For the better part of a century, extensive research has established that retinoic acid (RA), the major metabolic derivative of vitamin A, plays essential roles in early embryonic patterning and organogenesis in vertebrates. During vertebrate development, the levels of embryonic RA need to be tightly regulated, as congenital malformations in many organs result from excessive or decreased embryonic RA levels. Many recent reviews have extensively covered RA signaling components and their functions during development (Bastien and Rochette-Egly, 2004; Diez del Corral and Storey, 2004; Mark et al., 2006; Pan and Baker, 2007; Duester, 2008; Niederreither and Dolle, 2008; Pares et al., 2008; Mark et al., 2009; Pennimpede et al., 2010; Clagett-Dame and Knutson, 2011; Rhinn and Dolle, 2012; Samarut and Rochette-Egly, 2012; Schilling et al., 2012; Duester, 2013; Kedishvili, 2013; Das et al., 2014; Samarut et al., 2014). Therefore, the focus of this commentary is to highlight the importance of conserved canonical RA signaling feedback mechanisms to heart development and how new insights into improper RA signaling feedback may be cause for re-evaluation of congenital malformations due to aberrant canonical RA signaling. Although numerous recent studies have been insightful in elucidating non-canonical modes of RA signaling (reviewed in Al Tanoury et al., 2013), how these interesting modes of RA signaling impact heart development is currently not clear and is beyond the scope of this commentary. Here, we will first briefly review canonical RA signaling components and the progression of hypotheses regarding the role of RA signaling in early patterning of the vertebrate cardiac progenitors. We will then discuss the points of RA signaling feedback within the pathway and the consequences of improper feedback mechanisms to our understanding of normal development.
Components of the canonical RA signaling pathway
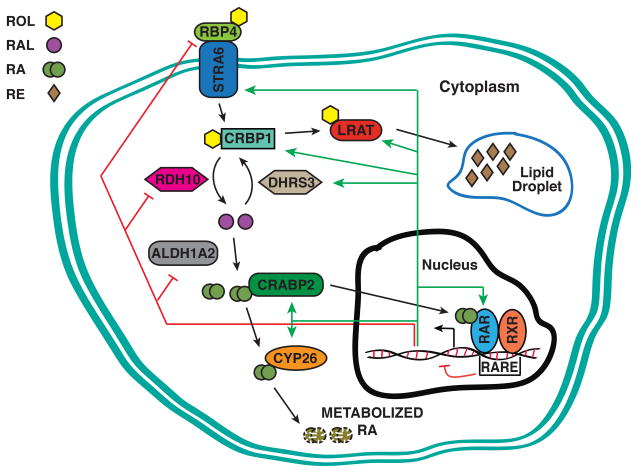
RA is the most active metabolic product of vitamin A. Vitamin A/retinol is provided in two dietary forms: preformed vitamin A, usually found in animal products, and carotenoids, the vitamin A precursor that is found primarily in plants (Bendich and Langseth, 1989). In amniotes, retinol is supplied from the mother through the circulation (D’Ambrosio et al., 2011). Retinol is stored in the liver through esterification by lecithin retinol acyltransferase (LRAT) (O’Byrne et al., 2005; Isken et al., 2007; Isken et al., 2008; Kim et al., 2008; Amengual et al., 2012), transported through the blood by Retinol binding protein 4 (RBP4) (Quadro et al., 2005; Isken et al., 2008; Amengual et al., 2014), and taken up into cells via Stimulated by retinoic acid-6 (STRA6) receptor (Fig. 1) (Blaner, 2007; Kawaguchi et al., 2007; Isken et al., 2008; Berry et al., 2013; Amengual et al., 2014). (NOTE: When referring to conserved genes and proteins from multiple species, we will follow the human nomenclature rules. However, when we refer to experiments from specific species, we will use the nomenclature rules for that organism.) In anamniotes, retinol is supplied to the embryo through the yolk (Albalat, 2009). In zebrafish lratb, stra6, and rbp4 and rbp4l are all expressed in the yolk syncytial layer and have specific expression within the early embryo (Isken et al., 2007; Isken et al., 2008; zfin.org). Despite the different maternal sources of retinol in the embryos, the retinol storage and production machinery is conserved in amniotes and anamniotes. Upon entering the cells of the target tissue cells, retinol is bound by cellular retinol-binding proteins (also referred to as RBPs, although we will use the more common acronym CRBPs to distinguish from the plasma RBP4) (Dolle et al., 1990; Ruberte et al., 1991; Ghyselinck et al., 1999). During development, retinol is oxidized into retinal primarily through microsomal short-chain dehydrogenases/reductases (SDRs) (Pares et al., 2008; Kumar et al., 2012; Kedishvili, 2013). Specifically, in vertebrates, a SDR retinol dehydrogenase 10 (RDH10) promotes retinal formation, while a related SDR, called DHRS3, converts retinal back to retinol for eventual storage (Fig. 1). Although it was previously thought that the production of retinal was essentially ubiquitous within the vertebrate embryo, we now understand that DHRS3 and RDH10 have specific embryonic expression patterns and are the major enzymes involved in embryonic retinol storage and retinal production (Sandell et al., 2007; Feng et al., 2010; Sandell et al., 2012; Billings et al., 2013). Following the production of retinal, it is then oxidized into RA by retinaldehyde dehydrogenases (ALDH1As; Fig. 1), members of the aldehyde dehydrogenase family of enzymes (Niederreither et al., 1997; Niederreither et al., 1999; Marchitti et al., 2008; Napoli, 2012). Upon the production of RA, it is bound by cellular retinoic acid binding proteins (CRABPs), which facilitate the distribution of RA to the retinoic acid receptors (RARs) and the RA degradation machinery (Dolle et al., 1990; Ruberte et al., 1991; Durand et al., 1992; Romand et al., 2000; Cai et al., 2012). Within the target tissue, RARs and retinoid X receptors (RXRs), members of the nuclear hormone receptor family, mediate the transcriptional actions of RA (Bastien and Rochette-Egly, 2004; Samarut and Rochette-Egly, 2012). RAR/RXR heterodimers bind to retinoic acid response elements (RAREs), which commonly consist of direct repeats of canonical 6 nucleotide motifs separated by 1, 2, or 5 nucleotides (Bastien and Rochette-Egly, 2004; Delacroix et al., 2010; Mahony et al., 2011; Samarut and Rochette-Egly, 2012). However, recent chromatin immunoprecipitation-sequencing (ChIP-seq) data suggests that there is likely considerably more flexibility of RAR/RXRs, which bind to a much greater range of RARE motifs than previously appreciated (Moutier et al., 2012). It is generally thought that upon binding of RA to RARs, the RAR/RXR heterodimers shed transcriptional repressive machinery and recruit transcriptional activation machinery to active target gene expression (Bastien and Rochette-Egly, 2004; Samarut and Rochette-Egly, 2012). Members of the cytochrome P450 superfamily (CYP26s) oxidize RA, which facilitates its degradation (Fig. 1) (Abu-Abed et al., 1998; Abu-Abed et al., 2001; Sakai et al., 2001; Niederreither et al., 2002; Pennimpede et al., 2010).
Figure 1. Schematic of feedback in the RA signaling pathway.
Retinol (ROL), bound to RBP4 binds to its receptor STRA6 in the cell membrane. Upon transport into the cell, CRBP1 binds ROL and delivers it to LRAT for storage as Retinyl esters (RE) or to RDH10, which oxides it to retinal (RAL). ALDH1A oxidizes RAL to RA or RAL can be reduced back to ROL via DHRS3 for storage. Upon conversion to RA, it binds to CRABP2, which can distribute RA to RARs or CYP26 enzymes for oxidation and degradation. Green arrows indicate where positive transcriptional feedback has been observed, while red bars indicate where negative transcriptional feedback has been observed. Although we depict that both positive and negative feedback is initiated through transcriptional activation, this is only a hypothesis. One mechanism of negative feedback could be through ligand-induced negative feedback at RAREs, indicated by the red bar emanating from the RARE. Overall, there is only limited data supporting that direct activation of the positively regulated RA signaling pathway genes is through RAREs in their promoters, while there is currently no data describing how the negatively regulated RA signaling pathway genes are suppressed.
History of RA in heart development
In humans, excess embryonic RA, from hypervitaminosis A and RA used in therapies of mothers (Lammer et al., 1985; Rizzo et al., 1991; Pan and Baker, 2007), can result in RA embryopathy, while RA deficiency can result from dietary maternal vitamin A deficiency, which is still common in developing countries (West and Mehra, 2010; Christian et al., 2013). Both excess and deficiency of RA result in a spectrum of development malformations in humans, including congenital heart defects. Animal models have demonstrated that the spectrum of congenital defects caused by inappropriate RA signaling is similar in all vertebrates (Heine et al., 1985; Stainier and Fishman, 1992; Lohnes et al., 1993; Lohnes et al., 1994; Mendelsohn et al., 1994; Yutzey et al., 1994; Niederreither et al., 1999; Hernandez et al., 2004; Emoto et al., 2005; Hernandez et al., 2007; Linville et al., 2009; Waxman and Yelon, 2009). Proper RA signaling is required for multiple aspects of vertebrate heart development, in particular the establishment of the cardiac progenitor fields (Pan and Baker, 2007; Duester, 2008; Niederreither and Dolle, 2008; Liu and Stainier, 2012; Schilling et al., 2012). Our understanding of how RA signaling affects early cardiac patterning has evolved considerably since the first studies to recognize the importance of proper RA signaling in normal heart development in the 1940s and ‘50s by Wilson and Warkany. These pioneering studies found that vitamin A deficient (VAD) rat embryos, achieved through limiting the maternal dietary supply of vitamin A, had pleiotropic developmental malformations, including ventricular outflow tract and aortic arch defects (Wilson and Warkany, 1949; Wilson and Warkany, 1950a; Wilson and Warkany, 1950b). Some time later, similar and actually more severe cardiac defects were found in VAD quail embryos, which had cardia bifida and hearts with a single enlarged ventricular chamber (Heine et al., 1985). Shortly after Warkany et al.’s initial studies, they also demonstrated that hypervitaminosis A, caused from excess dietary vitamin A, caused outflow tract and septal defects in mice (Kalter and Warkany, 1961), which are also found from hypervitaminosis A and RA embryopathy in humans (Pan and Baker, 2007).
While the experiments modulating vitamin A illustrated the importance of appropriate embryonic RA levels to heart development, it was an understanding that RA signaling is necessary and sufficient for proper anterior-posterior (A-P) patterning of the vertebrate embryo (Durston et al., 1989; Sive et al., 1990) that led to the hypothesis that the earliest requirement for RA signaling in heart development is to promote atrial cell identity at the expense of ventricular cell identity. The ventricular-atrial patterning hypothesis was based on the proposed relative anterior-posterior (A-P) positions, respectively, of the ventricular and atrial progenitors and the interpretations of some of the first experiments performed in chick and zebrafish embryos using exogenous RA (Osmond et al., 1991; Stainier and Fishman, 1992; Yutzey et al., 1994). Exogenous treatment of chick embryos with RA produces a variety of concentration dependent malformations, including smaller ventricles, enlarged atria, and expansion of cells expressing atrial myosin heavy chain (Osmond et al., 1991; Yutzey et al., 1994). Similarly, zebrafish treated with RA were reported to have smaller ventricles (Stainier and Fishman, 1992). Furthermore, the ventricular-atrial hypothesis gained greater traction due to the location of ALDH1A2 expression, the major embryonic RA producing enzyme, being posterior and adjacent to the cardiac progenitors in mouse and chick embryos and loss of function experiments using an RAR antagonist to inhibit RA signaling in chick embryos, which indicated that these embryos overtly had larger ventricles with smaller atria (Hochgreb et al., 2003).
Although the A-P model of atrio-ventricular patterning was initially an attractive model, the interpretations were based largely on cardiac chamber morphology, which is not a reliable indicator of cardiac progenitor specification and cardiomyocyte number in any model. Furthermore, the A-P model of cardiac patterning was derived prior to our understanding of the differential temporal contributions of progenitors from the first heart field (FHF) and second heart field (SHF), the availability of numerous tools and markers to assess chamber progenitor number, and a better understanding of the location of the atrial and ventricular progenitors within the anterior lateral plate mesoderm (ALPM). The first indication that RA signaling may not promote atrial specification at the expense of ventricular specification, despite its established role in A-P patterning, came from studies in zebrafish. A series of studies from the Yelon lab revealed, first, that RA signaling is required to limit the size of the cardiac progenitor field (Keegan et al., 2005) and, second, that RA signaling limits the specification of both atrial and ventricular cells (Waxman et al., 2008). In contrast to the interpretations of the previous studies, which was primarily based on morphological analysis, these experiments combined morphological analysis, counting individual cardiomyocytes using transgenic zebrafish embryos, and lineage tracing of the ALPM, which together revealed that RA signaling is required to limit the posterior boundary of both cardiac progenitors within the ALPM (Keegan et al., 2005; Waxman et al., 2008). Subsequent studies in Xenopus and mouse revealed similar observations. In Xenopus, RA signaling restricts cardiac specification, although the effects on chamber cell number were not examined (Collop et al., 2006). Analysis of Aldh1a2 knockout (KO) mice also revealed that RA signaling restricts the posterior limit of cardiac progenitors (Ryckebusch et al., 2008; Sirbu et al., 2008).
A second indication that RA signaling may not promote atrial specification at the expense of ventricular specification has come from gain of function experiments, also from the Yelon lab in zebrafish (Waxman and Yelon, 2009). Treating zebrafish embryos with different concentrations of RA, which cause various degrees of posteriorization, combined with counting the cardiomyocytes and chamber marker analysis indicated that there are dramatically different effects on cardiac progenitor specification that occur with increasing levels of embryonic RA (Waxman and Yelon, 2009). Specifically, treatment with high concentrations of RA, which strongly posteriorize embryos, can eliminate all cardiomyocyte specification. Importantly, RA treatment can eliminate cardiomyocytes in chicken embryos (Osmond et al., 1991; Yutzey et al., 1994), suggesting this effect on cardiomyocyte specification with high concentrations of RA is likely conserved in vertebrates. However, intermediate increases in RA, which retain cardiac specification, can produce independent effects on atrial and ventricular specification. Specifically, concentrations of RA causing a higher degree of posteriorization reduce ventricular specification without promoting atrial specification, while concentrations of RA causing a comparatively lower degree of posteriorization can promote atrial specification without affecting ventricular specification. Interestingly, more recently we found that treatment with very low increases in RA that cause a minimal degree of posteriorization can actually promote atrial and ventricular specification (D’Aniello et al., 2013). How might these different increases in embryonic RA cause such a great variety of different cardiac phenotypes? Given the correlation between the RA-induced degree of posteriorization and observed cardiac defects, we hypothesize that the differential effects on atrial and ventricular progenitor specification are due to a progressive flattening of the RA signaling gradient. Similar non-intuitive, differential effects on progenitor territories are observed with the modulation of other morphogens in early development (Dorsky et al., 2003; Tucker et al., 2008).
A final indicator that RA may not promote atrial specification at the expense of ventricular specification is that fate maps have revealed there is not a relative anterior-posterior relationship of the ventricular and atrial progenitors. In zebrafish embryos, lineage tracing indicated that within the blastula and the anterior lateral plate mesoderm (ALPM) the atrial and ventricular progenitors lie in medio-lateral orientation relative to each other (Keegan et al., 2004; Schoenebeck et al., 2007). Furthermore, a more medio-lateral orientation of ventricular and atrial progenitors has also been reported in a newer fate map of chick embryos that incorporate the SHF, the later differentiating cardiomyocytes (Abu-Issa and Kirby, 2008). Altogether, detailed cardiomyocyte counting, analysis of additional markers, and lineage tracing experiments, support a hypothesis that at the extremes of excess and deficiency of RA signaling it is necessary and sufficient for restricting both atrial and ventricular specification in vertebrates. Importantly, intermediate increases in RA signaling, through inducing varying degrees of posteriorization, can differentially and independently affect atrial and ventricular specification.
Requirements of RA pathway machinery on heart development
Within the vertebrate embryo, it is clear that an appropriate concentration of RA is critical during early heart development. Therefore, it is not surprising that animals with mutations, or deficiency, in many of the genes that control embryonic RA levels and signaling result in cardiac defects (Table 1). Beginning with the uptake of retinol in the embryo, mutations in STRA6 underlie Matthew-Wood syndrome (MWS) in humans, which includes a spectrum of congenital defects, including outflow tract, atrial and ventricular septal defects (Golzio et al., 2007; Pasutto et al., 2007). While mouse Stra6 KOs only have mild ocular defects (Amengual et al., 2014), Stra6 deficient zebrafish embryos have complex phenotypes indicative of decreases and increases in RA signaling (Isken et al., 2008). However, although the effects on heart development have not been analyzed in detail, overtly the cardiac phenotypes are reminiscent of increased RA signaling in zebrafish (Stainier and Fishman, 1992; Waxman and Yelon, 2009). Similarly, Lratb deficient zebrafish embryos have elevated embryonic retinoic acid and phenotypes consistent with increased RA signaling (Isken et al., 2007), although analysis of the hearts was not performed. While Lrat and Rbp4 mouse KO models do not have overt developmental defects (Quadro et al., 1999; Batten et al., 2004), interestingly Rbp4 KO and double Lrat; Rbp4 KO mutants are sensitive to modulation of vitamin A in their diets (Quadro et al., 2005; Kim et al., 2008), suggesting these proteins are necessary to help buffer RA signaling levels during development, which is discussed more below. However, Rbp4 KO mice do have subtle, transient cardiac developmental defects that include precocious cardiomyocyte differentiation and extracellular matrix deposition (Wendler et al., 2003). Of the enzymes required for retinol uptake and storage, DHRS3 seems to be the most important for development. Both Dhrs3 KO mice and Dhrs3a deficient zebrafish embryos have elevated levels of RA (Feng et al., 2010; Billings et al., 2013), while the Dhrs3 KO mice were also shown to have decreased retinyl esters, supporting their role in promoting RA storage. Importantly, Dhrs3 KO mice have ouflow tract and septal defects, reminiscent of RA treatment (Billings et al., 2013). Heart development was not examined in Dhrs3a deficient zebrafish embryos (Feng et al., 2010). However, compared to the Dhrs3 null mice, the overt phenotypes are not as strong, which could be due to redundancy with its paralog Dhrs3b (ZDB-GENE-041010-172; zfin.org).
Table 1.
| Affected Genes | Species | Heart Phenotypes | References |
|---|---|---|---|
| STRA6 | Human | Matthew-Wood sindrome: outflow tract-atrial and ventricular septal defects | (Golzio et al., 2007; Pasutto et al., 2007) |
| Zebrafish | Aberrant heart-reminiscent of increased RA signaling | (Isken et al., 2008) | |
| DHRS3 | Mouse | Outflow tract and septal defects | (Billings et al., 2013) |
| RDH10 | Mouse | Looping defects, enlarged hearts, and edema | (Sandell et al., 2012) |
| ALDH1A2 | Mouse | Looping defects, enlarged hearts, edema, increased cardiomyocyte specification | (Niederreither et al., 1999; Ryckebusch et al., 2008; Sirbu et al., 2008) |
| Zebrafish | Enlarged hearts due to increased cardiomyocyte specification | (Keegan et al., 2005) | |
| CYP26A1 | Human | DiGeorge and Klippel-feil; Outflow tract defects | (Pennimpede et al., 2010) |
| Mouse | Gross aberrant cardiac defects including looping defects | (Abu et al., 2001; Emoto et al., 2005; Ribes et al., 2007) | |
| CYP26A1; CYP26C1 | Mouse | Gross aberrant cardiac defects including looping defects | (Uehara et al., 2007) |
| Zebrafish | Abnormal heart with increased atrial CMs | (Rydeen et al., 2014) | |
| RARa2 | Chicken | Defects in cardiac inflow tract | (Romeih et al., 2003) |
| RARγ | Chicken | Defects in asymmetric orientation and looping morphogenesis | (Romeih et al., 2003) |
| RARal; RARβ | Mouse | Outflow tract and aortic arch abnormalities; persistent truncus arteriousus; dextroposed aorta; high ventricular septal defects; abnormal aortic arch pattern. | (Mendelsohn et al., 1994; Li et al., 2010) |
| RARal; RARγ | |||
| RARab1 | Zebrafish | Enlarged hearts with increased cardiomyocyte specification | (D’Aniello et al.,2013) |
The strongest congenital heart defects are observed in mouse KOs for the major embryonic proteins that promote the production of RA: RDH10 and ALDH1A2. Rdh10 mouse mutants were initially identified from a mutagenesis screen by Sandell et al (2007). While subsequent Rdh10 KO analysis revealed that the severity of the phenotype can vary with the genetic background (Rhinn et al., 2011; Sandell et al., 2012; Chatzi et al., 2013), the most severe developmental defects in the Rdh10 KO mice overtly resemble Aldh1a2 KO mice, including linear, dilated and unlooped hearts (Sandell et al., 2012). Detailed analysis of Aldh1a2 mutants, as mentioned above, has helped to support the hypothesis that a posterior source of RA signaling is necessary to restrict cardiac progenitor specification in vertebrates (Ryckebusch et al., 2008; Sirbu et al., 2008). Mouse Aldh1a2 KOs and zebrafish alh1a2 mutants both have overtly enlarged hearts (Niederreither et al., 1999; Keegan et al., 2005). The enlarged hearts in zebrafish aldh1a2 mutants are due to a surplus of atrial and ventricular cardiomyocyte specification (Keegan et al., 2005; Waxman et al., 2008). For the mouse Aldh1a2 KOs, although the cardiac phenotypes were initially interpreted as supporting the atrial-ventricular patterning model from cardiac morphology, revisiting the analysis of the cardiac defects with additional cardiac progenitor markers revealed that these mutants also display a posterior expansion of the cardiac progenitors (Ryckebusch et al., 2008; Sirbu et al., 2008). Studies initially in zebrafish and later in mice have suggested that the posterior expansion of the cardiac progenitors is potentially at the expense of neighboring forelimbs progenitors (Waxman et al., 2008; Zhao et al., 2009; Sorrell and Waxman, 2011; Cunningham et al., 2013). Despite the genetic data supporting a conserved requirement for RA in restricting cardiomyocyte specification, there are differences in interpretation as to whether or not there is strictly an expansion of the FHF, the earlier differentiating population of cardiomyocytes, and/or the second SHF, a later differentiating population of cardiomyocytes (Ryckebusch et al., 2008; Sirbu et al., 2008).
The transcriptional output of RA is mediated through RARs. In tetrapods, there are three RARs - α, β, and γ (Lohnes et al., 1994; Mendelsohn et al., 1994; Bastien and Rochette-Egly, 2004), while in zebrafish, there are four RARs-two α (RARαa and RARαb) and two γs (RARγa and RARγb), but no RARβ (Waxman and Yelon, 2007; Linville et al., 2009). Although numerous isoforms have been identified for each receptor in mice (Bastien and Rochette-Egly, 2004), only two additional isoforms have been identified in zebrafish (D’Aniello et al., 2013). There is considerable redundancy with respect to receptor function in mice, as the single mutants do not have significant cardiac defects. However, double KOs of RARα1−/− with RARβ −/− or -γ−/− have outflow tract defects (Mendelsohn et al., 1994). More recent studies indicate the outflow tract defects are due to inappropriate SHF specification (Li et al., 2010). Even though the double KOs do recapitulate defects observed with VAD in humans (Mendelsohn et al., 1994), it is interesting that they do not recapitulate the severity of the enlarged hearts with looping defects found in Rdh10 and Aldh1a2 KOs (Niederreither et al., 1999; Sandell et al., 2012), which are indicative of strong early patterning defects. One explanation for the differences between these phenotypes likely reflects that RA signaling plays multiple roles during cardiogenesis, with the RAR KOs revealing later requirements of these receptors. In contrast to the redundancy revealed in mice, avian embryos appear to have distinct and non-redundant roles during cardiac development: RARγ specifically regulates the asymmetric orientation and the normal looping morphogenesis of the heart, while RARα2 regulates cardiac inflow tract formation (Romeih et al., 2003). Using zebrafish, we have recently found that depletion of zebrafish RARαb1 causes enlarged hearts with increased cardiomyocyte specification. Surprisingly, the enlarged hearts appear to be due to effects of excessive RA feedback (D’Aniello et al., 2013), which will be discussed below.
Significant work has examined the function CYP26 genes during development. In vertebrates, there are three CYP26 genes: CYP26a1, -b1, and-c1. CYP26A1 is the most prevalent RA degrading enzyme in the vertebrate embryo, expressed primarily in the anterior of the embryo during early development (Abu-Abed et al., 2001; Kudoh et al., 2002; Dobbs-McAuliffe et al., 2004; Emoto et al., 2005). In humans, loss of CYP26A1 is associated with DiGeorge and Klipplel-Feil syndromes (Pennimpede et al., 2010), which both include outflow tract defects. While loss of CYP26A1 alone in mice and zebrafish results in a spectrum of RA signaling defects indicative of increased RA signaling (Abu-Abed et al., 2001; Sakai et al., 2001; Niederreither et al., 2002; Emoto et al., 2005; Hernandez et al., 2007; Uehara et al., 2007), the defects are somewhat mild compared to treatment with exogenous RA, suggesting that increases in embryonic RA that can occur from failure to degrade endogenous sources of RA are somewhat minimal. However, Cyp26a1 KO mouse and cyp26a1 mutant zebrafish embryos are extremely hypersensitive to treatment with RA (Hernandez et al., 2007; Ribes et al., 2007), indicating they are necessary to buffer embryonic RA levels against excessive RA. With respect to the heart, Cyp26a1 KO mice have looping defects (Abu-Abed et al., 2001). There is functional redundancy in the embryonic requirements for the Cyp26 enzymes during early development. Although Cyp26c1 KO mice do not have significant defects, the double Cyp26a1 and Cyp26c1 mutants are more posteriorized than the Cyp26a1 KOs alone, with more severe cardiac looping defects (Uehara et al., 2007).
Because the mechanisms underlying the cardiac defects were not really understood in vertebrates, we recently examined the roles of Cyp26 enzymes in zebrafish (Rydeen and Waxman, 2014). We found that while there was not evidence for significant early cardiac patterning defects in zebrafish deficient Cyp26a1 or Cyp26c1 alone, depletion of Cyp26a1 and Cyp26c1 together promotes an increase in atrial specification, due to an anterior shift in the cardiac progenitor field within the ALPM. Importantly, lineage tracing and marker analysis of the ALPM in the Cyp26 deficient embryos revealed that the increase in atrial specification was at the expense of the more anterior endothelial progenitors, and not at the expense of the ventricular progenitors (Rydeen and Waxman, 2014). These results suggest that endogenous increases in RA signaling, due to Cyp26a1 and Cyp26c1 loss, cause remarkably similar cardiac specification defects to what we observed with low-intermediate levels of posteriorization from treatments with exogenous RA (Waxman and Yelon, 2009). Moreover, these results provide additional evidence that RA signaling does not promote an increase in atrial cells at the expense of ventricular cells.
Feedback on RA pathway machinery
Within the early embryo, RA functions as a morphogen with sources and sinks (White et al., 2007; Cai et al., 2012; Schilling et al., 2012). Like other morphogens, RA signaling incorporates feedback to ensure robustness of the A-P RA signaling gradient, which is necessary for proper embryonic patterning. One of the most intriguing aspects of RA signaling is the thoroughness by which it incorporates positive and negative regulation of the RA pathway machinery (Fig. 1). The manner by which RA signaling regulates its own production machinery is somewhat intuitive. Within the embryo, RA positively regulates the expression of enzymes involved in retinol storage or RA degradation, while it negatively regulates the expression enzymes involved in the production of RA (Fig. 1). Thus, excess RA promotes the expression of STRA6, LRAT, DHRS3, CRBP1, CRABP2, and CYP26A1, while repressing RBP4, RDH10 and ALDH1A2 expression (Fig. 1) (Smith et al., 1991; Chazaud et al., 1996; Niederreither et al., 1997; Begemann et al., 2001; Cerignoli et al., 2002; Emoto et al., 2005; Strate et al., 2009; Feng et al., 2010; Sandell et al., 2012; Billings et al., 2013; D’Aniello et al., 2013; Kashyap et al., 2013; Amengual et al., 2014). Deficiency in RA promotes the opposite responses in these genes. The regulation at the transcript level is a conserved aspect of RA signaling in vertebrates, as these feedback responses are found in zebrafish through mammals. Currently, we do not understand many aspects of the conserved feedback, including its evolutionary origin. From an evolutionary perspective, in silico genomic studies and functional studies in mollusks support the hypothesis that retinoic acid (RA) signaling is an ancient molecular pathway that predates the origins of the bilaterian body plan (Albalat, 2009; Andre et al., 2014). Therefore, it would be particularly interesting if the mechanisms underlying RA feedback regulation were ancient, predating the common bilaterian ancestor. While the intracellular production and degradation components are predicted to be conserved in protostomes and invertebrate deuterostomes (Albalat, 2009; Albalat and Canestro, 2009; Andre et al., 2014), if RA signaling regulates the expression of these genes in a manner similar to that found in vertebrates has not been examined. However, it is known that RA positively regulates Cyp26 expression in chordate Ciona, but does not significantly affect Aldh1a expression (Ishibashi et al., 2003; Ishibashi et al., 2005), suggesting that at least the positive regulation of Cyp26 by RA predates a common chordate ancestor.
Although the conserved feedback on production and degradation machinery in vertebrates is clear, the level and conservation of RA signaling feedback on RAR expression is still poorly understood. Recent studies have indicated that RA signaling promotes RAR and RXR expression in mouse and zebrafish embryos (Begemann et al., 2001; Feng et al., 2010; Kashyap et al., 2013). However, we currently do not understand the mechanisms underlying this transcriptional activation for the majority of these genes. Of course, the explosion of ChIP-seq technology and the ever increasing ability to efficiently analyze cis-regulatory modules at a systems level will enhance the ability to assess the directness of RA regulation of these genes. It has long been recognized that there are canonical RAREs in the promoters of multiple RARs, with the mouse RARβ2 being responsive to RA, suggesting it is a direct target (de The et al., 1990; Loudig et al., 2000). However, RA regulation of the RARβ2 isoforms is likely not highly conserved in vertebrates as zebrafish do not have an RARβ2 and RA signaling does not regulate the Ciona RAR (Ishibashi et al., 2003; Ishibashi et al., 2005; Waxman and Yelon, 2007). Despite it becoming apparent that RA signaling promotes the expression of its own receptors, regardless of the directness, the purpose of the positive regulation of RARs by RA signaling in development is also not understood and necessitates investigation.
Another level of RAR feedback in development is compensatory expression of RARs, which we hypothesize could be a relatively unappreciated feedback response to loss of RA signaling. However, initial reports of RAR KO mice suggested that when RARs are lost there was not increased expression of the remaining RARs (Lohnes et al., 1993; Lufkin et al., 1993). Contradictory to these early experiments, more recent reports in mouse and zebrafish found instead that when RARs are depleted, compensatory expression of the remaining RARs is initiated (Manshouri et al., 1997; D’Aniello et al., 2013). The different results in these studies may be due to experimental differences, depletion of the RARs vs. the KOs. However, given there are more sensitive assays available now to assess expression compared to what was available at the initial time analysis of the mouse RAR KOs, it is likely worthwhile revisiting if there is compensatory RAR expression in these models. Although we do not yet understand the mechanisms by which compensative RAR expression is achieved, it would be interesting if transcriptional de-repression is one way it is initiated. It has been proposed that active repression of RARs is required for some developmental processes (Koide et al., 2001; Weston et al., 2003; Janesick et al., 2014). Therefore, perhaps RAREs in the promoter of RARs that promote their expression are also used to facilitate de-repression of their expression when other RARs are depleted. Alternatively, the compensatory expression of RARs could be activated through RAREs bound by the remaining, but depleted RARs, as suggested by Manshouri et al. (1997).
Feedback through direct transcriptional activation
One of the mechanisms by which RA positively regulates pathway components during development is through direct transcriptional regulation. A number of the positively regulated RA signaling pathway components, in addition to the aforementioned RARs, have RAREs in their promoters. Pathway components that have RAREs in their promoters besides RARs, include CYP26A1, CRBP1, and CRABP2 (Smith et al., 1991; Durand et al., 1992; Loudig et al., 2000; Loudig et al., 2005; Hu et al., 2008; Li et al., 2012). Of these, the CYP26A1 promoter has been the best studied, in part because of its responsiveness to RA in many developmental contexts. The responsiveness of the CYP26A1 promoter to RA is mediated by multiple direct repeat 5 (DR5) RAREs in humans, mice, rats, and zebrafish, with at least two of them being almost absolutely conserved (Loudig et al., 2000; Loudig et al., 2005; Hu et al., 2008; Zhang et al., 2010; Li et al., 2012). In rats, mice, and zebrafish, the multiple RAREs in the promoters have been shown to interact to enhance the responsiveness to RA. Although the RAREs for CRBP1and CRABP2 have been shown to be responsive to RA in vitro (Smith et al., 1991; Durand et al., 1992), if these RAREs mediate the crabp2a responsiveness to RA in zebrafish embryos has not been shown (Cai et al., 2012). Furthermore, if RAREs mediate the conserved RA responsiveness of LRAT and DHRS3 during development has not been shown (Cerignoli et al., 2002; Feng et al., 2010; D’Aniello et al., 2013; Kashyap et al., 2013). In addition to promoting the storage and degradation of RA, RA represses the expression of the major embryonic production enzymes RDH10 and ALDH1A2 (Niederreither et al., 1997; Emoto et al., 2005; Strate et al., 2009; Feng et al., 2010; Sandell et al., 2012; D’Aniello et al., 2013). How this conserved negative response to RA takes places has not been investigated. Thus, despite some inroads, we currently do not have a thorough understanding of the transcriptional mechanisms underlying the conserved feedback responses to RA signaling during development. A direct transcriptional mechanism of regulation for all these genes, or at least the positively regulated RA responsive genes, would seem to be the most efficient feedback mechanism, and one that we hypothesize might be favored evolutionarily. We expect that direct transcriptional regulation would allow for the fastest response to fluctuations in RA signaling. An even more intriguing question is what are the transcriptional mechanisms of RA negatively regulated genes? As increased RA is typically associated with transcriptional activation, one hypothesis would be that the regulation of these genes would have to be indirect. However, a fascinating hypothesis is that these might also be direct targets and repression of expression would occur through ligand-induced transcriptional inhibition at RAREs (Hu et al., 2004; Epping et al., 2005; Kumar and Duester, 2014).
Buffering the RA signaling gradient
A combination of elegant mathematical modeling studies and in vivo analysis in zebrafish has illustrated that self-induced negative feedback is necessary for robustness of the RA morphogen gradient and helps explain how RA signaling can maintain an appropriate gradient even with >20 fold fluctuations in RA (White et al., 2007; Schilling et al., 2012). Specifically, in zebrafish, experimental work from the Schilling lab has demonstrated that Cyp26a1 and Crabp2a are necessary for the robustness of the signaling gradient (White et al., 2007; Cai et al., 2012). While extremely informative, the mathematical models have not yet incorporated the majority of RA responsive machinery. We speculate, however, that all the levels of RA positive and negative feedback contribute to the robustness of the RA signaling gradient in vertebrate embryos. Incorporating what is known about the sensitivity of mouse and zebrafish models to modulation of RA signaling, it could be already argued that these components are necessary for the robustness. For instance, the lack of overt developmental defects in Lrat and Rbp4 KO mice suggests that in vitamin A and RA sufficient conditions these enzymes are largely dispensable for embryonic development (Quadro et al., 1999; Batten et al., 2004). However, Rbp4 KOs alone or Lrat;Rbp4 double KOs are sensitive to modulation of vitamin A (Quadro et al., 2005; Kim et al., 2008), suggesting that they are indispensible for embryonic development in RA insufficient conditions. Specifically, when mothers of Rbp4 KO mice are kept on a vitamin A deficient diet, the Rbp4 KO embryos can resemble Aldh1a2 KO embryos (Quadro et al., 2005). Furthermore, double Lrat; Rbp4 KO mice are even more sensitive to vitamin A deficiency, resulting in more severely affected embryos compared to Rbp4 KO mice alone (Kim et al., 2008). The sensitively of Rbp4 KO mice to vitamin A deficiency, and similarity of these phenotypes in mice to individuals with MWS, has led to the hypothesis that the severity of MWS defects, which harbor mutations in STRA6, may be due to nutritional deficiencies of the mothers (Quadro et al., 2005; Amengual et al., 2014).
In contrast to the sensitivity of Rbp4 mutants to vitamin A deficiency, experiments demonstrate that CRABP2 and CYP26A1 enzymes act as buffers against increased embryonic RA signaling. Zebrafish Crabp2a deficient embryos have relatively minor hindbrain defects (Cai et al., 2012). However, increases and decreases in crabp2a expression, which transports RA to Cyp26a1 (Kedishvili, 2013), leads to hypersensitivity to RA treatment and less sensitivity to loss of RA signaling in zebrafish (Cai et al., 2012). Although CYP26A1 mutants in zebrafish and mice have comparatively stronger phenotypes than deficiency of the other aforementioned enzymes (Abu-Abed et al., 2001; Sakai et al., 2001; Hernandez et al., 2007; Ribes et al., 2007; Uehara et al., 2007), as mentioned above, the CYP26A1 mouse and zebrafish mutants are hypersensitive to RA treatment, with RA treatment using sub-optimal concentrations that do not affect wild-type animals causing strong posteriorization of CYP26A1 mutant embryos (Hernandez et al., 2007; Ribes et al., 2007). Therefore, while phenotypes in the RA signaling pathway may be relatively mild in vitamin A/RA sufficient embryonic contexts, these molecules are necessary to buffer against RA deficiency or excess.
Consequences of inappropriate RA feedback
Incorporating our understanding of the known RA feedback mechanisms and embryonic phenotypes, it would seem intuitive that deficiency or excess of RA would rapidly initiate feedback mechanisms that are necessary to restore a balance in RA signaling and a proper signaling gradient in early vertebrate development. However, one conundrum with our current understanding of defects from improper RA signaling is that excess or deficiency of RA signaling in embryos affects similar organs and can even cause overtly similar phenotypes. Multiple recent studies have found that inappropriate embryonic RA signaling can cause seemingly contradictory phenotypes indicative of both increases and decreases of RA signaling. For instance, increases of embryonic RA in mice can induce, paradoxically, longer-term induction of RA deficiency and perturb renal development (Lee et al., 2012). This study shows that after the initial RA treatment there is maintained deficiency of embryonic RA levels, which is correlated with decreased Aldh1a2 and Rdh10 and increased Cyp26a1 and Cyp26b1 expression (Lee et al., 2012). The renal defects can be partially rescued with RA treatment (Lee et al., 2012), supporting a hypothesis that the renal phenotypes are due to the suppression of embryonic RA after the initial excess of RA. In light of these results in mice, we have observed similar phenotypes in zebrafish using a hyper-active RAR, which induced both brain defects associated with increased RA signaling and loss of RA responsive gene expression, including hoxb5b and dhrs3a (Waxman and Yelon, 2011). However, we did not assess other components of the RA machinery or the effect on cardiogenesis. Therefore, although we currently do not understand if the subsequent loss of RA responsive targets in this context is due to inappropriate maintenance of depressed RA signaling as in mice, we hypothesize this is the case.
In contrast to RA-induced loss of RA signaling, we have also found evidence for the opposite: RA deficiency-induced gain of RA signaling. We found that depletion of RARαb1 caused larger hearts with increased cardiac specification, but this was unexpectedly induced from increased RA signaling (D’Aniello et al., 2013). Furthermore, we found that hox gene and cyp26a1 expression were increased in RARαb1 deficient embryos, while aldh1a2 was downregulated, supporting the hypothesis that an increase in RA was triggered in the RARαb1 depleted embryos (D’Aniello et al., 2013). Importantly, depletion of RARαb2, the second RARαb isoform in zebrafish, or concurrent depletion of RARαb1 and RARαb2 also resulted in increased RA signaling, suggesting this effect was not specific to RARαb1 (D’Aniello et al., 2013). Additionally, we found that the expression of the other zebrafish RARs was increased when the RARαb isoforms were depleted. We also found that treatment with low concentrations of RA were able to mimic this effect, supporting that low levels of RA can also promote increased cardiomyocyte specification (D’Aniello et al., 2013). Therefore, in contrast to increases in RA signaling, modest decreases in RA can lead to inappropriate positive feedback that results in teratogenic levels of RA signaling. Moreover, dramatic losses of RA signaling, as occur in Aldh1a2 KO mice (Niederreither et al., 1999), would not be predicted to elicit phenotypes due to inappropriate feedback, as the lack of RA would cause a breakdown of the feedback loop.
Why might gain or loss of RA signaling result in inappropriate signaling due to excessive feedback that causes congenital defects? We hypothesize that the RA feedback mechanism is designed to rapidly buffer embryonic RA levels within a certain range and length of time. Inappropriate increases or decreases in RA signaling outside of this range or sustained moderate increases or decreases may cause prolonged feedback that ultimately exceeds the intent of returning the RA signaling to a normal level. In light of these new revelations regarding the consequences of inappropriate RA signaling due to feedback and the variability of cardiac defects that can be induced by exogenous RA treatment, it may be worthwhile to revisit the cause of some RA induced congenital heart defects, as has been done with kidney development (Lee et al., 2012). Future studies elucidating the transcriptional mechanisms underlying RA signaling feedback and the scenarios where inappropriate RA signaling feedback occurs will enhance our understanding of the teratogenic mechanisms underlying RA induced congenital heart defects, as well as the broad spectrum of tissues affected by inappropriate RA signaling. Since RA is commonly used to treat acne and cancers (Soprano et al., 2004; Pan and Baker, 2007), a greater understanding of the mechanisms underlying inappropriate RA signaling feedback may lead to new, rather unexpected treatments for the prevention of congenital defects and unwanted side effects.
Acknowledgments
Grant Support: March of Dimes-6-FY14-389, NIH-R01 HL112893-A1
References
- Abu-Abed S, Dolle P, Metzger D, Beckett B, Chambon P, Petkovich M. The retinoic acid-metabolizing enzyme, CYP26A1, is essential for normal hindbrain patterning, vertebral identity, and development of posterior structures. Genes & development. 2001;15:226–240. doi: 10.1101/gad.855001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Abed SS, Beckett BR, Chiba H, Chithalen JV, Jones G, Metzger D, Chambon P, Petkovich M. Mouse P450RAI (CYP26) expression and retinoic acid-inducible retinoic acid metabolism in F9 cells are regulated by retinoic acid receptor gamma and retinoid X receptor alpha. The Journal of biological chemistry. 1998;273:2409–2415. doi: 10.1074/jbc.273.4.2409. [DOI] [PubMed] [Google Scholar]
- Abu-Issa R, Kirby ML. Patterning of the heart field in the chick. Dev Biol. 2008;319:223–233. doi: 10.1016/j.ydbio.2008.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Tanoury Z, Piskunov A, Rochette-Egly C. Vitamin A and retinoid signaling: genomic and nongenomic effects. J Lipid Res. 2013;54:1761–1775. doi: 10.1194/jlr.R030833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albalat R. The retinoic acid machinery in invertebrates: ancestral elements and vertebrate innovations. Mol Cell Endocrinol. 2009;313:23–35. doi: 10.1016/j.mce.2009.08.029. [DOI] [PubMed] [Google Scholar]
- Albalat R, Canestro C. Identification of Aldh1a, Cyp26 and RAR orthologs in protostomes pushes back the retinoic acid genetic machinery in evolutionary time to the bilaterian ancestor. Chem Biol Interact. 2009;178:188–196. doi: 10.1016/j.cbi.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Amengual J, Golczak M, Palczewski K, von Lintig J. Lecithin:retinol acyltransferase is critical for cellular uptake of vitamin A from serum retinol-binding protein. J Biol Chem. 2012;287:24216–24227. doi: 10.1074/jbc.M112.353979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amengual J, Zhang N, Kemerer M, Maeda T, Palczewski K, Von Lintig J. STRA6 is critical for cellular vitamin A uptake and homeostasis. Hum Mol Genet. 2014;23:5402–5417. doi: 10.1093/hmg/ddu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre A, Ruivo R, Gesto M, Castro LF, Santos MM. Retinoid metabolism in invertebrates: When evolution meets endocrine disruption. Gen Comp Endocrinol. 2014;208C:134–145. doi: 10.1016/j.ygcen.2014.08.005. [DOI] [PubMed] [Google Scholar]
- Bastien J, Rochette-Egly C. Nuclear retinoid receptors and the transcription of retinoid-target genes. Gene. 2004;328:1–16. doi: 10.1016/j.gene.2003.12.005. [DOI] [PubMed] [Google Scholar]
- Batten ML, Imanishi Y, Maeda T, Tu DC, Moise AR, Bronson D, Possin D, Van Gelder RN, Baehr W, Palczewski K. Lecithin-retinol acyltransferase is essential for accumulation of all-trans-retinyl esters in the eye and in the liver. J Biol Chem. 2004;279:10422–10432. doi: 10.1074/jbc.M312410200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begemann G, Schilling TF, Rauch GJ, Geisler R, Ingham PW. The zebrafish neckless mutation reveals a requirement for raldh2 in mesodermal signals that pattern the hindbrain. Development. 2001;128:3081–3094. doi: 10.1242/dev.128.16.3081. [DOI] [PubMed] [Google Scholar]
- Bendich A, Langseth L. Safety of vitamin A. Am J Clin Nutr. 1989;49:358–371. doi: 10.1093/ajcn/49.2.358. [DOI] [PubMed] [Google Scholar]
- Berry DC, Jacobs H, Marwarha G, Gely-Pernot A, O’Byrne SM, DeSantis D, Klopfenstein M, Feret B, Dennefeld C, Blaner WS, Croniger CM, Mark M, Noy N, Ghyselinck NB. The STRA6 receptor is essential for retinol-binding protein-induced insulin resistance but not for maintaining vitamin A homeostasis in tissues other than the eye. J Biol Chem. 2013;288:24528–24539. doi: 10.1074/jbc.M113.484014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billings SE, Pierzchalski K, Butler Tjaden NE, Pang XY, Trainor PA, Kane MA, Moise AR. The retinaldehyde reductase DHRS3 is essential for preventing the formation of excess retinoic acid during embryonic development. FASEB J. 2013;27:4877–4889. doi: 10.1096/fj.13-227967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaner WS. STRA6, a cell-surface receptor for retinol-binding protein: the plot thickens. Cell Metab. 2007;5:164–166. doi: 10.1016/j.cmet.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Cai AQ, Radtke K, Linville A, Lander AD, Nie Q, Schilling TF. Cellular retinoic acid-binding proteins are essential for hindbrain patterning and signal robustness in zebrafish. Development. 2012;139:2150–2155. doi: 10.1242/dev.077065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerignoli F, Guo X, Cardinali B, Rinaldi C, Casaletto J, Frati L, Screpanti I, Gudas LJ, Gulino A, Thiele CJ, Giannini G. retSDR1, a short-chain retinol dehydrogenase/reductase, is retinoic acid-inducible and frequently deleted in human neuroblastoma cell lines. Cancer Res. 2002;62:1196–1204. [PubMed] [Google Scholar]
- Chatzi C, Cunningham TJ, Duester G. Investigation of retinoic acid function during embryonic brain development using retinaldehyde-rescued Rdh10 knockout mice. Dev Dyn. 2013;242:1056–1065. doi: 10.1002/dvdy.23999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chazaud C, Bouillet P, Oulad-Abdelghani M, Dolle P. Restricted expression of a novel retinoic acid responsive gene during limb bud dorsoventral patterning and endochondral ossification. Dev Genet. 1996;19:66–73. doi: 10.1002/(SICI)1520-6408(1996)19:1<66::AID-DVG7>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Christian P, Klemm R, Shamim AA, Ali H, Rashid M, Shaikh S, Wu L, Mehra S, Labrique A, Katz J, West KP., Jr Effects of vitamin A and beta-carotene supplementation on birth size and length of gestation in rural Bangladesh: a cluster-randomized trial. Am J Clin Nutr. 2013;97:188–194. doi: 10.3945/ajcn.112.042275. [DOI] [PubMed] [Google Scholar]
- Clagett-Dame M, Knutson D. Vitamin A in reproduction and development. Nutrients. 2011;3:385–428. doi: 10.3390/nu3040385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collop AH, Broomfield JA, Chandraratna RA, Yong Z, Deimling SJ, Kolker SJ, Weeks DL, Drysdale TA. Retinoic acid signaling is essential for formation of the heart tube in Xenopus. Dev Biol. 2006;291:96–109. doi: 10.1016/j.ydbio.2005.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham TJ, Zhao X, Sandell LL, Evans SM, Trainor PA, Duester G. Antagonism between retinoic acid and fibroblast growth factor signaling during limb development. Cell Rep. 2013;3:1503–1511. doi: 10.1016/j.celrep.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ambrosio DN, Clugston RD, Blaner WS. Vitamin A metabolism: an update. Nutrients. 2011;3:63–103. doi: 10.3390/nu3010063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Aniello E, Rydeen AB, Anderson JL, Mandal A, Waxman JS. Depletion of Retinoic Acid Receptors Initiates a Novel Positive Feedback Mechanism that Promotes Teratogenic Increases in Retinoic Acid. PLoS genetics. 2013;9:e1003689. doi: 10.1371/journal.pgen.1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das BC, Thapa P, Karki R, Das S, Mahapatra S, Liu TC, Torregroza I, Wallace DP, Kambhampati S, Van Veldhuizen P, Verma A, Ray SK, Evans T. Retinoic acid signaling pathways in development and diseases. Bioorg Med Chem. 2014;22:673–683. doi: 10.1016/j.bmc.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de The H, Vivanco-Ruiz MM, Tiollais P, Stunnenberg H, Dejean A. Identification of a retinoic acid responsive element in the retinoic acid receptor beta gene. Nature. 1990;343:177–180. doi: 10.1038/343177a0. [DOI] [PubMed] [Google Scholar]
- Delacroix L, Moutier E, Altobelli G, Legras S, Poch O, Choukrallah MA, Bertin I, Jost B, Davidson I. Cell-specific interaction of retinoic acid receptors with target genes in mouse embryonic fibroblasts and embryonic stem cells. Mol Cell Biol. 2010;30:231–244. doi: 10.1128/MCB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diez del Corral R, Storey KG. Opposing FGF and retinoid pathways: a signalling switch that controls differentiation and patterning onset in the extending vertebrate body axis. Bio Essays. 2004;26:857–869. doi: 10.1002/bies.20080. [DOI] [PubMed] [Google Scholar]
- Dobbs-McAuliffe B, Zhao Q, Linney E. Feedback mechanisms regulate retinoic acid production and degradation in the zebrafish embryo. Mech Dev. 2004;121:339–350. doi: 10.1016/j.mod.2004.02.008. [DOI] [PubMed] [Google Scholar]
- Dolle P, Ruberte E, Leroy P, Morriss-Kay G, Chambon P. Retinoic acid receptors and cellular retinoid binding proteins. I. A systematic study of their differential pattern of transcription during mouse organogenesis. Development. 1990;110:1133–1151. doi: 10.1242/dev.110.4.1133. [DOI] [PubMed] [Google Scholar]
- Dorsky RI, Itoh M, Moon RT, Chitnis A. Two tcf3 genes cooperate to pattern the zebrafish brain. Development. 2003;130:1937–1947. doi: 10.1242/dev.00402. [DOI] [PubMed] [Google Scholar]
- Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell. 2008;134:921–931. doi: 10.1016/j.cell.2008.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duester G. Retinoid signaling in control of progenitor cell differentiation during mouse development. Semin Cell Dev Biol. 2013;24:694–700. doi: 10.1016/j.semcdb.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Saunders M, Leroy P, Leid M, Chambon P. All-trans and 9-cis retinoic acid induction of CRABPII transcription is mediated by RAR-RXR heterodimers bound to DR1 and DR2 repeated motifs. Cell. 1992;71:73–85. doi: 10.1016/0092-8674(92)90267-g. [DOI] [PubMed] [Google Scholar]
- Durston AJ, Timmermans JP, Hage WJ, Hendriks HF, de Vries NJ, Heideveld M, Nieuwkoop PD. Retinoic acid causes an anteroposterior transformation in the developing central nervous system. Nature. 1989;340:140–144. doi: 10.1038/340140a0. [DOI] [PubMed] [Google Scholar]
- Emoto Y, Wada H, Okamoto H, Kudo A, Imai Y. Retinoic acid-metabolizing enzyme Cyp26a1 is essential for determining territories of hindbrain and spinal cord in zebrafish. Dev Biol (NY 1985) 2005;278:415–427. doi: 10.1016/j.ydbio.2004.11.023. [DOI] [PubMed] [Google Scholar]
- Epping MT, Wang L, Edel MJ, Carlee L, Hernandez M, Bernards R. The human tumor antigen PRAME is a dominant repressor of retinoic acid receptor signaling. Cell. 2005;122:835–847. doi: 10.1016/j.cell.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Feng L, Hernandez RE, Waxman JS, Yelon D, Moens CB. Dhrs3a regulates retinoic acid biosynthesis through a feedback inhibition mechanism. Dev Biol (NY 1985) 2010;338:1–14. doi: 10.1016/j.ydbio.2009.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghyselinck NB, Bavik C, Sapin V, Mark M, Bonnier D, Hindelang C, Dierich A, Nilsson CB, Hakansson H, Sauvant P, Azais-Braesco V, Frasson M, Picaud S, Chambon P. Cellular retinol-binding protein I is essential for vitamin A homeostasis. EMBO J. 1999;18:4903–4914. doi: 10.1093/emboj/18.18.4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC. Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6. Am J Hum Genet. 2007;80:1179–1187. doi: 10.1086/518177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine UI, Roberts AB, Munoz EF, Roche NS, Sporn MB. Effects of retinoid deficiency on the development of the heart and vascular system of the quail embryo. Virchows Arch B Cell Pathol Incl Mol Pathol. 1985;50:135–152. doi: 10.1007/BF02889897. [DOI] [PubMed] [Google Scholar]
- Hernandez RE, Putzke AP, Myers JP, Margaretha L, Moens CB. Cyp26 enzymes generate the retinoic acid response pattern necessary for hindbrain development. Development. 2007;134:177–187. doi: 10.1242/dev.02706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez RE, Rikhof HA, Bachmann R, Moens CB. vhnf1 integrates global RA patterning and local FGF signals to direct posterior hindbrain development in zebrafish. Development. 2004;131:4511–4520. doi: 10.1242/dev.01297. [DOI] [PubMed] [Google Scholar]
- Hochgreb T, Linhares VL, Menezes DC, Sampaio AC, Yan CY, Cardoso WV, Rosenthal N, Xavier-Neto J. A caudorostral wave of RALDH2 conveys anteroposterior information to the cardiac field. Development. 2003;130:5363–5374. doi: 10.1242/dev.00750. [DOI] [PubMed] [Google Scholar]
- Hu P, Tian M, Bao J, Xing G, Gu X, Gao X, Linney E, Zhao Q. Retinoid regulation of the zebrafish cyp26a1 promoter. Dev Dyn. 2008;237:3798–3808. doi: 10.1002/dvdy.21801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Chen Y, Farooqui M, Thomas MC, Chiang CM, Wei LN. Suppressive effect of receptor-interacting protein 140 on coregulator binding to retinoic acid receptor complexes, histone-modifying enzyme activity, and gene activation. J Biol Chem. 2004;279:319–325. doi: 10.1074/jbc.M307621200. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Nakazawa M, Ono H, Satoh N, Gojobori T, Fujiwara S. Microarray analysis of embryonic retinoic acid target genes in the ascidian Ciona intestinalis. Dev Growth Differ. 2003;45:249–259. doi: 10.1046/j.1524-4725.2003.694.x. [DOI] [PubMed] [Google Scholar]
- Ishibashi T, Usami T, Fujie M, Azumi K, Satoh N, Fujiwara S. Oligonucleotide-based microarray analysis of retinoic acid target genes in the protochordate, Ciona intestinalis. Dev Dyn. 2005;233:1571–1578. doi: 10.1002/dvdy.20486. [DOI] [PubMed] [Google Scholar]
- Isken A, Golczak M, Oberhauser V, Hunzelmann S, Driever W, Imanishi Y, Palczewski K, von Lintig J. RBP4 disrupts vitamin A uptake homeostasis in a STRA6-deficient animal model for Matthew-Wood syndrome. Cell Metab. 2008;7:258–268. doi: 10.1016/j.cmet.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isken A, Holzschuh J, Lampert JM, Fischer L, Oberhauser V, Palczewski K, von Lintig J. Sequestration of retinyl esters is essential for retinoid signaling in the zebrafish embryo. J Biol Chem. 2007;282:1144–1151. doi: 10.1074/jbc.M609109200. [DOI] [PubMed] [Google Scholar]
- Janesick A, Nguyen TT, Aisaki K, Igarashi K, Kitajima S, Chandraratna RA, Kanno J, Blumberg B. Active repression by RARgamma signaling is required for vertebrate axial elongation. Development. 2014;141:2260–2270. doi: 10.1242/dev.103705. [DOI] [PubMed] [Google Scholar]
- Kalter H, Warkany J. Experimental production of congenital malformations in strains of inbred mice by maternal treatment with hypervitaminosis A. Am J Pathol. 1961;38:1–21. [PMC free article] [PubMed] [Google Scholar]
- Kashyap V, Laursen KB, Brenet F, Viale AJ, Scandura JM, Gudas LJ. RARgamma is essential for retinoic acid induced chromatin remodeling and transcriptional activation in embryonic stem cells. J Cell Sci. 2013;126:999–1008. doi: 10.1242/jcs.119701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Yu J, Honda J, Hu J, Whitelegge J, Ping P, Wiita P, Bok D, Sun H. A membrane receptor for retinol binding protein mediates cellular uptake of vitamin A. Science. 2007;315:820–825. doi: 10.1126/science.1136244. [DOI] [PubMed] [Google Scholar]
- Kedishvili NY. Enzymology of retinoic acid biosynthesis and degradation. J Lipid Res. 2013;54:1744–1760. doi: 10.1194/jlr.R037028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan BR, Feldman JL, Begemann G, Ingham PW, Yelon D. Retinoic acid signaling restricts the cardiac progenitor pool. Science. 2005;307:247–249. doi: 10.1126/science.1101573. [DOI] [PubMed] [Google Scholar]
- Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- Kim YK, Wassef L, Hamberger L, Piantedosi R, Palczewski K, Blaner WS, Quadro L. Retinyl ester formation by lecithin: retinol acyltransferase is a key regulator of retinoid homeostasis in mouse embryogenesis. J Biol Chem. 2008;283:5611–5621. doi: 10.1074/jbc.M708885200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide T, Downes M, Chandraratna RA, Blumberg B, Umesono K. Active repression of RAR signaling is required for head formation. Genes & development. 2001;15:2111–2121. doi: 10.1101/gad.908801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudoh T, Wilson SW, Dawid IB. Distinct roles for Fgf, Wnt and retinoic acid in posteriorizing the neural ectoderm. Development. 2002;129:4335–4346. doi: 10.1242/dev.129.18.4335. [DOI] [PubMed] [Google Scholar]
- Kumar S, Duester G. Retinoic acid controls body axis extension by directly repressing Fgf8 transcription. Development. 2014;141:2972–2977. doi: 10.1242/dev.112367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sandell LL, Trainor PA, Koentgen F, Duester G. Alcohol and aldehyde dehydrogenases: retinoid metabolic effects in mouse knockout models. Biochim Biophys Acta. 2012;1821:198–205. doi: 10.1016/j.bbalip.2011.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammer EJ, Chen DT, Hoar RM, Agnish ND, Benke PJ, Braun JT, Curry CJ, Fernhoff PM, Grix AW, Jr, Lott IT, et al. Retinoic acid embryopathy. N Engl J Med. 1985;313:837–841. doi: 10.1056/NEJM198510033131401. [DOI] [PubMed] [Google Scholar]
- Lee LM, Leung CY, Tang WW, Choi HL, Leung YC, McCaffery PJ, Wang CC, Woolf AS, Shum AS. A paradoxical teratogenic mechanism for retinoic acid. Proc Natl Acad Sci USA. 2012;109:13668–13673. doi: 10.1073/pnas.1200872109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Hu P, Li K, Zhao Q. Identification and characterization of a novel retinoic acid response element in zebrafish cyp26a1 promoter. Anat Rec (Hoboken) 2012;295:268–277. doi: 10.1002/ar.21520. [DOI] [PubMed] [Google Scholar]
- Li P, Pashmforoush M, Sucov HM. Retinoic acid regulates differentiation of the secondary heart field and TGFbeta-mediated outflow tract septation. Dev Cell. 2010;18:480–485. doi: 10.1016/j.devcel.2009.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linville A, Radtke K, Waxman JS, Yelon D, Schilling TF. Combinatorial roles for zebrafish retinoic acid receptors in the hindbrain, limbs and pharyngeal arches. Dev Biol (NY 1985) 2009;325:60–70. doi: 10.1016/j.ydbio.2008.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Stainier DY. Zebrafish in the study of early cardiac development. Circ Res. 2012;110:870–874. doi: 10.1161/CIRCRESAHA.111.246504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohnes D, Kastner P, Dierich A, Mark M, LeMeur M, Chambon P. Function of retinoic acid receptor gamma in the mouse. Cell. 1993;73:643–658. doi: 10.1016/0092-8674(93)90246-m. [DOI] [PubMed] [Google Scholar]
- Lohnes D, Mark M, Mendelsohn C, Dolle P, Dierich A, Gorry P, Gansmuller A, Chambon P. Function of the retinoic acid receptors (RARs) during development (I). Craniofacial and skeletal abnormalities in RAR double mutants. Development. 1994;120:2723–2748. doi: 10.1242/dev.120.10.2723. [DOI] [PubMed] [Google Scholar]
- Loudig O, Babichuk C, White J, Abu-Abed S, Mueller C, Petkovich M. Cytochrome P450RAI(CYP26) promoter: a distinct composite retinoic acid response element underlies the complex regulation of retinoic acid metabolism. Mol Endocrinol. 2000;14:1483–1497. doi: 10.1210/mend.14.9.0518. [DOI] [PubMed] [Google Scholar]
- Loudig O, Maclean GA, Dore NL, Luu L, Petkovich M. Transcriptional co-operativity between distant retinoic acid response elements in regulation of Cyp26A1 inducibility. Biochem J. 2005;392:241–248. doi: 10.1042/BJ20050874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lufkin T, Lohnes D, Mark M, Dierich A, Gorry P, Gaub MP, LeMeur M, Chambon P. High postnatal lethality and testis degeneration in retinoic acid receptor alpha mutant mice. Proc Natl Acad Sci USA. 1993;90:7225–7229. doi: 10.1073/pnas.90.15.7225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony S, Mazzoni EO, McCuine S, Young RA, Wichterle H, Gifford DK. Ligand-dependent dynamics of retinoic acid receptor binding during early neurogenesis. Genome Biol. 2011;12:R2. doi: 10.1186/gb-2011-12-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manshouri T, Yang Y, Lin H, Stass SA, Glassman AB, Keating MJ, Albitar M. Downregulation of RAR alpha in mice by antisense transgene leads to a compensatory increase in RAR beta and RAR gamma and development of lymphoma. Blood. 1997;89:2507–2515. [PubMed] [Google Scholar]
- Marchitti SA, Brocker C, Stagos D, Vasiliou V. Non-P450 aldehyde oxidizing enzymes: the aldehyde dehydrogenase superfamily. Expert Opin Drug Metab Toxicol. 2008;4:697–720. doi: 10.1517/17425250802102627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Ghyselinck NB, Chambon P. Function of retinoid nuclear receptors: lessons from genetic and pharmacological dissections of the retinoic acid signaling pathway during mouse embryogenesis. Annu Rev Pharmacol Toxicol. 2006;46:451–480. doi: 10.1146/annurev.pharmtox.46.120604.141156. [DOI] [PubMed] [Google Scholar]
- Mark M, Ghyselinck NB, Chambon P. Function of retinoic acid receptors during embryonic development. Nuclear receptor signaling. 2009;7:e002. doi: 10.1621/nrs.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendelsohn C, Lohnes D, Decimo D, Lufkin T, LeMeur M, Chambon P, Mark M. Function of the retinoic acid receptors (RARs) during development (II). Multiple abnormalities at various stages of organogenesis in RAR double mutants. Development. 1994;120:2749–2771. doi: 10.1242/dev.120.10.2749. [DOI] [PubMed] [Google Scholar]
- Moutier E, Ye T, Choukrallah MA, Urban S, Osz J, Chatagnon A, Delacroix L, Langer D, Rochel N, Moras D, Benoit G, Davidson I. Retinoic acid receptors recognize the mouse genome through binding elements with diverse spacing and topology. J Biol Chem. 2012;287:26328–26341. doi: 10.1074/jbc.M112.361790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta. 2012;1821:152–167. doi: 10.1016/j.bbalip.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederreither K, Abu-Abed S, Schuhbaur B, Petkovich M, Chambon P, Dolle P. Genetic evidence that oxidative derivatives of retinoic acid are not involved in retinoid signaling during mouse development. Nat Genet. 2002;31:84–88. doi: 10.1038/ng876. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Dolle P. Retinoic acid in development: towards an integrated view. Nature reviews. Genetics. 2008;9:541–553. doi: 10.1038/nrg2340. [DOI] [PubMed] [Google Scholar]
- Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev. 1997;62:67–78. doi: 10.1016/s0925-4773(96)00653-3. [DOI] [PubMed] [Google Scholar]
- Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet. 1999;21:444–448. doi: 10.1038/7788. [DOI] [PubMed] [Google Scholar]
- O’Byrne SM, Wongsiriroj N, Libien J, Vogel S, Goldberg IJ, Baehr W, Palczewski K, Blaner WS. Retinoid absorption and storage is impaired in mice lacking lecithin:retinol acyltransferase (LRAT) J Biol Chem. 2005;280:35647–35657. doi: 10.1074/jbc.M507924200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmond MK, Butler AJ, Voon FC, Bellairs R. The effects of retinoic acid on heart formation in the early chick embryo. Development. 1991;113:1405–1417. doi: 10.1242/dev.113.4.1405. [DOI] [PubMed] [Google Scholar]
- Pan J, Baker KM. Retinoic acid and the heart. Vitam Horm. 2007;75:257–283. doi: 10.1016/S0083-6729(06)75010-5. [DOI] [PubMed] [Google Scholar]
- Pares X, Farres J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families: Medium-chain and short-chain dehydrogenases/reductases in retinoid metabolism. Cell Mol Life Sci. 2008;65:3936–3949. doi: 10.1007/s00018-008-8591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasutto F, Sticht H, Hammersen G, Gillessen-Kaesbach G, Fitzpatrick DR, Nurnberg G, Brasch F, Schirmer-Zimmermann H, Tolmie JL, Chitayat D, Houge G, Fernandez-Martinez L, Keating S, Mortier G, Hennekam RC, von der Wense A, Slavotinek A, Meinecke P, Bitoun P, Becker C, Nurnberg P, Reis A, Rauch A. Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation. Am J Hum Genet. 2007;80:550–560. doi: 10.1086/512203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennimpede T, Cameron DA, MacLean GA, Li H, Abu-Abed S, Petkovich M. The role of CYP26 enzymes in defining appropriate retinoic acid exposure during embryogenesis. Birth defects research. Part A, Clinical and molecular teratology. 2010;88:883–894. doi: 10.1002/bdra.20709. [DOI] [PubMed] [Google Scholar]
- Quadro L, Blaner WS, Salchow DJ, Vogel S, Piantedosi R, Gouras P, Freeman S, Cosma MP, Colantuoni V, Gottesman ME. Impaired retinal function and vitamin A availability in mice lacking retinol-binding protein. EMBO J. 1999;18:4633–4644. doi: 10.1093/emboj/18.17.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadro L, Hamberger L, Gottesman ME, Wang F, Colantuoni V, Blaner WS, Mendelsohn CL. Pathways of vitamin A delivery to the embryo: insights from a new tunable model of embryonic vitamin A deficiency. Endocrinology. 2005;146:4479–4490. doi: 10.1210/en.2005-0158. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Dolle P. Retinoic acid signalling during development. Development. 2012;139:843–858. doi: 10.1242/dev.065938. [DOI] [PubMed] [Google Scholar]
- Rhinn M, Schuhbaur B, Niederreither K, Dolle P. Involvement of retinol dehydrogenase 10 in embryonic patterning and rescue of its loss of function by maternal retinaldehyde treatment. Proc Natl Acad Sci U S A. 2011;108:16687–16692. doi: 10.1073/pnas.1103877108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribes V, Fraulob V, Petkovich M, Dolle P. The oxidizing enzyme CYP26a1 tightly regulates the availability of retinoic acid in the gastrulating mouse embryo to ensure proper head development and vasculogenesis. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:644–653. doi: 10.1002/dvdy.21057. [DOI] [PubMed] [Google Scholar]
- Rizzo R, Lammer EJ, Parano E, Pavone L, Argyle JC. Limb reduction defects in humans associated with prenatal isotretinoin exposure. Teratology. 1991;44:599–604. doi: 10.1002/tera.1420440602. [DOI] [PubMed] [Google Scholar]
- Romand R, Sapin V, Ghyselinck NB, Avan P, Le Calvez S, Dolle P, Chambon P, Mark M. Spatio-temporal distribution of cellular retinoid binding protein gene transcripts in the developing and the adult cochlea. Morphological and functional consequences in CRABP- and CRBPI-null mutant mice. Eur J Neurosci. 2000;12:2793–2804. doi: 10.1046/j.1460-9568.2000.00165.x. [DOI] [PubMed] [Google Scholar]
- Romeih M, Cui J, Michaille JJ, Jiang W, Zile MH. Function of RARgamma and RARalpha2 at the initiation of retinoid signaling is essential for avian embryo survival and for distinct events in cardiac morphogenesis. Dev Dyn. 2003;228:697–708. doi: 10.1002/dvdy.10419. [DOI] [PubMed] [Google Scholar]
- Ruberte E, Dolle P, Chambon P, Morriss-Kay G. Retinoic acid receptors and cellular retinoid binding proteins. II. Their differential pattern of transcription during early morphogenesis in mouse embryos. Development. 1991;111:45–60. doi: 10.1242/dev.111.1.45. [DOI] [PubMed] [Google Scholar]
- Ryckebusch L, Wang Z, Bertrand N, Lin SC, Chi X, Schwartz R, Zaffran S, Niederreither K. Retinoic acid deficiency alters second heart field formation. Proc Natl Acad Sci U S A. 2008;105:2913–2918. doi: 10.1073/pnas.0712344105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydeen AB, Waxman JS. Cyp26 enzymes are required to balance the cardiac and vascular lineages within the anterior lateral plate mesoderm. Development. 2014;141:1638–1648. doi: 10.1242/dev.105874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Meno C, Fujii H, Nishino J, Shiratori H, Saijoh Y, Rossant J, Hamada H. The retinoic acid-inactivating enzyme CYP26 is essential for establishing an uneven distribution of retinoic acid along the anterio-posterior axis within the mouse embryo. Genes & development. 2001;15:213–225. doi: 10.1101/gad.851501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarut E, Fraher D, Laudet V, Gibert Y. ZebRA: An overview of retinoic acid signaling during zebrafish development. Biochim Biophys Acta. 2014 doi: 10.1016/j.bbagrm.2014.05.030. [DOI] [PubMed] [Google Scholar]
- Samarut E, Rochette-Egly C. Nuclear retinoic acid receptors: conductors of the retinoic acid symphony during development. Mol Cell Endocrinol. 2012;348:348–360. doi: 10.1016/j.mce.2011.03.025. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Lynn ML, Inman KE, McDowell W, Trainor PA. RDH10 oxidation of Vitamin A is a critical control step in synthesis of retinoic acid during mouse embryogenesis. PLoS One. 2012;7:e30698. doi: 10.1371/journal.pone.0030698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Sanderson BW, Moiseyev G, Johnson T, Mushegian A, Young K, Rey JP, Ma JX, Staehling-Hampton K, Trainor PA. RDH10 is essential for synthesis of embryonic retinoic acid and is required for limb, craniofacial, and organ development. Genes & development. 2007;21:1113–1124. doi: 10.1101/gad.1533407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Nie Q, Lander AD. Dynamics and precision in retinoic acid morphogen gradients. Curr Opin Genet Dev. 2012;22:562–569. doi: 10.1016/j.gde.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirbu IO, Zhao X, Duester G. Retinoic acid controls heart anteroposterior patterning by down-regulating Isl1 through the Fgf8 pathway. Dev Dyn. 2008;237:1627–1635. doi: 10.1002/dvdy.21570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Draper BW, Harland RM, Weintraub H. Identification of a retinoic acid-sensitive period during primary axis formation in Xenopus laevis. Genes Dev. 1990;4:932–942. doi: 10.1101/gad.4.6.932. [DOI] [PubMed] [Google Scholar]
- Smith WC, Nakshatri H, Leroy P, Rees J, Chambon P. A retinoic acid response element is present in the mouse cellular retinol binding protein I (mCRBPI) promoter. EMBO J. 1991;10:2223–2230. doi: 10.1002/j.1460-2075.1991.tb07758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soprano DR, Qin P, Soprano KJ. Retinoic acid receptors and cancers. Annu Rev Nutr. 2004;24:201–221. doi: 10.1146/annurev.nutr.24.012003.132407. [DOI] [PubMed] [Google Scholar]
- Sorrell MR, Waxman JS. Restraint of Fgf8 signaling by retinoic acid signaling is required for proper heart and forelimb formation. Dev Biol (NY 1985) 2011;358:44–55. doi: 10.1016/j.ydbio.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier DY, Fishman MC. Patterning the zebrafish heart tube: acquisition of anteroposterior polarity. Dev Biol. 1992;153:91–101. doi: 10.1016/0012-1606(92)90094-w. [DOI] [PubMed] [Google Scholar]
- Strate I, Min TH, Iliev D, Pera EM. Retinol dehydrogenase 10 is a feedback regulator of retinoic acid signalling during axis formation and patterning of the central nervous system. Development. 2009;136:461–472. doi: 10.1242/dev.024901. [DOI] [PubMed] [Google Scholar]
- Tucker JA, Mintzer KA, Mullins MC. The BMP signaling gradient patterns dorsoventral tissues in a temporally progressive manner along the anteroposterior axis. Dev Cell. 2008;14:108–119. doi: 10.1016/j.devcel.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara M, Yashiro K, Mamiya S, Nishino J, Chambon P, Dolle P, Sakai Y. CYP26A1 and CYP26C1 cooperatively regulate anterior-posterior patterning of the developing brain and the production of migratory cranial neural crest cells in the mouse. Dev Biol (NY 1985) 2007;302:399–411. doi: 10.1016/j.ydbio.2006.09.045. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Keegan BR, Roberts RW, Poss KD, Yelon D. Hoxb5b acts downstream of retinoic acid signaling in the forelimb field to restrict heart field potential in zebrafish. Dev Cell. 2008;15:923–934. doi: 10.1016/j.devcel.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman JS, Yelon D. Comparison of the expression patterns of newly identified zebrafish retinoic acid and retinoid X receptors. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:587–595. doi: 10.1002/dvdy.21049. [DOI] [PubMed] [Google Scholar]
- Waxman JS, Yelon D. Increased Hox activity mimics the teratogenic effects of excess retinoic acid signaling. Developmental dynamics: an official publication of the American Association of Anatomists. 2009;238:1207–1213. doi: 10.1002/dvdy.21951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman JS, Yelon D. Zebrafish retinoic acid receptors function as context-dependent transcriptional activators. Dev Biol (NY 1985) 2011;352:128–140. doi: 10.1016/j.ydbio.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendler CC, Schmoldt A, Flentke GR, Case LC, Quadro L, Blaner WS, Lough J, Smith SM. Increased fibronectin deposition in embryonic hearts of retinol-binding protein-null mice. Circ Res. 2003;92:920–928. doi: 10.1161/01.RES.0000069030.30886.8F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West KP, Jr, Mehra S. Vitamin A intake and status in populations facing economic stress. J Nutr. 2010;140:201S–207S. doi: 10.3945/jn.109.112730. [DOI] [PubMed] [Google Scholar]
- Weston AD, Blumberg B, Underhill TM. Active repression by unliganded retinoid receptors in development: less is sometimes more. J Cell Biol. 2003;161:223–228. doi: 10.1083/jcb.200211117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RJ, Nie Q, Lander AD, Schilling TF. Complex regulation of cyp26a1 creates a robust retinoic acid gradient in the zebrafish embryo. PLoS biology. 2007;5:e304. doi: 10.1371/journal.pbio.0050304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JG, Warkany J. Aortic-arch and cardiac anomalies in the offspring of vitamin A deficient rats. The American journal of anatomy. 1949;85:113–155. doi: 10.1002/aja.1000850106. [DOI] [PubMed] [Google Scholar]
- Wilson JG, Warkany J. Cardiac and aortic arch anomalies in the offspring of vitamin A deficient rats correlated with similar human anomalies. Pediatrics. 1950a;5:708–725. [PubMed] [Google Scholar]
- Wilson JG, Warkany J. Congenital anomalies of heart and great vessels in offspring of vitamin A-deficient rats. American journal of diseases of children. 1950b;79:963. [PubMed] [Google Scholar]
- Yutzey KE, Rhee JT, Bader D. Expression of the atrial-specific myosin heavy chain AMHC1 and the establishment of anteroposterior polarity in the developing chicken heart. Development. 1994;120:871–883. doi: 10.1242/dev.120.4.871. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Zolfaghari R, Ross AC. Multiple retinoic acid response elements cooperate to enhance the inducibility of CYP26A1 gene expression in liver. Gene. 2010;464:32–43. doi: 10.1016/j.gene.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sirbu IO, Mic FA, Molotkova N, Molotkov A, Kumar S, Duester G. Retinoic acid promotes limb induction through effects on body axis extension but is unnecessary for limb patterning. Curr Biol. 2009;19:1050–1057. doi: 10.1016/j.cub.2009.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]