Abstract
The ubiquitin-like molecule ATG12 is required for the early steps of autophagy. Recently, we identified ATG3, the E2-like enzyme required for LC3 lipidation during autophagy, as an ATG12 conjugation target. Here, we demonstrate that cells lacking ATG12-ATG3 have impaired basal autophagic flux, accumulation of perinuclear late endosomes, and impaired endolysosomal trafficking. Furthermore, we identify an interaction between ATG12-ATG3 and the ESCRT-associated protein Alix (also known as PDCD6IP) and demonstrate that ATG12-ATG3 controls multiple Alix-dependent processes including late endosome distribution, exosome biogenesis, and viral budding. Lastly, similar to ATG12-ATG3, Alix is functionally required for efficient basal, but not starvation-induced, autophagy. Overall, these results identify a link between the core autophagy and ESCRT machineries and uncover a role for ATG12-ATG3 in late endosome function that is distinct from the canonical role of either ATG in autophagosome formation.
Introduction
Autophagy is a tightly regulated catabolic process important for cellular homeostasis and stress response1, 2. Autophagy is controlled by a set of conserved autophagy-related proteins (ATGs), among which several core ATGs function in two ubiquitin-like conjugation systems essential for autophagosome formation3–5. The first involves the ubiquitin-like molecule (UBL) ATG12, which is activated by the E1-like enzyme ATG7, transferred to the E2-like conjugating enzyme ATG10, and ultimately attached to ATG54, 6, 7. In the second, the UBL LC3 (ATG8 in yeast) is conjugated to the lipid phosphatidylethanolamine by ATG7 and the E2-like enzyme ATG33, 5, 7, 8.
In addition to the early steps of autophagy, ATGs in these pathways enable other functions9, 10. For example, although the principal substrate of ATG12 is ATG5, resulting in formation of the ATG12-ATG5 conjugate required for autophagy, we recently identified ATG3 as an additional ATG12 target. Surprisingly, although ATG12 and ATG3 are both core autophagy components, disrupting ATG12 conjugation to ATG3 did not compromise ATG3-mediated LC3 lipidation or starvation-induced autophagy. Rather, cells lacking ATG12-ATG3 displayed increased mitochondrial mass and reduced autophagosome targeting to mitochondria11. Interestingly, in those initial studies, we consistently observed increased numbers of autophagosomes in cells lacking ATG12-ATG3 under nutrient-rich conditions, but not during starvation11. These findings suggested that loss of ATG12-ATG3 affects basal autophagy, either by enhancing autophagosome formation or delaying autophagosome maturation. Here, by more carefully analyzing how ATG12-ATG3 impacts basal autophagy, we find that ATG12-ATG3 conjugation promotes autolysosome formation under nutrient-rich conditions.
In addition to basal autophagic flux defects, we demonstrate that cells lacking ATG12-ATG3 accumulate perinuclear multivesicular bodies (MVBs) and exhibit defects in late endosome to lysosome trafficking. The endosomal sorting complexes required for transport (ESCRT) components are a class of proteins required for MVB intralumenal vesicle formation and sorting of endocytosed proteins into MVBs for subsequent lysosomal degradation12. Although recent work indicates ESCRT function is also required for autophagosome maturation13–15, functional interactions between core autophagy and ESCRT components have not been established. Here, we identify an interaction between ATG12-ATG3 and the ESCRT-associated protein Alix (also known as PDCD6IP) and demonstrate that ATG12-ATG3 conjugation controls multiple Alix-mediated functions including MVB distribution, exosome biogenesis, and viral budding. Conversely, Alix deficiency specifically impairs basal autophagy, similarly to ATG12-ATG3. Overall, these results identify an interconnection between the core autophagy and ESCRT machineries that facilitates basal autophagic flux and multiple Alix-associated activities at the late endosome.
RESULTS
ATG12-ATG3 promotes basal autophagic flux
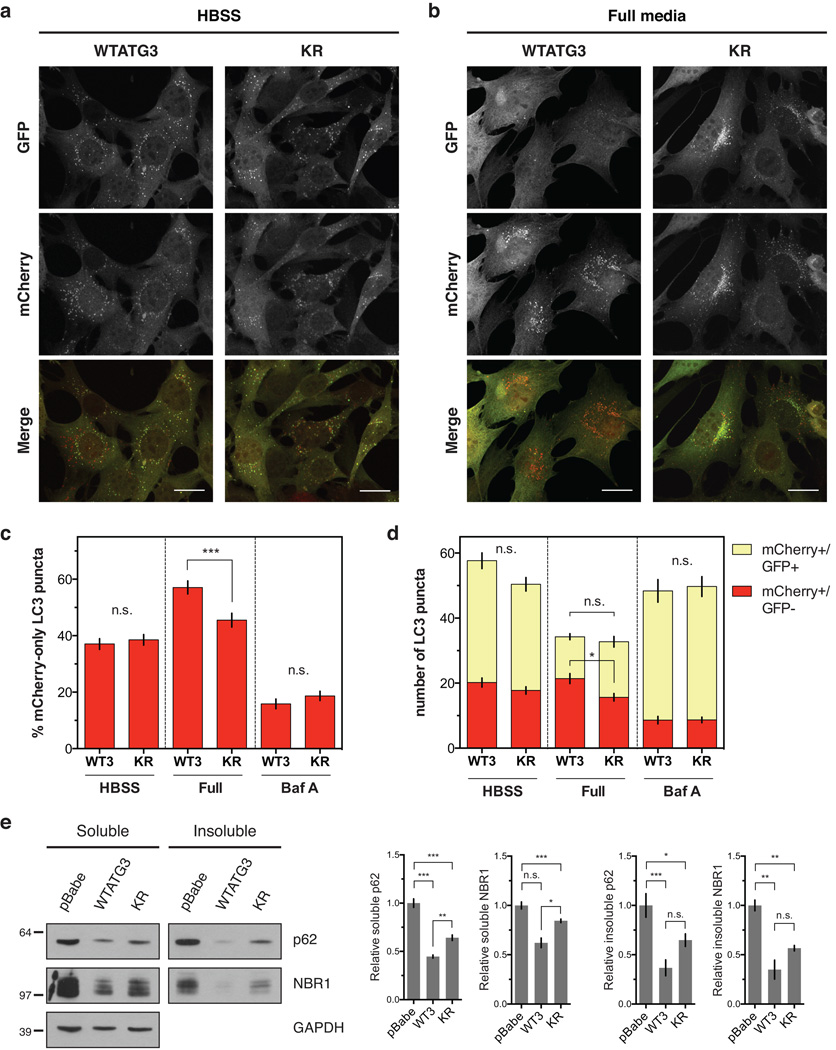
Cells lacking ATG12-ATG3 exhibit normal starvation- and rapamycin-induced autophagy, yet, under basal conditions, they exhibit significantly increased numbers of autophagosomes11. To determine whether this phenotype was due to increased autophagosome induction versus impaired maturation, we used a tandem mCherry-GFP-LC3 reporter assay. Since GFP is quenched in the acidic lysosome but mCherry remains fluorescent16, early autophagosomes correspond to double-positive mCherry+/GFP+ puncta whereas mature autolysosomes correspond to mCherry-only puncta. We reconstituted atg3−/− mouse embryonic fibroblasts (MEFs) with wild-type ATG3 (WTATG3), the K243R mutant of ATG3 (KR) that cannot be conjugated to ATG12, or an empty vector control (pBabe) (Supplementary Fig. 1a–c). WTATG3 reconstituted MEFs had similar levels of autophagic flux as wild-type atg3+/+ MEFs, while pBabe reconstituted MEFs were autophagy-deficient (Supplementary Fig. 1d–f). Consistent with previous results, WTATG3 and KR cells had no significant differences in the percentage or total number of mature autolysosomes following starvation in Hank’s buffered saline solution (HBSS) (Fig. 1a,c,d)11. However, in nutrient-rich conditions, KR cells exhibited reduced mature autolysosomes relative to WTATG3 cells (Fig. 1b–d), indicative of attenuated basal autophagic flux. Importantly, while KR cells had reduced autophagosome maturation under full media conditions, there were no differences in autophagosome induction as measured by total number of LC3 puncta (Fig. 1d). Treatment with the lysosomal inhibitor bafilomycin A (Baf A) further reduced autolysosomes in both WTATG3 and KR cells, indicating that KR cells exhibited a reduction, rather than complete block, in basal autophagy (Fig. 1c,d, Supplementary Fig. 1g). To confirm this defect in basal autophagy, we measured protein levels of the autophagy substrates p62 and NBR1 in the detergent-soluble and -insoluble fractions from reconstituted MEFs. KR cells had increased accumulation of soluble p62 and NBR1 relative to WTATG3 cells (Fig. 1e). We observed similar increases in the insoluble fraction, but due to variability in monitoring insoluble protein levels, these differences were not statistically significant (Fig. 1e). Overall, these results indicate that ATG12-ATG3 specifically promotes basal autophagosome maturation but does not affect starvation-induced autophagy.
Figure 1. ATG12-ATG3 promotes basal autophagic flux.
Stable pools of atg3−/− MEFs expressing an empty vector control (pBabe), wild-type mouse ATG3 (WTATG3) or the ATG3 K243R mutant unable to be conjugated to ATG12 (KR) were used for experiments as indicated. (a) Indicated cell types expressing mCherry-GFP-LC3 were HBSS starved for 2 h. Scale bar, 20 µm. (b) Indicated cell types expressing mCherry-GFP-LC3 were grown in full media. Scale bar, 20 µm. (c) Quantification of the percentage of mCherry-positive, GFP-negative (mCherry-only) LC3 puncta per cell as described in a–b and Supplementary Fig. 1g (mean ± SEM; n = 200 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001). (d) Quantification of the number of mCherry-only (mCherry+/GFP−) or double positive (mCherry+/GFP+) LC3 puncta per cell as described in a–b and Supplementary Fig. 1g (mean ± SEM; n = 200 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (*P < 0.05). (e) Indicated cell types were lysed in 1% Triton X-100, and triton-soluble and -insoluble fractions were immunoblotted for anti-p62, anti-NBR1, and anti-GAPDH. Right: Quantification of p62 and NBR1 protein levels (mean ± SEM; n = 5 independent experiments for p62, n = 3 independent experiments for NBR1). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test. (***P < 0.001, **P < 0.01, *P < 0.05). See also Supplementary Figure 1. Uncropped images of blots are shown in Supplementary Figure 6.
Cells lacking ATG12-ATG3 accumulate perinuclear late endosomes
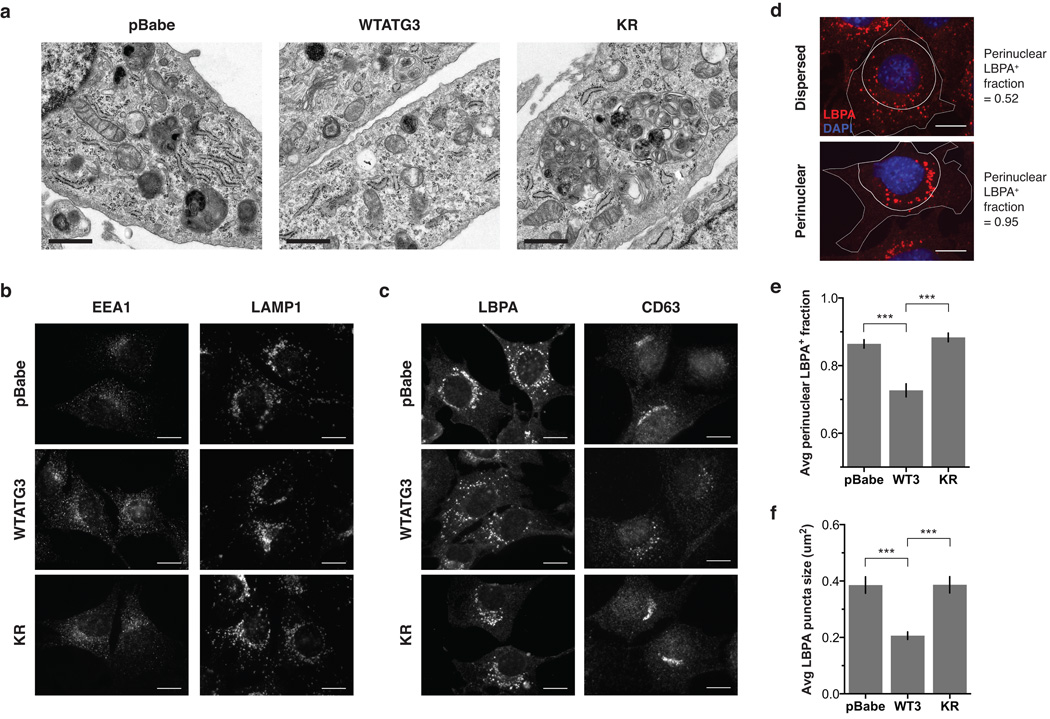
To further evaluate cells lacking ATG12-ATG3, we performed electron microscopy and observed enlarged multivesicular structures in KR cells, corresponding to late endosomes or MVBs (Fig. 2a). Hence, we immunostained for the early endosomal marker EEA1, the late endosomal markers lysobisphosphatidic acid (LBPA) and CD63, and the lysosomal marker LAMP1, to more precisely define the organelle alterations in cells lacking ATG12-ATG3. Whereas no obvious changes were seen in early endosomes or lysosomes (Fig. 2b), a striking difference in late endosome distribution and morphology was observed in cells lacking ATG12-ATG3. In pBabe and KR cells, MVBs were enlarged and clustered around the nucleus, while in WTATG3 cells they were smaller and distributed throughout the cytoplasm (Fig. 2c). To quantify MVB distribution, we measured the perinuclear LBPA+ fraction, defined as the fraction of LBPA-positive area located within 10 µm of the nucleus (Fig. 2d). Compared to WTATG3 cells, pBabe and KR cells exhibited a significantly increased perinuclear LBPA+ fraction, increased LBPA puncta size, and decreased LBPA puncta number (Fig. 2e,f, Supplementary Fig. 2a). CD63-positive late endosomes were also more tightly clustered around the nucleus in cells lacking ATG12-ATG3 (Supplementary Fig. 2b,c). Hence, ATG12-ATG3 controls MVB distribution and morphology.
Figure 2. Cells lacking ATG12-ATG3 accumulate enlarged perinuclear late endosomes.
(a) Indicated cell types were analysed by electron microscopy. Scale bar, 1 µm. (b) Indicated cell types were immunostained with anti-EEA1 or anti-LAMP1 to mark early endosomes and lysosomes, respectively. Scale bar, 10 µm. (c) Indicated cell types were immunostained with anti-lysobisphosphatidic acid (LBPA) or anti-CD63 to mark late endosomes. Scale bar, 10 µm. (d) Cells were immunostained with anti-LBPA and DAPI. The perinuclear LBPA+ fraction was defined as the fraction of LBPA area located within 10 µm of the nucleus. Cells with dispersed LBPA localization have a perinuclear LBPA+ fraction close to 0.5, whereas cells with completely clustered LBPA localization have a perinuclear LBPA+ fraction close to 1. Scale bar, 10 µm. (e) Quantification of the perinuclear LBPA+ fraction as described in d (mean ± SEM; n = 60 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001). (f) Quantification of the average number of LBPA puncta (mean ± SEM; n = 60 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001). See also Supplementary Figure 2.
ATG12-ATG3 promotes late endosome to lysosome trafficking
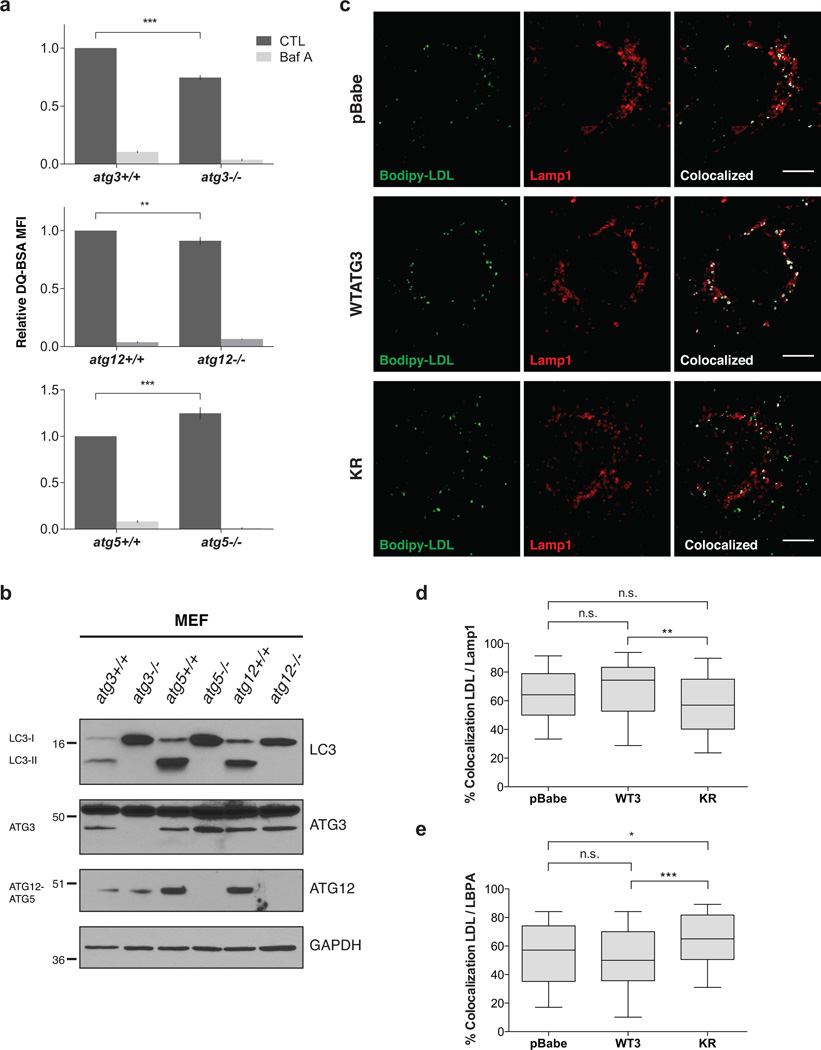
Since late endosome function is essential for autophagic flux14, we hypothesized that ATG12-ATG3 conjugation supports late endosome to lysosome trafficking. To evaluate endolysosomal trafficking, we used DQ Green BSA (DQ-BSA), a fluorogenic probe that traffics through the endosomal pathway and is ultimately dequenched following proteolytic cleavage in the lysosome. atg3−/− and atg12−/− MEFs, which both lack ATG12-ATG3, had decreased lysosomal degradation relative to wild-type controls. In contrast, atg5−/− MEFs, which lack ATG12-ATG5 but still produce ATG12-ATG3, had increased lysosomal degradation (Fig. 3a). Importantly, all three ATG-deficient lines were autophagy-incompetent, confirmed by their inability to form lipid-conjugated LC3 (LC3-II) (Fig. 3b). Thus, the effects of ATG12 or ATG3 genetic loss on endolysosomal trafficking are separable from the effects of ATG loss on autophagosome formation.
Figure 3. ATG12-ATG3 promotes late endosome to lysosome trafficking.
(a) Matched wild-type and atg-deficient MEFs were incubated with DQ-BSA for 30 min at 37 °C followed by a 2h chase in full media. Lysosomal degradation as measured by DQ-BSA dequenching was analysed by flow cytometry. When indicated, Baf A (50 nM) was used to block lysosome function. Data are presented as mean ± SEM relative fluorescence (n = 3 independent experiments for top graph, n = 7 independent experiments for bottom two graphs). Statistical significance calculated using unpaired two-tailed t test (***P < 0.001, **P < 0.01). (b) Indicated cell types were lysed and immunoblotted with anti-LC3, anti-ATG3, and anti-ATG12. (c) Stable pools of reconstituted atg3−/− MEFs were incubated with Bodipy-LDL for 15 min at 37 °C followed by a 2 h chase in full media. Cells were immunostained with anti-LAMP1 to mark lysosomes. Colocalized pixels were highlighted in white using the Colocalization plugin in ImageJ. Scale bar, 5 µm. (d) Quantification of the percentage of LDL puncta colocalized with LAMP1 as described in c. Data are presented as median (horizontal line), interquartile range (box), and 10–90th percentile (whiskers); n = 100 cells pooled from three independent experiments. Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (**P < 0.01). (e) Quantification of the percentage of LDL puncta colocalized with LBPA. Indicated cell types were incubated with Bodipy-LDL as in c and immunostained with anti-LBPA to mark late endosomes. Data are presented as median, interquartile range, and 10–90th percentile; n = 100 cells pooled from three independent experiments. Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001, *P < 0.05). See also Supplementary Figures 3 and 4. Uncropped images of blots are shown in Supplementary Figure 6.
To define the role of ATG12-ATG3 conjugation in endolysosomal trafficking, we incubated cells with fluorescent Bodipy-labeled LDL and monitored trafficking after a 120 min chase period. Following endocytosis, LDL is trafficked from early to late endosomes and ultimately to lysosomes for degradation. In WTATG3 cells, LDL primarily localized to EEA1-positive early endosomes at 0 min, throughout the endocytic pathway at 30 min, and to LBPA-positive late endosomes and LAMP1-positive lysosomes at 120 min (Supplementary Fig. 3a). Consistent with a requirement for ATG12-ATG3 in late endosome function, KR cells exhibited significantly decreased trafficking of LDL to the lysosome compared to WTATG3, with a 20% reduction in LDL colocalization with LAMP1 at 120 min (Fig. 3c,d). Baf A attenuated lysosomal trafficking in all cell types (Supplementary Fig. 3b). Moreover, at this same time point, KR cells had increased colocalization between LDL and LBPA relative to WTATG3 (Fig. 3e, Supplementary Fig 3c), but minimal colocalization between LDL and EEA1 (Supplementary Fig. 3d), indicating a specific block at late endosomes. Of note, LDL trafficking defects in autophagy-deficient pBabe cells were less pronounced than in KR cells, suggesting that complete autophagy ablation impacts endosomal trafficking independent of ATG12-ATG3 conjugation.
To assess whether impaired endolysosomal trafficking in KR cells was specific to late endosome function, we incubated cells with fluorescently labeled transferrin (Tfn). Tfn is taken up by receptor-mediated endocytosis, sorted to early endosomes, and recycled back to the plasma membrane. No differences were observed in either Tfn uptake or recycling (Supplementary Fig. 4a,b), confirming that early endosome function was intact in cells lacking ATG12-ATG3. Additionally, lysosomal mass, acidification, and cathepsin activity were unchanged among the cell types (Supplementary Fig. 4c,d). Thus, defects in endosomal trafficking in cells lacking ATG12-ATG3 are not secondary to impaired lysosomal acidification or proteolytic activity; rather, they result from defective trafficking from late endosomes to lysosomes.
ATG12-ATG3 interacts with Alix
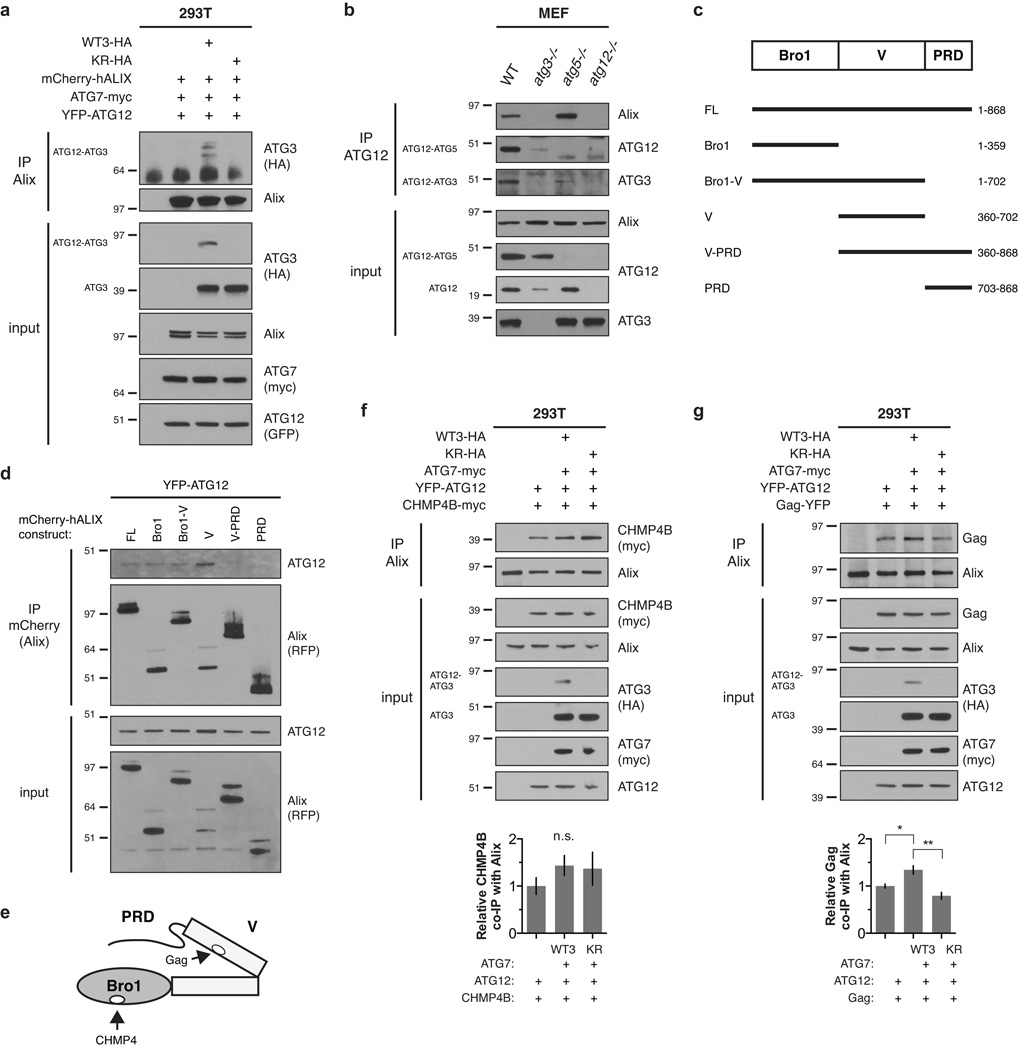
To further dissect the role of ATG12-ATG3 in late endosome function, we immunoprecipitated ATG12 from atg5−/− MEFs stably expressing tandem FLAG-HA-tagged ATG12 (FHA-ATG12) and performed liquid chromatography-tandem mass spectrometry. We identified the ESCRT-associated protein Alix (also known as PDCD6IP; expected value 7.1×10−8) as an interacting protein (Supplementary Fig. 5). Alix associates with LBPA-containing late endosomes and components of the ESCRT machinery to control topologically similar membrane events including MVB intralumenal vesicle biogenesis, exosome release, and viral budding17–21. To confirm the interaction between Alix and ATG12-ATG3, we transiently expressed WTATG3 or KR in HEK293T cells along with ATG7, ATG12, and Alix. Using this reconstitution system, the ATG12-ATG3 conjugate specifically co-immunoprecipitated with Alix, whereas unconjugated ATG3 from either WTATG3 or KR cells did not (Fig. 4a). We corroborated this interaction between Alix and the ATG12-ATG3 conjugate at endogenous protein levels; Alix co-immunoprecipitated with ATG12 in wild-type and atg5−/− MEFs, but not in atg3−/− MEFs, which lack ATG12-ATG3 (Fig. 4b).
Figure 4. ATG12-ATG3 interacts with Alix.
(a) HEK293T cells were transfected with mCherry-hALIX, Myc-tagged ATG7 (ATG7-myc), YFP-ATG12, and HA-tagged wild-type ATG3 (WTATG3-HA) or mutant K243R ATG3 (KR-HA) as indicated. Lysates were immunoprecipitated with anti-Alix. Immune complexes (IP Alix) were resolved by SDS-PAGE and immunoblotted with anti-HA and anti-Alix. (b) MEFs with the indicated genotypes were lysed and immunoprecipitated with anti-ATG12. Immune complexes (IP ATG12) were resolved by SDS-PAGE and immunoblotted with anti-Alix, anti-ATG12, and anti-ATG3. (c) Schematic representation of the conserved structural domains in Alix and truncation mutants used in this study. Numbers represent the N- and C-terminal amino acid residues of deletion mutants. (d) HEK293T cells were transfected with YFP-ATG12 and the indicated mCherry-hALIX truncation mutants. Lysates were immunoprecipitated with anti-mCherry. Immune complexes (IP mCherry) were resolved by SDS-PAGE and immunoblotted with anti-RFP and anti-ATG12. (e) Schematic representation of the conserved structural domains and protein binding sites in Alix. (f) HEK293T cells were transfected with CHMP4B-myc, ATG7-myc, YFP-ATG12, and HA-tagged wild-type ATG3 (WTATG3-HA) or mutant K243R ATG3 (KR-HA) as indicated. Lysates were immunoprecipitated with anti-Alix. Immune complexes (IP Alix) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Bottom: Quantification of co-immunoprecipitated CHMP4B (mean ± SEM; n = 5 independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test. (g) HEK293T cells were transfected with YFP-tagged murine leukemia virus Gag (Gag-YFP), ATG7-myc, YFP-ATG12, and HA-tagged wild-type ATG3 (WTATG3-HA) or mutant K243R ATG3 (KR-HA) as indicated. Lysates were immunoprecipitated with anti-Alix. Immune complexes (IP Alix) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Bottom: Quantification of co-immunoprecipitated MLV Gag (mean ± SEM; n = 3 independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (**P < 0.01, *P < 0.05). See also Supplementary Figure 5. Uncropped images of blots are shown in Supplementary Figure 6.
To define the interaction between ATG12-ATG3 and Alix, we performed co-immunoprecipitation studies with full-length Alix (FL) or Alix truncation mutants (Fig. 4c). Since unconjugated ATG3 did not co-immunoprecipitate with Alix in a HEK293T reconstitution system (Fig. 4a), we hypothesized that ATG12-ATG3 interacted with Alix via ATG12. Indeed, although Alix specifically co-immunoprecipitated with ATG12-ATG3 at endogenous protein levels (Fig. 4b), we observed interaction of free ATG12 with full-length Alix upon co-overexpressing both Alix and ATG12 (Fig. 4d).
Alix is comprised of three conserved structural domains. The N-terminal Bro1 domain binds ESCRT-III CHMP4 family members18, the central V domain binds YPXnL motifs present in retroviral Gag proteins and the adaptor syntenin17, 21, and the C-terminal proline rich domain (PRD) binds the ESCRT-I component TSG101 (Fig. 4c,e)22. The YPXnL-binding site of Alix is autoinhibited by intramolecular interaction of the PRD with the Bro1 and V domains (Fig. 4e)23, 24. In HEK293T cells co-expressing ATG12 and Alix truncation mutants, ATG12 co-immunoprecipitated with the Bro1, V, and combined Bro1-V domains. No interaction was detected between ATG12 and the PRD or V-PRD Alix mutants (Fig. 4d). Together, these data demonstrate that ATG12 interacts with the Bro1 and V domains of Alix, and suggest that the Bro1 domain is required for ATG12 binding to Alix mutants containing the PRD.
Based on these results, we postulated that ATG12-ATG3 displaces inhibitory PRD interactions in Alix, thereby promoting an active “open” conformation of Alix and enhancing the binding of V domain partners. To test this, we expressed either CHMP4B or the murine leukemia virus (MLV) Gag protein in the HEK293T reconstitution system. CHMP4B binds the Bro1 domain of Alix independently of PRD displacement18, 23, whereas Gag binding to the V domain requires PRD dissociation from Bro1-V (Fig. 4e)23, 24. Co-immunoprecipitation of CHMP4B with endogenous Alix was unaffected by ATG12-ATG3 conjugation (Fig. 4f). In contrast, co-immunoprecipitation of Gag with Alix was enhanced in cells expressing ATG12-ATG3 compared to those expressing ATG12 alone or the KR mutant (Fig. 4g). Overall, these results support the model that ATG12-ATG3 binding to Alix promotes its active “open” conformation, leading to enhanced binding of YPXnL-containing proteins such as Gag to its V domain.
ATG12-ATG3 promotes multiple Alix-mediated functions
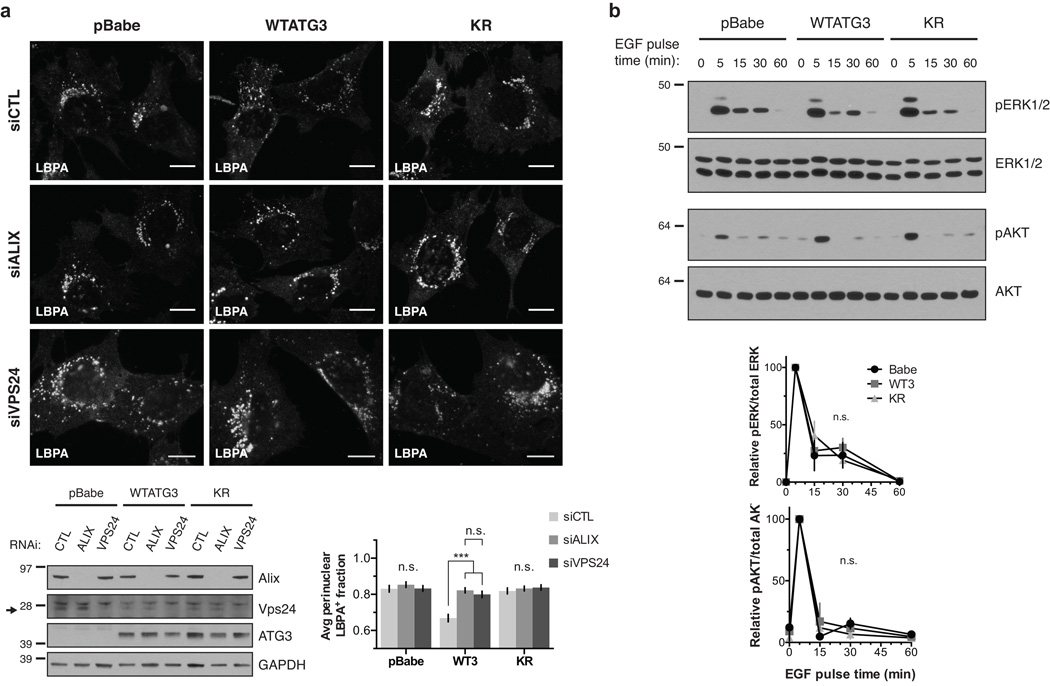
Given these interactions between ATG12-ATG3 and Alix, we reasoned that the late endosome localization and trafficking defects in KR cells were due to impaired Alix function. Previous studies have demonstrated that Alix depletion results in a perinuclear MVB distribution similar to KR cells25. These defects in Alix-depleted cells are likely mediated by changes in actin cytoskeletal organization, because the Alix V domain interacts with the actin-binding protein cortactin, and loss of Alix leads to accumulation of abnormal cortactin-positive actin structures25, 26. Indeed, we confirmed that Alix knockdown in WTATG3 cells led to perinuclear MVB accumulation similar to that observed in pBabe and KR cells. In contrast, Alix knockdown in pBabe and KR cells did not affect MVB localization, which remained perinuclear (Fig. 5a). Notably, Vps24 knockdown also resulted in the perinuclear accumulation of MVBs in WTATG3 cells, indicating that MVB localization was dependent upon ESCRT-III function (Fig. 5a). Despite causing this striking redistribution of late endosomes, loss of Alix does not affect canonical ESCRT-mediated lysosomal degradation of EGFR25, 27. Accordingly, we did not observe significant differences in EGF-induced ERK or AKT activation between pBabe, WTATG3, and KR cells (Fig. 5b).
Figure 5. Loss of ATG12-ATG3 conjugation phenocopies loss of Alix.
(a) Indicated cell types were transfected with siRNA against ALIX (siALIX), VPS24 (siVPS24), or a non-targeting control (siCTL) and immunostained with anti-LBPA to mark late endosomes. Scale bar, 10 µm. Right: Quantification of the perinuclear LBPA+ fraction as described in Fig. 2d (mean ± SEM; n = 60 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001). Matched lysates were immunoblotted for anti-Alix and anti-Vps24. (b) Indicated cell types were serum-starved overnight at 37 °C followed by treatment with 100 ng/mL EGF for the indicated times. Lysates were immunoblotted for anti-phosphorylated-ERK1/2 (pERK1/2), anti-ERK1/2, anti-phosphorylated-AKT (pAKT), and anti-AKT. Right: Quantification of phosphorylated ERK and AKT levels (mean SEM; n = 3 independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test. Uncropped images of blots are shown in Supplementary Figure 6.
Based on these results, we hypothesized that ATG12-ATG3 controls Alix functions mediated by partner-protein binding to the V domain, such as exosome release and viral budding. When MVBs fuse with the plasma membrane, the extracellular release of intralumenal vesicles gives rise to exosomes28, 29. During exosome biogenesis, Alix forms a complex with the scaffold protein syntenin, mediating loading of signaling cargo into exosomes and promoting exosome release. Viral budding involves a topologically similar process in which retroviral Gag proteins interact with Alix and other ESCRT proteins, such as TSG101, to hijack the membrane abscission machinery12, 30. Importantly, both syntenin and Gag interact directly with the V domain of Alix via conserved YPXnL motifs17, 21.
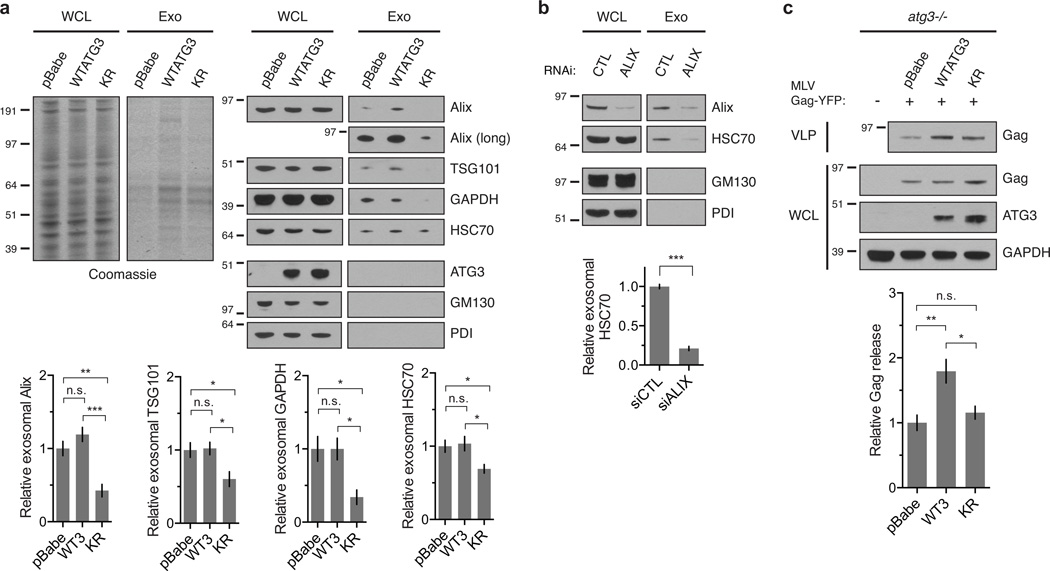
To test whether ATG12-ATG3 deficiency impaired exosome biogenesis, we isolated exosomal fractions from reconstituted MEFs by differential high-speed centrifugation. Purified exosomes contained exosomal markers including Alix and TSG101 but were absent of contamination from other membrane sources such as ER and Golgi (Fig. 6a). As a control, we confirmed that Alix knockdown impaired exosome production in wild-type MEFs (Fig. 6b). Strikingly, both total exosomal protein and specific exosomal marker proteins were decreased in KR versus WTATG3 cells (Fig. 6a), confirming reduced exosome biogenesis in cells lacking ATG12-ATG3. Notably, in contrast to KR cells, exosome release was not significantly impaired in pBabe cells. Because previous work demonstrates that autophagy inhibition enhances exosomal secretion of GAPDH31, we speculate that compensatory pathways in autophagy-deficient cells may bypass the requirement for ATG12-ATG3 in exosome biogenesis.
Figure 6. ATG12-ATG3 conjugation promotes multiple Alix functions.
(a) Whole cell lysates (WCL) and exosomal fractions (Exo) from indicated cell types were resolved by SDS-PAGE and stained by Coomassie (left) or immunoblotted for exosomal markers anti-Alix, anti-TSG101, anti-GAPDH, and anti-HSC70 (right) to measure relative exosome release. Bottom: Quantification of exosomal protein release (mean ± SEM; n = 7 independent experiments for Alix and GAPDH, n = 6 for HSC70, n = 5 for TSG101). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001, **P < 0.01, *P < 0.05). (b) Whole cell lysates (WCL) and exosomal fractions (Exo) from wild-type MEFs transfected with siRNA against Alix (ALIX) or a non-targeting control (CTL) were resolved by SDS-PAGE and immunoblotted for exosomal markers anti-Alix and anti-HSC70 to measure relative exosome release. Bottom: Quantification of exosomal HSC70 protein levels (mean ± SEM; n = 3 independent experiments). Statistical significance calculated using unpaired two-tailed t test (***P < 0.001). (c) Indicated cell types were transfected with YFP-tagged murine leukemia virus Gag (MLV Gag-YFP). Virus-like particles (VLPs) and whole cell lysates (WCL) were resolved by SDS-PAGE and immunoblotted for anti-Gag to measure relative viral budding. Bottom: Quantification of Gag release (mean ± SEM; n = 4 independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (**P < 0.01, *P < 0.05). Uncropped images of blots are shown in Supplementary Figure 6.
To assess viral budding, we isolated virus-like particles (VLPs) from cells transfected with YFP-tagged MLV Gag. Since Gag expression is sufficient for VLP production, this assay allows quantification of viral budding independent of infectivity and replication. Similar Alix deficiency, which attenuates MLV VLP production20, viral budding was decreased in pBabe and KR compared to WTATG3 cells, evidenced by reduced Gag levels in the VLP fraction (Fig. 6c). Overall, these data demonstrate that ATG12-ATG3 promotes multiple V domain-associated Alix functions, including late endosome distribution, exosome biogenesis, and viral budding.
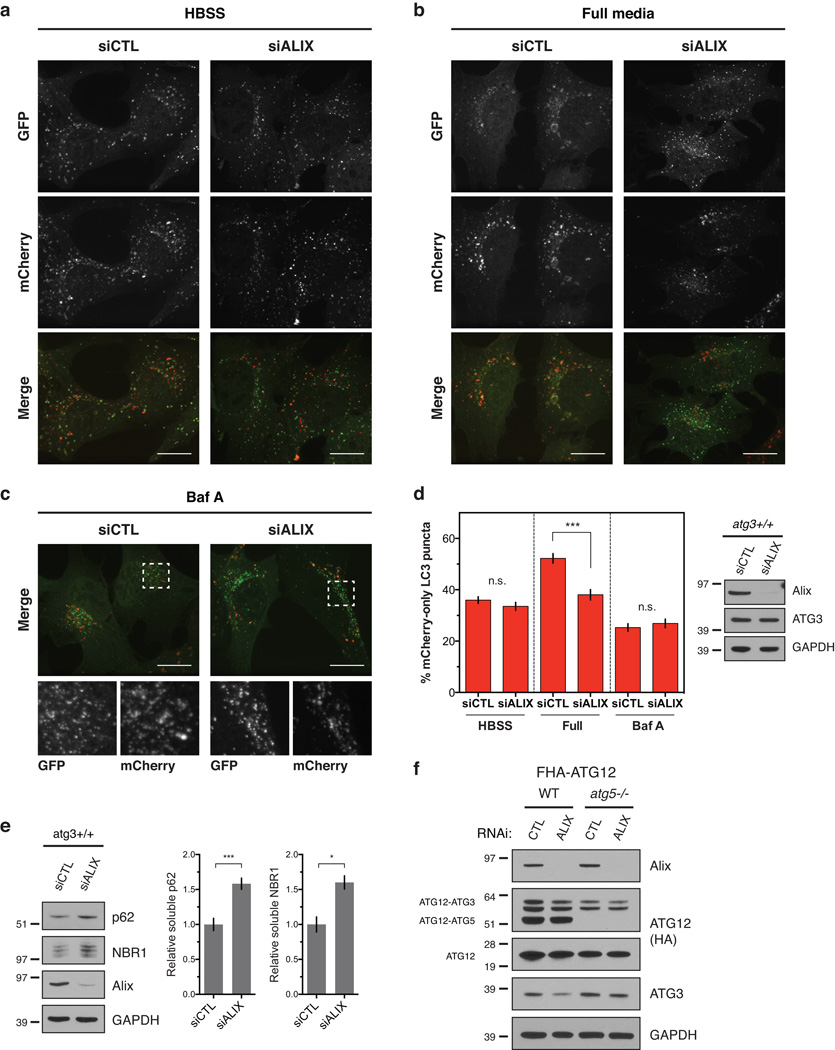
Finally, we asked whether Alix knockdown impacts macroautophagy. Consistent with previous data32, Alix depletion did not impact starvation-induced autophagy (Fig. 7a,d); rather, it specifically impaired basal autophagic flux in nutrient-rich conditions (Fig. 7b,d). Similar to loss of ATG12-ATG3, Alix depletion led to a reduction rather than complete block in basal autophagy, as Baf A treatment further reduced autolysosome formation (Fig. 7c,d). Moreover, Alix-depleted MEFs exhibited increased accumulation of p62 and NBR1 relative to cells expressing non-targeting control siRNA (Fig. 7e). This defect in basal autophagy following Alix depletion was not due to changes in ATG12-ATG3 conjugation, as Alix knockdown had no effect on ATG12-ATG3 levels (Fig. 7f). Overall, these results support that interactions between ATG12-ATG3 and Alix promote endolysosomal trafficking, exosome release, viral budding, and basal autophagy.
Figure 7. Loss of Alix specifically impairs basal autophagic flux.
(a) atg3+/+ MEFs expressing mCherry-GFP-LC3 were transfected with siRNA against ALIX (siALIX) or a non-targeting control (siCTL) and HBSS starved for 2 h. Scale bar, 15 µm. (b) atg3+/+ MEFs expressing mCherry-GFP-LC3 were transfected with siALIX or siCTL and grown in full media. Scale bar, 15 µm. (c) atg3+/+ MEFs expressing mCherry-GFP-LC3 were transfected with siRNA against ALIX (siALIX) or a non-targeting control (siCTL) and treated for 2 h with Baf A (50 nM) to block lysosome function. Scale bar, 15 µm. (d) Left: Quantification of the percentage of mCherry-positive, GFP-negative (mCherry-only) puncta per cell as described in a–c (mean ± SEM; n = 100 cells pooled from three independent experiments). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001). Right: Indicated cells were grown in full media, lysed, and immunoblotted for anti-Alix. (e) atg3+/+ MEFs were transfected with siALIX or a non-targeting control (siCTL), lysed in 1% Triton X-100, and immunoblotted with anti-p62, anti-NBR1, and anti-Alix. Right: Quantification of p62 and NBR1 protein levels (mean ± SEM; n = 5 independent experiments for p62, n = 3 for NBR1). Statistical significance calculated using ANOVA, followed by Tukey’s HSD test (***P < 0.001, *P < 0.05). (f) Stable pools of wild-type or atg5−/− MEFs expressing FLAG-HA-ATG12 (FHA-ATG12) were transfected with siRNA against Alix (ALIX) or a non-targeting control (CTL). Lysates were immunoblotted for anti-HA, anti-Alix, and anti-ATG3. Uncropped images of blots are shown in Supplementary Figure 6.
DISCUSSION
In this study, we identify an interaction between ATG12-ATG3 and Alix that enables multiple interconnected pathways, including basal autophagosome maturation, late endosome to lysosome trafficking of LDL, and Alix-dependent functions such as exosome biogenesis and viral budding. The endosomal and autophagy pathways are functionally connected; immature autophagosomes can fuse with MVBs prior to delivery to the lysosome33, 34. Multiple recent studies implicate components of the endosomal trafficking and vesicle fusion machineries in autophagosome maturation, including ESCRT complexes14, 15, SNAREs35–38, and Rab GTPases39–41. However, little is known about how individual ATGs functionally interact with the endosomal machinery. Here, we demonstrate that ATG12-ATG3 interacts with Alix and controls MVB morphology, distribution, and function. Moreover, these functions are distinct and separable from the established roles of either ATG in the early steps of autophagy.
Importantly, we show that both ATG12-ATG3 and Alix promote basal, but not starvation-induced autophagic flux. We speculate that signals activated during nutrient starvation bypass the requirement for these proteins in autolysosome formation. Previous studies support that homeostatic and stress-induced autophagosome maturation are functionally distinct processes. Similar to ATG12-ATG3, the histone deacetylase HDAC6 is required for basal autophagic flux but dispensable for starvation-induced autophagy. HDAC6 binds ubiquitinated protein aggregates and recruits cytoskeleton-remodeling machinery components to promote autophagosome-lysosome fusion at these sites. The HDAC6 substrate cortactin is required for this process42. In addition, in vitro studies demonstrate that autophagosomes isolated from starved hepatocytes can fuse normally with late endosomes and lysosomes following latrunculin treatment to block actin polymerization, whereas autophagosomes isolated from hepatocytes grown in full medium exhibit significantly decreased fusion42. These data suggest that basal autophagy is uniquely dependent upon actin remodeling for autophagosome maturation. Notably, Alix also interacts with cortactin and is required for normal cytoskeleton organization25, 26. Determining whether interactions between ATG12-ATG3 and Alix promote autophagosome maturation through similar actin-based mechanisms remains an important topic for future study. Moreover, starvation activates the transcription factors TFEB and TFE3, leading to coordinated induction of autophagy genes and lysosomal expansion in order to promote efficient autophagic flux43, 44. Hence, following starvation-induced lysosomal biogenesis, cells may no longer require ATG12-ATG3 or Alix to facilitate autophagic flux.
The identification of Alix as an ATG12-ATG3 binding partner led us to scrutinize the role of ATG12-ATG3 in Alix-dependent functions. Alix interacts with components of the ESCRT machinery to control multiple topologically similar membrane events: MVB intralumenal vesicle formation18, 19, exosome release17, and viral budding20, 21. At endogenous protein levels, Alix specifically co-immunoprecipitated with ATG12-ATG3, but not with ATG12-ATG5 or unconjugated ATG12. Intriguingly, recent work identified an amphipathic helix in ATG3 that recognizes membranes with high curvature, such as those at the growing autophagosome45. We speculate that ATG3-mediated targeting of ATG12-ATG3 to the highly curved membrane at sites of intralumenal vesicle formation, where Alix is localized, may facilitate Alix-dependent functions. Moreover, we demonstrate that the interaction between ATG12-ATG3 and Alix is mediated by binding of ATG12 to the Bro1 and V domains of Alix, and that ATG12-ATG3 promotes the co-immunoprecipitation of Gag with Alix. These data support a model in which ATG12-ATG3 binding relieves Alix auto-inhibition by its PRD domain, thereby promoting binding of partner proteins to its V domain. Consequently, loss of ATG12-ATG3 impairs Alix functions that require binding of YPXnL motif-containing proteins such as syntenin in exosome biogenesis and Gag proteins in viral budding.
Exosome biogenesis occurs when MVBs fuse with the plasma membrane, leading to intralumenal vesicle release as exosomes29. Exosomes have been identified as important signaling molecules in a wide variety of contexts, including the immune response and tumor priming of distant metastatic sites17, 46. Interestingly, studies in yeast implicate multiple ATGs in unconventional secretion of Acb1 through an exosome-like intermediate47. Our data identify the ATG12-ATG3 conjugate as a regulator of exosome release in mammalian cells. Our studies also demonstrate that ATG12-ATG3 promotes viral budding, a process mechanistically similar to exosome biogenesis in which viral proteins interact with Alix and other components of the membrane scission machinery30. Further understanding the physiological significance of ATG12-ATG3 and Alix in exosome-dependent processes such as immune signaling and metastasis and in viral infection are important topics for future study.
Defects in autophagic flux are a hallmark of various human diseases, most notably neurodegenerative conditions such as Parkinson’s and Alzheimer’s diseases48. Endolysosomal defects are also observed in Alzheimer’s disease, and ESCRT dysfunction is associated with other neuronal pathologies including frontotemporal dementia38, 49. We speculate that ATG12-ATG3 may be particularly important in non-dividing, terminally differentiated cells such as neurons that depend upon basal autophagy to degrade damaged organelles and proteins. Further dissecting the molecular basis and regulation of the interaction between ATG12-ATG3 and Alix may inform new therapeutic strategies for coordinately augmenting autophagosome maturation and late endosomal function in the treatment of such disorders.
METHODS
Reagents
Dr. Noboru Mizushima (University of Tokyo) provided atg5+/+ and atg5−/− MEFs and Dr. Masaaki Komatsu (Tokyo Metropolitan Institute) provided atg3+/+ and atg3−/− MEFs. Cells were cultured in DMEM with 10% FBS, penicillin, and streptomycin.
Commercial Antibodies
Antibodies used in this study include: anti-p62 (Progen GP62-C; 1:2,000), anti-NBR1 (Novus 71703; 1:1,000), anti-Atg3 (Sigma A3231; 1:500), anti-GAPDH (Millipore AB2302; 1:5,000), anti-EEA1 (Cell Signaling 2411; 1:100), anti-LAMP1 (BD 553792, clone 1D4B; 1:1,000), anti-LBPA (Echelon Z-SLBPA, clone 6C4; 1:100), anti-CD63 (Santa Cruz 15363; 1:50), anti-Atg12 (Cell Signaling 2011; 1:1,000), anti-Atg12 (Cell Signaling 4180, clone D88H11; 1:1,000), anti-HA (Covance HA.11, clone 16B12; 1:1,000), anti-HA (Cell Signaling 3724, clone C29F4; 1:2,000), anti-Alix (Cell Signaling 2171, clone 3A9, 1:1,000), anti-myc (Sigma 5546, clone 9E10; 1:1,000), anti-myc (Cell Signaling 2272; 1:2,000), anti-GFP (Santa Cruz 390394, clone C-2; 1:200), anti-RFP (Invitrogen R10367; 1:1,000), anti-mCherry (Molecular Probes M11217, clone 16D7; 1:100), anti-phospho-ERK1/2 (Biosource 44-680G; 1:2,000), anti-ERK1/2 (Zymed 13-6200, clone 7D8; 1:500), anti-phospho-AKT (Cell Signaling 2965, clone C31E5E; 1:500), anti-AKT (Cell Signaling 9272; 1:1,000), anti-Vps24 (Abcam 76333; 1:1,000), anti-TSG101 (Abcam 30871; 1:500), anti-HSC70 (Cell Signaling 8444, clone D12F2; 1:1,000), anti-GM130 (BD 610822, clone 35; 1:1,000), anti-PDI (Cell Signaling 2446; 1:500), and anti-Gag (Abcam 100970; 1:1,000). A rabbit polyclonal anti-LC3 antibody was created using a peptide corresponding to the conserved N-terminal sequence of human, rat, and mouse MAP1LC350 and is now commercially available (Millipore ABC232; 1:2,000).
cDNAs and generation of stably reconstituted MEFs
MLV-Gag-YFP (Plasmid 1813) and mCherry-hAlix (Plasmid 21504) constructs were obtained from Addgene51, 52. CHMP4B-myc was purchased from Origene (RC207637). Other cDNAs used in this study were previously described11 and have been deposited with Addgene. For retroviral infection, VSV-G pseudotyped retroviruses were generated, and MEFs were infected and selected as previously described11. Following selection, stable pools were used at a maximum of 6–8 passages to avoid clonal selection or drift.
RNAi
For siRNA-mediated Alix knockdown, siGenome SMARTpool siRNAs against mouse PDCD6IP/Alix (L-062173-01) and mouse CHMP3/Vps24 (L-062411-01) were purchased from Dharmacon RNA Technologies, and cells were transfected using an Amaxa nucleofactor apparatus (program U-020) and nucleofactor kit V according to manufacturer’s instructions.
Microscopy
Immunofluorescence imaging was performed using the 100× (1.3 NA) objective of a Zeiss Axiovert 200 microscope equipped with a Spot RT camera (Diagnostics Instruments) and mercury lamp; images were acquired using Metamorph software (Molecular Devices v6.0). Confocal analysis of mCherry-GFP-LC3 puncta in reconstituted atg3−/− MEFs was performed using the 60× (1.4 NA) objective of a Nikon C1si spectral confocal system equipped with an argon laser (488 line) and two solid-state diodes (405 and 546 lines); images were acquired using Nikon EZ-C1 software. All other confocal analysis was performed using the 100× (1.49 NA) objective of a Nikon inverted microscope (TE-2000 PFS) equipped with a CSU10 spinning-disk confocal unit (Yokogawa), solid-state 488 and 561 lasers, and a cooled charge-coupled device camera (Cool-SNAP-HQ2, Photometrics); images were acquired using NIS Elements software. Images were analysed in ImageJ (v1.44i).
Immunofluorescence
For endosomal/lysosomal immunostaining of EEA1, Lamp1, and LBPA, cells were fixed with 4% PFA for 10 min at room temperature and blocked/permeabilized for 30 min at room temperature in 0.05% saponin + 10% goat serum in PBS (saponin/GS). For CD63 immunostaining, cells were permeabilized with 0.05% saponin in PBS prior to fixation. Primary and secondary incubations were performed at room temperature in saponin/GS, and cells were mounted using Prolong Gold with or without DAPI. When representative images are shown, experiments were repeated a minimum of three times.
Immunoblotting
Unless otherwise indicated, cells were lysed in RIPA buffer (1% Triton X-100, 1% sodium deoxycholate, 0.1% SDS, 25 mM Tris, pH 7.6, 150 mM NaCl, 10 mM NaF, 10mM β-glycerophosphate, 1 mM Na3VO3, 10 nM calyculin A, 0.5 mM PMSF, 0.1 mM E-64-c, 10 µg/mL pepstatin A) plus protease inhibitors. Lysates were cleared by centrifugation for 15 min at 4°C, boiled in sample buffer, resolved by SDS-PAGE, and transferred to PVDF membrane. Membranes were blocked in 5% milk in PBS + 0.1% TWEEN 20 (PBST), incubated with primary antibodies overnight at 4°C in blocking buffer, washed in PBST, incubated with HRP-conjugated secondary antibodies, and analysed by enhanced chemiluminescence. When representative images are shown, experiments were repeated a minimum of three times.
mCherry-GFP-LC3 autophagic flux
MEFs expressing mCherry-GFP-LC3 were grown overnight on fibronectin-coated coverslips prior to HBSS starvation or treatment with 50 nM Baf A. Cells were fixed with 4% paraformaldehyde (PFA), washed with PBS, mounted using ProLong Gold (Life Technologies), and analysed by confocal microscopy as described above. Images were acquired randomly, and cells with intermediate total mCherry-GFP-LC3 fluorescence were chosen for further analysis. Cells were outlined manually, and each channel was independently autothresholded using the same settings across all images from each channel. mCherry and GFP puncta were quantified using the Analyze Particles plugin in ImageJ. Double-positive puncta were identified and counted using the Colocalization and Analyze Particles plugins in ImageJ.
Triton fractionation
Cells were lysed in 1% Triton X-100 in PBS plus 10 mM NaF, 10 mM β -glycerophosphate, 1 mM Na3VO3, 10 nM calyculin A, 0.5 mM PMSF, 0.1 mM E-64-c, 10 µg/mL pepstatin A, and protease inhibitors. Lysates were incubated on ice for 30 min, and soluble and insoluble fractions were separated by centrifugation at 4°C for 20 min at 16,000g. The soluble fraction was boiled in sample buffer, and the insoluble fraction was resuspended in sample buffer, sonicated, and boiled prior to immunoblotting. Protein levels were quantified by densitometry and normalized to GAPDH loading controls.
Electron microscopy
Cells were pelleted for 5 min at 4 °C at 600g, fixed on ice for 30 min in 0.1 M sodium cacodylate buffer (pH 7.4) plus 2% glutaraldehyde and 1% PFA, and pelleted at 3,000g for 10 min at 4 °C. Samples were submitted to the Gladstone Institute (UCSF) Electron Microscopy Core Facility for standard electron microscopy ultrastructural analyses. Representative images from n = 28–48 images acquired per condition, are shown.
Perinuclear LBPA and CD63 quantification
Cells were immunostained for LBPA or CD63 and DAPI and imaged by epifluorescence as described above. Images were autothresholded, and a 10 µm (for LBPA) or 5 µm (for CD63) region of interest (ROI) around the nucleus was selected using the wand tracing and enlarge selection tools in ImageJ. The perinuclear LBPA+ or CD63+ fractions were defined as the fraction of LBPA or CD63-positive integrated density within this ROI relative to the whole cell.
DQ Green BSA lysosomal degradation
Cells were incubated with 20 µg/mL DQ Green BSA (DQ-BSA; Life Technologies) for 30 min at 37 °C followed by a 2h chase in full media with or without 50 nM Baf A. Following treatment, cells were trypsinized, collected in cold PBS plus pepstatin A (10 µg/mL) and E-64-d (10 µg/mL) to block further degradation, and analysed by flow cytometry using a FACSCalibur (BD) and FlowJo software.
Bodipy-LDL trafficking
Cells were serum-starved in DMEM + 0.5% BSA + 20 mM HEPES for 1h at 37 °C, incubated with 10 µg/mL Bodipy FL LDL (LDL; Life Technologies) for 15 min at 37 °C in serum-free media, and chased for 120 min in full media. Cells were fixed in 4% PFA, immunostained for EEA1, LBPA, or LAMP1, and analysed by confocal microscopy as described above. Images were acquired randomly, and cells with at least five LDL puncta were chosen for further analysis. Cells were outlined manually, and each channel was independently autothresholded using the same settings across all images from each channel. Puncta were quantified using the Analyze Particles plugin in ImageJ. Colocalized puncta were identified and counted using the Colocalization and Analyze Particles plugins in ImageJ.
Transferrin uptake and recycling
Cells were serum-starved in DMEM + 0.5% BSA + 20 mM HEPES for 1h at 37 °C, incubated with 25 µg/mL fluorescently labeled transferrin (Tfn; Life Technologies) for 15 min at 37 °C in serum-free media, and chased in full media for up to 60 min. Cells were immunostained for EEA1 and analysed by confocal microscopy or trypsinized, fixed in 4% paraformaldehyde, and analysed by flow cytometry using a FACSCalibur (BD) and FlowJo software.
Lysosomal acidification and cathepsin activity
For lysosomal mass and acidification measurements, cells were incubated with 1 µM Lysosensor DND-153 or DND-189 (Life Technologies) in full media for 30 min at 37 °C, trypsinized, washed in PBS, and analysed by flow cytometry using a FACSCalibur (BD) and FlowJo software. For cathepsin activity assays, cells were trypsinized, washed in full media, incubated with Magic Red Cathepsin B or L substrates (ImmunoChemistry) for 30 min at 37 °C according to the manufacturer’s protocol, and analysed by flow cytometry using a FACSCalibur (BD) and FlowJo software.
Mass Spectrometry
Lysates were immunoprecipitated with monoclonal anti-HA conjugated to agarose and eluted with HA peptide (Sigma). The eluate was separated by SDS-PAGE, and Coomassie stained bands were excised, trypsin digested, and analysed on an LTQ-Orbitrap (Thermo Fisher Scientific) located in the UCSF Mass Spectroscopy Core. Fractions were separated on a 100 µm × 10 cm C18 column at flow rate 350 nL/min over a 60 min HPLC run. MS spectra were captured using one survey scan in FT at 30,000 resolution and six MS/MS events in the ion trap. Proteins were identified using Protein Prospector against the SwissProt database.
Co-immunoprecipitation
Cells were lysed in non-denaturing lysis buffer (1% Triton X-100, 50 mM Tris·Cl pH 7.4, 300 mM NaCl, 5 mM EDTA, 0.02% sodium azide, 10 mM NaF, 10mM β-glycerophosphate, 1 mM Na3VO3, 10 nM calyculin A, 0.5 mM PMSF, 0.1 mM E-64-c, 10 µg/mL pepstatin A) plus protease inhibitors, incubated on ice for 20 min, and cleared by centrifugation for 15 min at 4°C. Lysates were pre-cleared with protein A/G agarose beads (Santa Cruz) and incubated with primary antibody overnight at 4 °C followed by incubation with 20 µL A/G beads for 2 h at 4 °C. Lysates were washed 5 times with 1 mL of PBS, eluted in sample buffer, and analysed by immunoblotting as described above. For CHMP4B and Gag co-immunoprecipitations with Alix in Figure 5, protein levels were quantified by densitometry and normalized to immunoprecipitated Alix. When representative images are shown, experiments were repeated a minimum of three times.
EGF signaling
Cells were serum-starved in DMEM + 0.5% BSA + 20 mM HEPES for overnight at 37 °C, treated with 100 ng/mL EGF for up to 60 min at 37 °C in serum-free media, and analyzed by immunoblotting as described above. Levels of phosphorylated ERK and AKT were quantified by densitometry and normalized to total ERK/AKT levels.
Exosome isolation
For exosome biogenesis assays, equal numbers of cells were seeded; upon reaching 80% confluence, cells were washed and incubated in equal volumes of DMEM containing 10% exosome-depleted FBS. Exosome isolation was performed 24h later by differential centrifugation as described previously53. The corresponding cells were lysed in RIPA as described above. Exosomal and whole cell lysate (WCL) fractions were resolved by SDS-PAGE and stained by Coomassie to visualize total protein or immunoblotted for exosomal and membrane markers. Exosomal protein levels were quantified by densitometry.
Virus-like particle (VLP) isolation
For VLP budding assays, equal numbers of cells were seeded; upon reaching 80% confluence, cells were washed and incubated in equal volumes of full media. VLP isolation was performed 24h later by sequential centrifugation at 4 °C. Conditioned media was spun for 5 min at 200 g to remove cells, filtered through a 0.45 µm pre-wet filter to remove cellular debris, overlaid on a 20% sucrose cushion and spun for 2h at 130,000g to pellet VLPs. The corresponding cells were lysed in RIPA, and VLP and WCL fractions were resolved by SDS-PAGE and immunoblotted with anti-Gag. Levels of VLP Gag were quantified by densitometry and normalized to levels of Gag in the whole cell lysate.
Statistics
Experimental groups were compared using unpaired t test or ANOVA followed by Tukey’s HSD test for multiple comparisons as indicated.
Supplementary Material
Acknowledgements
Confocal microscopy was performed in the Biological Imaging Development Center at UCSF. Grant support to J.D. includes the NIH (CA126792, CA188404) and a Howard Hughes Medical Institute Physician-Scientist Early Career Award. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship to L.M. under grant DGE-1144247.
Footnotes
Author contributions
L.M. and J.D. conceived the study and designed the experiments. L.M. performed the experiments and analysed the data. R.M. performed the mass spectrometry analysis. L.M. and J.D. wrote the manuscript.
Competing financial interest
The authors declare no competing financial interests.
References
- 1.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annual Review of Pathology: Mechanisms of Disease. 2013;8:105–137. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ichimura Y, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 4.Mizushima N, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 5.Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nature Reviews Molecular Cell Biology. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- 6.Nemoto T, et al. The mouse APG10 homologue, an E2-like enzyme for Apg12p conjugation, facilitates MAP-LC3 modification. Journal of Biological Chemistry. 2003;278:39517–39526. doi: 10.1074/jbc.m300550200. [DOI] [PubMed] [Google Scholar]
- 7.Tanida I, et al. Apg7p/Cvt2p: a novel protein-activating enzyme essential for autophagy. Molecular biology of the cell. 1999;10:1367–1379. doi: 10.1091/mbc.10.5.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. Journal of Biological Chemistry. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- 9.Bestebroer J, V'kovski P, Mauthe M, Reggiori F. Hidden behind autophagy: the unconventional roles of ATG proteins. Traffic. 2013;14:1029–1041. doi: 10.1111/tra.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Subramani S, Malhotra V. Non-autophagic roles of autophagy-related proteins. EMBO reports. 2013;14:143–151. doi: 10.1038/embor.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radoshevich L, et al. ATG12 conjugation to ATG3 regulates mitochondrial homeostasis and cell death. Cell. 2010;142:590–600. doi: 10.1016/j.cell.2010.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raiborg C, Stenmark H. The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature. 2009;458:445–452. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 13.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. The Journal of cell biology. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee J-A, Beigneux A, Ahmad ST, Young SG, Gao F-B. ESCRT-III dysfunction causes autophagosome accumulation and neurodegeneration. Current biology. 2007;17:1561–1567. doi: 10.1016/j.cub.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 15.Rusten TE, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Current biology. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 16.Kimura S, Noda T, Yoshimori T. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 2007;3:452–460. doi: 10.4161/auto.4451. [DOI] [PubMed] [Google Scholar]
- 17.Baietti MF, et al. Syndecan-syntenin-ALIX regulates the biogenesis of exosomes. Nature cell biology. 2012;14:677–685. doi: 10.1038/ncb2502. [DOI] [PubMed] [Google Scholar]
- 18.Katoh K, et al. The ALG-2-interacting protein Alix associates with CHMP4b, a human homologue of yeast Snf7 that is involved in multivesicular body sorting. Journal of Biological Chemistry. 2003;278:39104–39113. doi: 10.1074/jbc.M301604200. [DOI] [PubMed] [Google Scholar]
- 19.Matsuo H, et al. Role of LBPA and Alix in multivesicular liposome formation and endosome organization. Science. 2004;303:531–534. doi: 10.1126/science.1092425. [DOI] [PubMed] [Google Scholar]
- 20.Segura-Morales C, et al. Tsg101 and Alix interact with murine leukemia virus Gag and cooperate with Nedd4 ubiquitin ligases during budding. Journal of Biological Chemistry. 2005;280:27004–27012. doi: 10.1074/jbc.M413735200. [DOI] [PubMed] [Google Scholar]
- 21.Strack B, Calistri A, Craig S, Popova E, Göttlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003;114:689–699. doi: 10.1016/s0092-8674(03)00653-6. [DOI] [PubMed] [Google Scholar]
- 22.von Schwedler UK, et al. The protein network of HIV budding. Cell. 2003;114:701–713. doi: 10.1016/s0092-8674(03)00714-1. [DOI] [PubMed] [Google Scholar]
- 23.Zhai Q, et al. Activation of the retroviral budding factor ALIX. Journal of virology. 2011;85:9222–9226. doi: 10.1128/JVI.02653-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, et al. The HIV-1 p6/EIAV p9 docking site in Alix is autoinhibited as revealed by a conformation-sensitive anti-Alix monoclonal antibody. Biochem. J. 2008;414:215–220. doi: 10.1042/BJ20080642. [DOI] [PubMed] [Google Scholar]
- 25.Cabezas A, Bache KG, Brech A, Stenmark H. Alix regulates cortical actin and the spatial distribution of endosomes. Journal of cell science. 2005;118:2625–2635. doi: 10.1242/jcs.02382. [DOI] [PubMed] [Google Scholar]
- 26.Pan S, et al. Involvement of the conserved adaptor protein Alix in actin cytoskeleton assembly. Journal of Biological Chemistry. 2006;281:34640–34650. doi: 10.1074/jbc.M602263200. [DOI] [PubMed] [Google Scholar]
- 27.Bowers K, et al. Degradation of endocytosed epidermal growth factor and virally ubiquitinated major histocompatibility complex class I is independent of mammalian ESCRTII. Journal of Biological Chemistry. 2006;281:5094–5105. doi: 10.1074/jbc.M508632200. [DOI] [PubMed] [Google Scholar]
- 28.Fevrier B, Raposo G. Exosomes: endosomal-derived vesicles shipping extracellular messages. Current opinion in cell biology. 2004;16:415–421. doi: 10.1016/j.ceb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nature Reviews Immunology. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 30.Martin-Serrano J, Neil SJ. Host factors involved in retroviral budding and release. Nature Reviews Microbiology. 2011;9:519–531. doi: 10.1038/nrmicro2596. [DOI] [PubMed] [Google Scholar]
- 31.Sahu R, et al. Microautophagy of cytosolic proteins by late endosomes. Developmental cell. 2011;20:131–139. doi: 10.1016/j.devcel.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petiot A, et al. Alix differs from ESCRT proteins in the control of autophagy. Biochemical and biophysical research communications. 2008;375:63–68. doi: 10.1016/j.bbrc.2008.07.136. [DOI] [PubMed] [Google Scholar]
- 33.Berg TO, Fengsrud M, Stromhaug PE, Berg T, Seglen PO. Isolation and characterization of rat liver amphisomes - Evidence for fusion of autophagosomes with both early and late endosomes. Journal of Biological Chemistry. 1998;273:21883–21892. doi: 10.1074/jbc.273.34.21883. [DOI] [PubMed] [Google Scholar]
- 34.Fader CM, Sanchez D, Furlan M, Colombo MI. Induction of autophagy promotes fusion of multivesicular bodies with autophagic vacuoles in k562 cells. Traffic. 2008;9:230–250. doi: 10.1111/j.1600-0854.2007.00677.x. [DOI] [PubMed] [Google Scholar]
- 35.Moreau K, Ravikumar B, Renna M, Puri C, Rubinsztein DC. Autophagosome precursor maturation requires homotypic fusion. Cell. 2011;146:303–317. doi: 10.1016/j.cell.2011.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nair U, et al. SNARE proteins are required for macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Zhang Z, Sun D, Sweeney ST, Gao F-B. Syntaxin 13, a Genetic Modifier of Mutant CHMP2B in Frontotemporal Dementia, Is Required for Autophagosome Maturation. Molecular cell. 2013;52:264–271. doi: 10.1016/j.molcel.2013.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gutierrez MG, Munafó DB, Berón W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. Journal of cell science. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- 40.Jäger S, et al. Role for Rab7 in maturation of late autophagic vacuoles. Journal of cell science. 2004;117:4837–4848. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- 41.Ganley IG, Wong P-M, Gammoh N, Jiang X. Distinct autophagosomal-lysosomal fusion mechanism revealed by thapsigargin-induced autophagy arrest. Molecular cell. 2011;42:731–743. doi: 10.1016/j.molcel.2011.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee JY, et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. The EMBO journal. 2010;29:969–980. doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martina JA, et al. The Nutrient-Responsive Transcription Factor TFE3 Promotes Autophagy, Lysosomal Biogenesis, and Clearance of Cellular Debris. Science signaling. 2014;7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Settembre C, et al. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nath S, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nature cell biology. 2014 doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bobrie A, Colombo M, Raposo G, Théry C. Exosome secretion: molecular mechanisms and roles in immune responses. Traffic. 2011;12:1659–1668. doi: 10.1111/j.1600-0854.2011.01225.x. [DOI] [PubMed] [Google Scholar]
- 47.Duran JM, Anjard C, Stefan C, Loomis WF, Malhotra V. Unconventional secretion of Acb1 is mediated by autophagosomes. The Journal of cell biology. 2010;188:527–536. doi: 10.1083/jcb.200911154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheung ZH, Ip NY. Autophagy deregulation in neurodegenerative diseases – recent advances and future perspectives. Journal of neurochemistry. 2011;118:317–325. doi: 10.1111/j.1471-4159.2011.07314.x. [DOI] [PubMed] [Google Scholar]
- 49.Nixon RA. Endosome function and dysfunction in Alzheimer's disease and other neurodegenerative diseases. Neurobiology of aging. 2005;26:373–382. doi: 10.1016/j.neurobiolaging.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 50.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of autophagy during extracellular matrix detachment promotes cell survival. Molecular biology of the cell. 2008;19:797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sherer NM, et al. Visualization of retroviral replication in living cells reveals budding into multivesicular bodies. Traffic. 2003;4:785–801. doi: 10.1034/j.1600-0854.2003.00135.x. [DOI] [PubMed] [Google Scholar]
- 52.Lee HH, Elia N, Ghirlando R, Lippincott-Schwartz J, Hurley JH. Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science. 2008;322:576–580. doi: 10.1126/science.1162042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Current Protocols in Cell Biology. 2006:3.22. 21–23.22. 29. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.