Abstract
Visualization of hormonal signaling input and output is of key importance for understanding regulation of multicellular development. The plant signaling molecule auxin triggers many growth and developmental responses, but current tools lack sensitivity or precision to visualize these. We developed a set of novel fluorescent reporters that allow sensitive and semi-quantitative readout of auxin responses at cellular resolution in Arabidopsis. These generic tools are suitable for any transformable plant species.
The plant signaling molecule auxin plays a fundamental role in plant development. Gene expression responses to auxin mediate most patterning processes1, but also underlie differential growth in response to light or gravity2. The ability to visualize sites of auxin response in a dynamic and quantitative manner is therefore of great importance in understanding mechanisms and dynamics of auxin-controlled plant development.
Auxin response leading to gene expression changes starts with auxin binding to the nuclear auxin receptor TRANSPORT INHIBITOR RESISTANT1/AUXIN F-BOX(TIR1/AFB) in SKP1-CULLIN1-F-BOX (SCF) ubiquitin ligase complexes3, 4. This binding increases affinity between SCF(TIR1/AFB) ubiquitin ligase complexes and their substrates, AUXIN/INDOLE-3-ACETIC ACIDs (Aux/IAAs)5, which act as inhibitors of AUXIN RESPONSE FACTORs (ARFs)6. ARFs are transcription factors that recognize AUXIN RESPONSE ELEMENTs (AuxREs)7 in promoter regions and regulate downstream gene activities8. Degradation of ubiquitin-modified Aux/IAA proteins releases ARFs from inhibition, allowing activation or repression of auxin responding genes (reviewed in ref. 9). A widespread reporter of auxin response, the synthetic DR5 promoter, which consists of 7-9 AuxRE repeats, marks sites of transcriptional auxin response by activating fused reporter genes such as β-glucuronidase10, fluorescent proteins11, or luciferase12. While DR5 marks many auxin-dependent processes10, several others are not accompanied by its activity13, 14. Notably, computational modeling of auxin accumulation patterns based on the topology and dynamics of the auxin transport network predicted auxin gradients in the root tip, but these cannot be directly visualized15, and thus the reported DR5 expression sites are often referred to as auxin “maxima”. The AuxRE in the DR5 promoter was first identified through deletion analysis of a single auxin-responsive soybean promoter10. No exhaustive analysis of DNA-binding specificity of the ARFs had been performed, and it has remained unclear whether the canonical AuxRE is a high-affinity binding site until recently. We have solved crystal structures of 2 functionally divergent ARFs and systematically determined binding sites through Protein Binding Microarrays. This analysis revealed that the AuxRE in DR5 is not a high-affinity binding site and identified another site (TGTCGG) that showed higher affinity16. It is thus conceivable that the limited sensitivity of DR5 reporters is due to its element being medium ARF affinity.
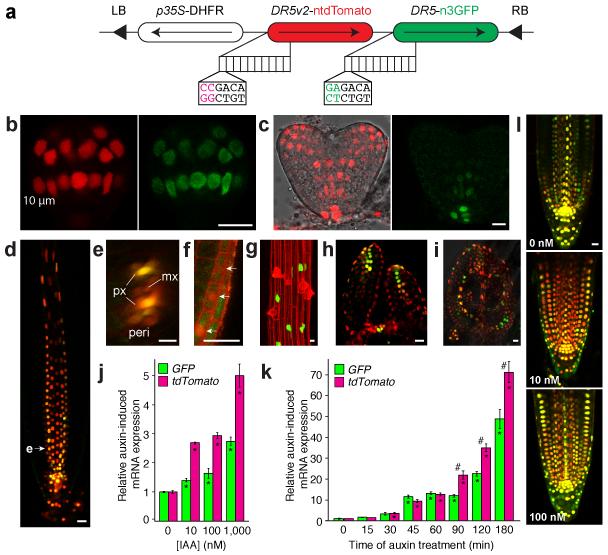
To address this question, we engineered DR5 reporters by replacing the 9 original AuxREs in the DR5-rev promoter10 with this novel binding site and named the new reporter DR5v2. To directly compare the two reporters without confounding effects of transgene integration site and expression level, we fused each to a different nuclear localized fluorescent protein and expressed both reporters from a single transgene in Arabidopsis thaliana (Fig. 1a). Analysis of these lines showed activity of DR5v2 in extended domains compared to DR5. During embryogenesis, DR5v2 expression is comparable to DR5 until the early globular stage (Fig. 1b). From transition stage onward, additional expression domains of DR5v2 become more distinct in the incipient cotyledon and vasculature (Fig. 1c) where proper auxin response is required for normal development17. In the post-embryonic root, while DR5 and DR5v2 mark quiescent center, columella root cap, and protoxylem (Fig. 1d,e), DR5v2 was additionally expressed in metaxylem, pericycle (Fig. 1e), lateral root cap (Fig. 1f), and epidermal cells (Fig. 1g). Strikingly, the epidermal cells expressing DR5v2 were trichoblasts (Fig. 1g), which require auxin response for normal root hair development13. In the (first rosette) leaf primordia, both DR5v2 and DR5 report auxin maxima in the most distal domain and incipient leaf vein; however, DR5v2 also shows expression in surrounding cells and the L1 layer (Fig. 1h,i). These additional DR5v2 expression domains match the predicted auxin accumulation sites based on convergence of polar auxin transporter localization14. To exclude that differences in expression were due to different fluorophores, we also compared separate DR5v2 and DR5 reporter lines both driving the same fluorescent protein, which confirmed extended expression of DR5v2 (Supplementary Fig. 1). This suggests that the new DR5v2 marker is visualizing previously predicted sites of auxin response. Importantly, DR5v2 reported low-level activity in most cells in dividing areas in embryo, leaf and shoot meristem. This extended domain is consistent with the known involvement of auxin in cell division and elongation18, and suggests that the DR5v2 reporter is sufficiently sensitive to detect these more generic auxin reponses. Interestingly, while all sites of DR5 activity are also marked by DR5v2, the relative intensity across cell types is not identical. For example, in roots DR5 has the highest expression in the QC, while DR5v2 has increased response in subtending columella cells (Fig. 1d). This presumably reflects a difference in binding affinity towards TGTCTC (DR5) and TGTCGG (DR5v2) by the ARFs that act in each cell type.
Figure 1. DR5v2 sensitively reports auxin response.
(a) Schematic of DR5v2-ntdTomato/DR5-n3GFP double reporter. 10 repeats of either reverse TGTCTC (DR5) or TGTCGG (DR5v2) are positioned upstream of a minimal promoter and either nuclear 3×eGFP (n3GFP) or nuclear tandem Tomato (ntdTomato). LB/RB, Left/Right Border; DHFR, Methotrexate resistance gene. (b-j) DR5v2 (red) and DR5 (Green) activity in early globular (b) and heart stage (c) embryos, root tip (d, longitudinal section; e, transverse section along plane at arrowhead in d; px, protoxylem; mx, metaxylem; peri, pericycle), lateral root cap (f), root epidermis (g, here shown in a DR5v2::n3eGFP root), SAM (h) and young leaf (i). (j,k) Relative qRT-PCR GFP and tdTomato transcript level in DR5v2-ntdTomato/DR5-n3GFP seedlings after 12 hour pre-treatment with 10 μM NPA followed by 2 hour treatment with auxin (IAA) concentrations as indicated (j), or treated with 1 μM IAA for the indicated times without NPA pre-treatment (k). Expression in mock treatments in are set to 1. Bars indicate standard error from the mean (n= 3). Asterisks (*) indicate significantly different expression compared to untreated control, while number signs (#) indicate significant difference between DR5 and DR5v2 (Two-tailed t-test; p<0.05). (l) Visualization of DR5v2 (red)-DR5 (Green) double reporter activity in root tips after 12-hour co-treatment of 10 μM NPA with indicated IAA concentrations. Scale bar in panels (b-i, l) is 10 μm.
We next tested if the extended domain of DR5v2 expression correlates with increased sensitivity to auxin. We treated DR5-n3EGFP/DR5v2-ntdTomato double reporter seedlings with a range of exogenous auxin concentrations and monitored gene activation using both qRT-PCR (Fig. 1j,k; Supplementary Fig 2) and microscopy (Fig. 1l; Supplementary Fig. 3). While both DR5 and DR5v2 responded to concentrations as low as 3 nM (Supplementary Fig. 3), the amplitude of response was much higher for DR5v2 at all concentrations tested. To exclude that folding and/or stability of fluorescent proteins contributed to differential signal intensity, we performed qRT-PCR analysis on GFP and tdTomato transcripts. In this analysis, DR5v2-ntdTomato indeed responded to lower auxin concentrations than DR5-n3EGFP (Fig. 1j), confirming its increased sensitivity. Likewise, when treated with the same concentration of auxin, the increased amplitude of response of DR5v2 is distinctly visible after prolonged auxin treatment (Fig. 1k,l). Thus, while both reporters respond to the same range of auxin concentrations (Supplementary Fig. 3), the increased amplitude of DR5v2 response allows in vivo detection of 10-fold lower auxin concentrations (Fig. 1l). Importantly however, neither of the reporters shows a linear response to auxin concentrations (Fig. 1j) or treatment duration (Fig. 1k), and hence can not be used to infer actual auxin levels. Yet, the highly sensitive DR5v2 reporter enables visualization of novel and weaker auxin responses.
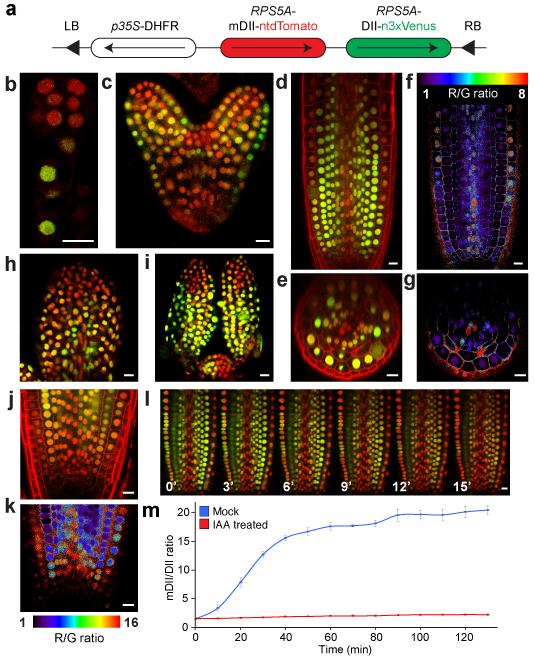
Any novel auxin response site, as marked by DR5v2, would benefit from being confirmed by an independent reporter. Recently, a conceptually different reporter was developed: in lines that carry DII-VENUS, a fusion of the auxin-dependent degradation domain II of an Aux/IAA protein to Venus fluorescent protein, absence of fluorescence marks auxin accumulation19. Comparison with lines in which the residues responsible for auxin-dependent degradation are mutated (mDII-Venus) reveals sites in which auxin promotes degradation (here termed “auxin input”). Importantly, such a reporter has the potential of semi-quantitative measurement of auxin input as it omits gene regulation. In addition, the reporter can also be used to report auxin accumulation that does not lead to gene activation. However, the 35S promoter used is not ideal for several developmental processes, such as embryogenesis20. Therefore, we generated DII-Venus and mDII-Venus versions expressed from the RPS5A promoter that is active in most dividing cells21, and thus encompasses most sites of auxin response in growth and development. While these lines allow observing auxin activity in embryos and meristems (Supplementary Fig. 4), quantification and comparison of signals is extremely challenging without an internal reference for the theoretical activity in the absence of auxin. We therefore designed a novel reporter that would allow ratiometric analysis of the two fluorescent proteins, the utility of which has recently been demonstrated22, 23. This reporter, named R2D2 (Ratiometric version of 2 D2’s), combines RPS5A-driven DII fused to n3×Venus and RPS5A-driven mDII fused to ntdTomato on a single transgene (Fig. 2a). Auxin accumulation will be visible as reduction of yellow signal relative to the red signal. Indeed, observation of untreated root tips showed results qualitatively similar to separate DII and mDII lines (Supplementary Fig. 4), yet allow comparison of the two signals in every cell (Fig. 2b-k). We implemented a simple image analysis algorithm (see Materials and Methods) to infer relative auxin distribution from the yellow/red ratio. Following background subtraction, yellow/red ratio of each pixel was calculated and visualized into a false-color scale in real time. Here, as low yellow/red ratio correlates with higher auxin levels, we plot the inverse ratio such that increased ratio corresponds to higher auxin (Fig. 2f). We used this analysis to image auxin input in various developmental processes. During early embryogenesis, auxin input detected by R2D2 is consistent with auxin response presented by DR5 and DR5v2 activity; high auxin activity in embryo proper until globular stage11 (Fig. 2b), and confined to incipient cotyledon and vasculature, and hypophysis and its daughter cells in heart stage (Fig. 2c). From heart stage, however, an additional domain of auxin input in the shoot apical meristem (SAM) is detected only by R2D2 (Fig. 2c; Supplementary Fig. 5). This finding is consistent with the expression of several key auxin biosynthesis enzymes24 in these cells, which does not translate to auxin response, as was also predicted and demonstrated for the post-embryonic shoot meristem25. In post-embryonic root (Fig. 2d-g), young leaves and leaf primordia (Fig. 2h,i), and shoot apical meristem (Fig. 2i), in addition to confirming auxin response shown by DR5v2 (Fig. 1), R2D2 revealed quantitative properties of early auxin signaling.
Figure 2. R2D2, a semi-quantitative and rapid auxin input reporter.
(a) Schematic of R2D2. LB/RB, Left/Right Border; DHFR, Methothrexate resistance gene. (b-k) ntdTomato (red) and n3×Venus (green) fluorescence signal overlays (b-e, h-j) and inverse n3×Venus/ ntdTomato signal ratio (f,g,k; false color) in pre-globular (b) and heart stage (c) embryos, root tip (longitudinal section in d,e; radial section in e,g; detail in j,k), young leaf (h) and SAM (i). Note the descending gradient of auxin input in RAM in (j,k). (l) Successive images of R2D2 root tips treated with 1 μM IAA for the indicated time. (m) Whole-frame quantification of inverse n3×Venus/ ntdTomato signal ratio after treatment with 1 μM IAA and untreated mock control. Bars indicate standard error from the mean (n=3). Scale bar in panels (b-l) is 10 μm.
We used R2D2 to address whether auxin gradients, as predicted by simulations of accumulation based on the transport network15, and as inferred from comparison of DII-Venus and mDII-Venus roots19, could be directly visualized. We noticed that a steep auxin gradient could be observed in the cells closest to the quiescent center in the root tip. In all cell files except epidermis and xylem cells, auxin input levels decreased from maximum to background over a range of 6-7 nuclei (Fig. 2j,k; Supplementary Fig. 6). This, to our interpretation, clearly marks an auxin gradient that is entirely consistent with computational predictions15. Importantly, while average gradients could be inferred from comparison of DII-Venus and mDII-Venus lines19, their accurate quantitative analysis requires a dedicated ratiometric tool such as R2D2.
In addition to its semi-quantitative property to detect auxin input, R2D2 should also allow observation of rapid changes in auxin accumulation due to the omission of the time required for transcription, translation and fluorophore maturation19. Indeed, treatment of R2D2 seedlings with exogenous auxin led to a rapid and uniform loss of yellow signal without appreciable effect on the red signal (Fig. 2l,m; Supplementary Movies 1 and 2). This capability to rapidly monitor auxin accumulation makes it possible to measure auxin input at cellular resolution in real-time.
Understanding of the developmental roles and dynamic regulation by signaling molecules is greatly accelerated by the ability to visualize its activity at high resolution. Here, we have developed a set of novel tools that allow sensitive and semi-quantitative detection of auxin signaling and responses in plants. As the AuxRE is a generic ARF binding site16, DR5v2 is likely to be functional in any genetically transformable plant species, and dual-color imaging of high and medium affinity ARF binding sites allows simultaneously visualizing both an extended range of auxin responses and the maxima. Likewise, R2D2 has the potential of being a generic reporter for transformable plant species, although the choice of the promoter has to be adapted for specific tissues and cell types. Finally, combining the high-affinity DR5v2 reporter and R2D2 in a single-transgene, triple-color marker will enable correlating auxin input and output at high resolution, and detect sites where auxin accumulation does not elicit a response. We expect that these novel tools will be of crucial importance in defining and quantifying responses to the pivotal auxin molecule.
Online Methods
Plant material
DR5v2 was designed by replacing the 9 AuxRE’s in DR5 (TGTCTC), with TGTCGG, and synthesized to generate cloning vector pUC57/DR5v2 (GenScript). Double reporter pGIIM/DR5v2::ntdTomato-DR5::n3eGFP was created in two steps. DR5 reporter cassette from pGIIK/DR5::n3eGFP was first excised by BamHI and EcoRI digestion and cloned into BamHI and EcoRI digested pGIIM/LIC_SwaI-ntdTomato-LIC_HpaI-n3EGFP (a kind gift from Thomas Laux, Freiburg) to create pGIIM/LIC_SwaI-ntdTomato- DR5::n3eGFP. DR5v2 reporter cassette amplified from pUC57/DR5v2 using primer set “DR5v2” was then cloned into SwaI-digested pGIIM/LIC_SwaI-ntdTomato- DR5::n3eGFP via Ligation Independent Cloning 26. pGIIM/LIC_SwaI-ntdTomato- DR5v2::n3eGFP and pGIIM/LIC_SwaI-ntdTomato- DR5v2::n3VENUS were created by excising DR5v2 reporter cassette from pUC57/DR5v2 via BamHI and EcoRI digestion followed by cloning into BamHI and EcoRI digested pGIIM/LIC_SwaI-ntdTomato- LIC_HpaI-n3eGFP or pGIIM/LIC_SwaI-ntdTomato- LIC_HpaI-n3Venus.
The pRPS5A::DII-Venus and pRPS5A::mDII-Venus binary vectors were constructed using the multisite Gateway technology (Invitrogen) and following the provider instructions. To do so, the RPS5A promoter was cloned in pDONR P4-P1R using primers listed in Supplementary Table 1. This vector was used together with the previously described DII/mDII cloned in pDONR221 and Venus fused to the N7 nuclear localization signal cloned in pDONR P2R-P319 for recombination in the binary gateway vector pH7m34GW (http://gateway.psb.ugent.be/).
R2D2 in pGIIM/ RPS5A:: mDII: ntdTomato- RPS5A:: DII: n3Venus was created through two subsequent Ligation Independent Cloning events. First, RPS5A:: DII reporter cassette amplified from genomic DNA of pRPS5a:: DII: Venus using primer set “pRPS5a:: DII” was cloned into HpaI digested pGIIM/LIC_SwaI-ntdTomato- LIC_HpaI-n3Venus (a kind gift from Thomas Laux, Freiburg) to create pGIIM/LIC_SwaI-ntdTomato- RPS5A:: DII:n3Venus. RPS5A:: mDII control cassette amplified from genomic DNA of pRPS5a:: mDII: Venus control line using primer set “pRPS5a:: mDII” was then cloned into SwaI digested pGIIM/LIC_SwaI-ntdTomato- RPS5A:: DII:n3Venus to create R2D2. Sequences of primers used for cloning aforementioned constructs are listed in Supplementary Table 1. All transgenic lines were first created in Arabidopsis Col-0 ecotype.
Plant Growth condition
Arabidopsis plants were grown at 22°C in 16 hours light/ 8 hours dark cycle for every experiments. All seeds were surface sterilized, sown on half-strength Murashige and Skoog medium with 0.8% Daichin agar (Duchefa) (1/2 MS plate) if not mentioned otherwise, and vernalized at 4°C for 2 days. For microscopic analysis of root, seedlings were grown vertically for five days after transfer to growth chamber, while this period was decreased to three or four days for microscopic analysis in shoot. Methotrexate (MTX) selection was conducted by growing sterilized seeds on 1/2 MS plates containing 0.1 mg/L MTX (Sigma; A6770).
For DR5/DR5v2 auxin sensitivity analysis via qRT-PCR, surface sterilized seeds were sown on sterilized nylon mesh placed on 1/2 MS plates after stratification and grown in growth chamber for four days then transferred to 1/2 MS plates with 0.11% DMSO and 10 μM N-1-Naphthylphthalamidic acid (NPA; Chem Service) to inhibit auxin transport. After incubation for 12 hours, seedlings were transferred to plates containing 0.11% DMSO and 10 μM NPA with 0.01, 0.1, or 1.0 μM Indole 3-Acetic Acid (IAA; Duchefa) for treatments, 0.11% DMSO and 10 μM NPA for control for two hours before collection for RNA isolation.
For DR5/DR5v2 auxin sensitivity analysis via confocal microscopy, surface sterilized seeds were sown on sterilized nylon mesh placed on 1/2 MS plates after stratification and grown in growth chamber for four days then transferred to 1/2 MS plates with 0.11% DMSO and 10 μM NPA with 0.0001, 0.000316, 0.001, 0.00316, 0.01, 0.0316, 0.1, 0.316, or 1.0 μM IAA for treatments, 0.11% DMSO and 10 μM NPA for control for 12 hours before collection for imaging.
For temporal DR5/DR5v2 auxin response analysis, surface sterilized seeds were sown on sterilized nylon mesh placed on 1/2 MS plates after stratification and grown in growth chamber for five days then transferred to 1/2 MS plates with 0.01% DMSO and 1.0 μM IAA as treatment or 1/2 MS plates with 0.01% DMSO as control for given time before collection for RNA isolation.
Microscopic analysis
Images were acquired as 8-bit format using a Leica TCS SP5II confocal laser scanning microscope with 20× NA=0.75 and 63× NA=1.20 water-immersion objective and pinhole equivalent to 1.0× the Airy disk diameter. EGFP and VENUS were excited by argon ion laser while tdTomato and propidium iodide were excited using diode laser, and their emissions were detected sequentially with Leica HyD in standard mode to prevent cross-talks between fluorophores. Excitation and detection of fluorophores were configured as below, eGFP was excited at 488 nm and detected 498-530 nm; Venus was excited at 514 nm and detected 524-540 nm; tdTomato was excited at 561 nm and detected 571-630 nm; propidium iodide was excited at excited at 561 nm and detected 571-700 nm.
In comparisons of eGFP and tdTomato fluorescence in the same line, the highest fluorescence signal in reference cells listed below in each channel was used to set the upper limit of pixel intensity. Reference cells used to setup the upper limit of pixel intensity are lens shape cell of early heart stage when imagining embryos, quiescent center cells when imaging roots, and distal domain of leaf primordia when imaging shoot meristem and leaf primordial.
In comparisons of VENUS and tdTomato fluorescence in R2D2 line, the highest fluorescence signal in reference cells listed below in each channel was used to set the upper limit of pixel intensity. Reference cells used to setup the upper limit of pixel intensity are suspensor cells of early globular stage when imagining embryos, cortex cells when imaging roots, and trichome cells when imaging shoot meristem and leaf primordial.
Embryos were mounted in 1× phosphate solution saline (PBS) containing 4% paraformaldehyde and 5% glycerol as described27, and seedlings were mounted in demineralized water unless mentioned otherwise with 10 μg/mL propidium iodide4 for roots and without propidium iodide for shoot meristem, leaf primordial, DR5v2 auxin sensitivity analysis, and R2D2 auxin treatment live imaging.
Seedlings for live imaging were mounted in modified devices described28. The original plastic mask was replaced by a 15.5 mm × 21.0 mm× 0.5 mm frame made of Bioplastic with 10.0 mm × 15.0 mm opening in the center at where was covered with 0.4 μm PTFE mesh. Only one plastic frame was used, but agarose and culture medium were also omitted. Five five-day-after-germination seedlings were placed in a two chamber coverglass containing 100 μL demineralized water then covert by PTFE frame with 0.4 μm PTFE mesh facing to seedlings followed by adding 900μL of demineralized water to cover the roots. This device allows imaging multiple roots at identical condition at once via confocal microscope. Time interval and coordinates of regions of interest were first defined, and images referred as “0 sec” were taken before adding 111 μL of 0.1% DMSO or 10 μM IAA in 0.1% DMSO.
Virtual ratio images of R2D2 were generated by “Calcium Imaging Calculator” built in Leica LAS AF lite v2.6.3 or v 3.7.x through calculating ratios between signal intensities of each pixel from two channels after subtracting noise, which was defined as the average signal intensity of six 2.5-3.5 μm2 area in the cytoplasm of six epidermal cells from a single image and used for the rest of images taken with the same laser/detection configuration.
To monitor auxin gradient in the root apical meristem by R2D2, images from three z-stacks with 2.0 μm interval were acquired. The maximum projection of three images was examined to assure the section of region of interest contains only single cell layer. Approximate 10 μm2 area in nucleus of cell of interest was selected via the ROI tool, and the ratio of red/yellow signal ratio was calculated by “Calcium Imaging Calculator” built in Leica LAS AF lite v2.6.3 or v 3.7.x after noise subtraction. Red/yellow ratio of the first 7-10 continuous cells (depending on number of cells in each frame of images due to the different cell size of each tissue) from the quiescent center was acquired form 32 roots of R2D2 line. Cells from both sides of roots were used if possible to generate 32 to 47 data sets of each tissue.
Quantitative RT-PCR Analysis
Over 100 roots from treatments were collected and RNA was extracted with Plant RNeasy kit (QIAGEN). Poly(dT) cDNA was prepared from 600 ng total RNA with an iScript cDNA Synthesis Kit (Biorad). Primer pairs were designed with Beacon Designer 8.0 (Premier Biosoft International). Although the fluorescent proteins are tandem repeats (tandem dimer Tomato and 3×eGFP), we designed primers that generate a single amplicon per transcript. Primers were tested in qRT-PCR using serial diluted pGIIM/DR5v2::ntdTomato-DR5::n3eGFP plasmid as template to validate the correspondence between amount of amplicons and actual templates (See Supplementary Fig.2). qRT-PCR was conducted with iQ SYBR Green Supermix (BioRad) on CFX384 Real-Time PCR detection system (BioRad) according to the manufacturer’s instructions. Efficiency of primers for subjected cDNA, ntdTomato and n3eGFP, and concentrations of subjected cDNA in all samples have been tested in advance to ensure expressions of n3eGFP and ntdTomato are comparable. All individual reactions were done in triplicate with two biological replicates. Data were analyzed with qBase5. Expression levels were normalized to those of EEF1α4, GAPC, and iEF4A. Sequences of primers used for qRT-PCR were listed in Supplementary Table 1.
To compare qRT-PCR results, normalized data sets acquired form qBase were subjected to two-tailed Student’s t-test with threshold (alpha level) of 0.05 to determine if significance of the difference between each treatment.
Supplementary Material
Acknowledgements
The authors would like to thank Thomas Laux (Institut für Biologie III, Universität Freiburg) for plasmids and Bert de Rybel for helpful comments on the manuscript. This work was supported by grants from the European Research Council (ERC; CELLPATTERN; Contract number 281573) and the Netherlands Organization for Scientific Research (NWO; ALW-820.02.019) to D.W. and Human Frontier Science Program (HFSP; research grant RGP0054-2013) and Agence Nationale de la Recherche (ANR; AuxiFlo; Grant ANR-12-BSV6-0005) to T.V..
Footnotes
Competing Financial Interests statement
None of the authors has competing financial interests.
Accession codes
Plasmids and seeds described in this study have been deposited in Addgene (www.addgene.org; Deposit 71550) and the Nottingham Arabidopsis Stock Center (www.arabidopsis.info), respectively.
References
- 1.Lokerse AS, Weijers D. Curr. Opin. Plant Biol. 2009;12:520–526. doi: 10.1016/j.pbi.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 2.Muday GK. J. Plant Growth Regul. 2001;20:226–243. doi: 10.1007/s003440010027. [DOI] [PubMed] [Google Scholar]
- 3.Dharmasiri N, Dharmasiri S, Estelle M. Nature. 2005;435:441–445. doi: 10.1038/nature03543. [DOI] [PubMed] [Google Scholar]
- 4.Kepinski S, Leyser O. Nature. 2005;435:446–451. doi: 10.1038/nature03542. [DOI] [PubMed] [Google Scholar]
- 5.Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M. Nature. 2001;414:271–276. doi: 10.1038/35104500. [DOI] [PubMed] [Google Scholar]
- 6.Tan X, et al. Nature. 2007;446:640–645. doi: 10.1038/nature05731. [DOI] [PubMed] [Google Scholar]
- 7.Ulmasov T, Hagen G, Guilfoyle TJ. Science. 1997;276:1865–1868. doi: 10.1126/science.276.5320.1865. [DOI] [PubMed] [Google Scholar]
- 8.Ulmasov T, Hagen G, Guilfoyle TJ. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:5844–5849. doi: 10.1073/pnas.96.10.5844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R, Estelle M. Curr. Opin. Plant Biol. 2014;21:51–58. doi: 10.1016/j.pbi.2014.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ. Plant Cell. 1997;9:1963–1971. doi: 10.1105/tpc.9.11.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Friml J, et al. Nature. 2003;426:147–153. doi: 10.1038/nature02085. [DOI] [PubMed] [Google Scholar]
- 12.Moreno-Risueno MA, et al. Science. 2010;329:1306–1311. doi: 10.1126/science.1191937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones AR, et al. Nat. Cell Biol. 2009;11:78–84. doi: 10.1038/ncb1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scarpella E, Marcos D, Friml J, Berleth T. Genes and Development. 2006;20:1015–1027. doi: 10.1101/gad.1402406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grieneisen VA, Xu J, Marée AFM, Hogeweg P, Scheres B. Nature. 2007;449:1008–1013. doi: 10.1038/nature06215. [DOI] [PubMed] [Google Scholar]
- 16.Boer DR, et al. Cell. 2014;156:577–589. doi: 10.1016/j.cell.2013.12.027. [DOI] [PubMed] [Google Scholar]
- 17.Hardtke CS, et al. Development. 2004;131:1089–1100. doi: 10.1242/dev.00925. [DOI] [PubMed] [Google Scholar]
- 18.Perrot-Rechenmann C. Cold Spring Harbor perspectives in biology. 2010;2:a001446. doi: 10.1101/cshperspect.a001446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brunoud G, et al. Nature. 2012;482:103–106. doi: 10.1038/nature10791. [DOI] [PubMed] [Google Scholar]
- 20.Völker A, Stierhof YD, Jürgens G. J. Cell Sci. 2001;114:3001–3012. doi: 10.1242/jcs.114.16.3001. [DOI] [PubMed] [Google Scholar]
- 21.Weijers D, et al. Development. 2001;128:4289–4299. doi: 10.1242/dev.128.21.4289. [DOI] [PubMed] [Google Scholar]
- 22.Federici F, Dupuy L, Laplaze L, Heisler M, Haseloff J. Nat. Methods. 2012;9:483–485. doi: 10.1038/nmeth.1940. [DOI] [PubMed] [Google Scholar]
- 23.Wend S, et al. Scientific reports. 2013;3:2052. doi: 10.1038/srep02052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robert HS, et al. Curr. Biol. 2013;23:2506–2512. doi: 10.1016/j.cub.2013.09.039. [DOI] [PubMed] [Google Scholar]
- 25.Vernoux T, et al. Mol. Syst. Biol. 2011;7 doi: 10.1038/msb.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 26.de Rybel BD, et al. Plant Physiol. 2011;156:1292–1299. doi: 10.1104/pp.111.177337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Llavata-Peris C, Lokerse A, Möller B, De Rybel B, Weijers D. Methods in Molecular Biology. Vol. 959. Humana Press; Clifton, N.J.: 2013. pp. 137–148. [DOI] [PubMed] [Google Scholar]
- 28.Daghma DS, Kumlehn J, Hensel G, Rutten T, Melzer M. J. Exp. Bot. 2012;63:6017–6021. doi: 10.1093/jxb/ers254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Den Berg C, Willemsen V, Hage W, Weisbeek P, Scheres B. Nature. 1995;378:62–65. doi: 10.1038/378062a0. [DOI] [PubMed] [Google Scholar]
- 30.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. Genome biology. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.