman S. Further extracellular Darwinian experiments with replicating RNA molecules: diverse variants isolated under different selective conditions. Proc Natl Acad Sci U S A. 1969;63:805–811. doi: 10.1073/pnas.63.3.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kacian DL, Mills DR, Kramer FR, Spiegelman S. A replicating RNA molecule suitable for a detailed analysis of extracellular evolution and replication. Proc Natl Acad Sci U S A. 1972;69:3038–3042. doi: 10.1073/pnas.69.10.3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith G. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 6.Brannigan JA, Wilkinson AJ. Protein engineering 20 years on. Nat Rev Mol Cell Biol. 2002;3:964–970. doi: 10.1038/nrm975. [DOI] [PubMed] [Google Scholar]
- 7.Tobin MB, Gustafsson C, Huisman GW. Directed evolution: the ‘rational’ basis for ‘irrational’ design. Curr Opin Struct Biol. 2000;10:421–427. doi: 10.1016/s0959-440x(00)00109-3. [DOI] [PubMed] [Google Scholar]
- 8.Zhao H, Chockalingam K. Directed evolution of enzymes and pathways for industrial biocatalysis. Curr Opin Biotechnol. 2002;13:104–110. doi: 10.1016/s0958-1669(02)00291-4. [DOI] [PubMed] [Google Scholar]
- 9.Arnold FH, Volkov aa. Directed evolution of biocatalysts. Curr Opin Chem Biol. 1999;3:54–59. doi: 10.1016/s1367-5931(99)80010-6. [DOI] [PubMed] [Google Scholar]
- 10.Arnold FH. Combinatorial and computational challenges for biocatalyst design. Nature. 2001;409:253–257. doi: 10.1038/35051731. [DOI] [PubMed] [Google Scholar]
- 11.Chartrain M, Salmon PM, Robinson DK, Buckland BC. Metabolic engineering and directed evolution for the production of pharmaceuticals. Curr Opin Biotechnol. 2000;11:209–214. doi: 10.1016/s0958-1669(00)00081-1. [DOI] [PubMed] [Google Scholar]
- 12.Rohlin L, Oh MK, Liao JC. Microbial pathway engineering for industrial processes: evolution, combinatorial biosynthesis and rational design. Curr Opin Microbiol. 2001;4:330–335. doi: 10.1016/s1369-5274(00)00213-7. [DOI] [PubMed] [Google Scholar]
- 13.Zhao H. Directed evolution of novel protein functions. Biotechnol Bioeng. 2007;98:313–317. doi: 10.1002/bit.21455. [DOI] [PubMed] [Google Scholar]
- 14.Chen K, Arnold FH. Tuning the activity of an enzyme for unusual environments: sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc Natl Acad Sci U S A. 1993;90:5618–5622. doi: 10.1073/pnas.90.12.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stemmer WP. DNA shuffling by random fragmentation and reassembly: in vitro recombination for molecular evolution. Proc Natl Acad Sci U S A. 1994;91:10747–10751. doi: 10.1073/pnas.91.22.10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stemmer WP. Rapid evolution of a protein in vitro by DNA shuffling. Nature. 1994;370:389–391. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 17.Rubin-Pitel S, Cho CM-H, Chen W, Zhao H. Bioprocessing for Value-Added Products from Renewable Resources: New Technologies and Applications. New York: Elsevier; 2006. Directed evolution tools in bioproduct and bioprocess development; pp. 49–72. [Google Scholar]
- 18.Zhao H, Giver L, Shao Z, Affholter JA, Arnold FH. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat Biotechnol. 1998;16:258–261. doi: 10.1038/nbt0398-258. [DOI] [PubMed] [Google Scholar]
- 19.Zhao H, Arnold FH. Directed evolution converts subtilisin E into a functional equivalent of thermitase. Protein Eng. 1999;12:47–53. doi: 10.1093/protein/12.1.47. [DOI] [PubMed] [Google Scholar]
- 20.Keefe AD, Szostak JW. Functional proteins from a random-sequence library. Nature. 2001;410:715–718. doi: 10.1038/35070613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Maille PE, Bakhtina M, Tsai M-D. Structure-based combinatorial protein engineering (SCOPE) J Mol Biol. 2002;321:677–691. doi: 10.1016/s0022-2836(02)00675-7. [DOI] [PubMed] [Google Scholar]
- 22.Voigt CA, Martinez C, Wang Z-G, Mayo SL, Arnold FH. Protein building blocks preserved by recombination. Nat Struct Mol Biol. 2002;9:553–558. doi: 10.1038/nsb805. [DOI] [PubMed] [Google Scholar]
- 23.Dalby PA. Strategy and success for the directed evolution of enzymes. Curr Opin Struct Biol. 2011;21:473–480. doi: 10.1016/j.sbi.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Cobb RE, Si T, Zhao H. Directed evolution: an evolving and enabling synthetic biology tool. Curr Opin Chem Biol. 2012;16:285–291. doi: 10.1016/j.cbpa.2012.05.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chockalingam K, Chen Z, Katzenellenbogen JA, Zhao H. Directed evolution of specific receptor–ligand pairs for use in the creation of gene switches. Proc Natl Acad Sci U S A. 2005;102:5691–5696. doi: 10.1073/pnas.0409206102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reetz MT, Carballeira JD, Vogel A. Iterative saturation mutagenesis on the basis of B factors as a strategy for increasing protein thermostability. Angew Chem Int Ed Engl. 2006;45:7745–7751. doi: 10.1002/anie.200602795. [DOI] [PubMed] [Google Scholar]
- 27.Fasan R, Chen MM, Crook NC, Arnold FH. Engineered alkane-hydroxylating cytochrome P450BM3 exhibiting nativelike catalytic properties. Angew Chem. 2007;119:8566–8570. doi: 10.1002/anie.200702616. [DOI] [PubMed] [Google Scholar]
- 28.Savile CK, Janey JM, Mundorff EC, et al. Biocatalytic asymmetric synthesis of chiral amines from ketones applied to sitagliptin manufacture. Science. 2010;329:305–309. doi: 10.1126/science.1188934. [DOI] [PubMed] [Google Scholar]
- 29.Park H-S, Nam S-H, Lee JK, et al. Design and evolution of new catalytic activity with an existing protein scaffold. Science. 2006;311:535–538. doi: 10.1126/science.1118953. [DOI] [PubMed] [Google Scholar]
- 30.Röthlisberger D, Khersonsky O, Wollacott AM, et al. Kemp elimination catalysts by computational enzyme design. Nature. 2008;453:190–195. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 31.Karanicolas J, Corn JE, Chen I, et al. A de novo protein binding pair by computational design and directed evolution. Mol Cell. 2011;42:250–260. doi: 10.1016/j.molcel.2011.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Du J, Shao Z, Zhao H. Engineering microbial factories for synthesis of value-added products. J Ind Microbiol Biotechnol. 2011;38:873–890. doi: 10.1007/s10295-011-0970-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Du J, Yuan Y, Si T, Lian J, Zhao H. Customized optimization of metabolic pathways by combinatorial transcriptional engineering. Nucleic Acids Res. 2012:1–10. doi: 10.1093/nar/gks549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chatterjee R, Yuan L. Directed evolution of metabolic pathways. Trends Biotechnol. 2006;24:28–38. doi: 10.1016/j.tibtech.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Crameri A, Dawes G, Rodriguez E, Jr, Silver S, Stemmer WPC. Molecular evolution of an arsenate detoxification pathway by DNA shuffling. Nat Biotech. 1997;15:436–438. doi: 10.1038/nbt0597-436. [DOI] [PubMed] [Google Scholar]
- 36.Stutzman-Engwall K, Conlon S, Fedechko R, et al. Semi-synthetic DNA shuffling of aveC leads to improved industrial scale production of doramectin by Streptomyces avermitilis. Metab Eng. 2005;7:27–37. doi: 10.1016/j.ymben.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Wang C-W, Oh M-K, Liao JC. Directed evolution of metabolically engineered Escherichiacoli for carotenoid production. Biotechnol Prog. 2000;16:922–926. doi: 10.1021/bp000124f. [DOI] [PubMed] [Google Scholar]
- 38.Schmidt-Dannert C, Umeno D, Arnold FH. Molecular breeding of carotenoid biosynthetic pathways. Nat Biotechnol. 2000;18:750–753. doi: 10.1038/77319. [DOI] [PubMed] [Google Scholar]
- 39.Dietrich JA, Shis DL, Alikhani A, Keasling JD. Transcription factor-based screens and synthetic selections for microbial small-molecule biosynthesis. ACS Synth Biol. 2012 doi: 10.1021/sb300091d. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokobayashi Y, Weiss R, Arnold FH. Directed evolution of a genetic circuit. Proc Natl Acad Sci U S A. 2002;99:16587–16591. doi: 10.1073/pnas.252535999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16–e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA, Smith HO. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Meth. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 43.Zhang YX, Perry K, Vinci VA, Powell K, Stemmer WP, del Cardayre SB. Genome shuffling leads to rapid phenotypic improvement in bacteria. Nature. 2002;415:644–646. doi: 10.1038/415644a. [DOI] [PubMed] [Google Scholar]
- 44.Patnaik R, Louie S, Gavrilovic V, et al. Genome shuffling of Lactobacillus for improved acid tolerance. Nat Biotechnol. 2002;20:707–712. doi: 10.1038/nbt0702-707. [DOI] [PubMed] [Google Scholar]
- 45.Kuyper M, Toirkens MJ, Diderich JA, Winkler AA, van Dijken JP, Pronk JT. Evolutionary engineering of mixed-sugar utilization by a xylose-fermenting Saccharomyces cerevisiae strain. FEMS Yeast Res. 2005;5:925–934. doi: 10.1016/j.femsyr.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 46.Liu ZL, Slininger PJ, Gorsich SW. Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl Biochem Biotechnol. 2005;05:451–460. [PubMed] [Google Scholar]
- 47.Wang HH, Isaacs FJ, Carr Pa, et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature. 2009;460:894–898. doi: 10.1038/nature08187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carr PA, Wang HH, Sterling B, et al. Enhanced multiplex genome engineering through co-operative oligonucleotide co-selection. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang HH, Kim H, Cong L, Jeong J, Bang D, Church GM. Genome-scale promoter engineering by coselection MAGE. Nat Meth. 2012;9:591–593. doi: 10.1038/nmeth.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Isaacs FJ, Carr PA, Wang HH, et al. Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science. 2011;333:348–353. doi: 10.1126/science.1205822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang HH, Huang P-Y, Xu G, et al. Multiplexed in vivo His-tagging of enzyme pathways for in vitro single-pot multienzyme catalysis. ACS Synth Biol. 2012;1:43–52. doi: 10.1021/sb3000029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LB, Gill RT. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol. 2010;28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 53.Miyazaki K, Arnold FH. Exploring nonnatural evolutionary pathways by saturation mutagenesis: rapid improvement of protein function. J Mol Evol. 1999;49:716–720. doi: 10.1007/pl00006593. [DOI] [PubMed] [Google Scholar]
- 54.Gumulya Y, Sanchis J, Reetz MT. Many pathways in laboratory evolution can lead to improved enzymes: how to escape from local minima. ChemBioChem. 2012;13:1060–1066. doi: 10.1002/cbic.201100784. [DOI] [PubMed] [Google Scholar]
- 55.Peisajovich SG, Tawfik DS. Protein engineers turned evolutionists. Nat Methods. 2007;4:991–994. doi: 10.1038/nmeth1207-991. [DOI] [PubMed] [Google Scholar]
- 56.Tawfik OK, Tawfik DS. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu Rev Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 57.Aharoni A, Gaidukov L, Khersonsky O, Gould SM, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 58.Bloom JD, Arnold FH. In the light of directed evolution: pathways of adaptive protein evolution. Proc Natl Acad Sci U S A. 2009;106:9995–10000. doi: 10.1073/pnas.0901522106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arnold FH, Wintrode PL, Miyazaki K, Gershenson A. How enzymes adapt: lessons from directed evolution. Trends Biochem Sci. 2001;26:100–106. doi: 10.1016/s0968-0004(00)01755-2. [DOI] [PubMed] [Google Scholar]
- 60.Johannes TW, Zhao H. Directed evolution of enzymes and biosynthetic pathways. Current Opin Microbiol. 2006;9:261–267. doi: 10.1016/j.mib.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 61.Cobb RE, Sun N, Zhao H. Directed evolution as a powerful synthetic biology tool. Methods. 2012 doi: 10.1016/j.ymeth.2012.03.009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uhlen M, Svahn HA. Lab on a chip technologies for bioenergy and biosustainability research. Lab Chip. 2011;11:3389–3393. doi: 10.1039/c1lc90063c. [DOI] [PubMed] [Google Scholar]
- 63.Guo MT, Rotem A, Heyman Ja, Weitz Da. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12:2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 64.Wingler LM, Cornish VW. Reiterative recombination for the in vivo assembly of libraries of multigene pathways. Proc Natl Acad Sci U S A. 2011;108:15135–15140. doi: 10.1073/pnas.1100507108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nature Biotechnol. 2006;24:1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 66.Brenner K, You L, Arnold FH. Engineering microbial consortia: a new frontier in synthetic biology. Trends Biotechnol. 2008;26:483–489. doi: 10.1016/j.tibtech.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 67.Heinemann M, Panke S. Synthetic biology-putting engineering into biology. Bioinformatics. 2006;22:2790–2799. doi: 10.1093/bioinformatics/btl469. [DOI] [PubMed] [Google Scholar]
- 68.Peisajovich SG. Evolutionary synthetic biology. ACS Synthetic Biology. 2012;1:199–210. doi: 10.1021/sb300012g. [DOI] [PubMed] [Google Scholar]
- 69.Baker D. An exciting but challenging road ahead for computational enzyme design. Protein Sci. 2010;19:1817–1819. doi: 10.1002/pro.481. [DOI] [PMC free article] [PubMed] [Google Scholar]