Abstract
Rationale
Treatment of sinus node disease with regenerative or cell-based therapies will require a detailed understanding of gene regulatory networks in cardiac pacemaker cells (PCs).
Objective
To characterize the transcriptome of PCs using RNA sequencing, and to identify transcriptional networks responsible for PC gene expression.
Methods and Results
We used laser capture micro-dissection (LCM) on a sinus node reporter mouse line to isolate RNA from PCs for RNA sequencing (RNA-Seq). Differential expression and network analysis identified novel SAN-enriched genes, and predicted that the transcription factor Islet-1 (Isl1) is active in developing pacemaker cells. RNA-Seq on SAN tissue lacking Isl1 established that Isl1 is an important transcriptional regulator within the developing SAN.
Conclusions
(1) The PC transcriptome diverges sharply from other cardiomyocytes; (2) Isl1 is a positive transcriptional regulator of the PC gene expression program.
Keywords: Sinoatrial node, laser capture microdissection, Isl1, Hcn4, transcription factors, mouse heart development, RNA-Seq
INTRODUCTION
Pacemaker cells (PCs) within the sinoatrial node (SAN) generate the electrical impulse that initiates each heartbeat. Sinus node dysfunction (SND), characterized by loss of PCs and SAN fibrosis, is a common but poorly understood disease. With the advent of cellular reprogramming technology,1, 2 there is increasing interest in understanding gene regulatory networks in PCs, both to improve our understanding of SND and to facilitate novel therapies such as biological pacemakers and SAN regeneration.
Transcription factors important for SAN development and function have been identified, including Tbx18,3 Tbx3,4 Shox2,5 and Isl1.6, 7 Efforts to reprogram non-PCs into PCs have employed some of these SAN transcription factors,8–10 even though their downstream transcriptional networks have not been fully elucidated.
Although these efforts are highly promising, there have been no genome-wide transcriptome comparisons of putatively reprogrammed PC-like cells with bona fide PCs, largely because of the absence of PC transcriptome data. Transcriptional profiling of PCs presents special challenges because of the lack of specific molecular markers, the small size of the developing SAN, and the interdigitation of PCs with non-PCs in the SAN. At present, the lack of transcriptome data from PCs remains a barrier to further progress in understanding SAN biology, and to assessing and improving the fidelity of PC reprogramming technology.
METHODS
Laser capture microdissection
Embryos or hearts were removed intact, washed with cold PBS, and immediately embedded in OCT and stored at −20 °C until sectioning. Tissue was sectioned at a thickness of 8 microns onto membrane-coated slides (MembraneSlide NF 1.0 PEN, Zeiss Microscopy, Gottingen, Germany). For laser capture, slides were thawed to room temperature until evaporation of moisture (approximately 1 minute), placed on the microscope stage of a PALM Micro-Beam inverted microscope with LCM capability (Zeiss). The sinus node tissue was identified visually and outlined manually with the microscope user interface (Online Video II). Laser power and catapult energy were optimized prior to the experiment and varied from experiment-to-experiment. After each experiment, sections were stained with DAPI and anti-Hcn4, mounted, and visualized to confirm accurate dissection of the region of interest.
A detailed methods section is available online.
RESULTS
Laser capture micro-dissection of cardiac pacemaker cells for RNA sequencing
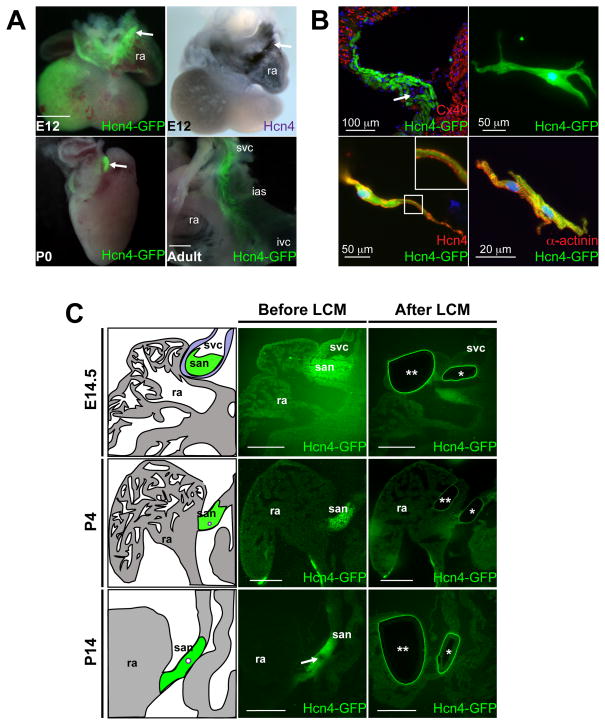
Hcn4-GFP BAC transgenic mice exhibited high-level GFP expression in the sinoatrial node (SAN) and venous pole at E12.5, with further restriction of expression to the SAN at P0 and in the adult heart (Figure 1A).11 GFP expression was complementary to Connexin-40, a marker of working atrial myocytes, and isolated adult GFP+ cells showed morphology typical of PCs and exhibited spontaneous beating (Figure 1B, Online Video I). We isolated SAN tissue and right atrial (RA) myocardium by laser capture micro-dissection (LCM) on unfixed, unstained cryosections from flash frozen Hcn4-GFP hearts harvested at E14.5, P4, and P14 (Figure 1C, Online Video II). The SAN is heterogeneous and includes non-PC cells such as fibroblasts, endothelial cells, and atrial cardiomyocytes. To optimize enrichment for PCs, we selected regions of interest for LCM within the SAN head (near the SAN artery) that contained purer populations of PCs (Online Figure I). Such regions within the SAN, as well as neighboring RA regions, were excised from 6 consecutive embryonic heart sections and pooled to generate total RNA. We repeated the LCM experiment for three different embryos for each time point to generate 18 independent RNA samples, reflecting 3 biological replicates for each tissue and time point (Online Table I). Each sample was processed separately for RNA-sequencing (RNA-seq) as described in the online methods section.
Figure 1. Laser Capture Micro-Dissection of Sinoatrial Node Pacemaker Cells from Hcn4-GFP BAC Transgenic.
(A) Whole-mount images of embryonic day (E) 12.5 hearts (top, left), post-natal day 0 (P0, bottom, left), and adult right atrium (bottom right) showed expression of Hcn4-GFP, compared to in-situ hybridization for Hcn4 (top, right). Arrows denote sinoatrial node (SAN). At the adult stage, GFP+ cells were present in the mature SAN, located in the inter-caval groove between the inter-atrial septum (ias) and right atrium (ra). (B) Immunohistochemistry on adult SAN from Hcn4-GFP hearts showing complementary expression of GFP and Connexin-40 (Cx40) (arrow denotes SAN artery). Isolated Hcn4-GFP+ cells (top, right), were Hcn4+ by immunocytochemistry (bottom, left – high magnification inset shows Hcn4 immunostaining at cell membrane) and were positive for the myocyte marker α-actinin (bottom, right). (C) Unfixed, unstained sections from Hcn4-GFP hearts were used for laser capture microdissection (LCM) of SAN and RA at time points indicated. Schematic depictions of the SAN and RA are shown for corresponding sections before or after LCM. Arrows denote SAN artery; ‘*’ denotes the excised SAN region and ‘**’ denotes excised right atrial tissue; svc, superior vena cava; ivc, inferior vena cava. Scalebars = 200 μM unless otherwise indicated.
Differential expression analysis
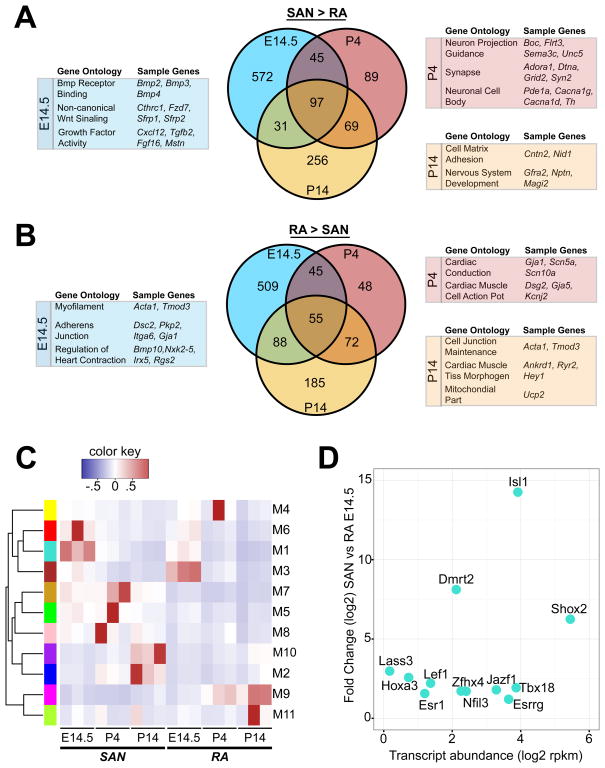
Differential expression (DE) analysis of RNA-seq data between SAN and RA tissue samples revealed hundreds of DE genes at each time point (Online Table II). Known SAN-associated genes Hcn4, Tbx3, Shox2, Tbx18, and Bmp4 were all enriched in the SAN tissue, while RA-associated genes Nkx2-5, Nppa, and Gja1 were enriched in the RA samples (Online Figure I), demonstrating the fidelity of tissue isolation. Of note, the core cardiac transcription factors Gata4, Mef2c and Tbx5, were not differentially expressed between SAN and RA. We also found numerous genes enriched in the SAN tissue that had not been previously associated with PCs (Figure 2A, Online Table II). Gene Ontology terms (GO) associated with SAN-enriched genes included Bmp and Wnt signaling pathways at E14.5, and neuronal development and function at later time points (Figure 2A, Online Table III). Conversely, RA-enriched genes were associated with GO terms that included conduction, contractile apparatus, and cell junction formation (Figure 2B, Online Table III). While a core set of genes in each tissue type exhibited DE at all time points examined (Online Table IV), there was considerable change over time within the DE gene set, highlighting the dynamic nature of expression during SAN development. Hierarchical clustering of SAN and RA samples revealed that biological replicates clustered together, and that, as differentiation progressed, SAN samples were more similar to each other than they were to the RA samples (Online Figure II).
Figure 2. Comparative Expression Analysis of SAN and RA.
(A) Venn diagram showing overlap of SAN-enriched genes at E14.5, P4, and P14 using a false discovery rate cutoff of 0.05. Selected enriched gene ontology terms with example genes are shown for each time point. (B) Similar Venn diagram as (A), but for RA-enriched genes. (C) Weighted gene correlation network analysis (WGCNA) identified 11 highly correlated modules within SAN/RA data set. Module eigengene activity was computed for each module for each of the 18 SAN/RA expression time points. Color key indicates activity scale. (D) Expression of SAN-enriched transcription factors within Module 1 at E14.5. Transcript levels are shown on the X-axis and differential expression (SAN vs RA) on the Y-axis. Abbreviations: SAN, sinoatrial node; RA, right atrium.
Network analysis
We employed weighted gene correlation network analysis (WGCNA) to partition the RA and SAN transcriptomes into modules exhibiting correlated gene expression (Online Figure III, Online Table V). Most time points of specific tissue were associated with at least one highly active module. Module 1 (M1) exhibited highest activity at E14.5, a critical period for SAN morphogenesis and PC differentiation (Figure 2C). M1 contained several previously identified SAN-enriched transcription factors, including Tbx18, Shox2, and Isl1, while Tbx3 clustered with Hcn4 and Hcn1 in M7. Of the factors in M1, Isl1 had the highest level of DE as well as high transcript abundance early in SAN development (Figure 2D).
Conditional selection of Isl1 after second heart field differentiation
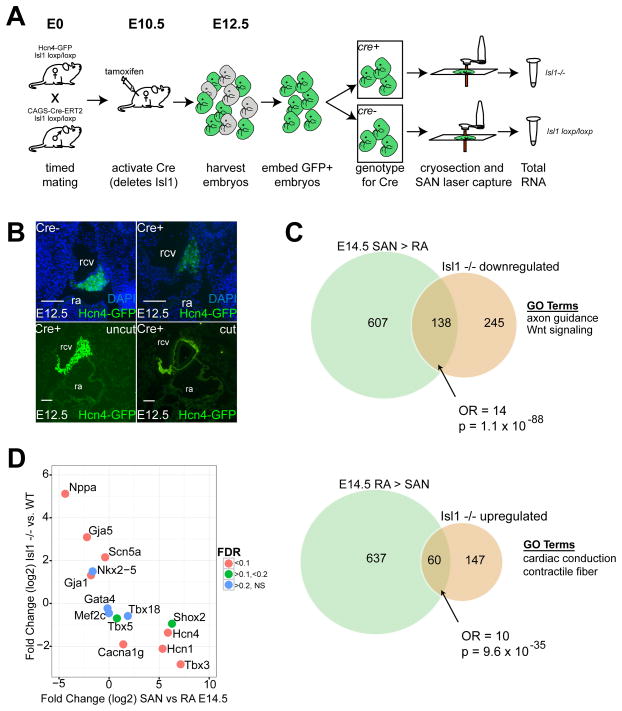
To test for a requirement of Isl1 in the PC gene expression program, we crossed Isl1loxp/loxp; CAG-Cre/Esr1 with Isl1loxp/loxp; Hcn4-GFP and injected intra-peritoneal tamoxifen at E10.5 (Figure 3A). This strategy generated a global deletion of Isl1 after second heart field differentiation, which circumvented the early embryonic lethality associated with Isl1 loss of function. Cre+;Hcn4-GFP+ embryos were recovered at Mendelian ratios at E12.5, indicating that loss of Isl1 after second heart field differentiation did not lead to rapid embryonic demise. SAN Hcn4-GFP expression was reduced but readily detectable in Cre+ embryos (Figure 3B).
Figure 3. SAN Transcriptome After Isl1 Deletion.
(A) Strategy to obtain RNA from SAN tissue lacking Isl1. (B). Top row: Immunohistochemistry of E12.5 SAN in Cre− embryos, with intact Isl1 expression (left) in comparison to Cre+ embryos (right) which lack Isl1. DAPI staining indicates nuclei. Bottom row: adjacent sections from Cre+ E12.5 SAN lacking Isl1. Left, uncut, and right, after laser capture microdissection. Scalebars = 100 μM (C) Venn diagrams showing statistically significant overlap of genes enriched in SAN (top) or RA (bottom) at E14.5 with genes that were downregulated (top) or upregulated (bottom) after Isl1 deletion. (D) Scatterplot showing that Isl1 results in downregulation of SAN-enriched genes and upregulation of RA-enriched genes. Abbreviations: rcv, right cardinal vein; ra, right atrium; NS, non-significant.
We then performed LCM on Cre+ and Cre− embryonic SAN at E12.5. Each sample was pooled from approximately 9 SAN sections per embryo, and we used 7 different embryos (3 for Cre+ group and 4 for Cre− group, Figure 3B). Quantitative PCR performed on amplified cDNA prior to RNA-Seq library preparation could not detect Isl1 transcript in the samples from Cre+ embryos, a finding confirmed by the absence of RNA-Seq reads mapped to the floxed portion of Isl1 (Online Figure IV).
SAN transcriptome in the absence of Isl1
590 genes exhibited DE between Cre+ and Cre− RNA samples, of which 65% were downregulated in SAN tissue lacking Isl1 (Figure 3C, Online Table VI). Among the genes enriched in SAN tissue vs. RA at E14.5, 18% were downregulated after deletion of Isl1 (odds ratio (OR) = 14, p=1.1 × 10−88, compared to non-DE genes), indicating a positive role for Isl1 in the PC-specific gene expression program. Of genes with established roles in SAN development and PC function, Tbx3, Hcn4, Hcn1, and Cacna1g were all significantly decreased after deletion of Isl1, while expression of other cardiac and SAN TFs including Nkx2.5, Gata4, Mef2C, Tbx5, and Tbx18 were not significantly changed (Figure 3D). Conversely, 9% of RA-enriched genes (including Nppa, Gja1, Gja5, and Scn5a) were upregulated in SAN after loss of Isl1 (OR = 10, p=9.6 × 10−35), suggesting a secondary role for Isl1 in repressing a differentiation program associated with working myocardium.
Network and motif analysis on dysregulated genes
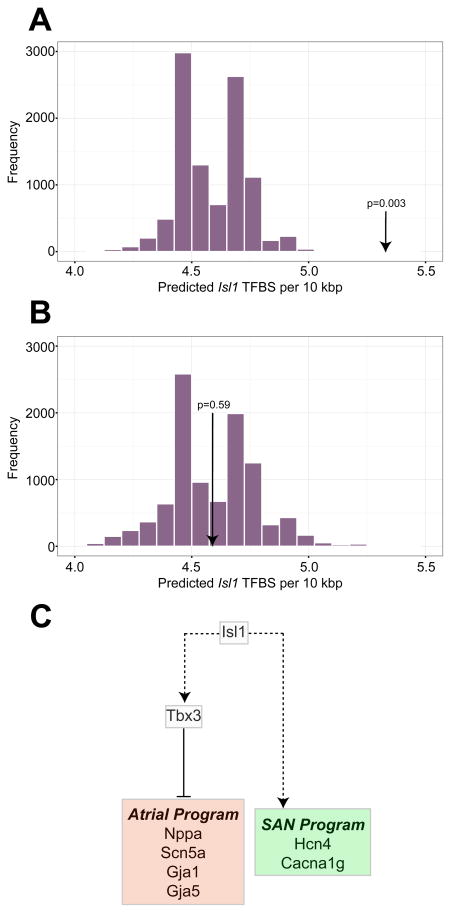
Assessment of individual network activity after loss of Isl1 function showed that M7 (containing Hcn4 and Tbx3), followed by M1 (containing Tbx18, Shox2 and Isl1), were the most reduced in overall activity (Online Figure VI). Conversely, the only modules that exhibited increased activity after loss of Isl1, M3 and M9, were associated with increased activity in RA. To relate these global changes in expression to loss of Isl1 function, we searched for Isl1 binding sites within introns, untranslated regions, and 10 kb upstream of the transcriptional start sites of genes that were downregulated after loss of Isl1, and compared the frequency of occurrence of these sites to genes that were unaffected by Isl1 deletion. This analysis showed highly significant enrichment (p = 0.0003) of Isl1 binding sites among the genes downregulated after Isl1 deletion, suggesting a role for Isl1 as a direct transcriptional activator in the developing SAN (Figure 4A, Online Figure VI). Examples of putative Isl1 binding sites in Tbx3 and Hcn4 regulatory regions are shown in Online Table VII. Because of the limiting number of pacemaker cells in each embryonic heart, it was not possible to test these sites directly using chromatin immunoprecipitation. Conversely, we did not observe enrichment of Isl1 binding sites near the genes upregulated after Isl1 deletion, suggesting that Isl1 may not function primarily as a direct transcriptional repressor of these genes, but may suppress the atrial gene expression program indirectly (Figure 4B, Online Figure V).
Figure 4. Inferring Isl1 Function in Cardiac Pacemaker Cells.
(A) For motif analysis on Isl1 binding sites, regions up to 10kb upstream of the transcriptional start site, introns, and untranslated regions for genes downregulated after Isl1 deletion were included in the query set. A null set was defined as genes that were not differentially expressed. The histogram shows a distribution of medians of 10,000 samples drawn from the null set by bootstrap sampling. The arrow denotes the observed frequency of Isl1 binding sites among genes downregulated after Isl1 deletion. (B) Similar motif analysis to (A), where the arrow denotes the observed frequency of Isl1 binding sites at genes upregulated after loss of Isl1. (C) A model for Isl1 transcriptional activity in the SAN. A solid line indicates a direct transcriptional relation; dashed lines indicate hierarchical relationships based on expression analysis only (may be direct or indirect).
Sensitivity analysis
Because inadvertent contamination of a Cre+ SAN sample with non-PC myocytes would bias our analysis towards the observed effects, we assessed our SAN samples for the presence of outliers. We found that one of the three Cre+ samples consistently exhibited more dramatic changes in gene expression at more diverse loci than the other two samples, raising the possibility of either contamination by non-PC myocytes or simply biological variation. To insure the robustness of our findings, we repeated our differential expression analysis and motif analysis after excluding this outlier sample and observed no significant changes in our results (Online Table VII, Online Figure VI).
DISCUSSION
Here, we present a comprehensive transcriptional analysis of mammalian PCs by deep sequencing SAN tissue obtained with LCM. While PCs and RA myocytes share core transcriptional machinery, mature PCs have adopted a distinct transcriptional program, consistent with morphological and electrical phenotypes exhibited by PCs. Our dataset highlights dynamic changes in PC gene expression associated with differentiation and maturation, and reveals a central role of Isl1 in establishing the PC gene program.
Establishment and utility of SAN-specific gene networks
Knowledge of the SAN transcriptome is a prerequisite to understanding the unique properties of this specialized cell type and its disruption in the setting of disease. In addition, the description of the major regulators and networks activated and repressed in the SAN provides a foundation for studies aimed at regulating SAN cell fate. Because of the difficulty of isolating pure PC RNA, there has been no dataset to use for comparison of putatively reprogrammed PCs with endogenous PCs. Because some non-PC cardiomyocytes can exhibit automaticity with PC-like action potentials,12, 13 and many SAN-associated genes such as Hcn4 exhibit dynamic expression outside the SAN,14 it is ideal to have a characteristic transcriptome as a means to determine the degree of fidelity during reprogramming.15
Our SAN expression profile can provide such a benchmark for investigators attempting to derive PCs from non-PCs and will permit more precise characterization of putatively reprogrammed cells. Analysis of specific gene sets with discordant expression between bona fide PCs and reprogrammed PCs may lead to targeted refinements of current methods. In addition, by quantifying changes in SAN gene expression during development, our dataset may suggest ways to promote maturation of incompletely reprogrammed cells.
Role of Isl1 in establishing the SAN gene network
Isl1 is required for second heart field development in mice16 and for SAN development in zebrafish.6 Although Isl1 expression and function in mouse PCs has been described previously,7, 17 its transcriptional role in the SAN remains unknown. Our loss of function data demonstrated that SAN-enriched genes were strongly overrepresented among the genes downregulated after Isl1 disruption, suggesting that Isl1 is required to generate the SAN-specific gene expression program. Furthermore, we observed significant enrichment of Isl1 binding sites within and around downregulated genes, providing strong evidence that Isl1 is a direct transcriptional activator of gene expression in PCs.
We also observed upregulation of genes associated with RA myocardium in the SAN after deletion of Isl1, including Nppa, Scn5a, Gja1, and Gja5. However, we did not observe enrichment of Isl1 binding sites near the upregulated genes, suggesting that Isl1 functions indirectly to block the RA myocyte gene expression in the SAN. Tbx3 is an established negative regulator of the atrial myocyte gene expression program4 and was found in our analysis to be highly downregulated after deletion of Isl1. One possibility is that upregulation of RA genes after Isl1 deletion is mediated in part by downregulation of Tbx3 (Figure 4C). This model situates Isl1 as a key transcriptional activator in PCs with a major role in establishing the SAN-specific gene expression program. As the scope of the present study is limited to expression analysis, further work will be required to delineate the functional and developmental roles of Isl1 in sinoatrial node, and to validate putative direct transcriptional targets. In addition, the potential of Isl1 to augment PC derivation or reprogramming has yet to be fully explored. Our analysis suggests that multifactor reprogramming using Isl1 in combination with other cardiac and/or SAN transcription factors may be a fruitful approach.
Novelty and Significance.
What Is Known?
Cardiac pacemaker cells (PCs) in the sinoatrial node (SAN) have a distinct gene expression program that underlies their unique functional properties, but are difficult to isolate in sufficient quantity and with adequate purity required for expression profiling.
Non-PC myocytes can be reprogrammed to exhibit PC-like behavior using SAN transcription factors, with varying efficiency and uncertain fidelity.
What New Information Does This Article Contribute?
Pure SAN tissue was isolated at mid-development, the perinatal period, and after maturation for comprehensive expression profiling using RNA sequencing.
Differential expression analysis of SAN tissue and right atrial tissue uncovered numerous SAN-enriched genes not previously associated with PCs.
RNA sequencing of SAN tissue in the absence of the transcription factor Islet-1 (Isl1) showed that Isl1 plays an important role in establishing the PC gene expression program during heart development.
The regeneration of sinoatrial node (SAN) or the creation of biological pacemakers using cellular reprogramming are promising new approaches to the treatment of sinus node disease (SND). Further development of these therapies will require a detailed understanding of transcriptional networks in specialized cardiac pacemaker cells (PC). However, these cells are difficult to characterize because they are few in number, they lack of specific molecular markers, and they are interspersed among non-PC cells. In this study, we used laser capture microdissection to isolate purer populations of PCs from embryonic, perinatal, and postnatal SAN for RNA sequencing. Analysis of RNA sequencing data uncovered numerous genes expressed in SAN that were not previously associated with PCs. Based on high differential expression, high absolute expression, and a weighted gene correlation network analysis, the transcription factor Isl1 was predicted to regulate PC-specific gene expression. RNA-Seq on SAN tissue lacking Isl1 confirmed this prediction and established Isl1 as an important transcriptional regulator within the developing SAN. The results of this study could from the basis of further mechanistic investigations into the gene regulatory networks active in the SAN and future translational work on SND.
Acknowledgments
We thank the Gladstone Genomics Core, the UCSF Center for Advanced Technology, and the Gladstone Bioinformatics Core for assistance with RNA library preparation, RNA sequencing, and initial sequence analysis, respectively. LCM was performed at the UCSF Laboratory for Cell Analysis.
SOURCES OF FUNDING
The authors were funded by the NIH/NHLBI (K08 HL101989 to V.V.; K08 HL093861, DP2 HL123228, and U01 HL107440-03 to R.C.D.; R01 HL057181, P01 HL089707 U01 HL100406, and U01 HL098179 to D.S.), the AHA (14BGIA20490269 to V.V.), the Younger Family Foundation, the Roddenberry Foundation, and the California Institute for Regenerative Medicine (to D.S.).
Nonstandard Abbreviations and Acronyms
- SAN
sinoatrial node
- RA
right atrium
- PC
pacemaker cell
- LCM
laser capture micro-dissection
- GFP
green fluorescent protein
- M
gene expression module
- Cre
Cre Recombinase
- DE
Differential expression
- GO
Gene ontology
Footnotes
DISCLOSURES
None.
References
- 1.Qian L, Srivastava D. Direct cardiac reprogramming: from developmental biology to cardiac regeneration. Circ Res. 2013;113:915–921. doi: 10.1161/CIRCRESAHA.112.300625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takahashi K. Cellular reprogramming. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a018606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiese C, Grieskamp T, Airik R, Mommersteeg MT, Gardiwal A, de Gier-de Vries C, Schuster-Gossler K, Moorman AF, Kispert A, Christoffels VM. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ Res. 2009;104:388–397. doi: 10.1161/CIRCRESAHA.108.187062. [DOI] [PubMed] [Google Scholar]
- 4.Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LY, Bakker ML, Clout DE, Wakker V, Barnett P, Ravesloot JH, Moorman AF, Verheijck EE, Christoffels VM. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaschke RJ, Hahurij ND, Kuijper S, Just S, Wisse LJ, Deissler K, Maxelon T, Anastassiadis K, Spitzer J, Hardt SE, Scholer H, Feitsma H, Rottbauer W, Blum M, Meijlink F, Rappold G, Gittenberger-de Groot AC. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation. 2007;115:1830–1838. doi: 10.1161/CIRCULATIONAHA.106.637819. [DOI] [PubMed] [Google Scholar]
- 6.Tessadori F, van Weerd JH, Burkhard SB, Verkerk AO, de Pater E, Boukens BJ, Vink A, Christoffels VM, Bakkers J. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE. 2012;7:e47644. doi: 10.1371/journal.pone.0047644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weinberger F, Mehrkens D, Friedrich FW, Stubbendorff M, Hua X, Muller JC, Schrepfer S, Evans SM, Carrier L, Eschenhagen T. Localization of Islet-1-positive cells in the healthy and infarcted adult murine heart. Circ Res. 2012;110:1303–1310. doi: 10.1161/CIRCRESAHA.111.259630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bakker ML, Boink GJ, Boukens BJ, Verkerk AO, van den Boogaard M, den Haan AD, Hoogaars WM, Buermans HP, de Bakker JM, Seppen J, Tan HL, Moorman AF, t Hoen PA, Christoffels VM. T-box transcription factor TBX3 reprogrammes mature cardiac myocytes into pacemaker-like cells. Cardiovasc Res. 2012;94:439–449. doi: 10.1093/cvr/cvs120. [DOI] [PubMed] [Google Scholar]
- 9.Kapoor N, Liang W, Marban E, Cho HC. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat Biotechnol. 2013;31:54–62. doi: 10.1038/nbt.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu YF, Dawkins JF, Cho HC, Marban E, Cingolani E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci Transl Med. 2014;6:245ra294. doi: 10.1126/scitranslmed.3008681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nam YJ, Lubczyk C, Bhakta M, Zang T, Fernandez-Perez A, McAnally J, Bassel-Duby R, Olson EN, Munshi NV. Induction of diverse cardiac cell types by reprogramming fibroblasts with cardiac transcription factors. Development. 2014;141:4267–4278. doi: 10.1242/dev.114025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yasui K, Liu W, Opthof T, Kada K, Lee JK, Kamiya K, Kodama I. I(f) current and spontaneous activity in mouse embryonic ventricular myocytes. Circ Res. 2001;88:536–542. doi: 10.1161/01.res.88.5.536. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Q, Timofeyev V, Qiu H, Lu L, Li N, Singapuri A, Torado CL, Shin HS, Chiamvimonvat N. Expression and roles of Cav1.3 (alpha1D) L-type Ca(2)+ channel in atrioventricular node automaticity. J Mol Cell Cardiol. 2011;50:194–202. doi: 10.1016/j.yjmcc.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang X, Wang G, Lin L, Lowe J, Zhang Q, Bu L, Chen Y, Chen J, Sun Y, Evans SM. HCN4 dynamically marks the first heart field and conduction system precursors. Circ Res. 2013;113:399–407. doi: 10.1161/CIRCRESAHA.113.301588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morris SA, Cahan P, Li H, Zhao AM, San Roman AK, Shivdasani RA, Collins JJ, Daley GQ. Dissecting engineered cell types and enhancing cell fate conversion via CellNet. Cell. 2014;158:889–902. doi: 10.1016/j.cell.2014.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cai CL, Liang X, Shi Y, Chu PH, Pfaff SL, Chen J, Evans S. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev Cell. 2003;5:877–889. doi: 10.1016/s1534-5807(03)00363-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann S, Berger IM, Glaser A, Bacon C, Li L, Gretz N, Steinbeisser H, Rottbauer W, Just S, Rappold G. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res Cardiol. 2013;108:339. doi: 10.1007/s00395-013-0339-z. [DOI] [PMC free article] [PubMed] [Google Scholar]