Abstract
Background
The S. cerevisiae carbamylphosphate synthetase – aspartate transcarbamylase multifunctional protein catalyses the first two reactions of the pyrimidine pathway. In this organism, these two reactions are feedback inhibited by the end product UTP. In the present work, the mechanisms of these integrated inhibitions were studied.
Results
The results obtained show that the inhibition is competitive in the case of carbamylphosphate synthetase and non-competitive in the case of aspartate transcarbamylase. They also identify the substrate whose binding is altered by this nucleotide and the step of the carbamylphosphate synthetase reaction which is inhibited. Furthermore, the structure of the domains catalyzing these two reactions were modelled in order to localize the mutations which, specifically, alter the aspartate transcarbamylase sensitivity to the feedback inhibitor UTP. Taken together, the results make it possible to propose a model for the integrated regulation of the two activities of the complex. UTP binds to a regulatory site located in the vicinity of the carbamylphosphate synthetase catalytic subsite which catalyzes the third step of this enzyme reaction. Through a local conformational change, this binding decreases, competitively, the affinity of this site for the substrate ATP. At the same time, through a long distance signal transmission process it allosterically decreases the affinity of the aspartate transcarbamylase catalytic site for the substrate aspartate.
Conclusion
This investigation provides informations about the mechanisms of allosteric inhibition of the two activities of the CPSase-ATCase complex. Although many allosteric monofunctional enzymes were studied, this is the first report on integrated allosteric regulation in a multifunctional protein. The positions of the point mutations which specifically abolish the sensitivity of aspartate transcarbamylase to UTP define an interface between the carbamylphosphate synthetase and aspartate transcarbamylase domains, through which the allosteric signal for the regulation of aspartate transcarbamylase must be propagated.
Background
Although numerous allosteric enzymes were studied, much less information is available concerning the coordinated regulation of activities in multienzymatic complexes. Two feedback inhibited multienzyme complexes were studied in Saccharomyces cerevisiae, the N-acetylglutamate synthase/N-acetyl glutamate kinase [1] and the carbamylphosphate synthetase – aspartate transcarbamylase [2] complexes.
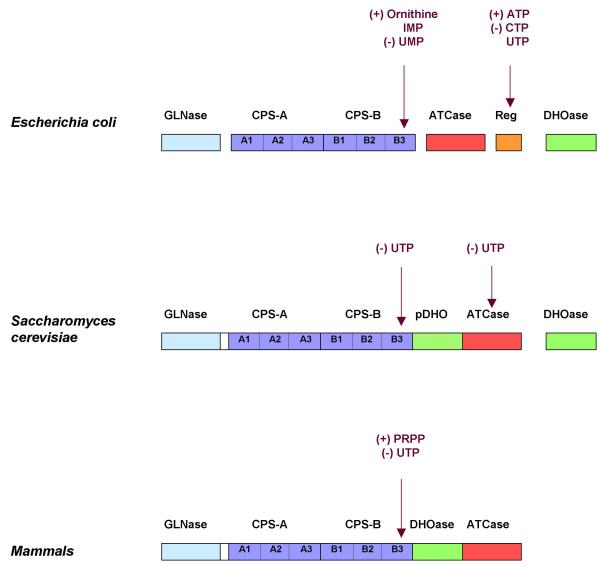
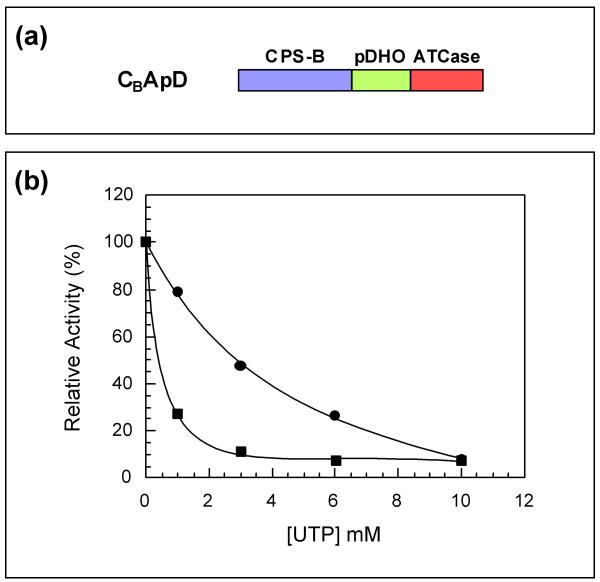
In procaryotes the first three reactions of the pyrimidine biosynthetic pathway are catalyzed by independent enzymes namely carbamylphosphate synthetase (CPSase, EC 2.7.2.9), aspartate transcarbamylase (ATCase, EC 2.1.3.2), and dihydroorotase (DHOase, EC 3.5.2.3). Escherichia coli CPSase is feedback inhibited by UMP [3] and ATCase by CTP and UTP [4] (Figure 1). In contrast, in mammals these three reactions are catalyzed by an hexameric 240-kDa multifunctional protein, the protein CAD, made of covalently linked domains, each one catalyzing one of these reactions [5-7]. In this case, only CPSase is regulated, being feedback inhibited by UTP [8]. An interesting intermediary organisation is observed in S. cerevisiae. In this case a 240-kDa bifunctional protein encoded by the URA2 locus possesses the CPSase and ATCase activities [2,9,10] but lacks the DHOase activity, although it contains an inactive pseudo-DHOase (pDHO) domain [10,11] homologous to the functional DHOases [5-7]. In this organism both CPSase and ATCase are feedback inhibited by UTP [12,13] (Figure 1). Similarly to what is observed in CAD, this yeast multifunctional protein is organized into four major functional domains:
Figure 1.
Organization and allosteric regulatory properties of the enzymes catalyzing the first three reactions of the pyrimidine pathway in E. coli, S. cerevisiae and mammals. This scheme shows the functional domains that catalyze the first steps in the de novo pyrimidine biosynthetic pathway, the amidotransferase or glutaminase domain (GLNase), the CPSase synthetase domain consisting of two subdomains (CPS-A and CPS-B), the dihydroorotase domain (DHOase), and the ATCase domain. The activities are associated with separate polypeptide chains in E. coli, whereas in mammals (CAD) all are consolidated on a single multifunctional protein. In yeast, the CPSase and ATCase domains are carried by a single polypeptide, but in this case the active DHOase that is encoded by a separate gene is replaced with an inactive DHOase homologue (pDHO). Eucaryotes ATCases lack the regulatory subunit (Reg) found in the E. coli protein. The scheme also shows the allosteric effectors that regulate the activity of these proteins and the localization of the regulatory sites represented by arrows.
-The glutaminase (GLNase) domain which hydrolyzes glutamine and transfers ammonia to the carbamylphosphate synthetase domain [10].
-The CPSase domain which catalyzes the synthesis of carbamylphosphate from two molecules of ATP, bicarbonate and ammonia in a stepwise fashion that involves three partial reactions: the activation of bicarbonate by ATP, the reaction of the activated species, carboxyphosphate, with ammonia to form carbamate and the ATP-dependent phosphorylation of carbamate to form carbamylphosphate [3,14]:
(1) ATP-Mg + HCO32- 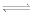 -OCOOPO32- + ADP-Mg
-OCOOPO32- + ADP-Mg
(2) -OCOOPO32- + NH3 (Gln) 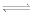 NH2COO- + Pi + (Glu)
NH2COO- + Pi + (Glu)
(3) NH2COO- + ATP-Mg 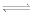 NH2COOPO32- + ADP-Mg
NH2COOPO32- + ADP-Mg
-The inactive pDHO domain.
-The ATCase domain which catalyzes the reaction of carbamylphosphate and aspartate to form carbamylaspartate.
The N- and C-halves of CPSases from all organisms examined so far, show a significant degree of sequence similarity [10,15-20], an observation which was interpreted to mean that the genes coding for these enzymes evolved through a process of gene duplication, fusion, and differentiation [15,16]. The two domains corresponding to these two halves are called CPS-A and CPS-B. Unexpectedly, it was discovered that each of these two domains of the mammalian CAD CPSase are able to independently catalyze the formation of carbamylphosphate provided that they dimerize [21,22]. In the same way, a truncated yeast bifunctional protein lacking the GLNase and CPS-A domains (CBApD) was shown to possess the CPSase activity regulated by UTP [23]. In contrast, the ATCase domain was no longer sensitive to this nucleotide, indicating that the two catalytic activities are controlled by distinct mechanisms [23].
In order to identify amino acid residues implicated in the feedback inhibition by UTP, genetics were used to positively select in vivo and characterize missense mutations in the URA2 gene, which specifically affect the feedback – inhibition of ATCase [24]. In these mutants ATCase is no longer inhibited by UTP although CPSase retains full sensitivity to this effector, indicating again that UTP affects the activities of CPSase and ATCase by different mechanisms [25].
In the present work the use of S. cerevisiae mutants in which single amino acid replacements abolish the sensitivity of ATCase to UTP allowed to study specifically the process of feedback inhibition of the CPSase domain by this nucleotide. In addition, the reaction step affected by the feedback inhibitor UTP was identified in both the entire complex and in the truncated protein (CBApD). Moreover a computational approach was used to predict the structures of the CPSase and ATCase domains. The results obtained provide informations about the integrated allosteric regulation of the two enzymatic activities of the complex, and indicate that the regulatory site is located in the CPS-B domain. The modelling defines an interface between the CPSase and ATCase domains for the transmission of the allosteric signal.
Results
Inhibition of the CPSase and ATCase activities of the complex
UTP inhibition of the coupled reaction
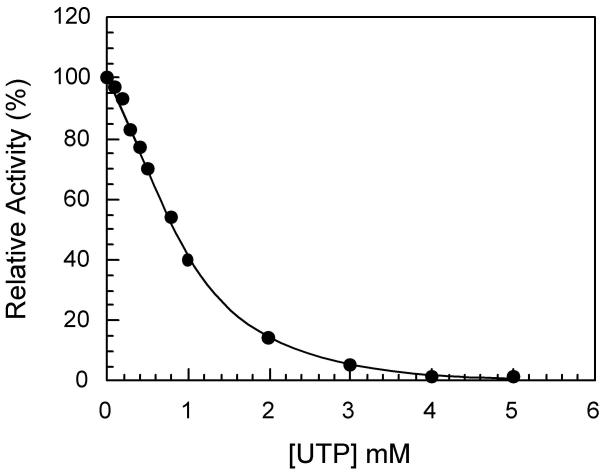
The influence of the feedback inhibitor UTP on the S. cerevisiae coupled CPSase-ATCase reaction catalyzed by the complex was determined using the wild type complex present in the URA2-EK1104 strain, and is shown in Figure 2. Under the conditions used, complete inhibition is observed for UTP concentrations superior to 3 mM, and half inhibition of the overall reaction is observed in the presence of 0.8 mM. The specific inhibition by UTP of each of the two individual reactions was then investigated.
Figure 2.
UTP inhibition of the S. cerevisiae CPSase-ATCase coupled reaction. The influence of UTP on the coupled CPSase-ATCase reaction was determined as indicated in the Methods using the dialyzed crude extract of the E. coli EK1104 strain transformed by pC4-URA2, in the presence of 50 mM aspartate.
UTP inhibition of ATCase
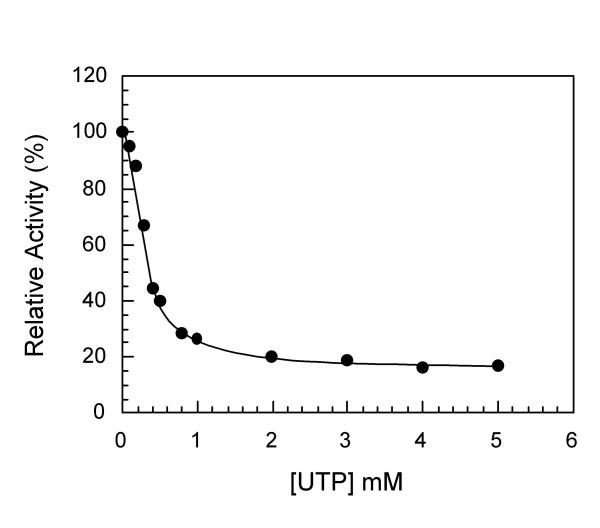
The ATCase activity of the complex can be specifically tested without interference of the CPSase activity by providing carbamylphosphate and aspartate to the complex. The influence of UTP on this activity was determined using the same strain than above. The result obtained is shown in Figure 3. Complete inhibition cannot be obtained and under the conditions used (50 mM aspartate) one observes a 20% residual activity. Similar results were previously reported [12,25,26], the value of the plateau of inhibition varying with the aspartate concentration [26]. Such a pattern is characteristic of a process of partial competitive inhibition in which the binding of the inhibitor to a site distinct from the catalytic site decreases the affinity of the active site for the substrate through a conformational change. This result is consistent with the previously reported observation that towards carbamylphosphate the inhibition by UTP is of uncompetitive type [26].
Figure 3.
UTP inhibition of the S. cerevisiae ATCase reaction of the CPSase-ATCase complex. The influence of UTP on the ATCase reaction of the bifunctional complex was determined as indicated in the Methods in the presence of 50 mM aspartate and 10 mM carbamylphosphate using dialyzed crude extract of the E. coli EK1104 strain transformed by pC4-URA2.
UTP inhibition of CPSase
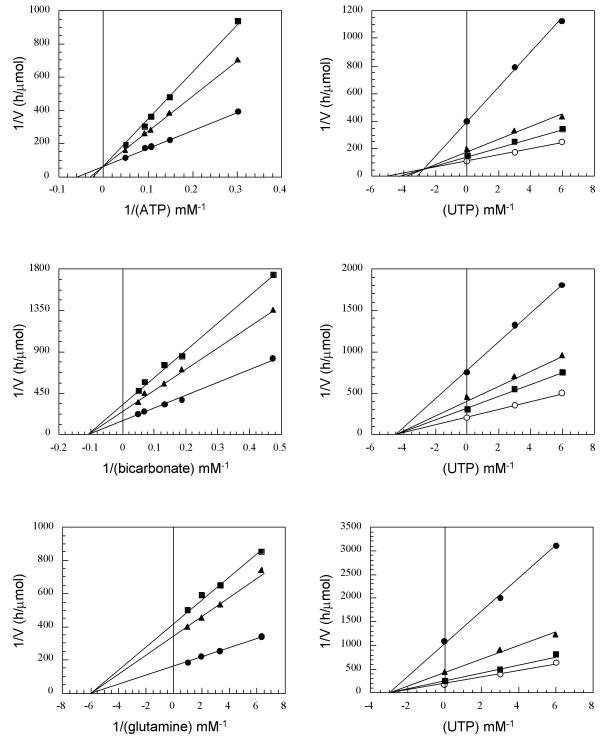
Since the CPSase assay involves its coupled reaction with ATCase, in order to study specifically the influence of the feedback inhibitor on the CPSase reaction, it was necessary to use mutants in which the ATCase reaction is insensitive to this effector. Such mutants were previously described [25] and it was shown that this specific desensitization could result from single missense mutations located either in the ATCase domain or in the CPSase domain of the complex [25]. Three of these mutants were used in this study: Asn1094→Asp located in the CPSase domain; Glu1933→Lys and Glu2182→Lys located in the ATCase domain [25] (Table 1). Complete inhibition of the CPSase reaction by UTP was observed in the three cases, in accordance with the complete inhibition of the coupled reaction reported above. In order to investigate the nature of this inhibition and to identify the substrate(s) whose binding or utilization is altered by the effector, the influence of UTP was measured in the presence of varying concentrations of the three substrates of CPSase: ATP, glutamine, and bicarbonate. The three mutants provide the same results which are presented here in the case of Glu2182→Lys. These results are shown in Figure 4, where double reciprocal plots are presented on the left, and Dixon plots on the right. Taken together, these results show unambiguously that UTP acts as a competitive inhibitor towards ATP and as a non competitive inhibitor towards bicarbonate and glutamine, a pattern which indicates that UTP acts by decreasing specifically the apparent affinity of the catalytic site for ATP with a KI of 3 ± 1 mM. These results indicate a process of absolute competitive inhibition toward ATP.
Table 1.
Position of the missense mutations affecting the sensitivity of ATCase to UTP.
| Mutants | Domain |
| Arg1076 →Ser | |
| Asn1094 →Asp | |
| Tyr1096 →Cys | CPSase |
| Trp1112 →Leu | |
| Asp1220 →Tyr | |
| Phe1924 →Val | |
| Glu1933 →Lys | |
| Glu1933 →Gly | |
| Glu2052 →Lys | |
| Gly2055 →Ser | ATCase |
| Asn2058 →Asp | |
| Asn2157 →Lys | |
| Glu2182 →Lys | |
| Asp2186 →Asn | |
All the mutants carry missense mutations leading to a single amino acids replacement. Five of them are located in the CPSase domains while the others are in the ATCase domain. In these mutants, ATCase is no longer inhibited by UTP although CPSase retains full sensitivity to this effector.
Figure 4.
Substrate saturation curves of the CPSase from the mutant Glu 2182→Lys in presence or absence of UTP. The CPSase activity was measured as indicated in the Methods. On the left are represented the double reciprocal plots in the presence of (●) 0 mM, (◆) 3 mM, and (■) 6 mM UTP. On the right are presented the corresponding Dixon plots. The concentrations used were (●) 6.7 mM, (◆) 9.3 mM, (■) 10.7 mM, and (○) 33 mM in the case of ATP; (●) 2.1 mM, (◆) 5.3 mM, (■) 7.5 mM, and (○) 20 mM in the case of bicarbonate; (●) 0.03 mM, (◆) 0.07 mM, (■) 0.3 mM, and (○) 1 mM in the case of glutamine.
The UTP inhibition curves of the complexe and of the individual CPSase and ATCase reactions exhibit a slight cooperativity with a Hill number of 1.2 ± 0.1. This is shown in Figure 5 in the case of the Glu2182→Lys mutant.
Figure 5.
UTP inhibition curve of the CPSase from the mutant Glu 2182→Lys. The percentage of inhibition by UTP was tested under the standard conditions as indicated in the Methods in the presence of increasing concentrations of UTP. The kinetic data were fit to the Hill equation : 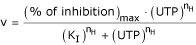
Determination of the CPSase reaction step influenced by the feedback inhibitor UTP
Wild type complex
As indicated in the Background section, the reaction catalyzed by CPSase is complex and consists in three partial reactions. Two of them involve a molecule of ATP and these two steps, (1) and (3), can be specifically tested: step (1) on the basis of the bicarbonate dependent ATP/ADP exchange and step (3) in the presence of carbamylphosphate and ADP measuring the ATP production using a coupled reaction with hexokinase and glucose 6-phosphate dehydrogenase (see Methods). The influence of UTP on these two partial reactions was investigated and the results obtained are shown in Figure 6, which shows clearly that it is strictly the third partial reaction catalyzed by the CPS-B domain, which is specifically inhibited by UTP.
Figure 6.
Influence of UTP on the partial reactions 1 and 3 of the CPSase reaction of the wild type complex. The influence of UTP on steps 1 and 3 was determined as indicated in the Methods using dialyzed crude extract of the E. coli EK1104 strain transformed by pC4-URA2. (●) partial reaction 1, and (■) partial reaction 3.
CBApD mutant
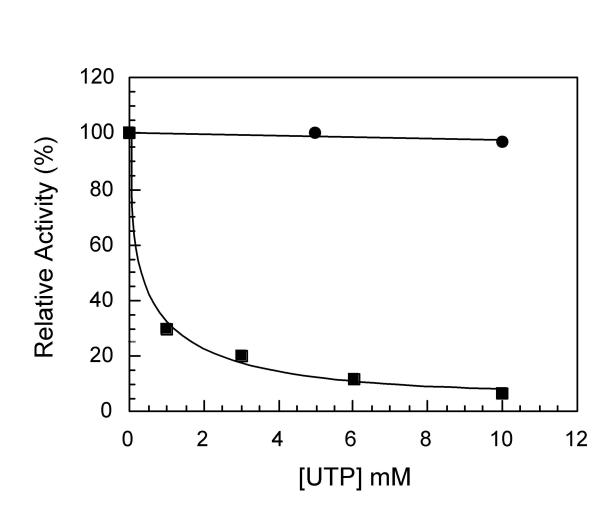
In the CBApD mutant it is a dimer of CPS-B domain (see Background and Fig. 7a) which catalyzes the three steps of the CPSase reaction [23]. Since in the wild type complex it is only the CPS-B domain which is sensitive to UTP, it was interesting to determine whether in this mutant the step 1 catalyzed by this domain becomes also sensitive to the feedback inhibitor. Thus, the putative influence of UTP on the partial reaction 1 was investigated in comparison with the partial reaction 1 catalyzed by CPS-A in the wild type complex. The results obtained are shown in Figure 7b were it appears that in this mutant, both steps 1 and 3 are feedback inhibited by UTP, indicating that the regulatory site for UTP is effective for the inhibition of step 1 when catalyzed by the CPS-B where it is located. This observation is consistent with the lack of a regulatory site in the CPS-A domain [27,28].
Figure 7.
UTP influence on the partial reactions 1 and 3 of the CPSase reaction catalyzed by CBApD. (a) The truncated complex CBApD possess CPS-B, pDHO and ATCase domains. (b) The influence of UTP on steps 1 and 3 was determined as indicated in the Methods using dialyzed crude extract. (●) partial reaction 1, and (■) partial reaction 3.
Modelling of the CPSase and ATCase domains and localization of the missense mutations affecting the sensitivity of ATCase to UTP
Amino acid sequences alignment
In order to model the CPSase domain of the complex, we used FASTP to compare the amino acid sequences of E. coli CPSase and the S. cerevisiae CPSase domain of the CPSase-ATCase complex (Fig. 8). These two homologous proteins have approximately the same size, and show very similar sequences with few insertions and deletions. The degree of identity between the E. coli enzyme (residues 2–1073) and the yeast CPSase domain (434–1502) is 42%. This alignment involves 10 small deletions and 6 insertions in the yeast sequence (Fig. 8). None of these insertions and deletions occur in highly conserved regions or within regions of well-defined secondary structure (not shown). Moreover, all the amino acid residues of the catalytic site which were identified in the X-ray structure of the E. coli CPSase are conserved in the yeast sequence. The same method was used to align the sequences of the ATCase domain of the S. cerevisiae complex and the ATCase catalytic chain of E. coli. The result obtained is identical to that previously reported from the use of the Needleman and Wunsch algorithm [29,30]. The degree of identity is 42% and that involves the presence of 4 minor deletions and 3 insertions. Here also all the amino acid residues which were previously shown to interact with the substrates in the catalytic site of E. coli ATCase are present in S. cerevisiae [30,31].
Figure 8.
Sequence alignment of the CPSase(s) from E. coli and S. cerevisiae. The amino acids sequences of the CPSase(s) from E. coli and S. cerevisiae were aligned as indicated in the Methods. CARB-E : E. coli; PYR1-Y: S. cerevisiae. ([ ]), limits of the sequence used for the modeling process. (↑), limit between A and B domains. (◆), point mutations which abolish the sensitivity of the ATCase domain to UTP (see table 1). (*) lysine992 and serine948 residues located in the UMP binding site of the E. coli CPSase.
Modelling of the S. cerevisiae CPSase and ATCase domains
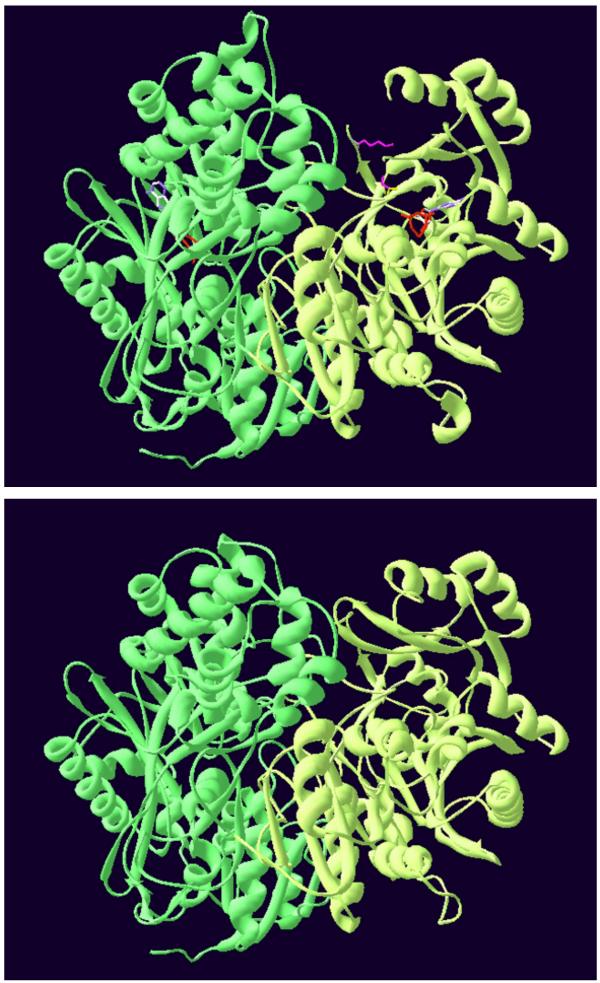
The similarity of the S. cerevisiae CPSase domain and the E. coli CPSase in sequence and kinetic properties prompted us to model the structure of this yeast domain using the X-ray coordinates of the E. coli CPSase [32] as a tertiary template as indicated in Methods. The model obtained is shown as a Swiss-PdbViewer representation in Figure 9. It appears clearly that these two structures are very similar. In the same way and for the sake of homogeneity, the same procedure was used to model the S. cerevisiae ATCase domain on the basis of the cristallographic structure of the homologous E. coli enzyme [33]. Again, the two structures appear to be strikingly identical (not shown) in accordance with the observation previously made using a different procedure [30].
Figure 9.
Modelled structure of the S. cerevisiae CPSase domain of the CPSase-ATCase complex. Top: cristallographic structure of the E. coli CPSase [32]. Bottom: modelled structure of the S. cerevisiae CPSase domain of the bifunctional protein obtained as indicated in the Methods. The CPS-A and CPS-B domains are in green and light-green respectively. The two catalytic subsites of the E. coli CPSase are indicated by two molecules of ADP. Lysine992 and serine948 located in the UMP binding site of the E. coli CPSase are in magenta.
Localization of the ATCase desensitizing mutations
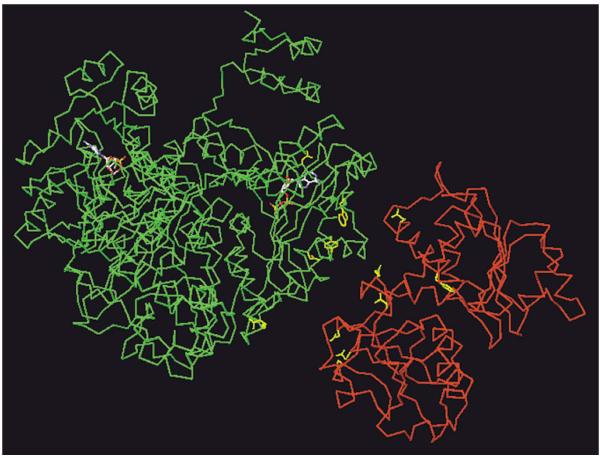
As reported above a series of missense mutants of the S. cerevisiae complex were obtained in which the ATCase activity is insensitive to the feedback inhibitor UTP, the CPSase activity remaining fully sensitive to this nucleotide [25]. Interestingly, these point mutations are located either in the ATCase or in the CPSase domains (Table 1). Thus, it was of interest to localize these point mutations in the modelled structures of the two domains. The result is shown in Figure 10. Most interestingly, it appears that these mutants are clustered in a particular region of the surface of the two domains, suggesting that these two regions constitute an interface between the two domains, through which the regulatory signal from the allosteric UTP binding site located in the CPSase domain to the catalytic site of the ATCase domain should be transmitted. However one of these mutations (Phe1924→Val) is located close to the catalytic site of ATCase suggesting that this residue would be rather involved in the transmission of the regulatory signal from the interface to the ATCase catalytic site.
Figure 10.
Probable interface between the CPSase and ATCase domains of the S. cerevisiae bifunctional complex. In green: the yeast CPSase domain. In red: the yeast ATCase domain. In yellow: point mutations which specifically abolish the sensitivity of the ATCase activity to UTP (see Table 1). From top to bottom: Asp1220, Trp1112, Tyr1096, Asn1094, Arg1076 in the CPSase domain; Glu1933, Glu2052, Phe1924, Asn2058, Asp2186, Glu2182, Asn2157 in the ATCase domain. The CPSase catalytic subsites are indicated by two molecules of ADP.
Discussion
In the S. cerevisiae CPSase-ATCase complex the two activities are feedback inhibited by the end-product UTP [12,13]. In the case of CPSase, the results reported here indicate that the substrate whose binding is altered by this effector is ATP as shown previously in the case of E. coli CPSase [34]. In the case of yeast ATCase, UTP decreases the affinity for the substrate aspartate [26]. Taken together with previously published observations, the results reported here show that the feedback inhibition of the yeast complex is of absolute competitive nature in the case of CPSase and of partial competitive nature in the case of ATCase. This last behavior is characteristic of a process of allosteric inhibition in which the effector binds to a regulatory site distinct from the catalytic site. In the case of CPSase the absolute competition indicates that UTP binds close enough to directly prevent the binding of ATP to this site through a local conformational change.
Several lines of evidence show that the regulatory site where UMP (procaryote CPSases) or UTP (enkaryote CPSases) binds is localized in the B3 subdomain (Fig. 1) [27,28,35]. Among the two steps of the CPSase reaction which, each, use a molecule of ATP (steps 1 and 3) only the third one is inhibited by UTP (Fig. 6). This partial reaction is specifically catalyzed by the B2 subdomain. Thus, it appears that the UTP binding site is located in the CPS-B domain which catalyzes the partial reaction which is specifically inhibited by this nucleotide, a feature which relates to the absolute competitive inhibition reported above. In the E. coli enzyme this regulatory site and the CPS-B catalytic site are distant by approximately 20 Å [36]. However, in the yeast complex this distance might be lower as the result of interactions with the ATCase and pDHOase domains. Alternatively, UTP might act through a different mechanism in the yeast complex and might provoke the competitive inhibition through binding to the CPS-B ATP binding site.
The first partial reaction catalyzed by the domain A is not affected by the presence of UTP. It was shown previously that the dimer of the isolated B domain is able to catalyze the synthesis of carbamylphosphate [23]. Interestingly, in this case, the first step of the CPSase reaction becomes sensitive to UTP (Fig. 7). Thus, the reaction normally catalyzed by the A domain becomes sensitive to UTP when it is catalyzed by the B domain which contains the allosteric UTP binding site. Taken together, these observations indicate that although it is homologous to the B3 domain, the A3 subdomain is unable to bind UTP.
As far as the feedback inhibition of the ATCase activity is concerned, several lines of evidence show that the presence of the CPSase domain of the S. cerevisiae complex is necessary for the ATCase domain to be sensitive to UTP, and that a single UTP binding site is located in the CPSase domain. Separation of the CPSase and ATCase domains by limited proteolysis [37] or genetic engeneering [23-25,38] leads to the desensitization of the ATCase domain. Taken together with the results reported here concerning the partial competitive character of the ATCase feedback inhibition, the requirement of the CPSase domain strongly suggests that the CPSase and the ATCase catalytic sites are both under the influence of the UTP binding regulatory site located in the B3 subdomain.
Conclusions
The results obtained provide informations about the mechanisms of allosteric inhibition of the two activities of the CPSase-ATCase complex. It is of particular interest that the mutations which specifically abolish the sensitivity to UTP of the ATCase reaction, without altering the CPSase inhibition, are clustered on the surface of either the CPS-B domain or the ATCase domain. This observation strongly suggests that these two regions constitute the interface between the two domains, interface through which the regulatory signal must be transmitted from the CPSase regulatory site to the catalytic site of the ATCase domain. This transmission could involve either a specific path between this interface and the catalytic site of ATCase, or a more global conformational change of this ATCase domain leading to a decrease of the affinity of this site for aspartate.
This is an original example of integrated allosteric regulation in a multifunctional complex that the catalytic domain of one activity is the allosteric site for the other activity.
Methods
Plasmids and strains
The 14.0-kb plasmid pC4-URA2 contains the yeast ura2 gene encoding the bifunctionnal CPSase-ATCase complex [25]. The pSV-CBApD recombinant plasmid encodes a protein CBApD, that possesses the C-terminal half of CPSase (CB), linked to the pDHO (pD) and the ATCase (A) domains [23].
The S. cerevisiae LJ5 strain was transformed by pC4 carrying a wild-type or mutated ura2 allele [25]. The LJ5 recipient strain was chosen because it is devoid of endogenous CPSase and ATCase activities.
The E. coli mutant L673 strain [39], defective in carA and carB, as well as the Lon-protease, was a gift from Dr. Carol Lusty (Public Health Research Institute of the City of New York). The genes carA and carB encode the small and large subunits of E. coli carbamoyl phosphate synthetase, respectively. The E. coli host strain EK1104 [40] lacks the pyrB1 genes. E. coli EK1104 and L673 cells were transformed with pC4-URA2 and pSV-CBApD respectively.
Cell growth and preparation of cell-free extracts
S. cerevisiae LJ5 strain harboring the recombinant plasmids was grown on YNB (6.7 g yeast nitrogen base / 1.2% glucose) at 30°C. Supplements were added to a 50 mg/ml final concentration. E. coli EK1104 and L673 cells harboring the recombinant plasmids were routinely grown from a single colony in 2xYT media supplemented with 100 μg/ml ampicillin. For induction of recombinant proteins under control of the pyrB1 promoter, the EK1104 and L673 cells were grown in a minimal media consisting of 6 g/l Na2HPO4, 3 g/l KH2PO4, 1 g/l NH4Cl, 5 g/l casamino acids, 4 g/l glucose, 0.5 mg/l ZnSO4.7H2O, 0.1 mM CaCl2, 1 mM MgSO4.7H2O, 10 mg/l tryptophan, supplemented with 12 μg/ml uracil and 100 μg/ml ampicillin. Under these conditions, there is sufficient uracil to sustain growth for about 19 to 21 hours, after which time, uracil is exhausted, growth is slowed, and the recombinant protein is expressed. Growth was monitored spectrophotometrically at 600 nm. The cells were harvested in late exponential phase or early stationary phase, by centrifugation at 3000 g for 30 minutes in a Centrikon T-124 centrifuge. The cells were resuspended in 50 mM Tris-HCl (pH 7.5), 1 mM EDTA, 1 mM DTT, and disrupted by sonication three times for one minute on ice, using a Biosonik III sonifier set at 20,000 kHz. The sonicate was cleared by centrigugation at 12,000 g for 30 minutes at 4°C. These extracts were dialyzed in order to eliminate all the metabolites, including nucleotides, which might interfere with enzyme assays. Protein concentrations were assayed by the Lowry method [41].
Enzymatic activities
Enzymatic activities were tested on crude dialyzed extracts as described by Penverne & Hervé [26]. The ATCase activity was tested as described by Denis-Duphil et al. [37]. The standard conditions used were 30 mM (14C)aspartate (0.03 μCi/μmol), 10 mM carbamylphosphate, and 50 mM Tris-HCl, pH 7.5. The assays were conducted at 30°C for 10 minutes. The CPSase activity of the yeast CPSase-ATCase and the CBApD complexes were assayed in the presence of 5 μg of E. coli ATCase catalytic subunits to efficiently trap all the unstable carbamylphosphate formed. The standard conditions used were 50 mM Tris-Ac, pH 7.5, 100 mM KCl, 100 mM NH4Cl, 150 mM (14C)sodium bicarbonate (0.168 μCi/μmol), 20 mM magnesium acetate, 10 mM ATP, and 50 mM aspartate. The assays were conducted at 30°C for 30 minutes. The extracts samples were extensively dialyzed immediately before the enzymatic tests in order to eliminate all the metabolites (including nucleotides) potentially able to interfere with the activities.
The overall carbamylphosphate synthetase – aspartate transcarbamylase activity was tested as described by Penverne et al. [42] without the addition of E. coli ATCase catalytic subunits. The partial reaction 1 of CPSase was assayed at 25°C by coupling the production of MgADP to the oxydation of NADH through the inclusion of pyruvate kinase, PEP, and lactate dehydrogenase in the assay mixture. Disappearance of NADH was followed continuously by monitoring the decrease in absorbance at 340 nm with a strip chart recorder. Each cuvette contained in a final volume of 1.0 ml the following: 50 mM Tris-Ac, pH 7.5, 100 mM KCl, 20 mM MgCl2, 0.2 mM NADH, 1 mM PEP, 50 mM NaHCO3, 10 mM L-glutamine, 10 mM MgATP, 0.1 mg of pyruvate kinase, and 0.15 mg of lactate dehydrogenase. The inhibitor UTP, if present, was 10 mM. The reaction was started with the addition of 100 μl of dialyzed crude extract. The partial reaction 3 of CPSase was measured at 25°C by coupling MgATP production with the reduction of NADP with hexokinase, glucose, and glucose-6-phosphate dehydrogenase. All cuvettes contained 50 mM Tris-Ac, pH 7.5, 100 mM KCl, 20 mM MgCl2, 1 mM NADP, 10 mM glucose, 1 unit each of hexokinase and glucose-6-phosphate dehydrogenase, 6 mM of MgADP (≈ Km), and 10 mM carbamoyl phosphate. The inhibitor UTP, if present, was 10 mM. The reaction was started by the addition of 100 μl of dialyzed crude extract. It was verified that the CPSase reaction was fully dependent on the presence of the three substrates. In the case of the L673 strain these controls were already published [23]. The same full requirement was observed in the case of the EK1104 strain. The bicarbonate ATPase-dependent reaction was undetectable in absence of bicarbonate.
Assay for UTP inhibition
The sensitivity of CPSase and ATCase to the feedback inhibitor UTP was assayed under the standard conditions described above in the presence of varying concentrations of this effector.
Sequence alignments
Sequences of E. coli CPSase (CARB, SwissProt P00968), E. coli ATCase catalytic chain (PYRB, SwissProt P00479) and S. cerevisiae CPSase-ATCase complex (PYR1, SwissProt P07259) were used. The sequences were aligned using the BIONET program FASTP (BLOSUM 50 matrix, ktup = 2) and Protein Information Ressource program ALIGN. Input parameters were chosen empirically.
Homology modelling of the S. cerevisiae CPSase and ATCase domains
The three-dimensional structure of the S. cerevisiae CPSase and ATCase domains were modelled by comparative protein modelling methods and energy minimization using the program SWISS-MODEL [43] in the optimized mode. The 2.10 Å coordinate set for the CPSase from E. coli [32] was used as the template for modelling the yeast CPSase monomer. The 2.5 Å structure of the E. coli ATCase complexed with the bisubstrate analogue N-(phosphonoacetyl)-L-aspartate [33] was used as the template for modelling the yeast ATCase monomer. Swiss-PdbViewer 3.5 [43] was used to analyse and visualize the structures.
List of abbreviations
The abbreviations used are: CPSase, carbamylphosphate synthetase; ATCase, aspartate transcarbamylase; GLNase: glutamine amidotransferase; DHOase, dihydroorotase; pDHO, the yeast domain that exibits sequence similarity to functional DHOases but which lacks activity; CBApD, the truncated yeast complex consisting of the CPS-B domain fused to the ATCase domain via the pDHO domain; CPS-A, the subdomain corresponding to the amino half of the CPSase synthetase domain or subunit; CPS-B and CB, the subdomain corresponding to the carboxy half of the CPSase synthetase domain or subunit.
Authors' contributions
VS performed the kinetic analysis with the participation of BP, the molecular modelling and the construction of the truncated CBApD protein in collaboration with HG and DE. SP and JLS provided the mutants. GH coordinated the investigations. All authors read and approved the final manuscript.
Acknowledgments
Acknowledgments
Our research was supported by the CNRS, Université Paris 6, and Université Paris 7.
Contributor Information
Valérie Serre, Email: serre@ccr.jussieu.fr.
Bernadette Penverne, Email: penverne@ccr.jussieu.fr.
Jean-Luc Souciet, Email: souciet@gem.u-strasbg.fr.
Serge Potier, Email: potier@gem.u-strasbg.fr.
Hedeel Guy, Email: hguy@cmb.biosci.wayne.edu.
David Evans, Email: devans@cmb.biosci.wayne.edu.
Patrick Vicart, Email: vicart@ext.jussieu.fr.
Guy Hervé, Email: gherve@ccr.jussieu.fr.
References
- Pauwels K, Abadjieva A, Hilven P, Stankiewicz A, Crabeel M. The N-acetylglutamate synthase/N-acetylglutamate kinase metabolon of Saccharomyces cerevisiae allows co-ordinated feedback regulation of the first two steps in arginine biosynthesis. Eur J Biochem. 2003;270:1014–1024. doi: 10.1046/j.1432-1033.2003.03477.x. [DOI] [PubMed] [Google Scholar]
- Herve G, Nagy M, Le Gouar M, Penverne B, Ladjimi M. The carbamoyl phosphate synthetase-aspartate transcarbamoylase complex of Saccharomyces cerevisiae: molecular and cellular aspects. Biochem Soc Trans. 1993;21:195–198. doi: 10.1042/bst0210195. [DOI] [PubMed] [Google Scholar]
- Meister A. Mechanism and regulation of the glutamine-dependent carbamyl phosphate synthetase of Escherichia coli. Adv Enzymol Relat Areas Mol Biol. 1989;62:315–374. doi: 10.1002/9780470123089.ch7. [DOI] [PubMed] [Google Scholar]
- Wild JR, Loughrey-Chen SJ, Corder TS. In the presence of CTP, UTP becomes an allosteric inhibitor of aspartate transcarbamoylase. Proc Natl Acad Sci U S A. 1989;86:46–50. doi: 10.1073/pnas.86.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoaf WT, Jones ME. Uridylic acid synthesis in Ehrlich ascites carcinoma. Properties, subcellular distribution, and nature of enzyme complexes of the six biosynthetic enzymes. Biochemistry. 1973;12:4039–4051. doi: 10.1021/bi00745a004. [DOI] [PubMed] [Google Scholar]
- Mori M, Ishida H, Tatibana M. Aggregation states and catalytic properties of the multienzyme complex catalyzing the initial steps of pyrimidine biosynthesis in rat liver. Biochemistry. 1975;14:2622–2630. doi: 10.1021/bi00683a010. [DOI] [PubMed] [Google Scholar]
- Coleman PF, Suttle DP, Stark GR. Purification from hamster cells of the multifunctional protein that initiates de novo synthesis of pyrimidine nucleotides. J Biol Chem. 1977;252:6379–6385. [PubMed] [Google Scholar]
- Levine RL, Kretchmer N. Conversion of carbamoyl phosphate to hydroxyurea. An assay for carbamoylphosphate synthetase. Anal Biochem. 1971;42:324–337. doi: 10.1016/0003-2697(71)90044-3. [DOI] [PubMed] [Google Scholar]
- Lue PF, Kaplan JG. The aspartate transcarbamylase and carbamoyl phosphate synthetase of yeast: a multi-functional enzyme complex. Biochem Biophys Res Commun. 1969;34:426–433. doi: 10.1016/0006-291x(69)90399-4. [DOI] [PubMed] [Google Scholar]
- Souciet JL, Nagy M, Le Gouar M, Lacroute F, Potier S. Organization of the yeast URA2 gene: identification of a defective dihydroorotase-like domain in the multifunctional carbamoylphosphate synthetase-aspartate transcarbamylase complex. Gene. 1989;79:59–70. doi: 10.1016/0378-1119(89)90092-9. [DOI] [PubMed] [Google Scholar]
- Williams NK, O'Donoghue S, Christopherson RI. Homology and mutagenesis studies of hamster dihydroorotase. Adv Exp Med Biol. 1994;370:597–601. doi: 10.1007/978-1-4615-2584-4_124. [DOI] [PubMed] [Google Scholar]
- Kaplan JG, Duphil M, Lacroute F. A study of the aspartate transcarbamylase activity of yeast. Arch Biochem Biophys. 1967;119:541–551. doi: 10.1016/0003-9861(67)90489-4. [DOI] [PubMed] [Google Scholar]
- Aitken DM, Lue PF, Kaplan JG. Kinetics and reaction mechanism of the carbamylphosphate synthetase of a multienzyme aggregate from yeast. Can J Biochem. 1975;53:721–730. doi: 10.1139/o75-099. [DOI] [PubMed] [Google Scholar]
- Anderson PM, Meister A. Evidence for an activated form of carbon dioxide in the reaction catalyzed by Escherichia coli carbamyl phosphate synthetase. Biochemistry. 1965;4:2803–2809. doi: 10.1021/bi00888a034. [DOI] [PubMed] [Google Scholar]
- Lusty CJ, Widgren EE, Broglie KE, Nyunoya H. Yeast carbamyl phosphate synthetase. Structure of the yeast gene and homology to Escherichia coli carbamyl phosphate synthetase. J Biol Chem. 1983;258:14466–14477. [PubMed] [Google Scholar]
- Nyunoya H, Lusty CJ. The carB gene of Escherichia coli: a duplicated gene coding for the large subunit of carbamoyl-phosphate synthetase. Proc Natl Acad Sci U S A. 1983;80:4629–4633. doi: 10.1073/pnas.80.15.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyunoya H, Broglie KE, Lusty CJ. The gene coding for carbamoyl-phosphate synthetase I was formed by fusion of an ancestral glutaminase gene and a synthetase gene. Proc Natl Acad Sci U S A. 1985;82:2244–2246. doi: 10.1073/pnas.82.8.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freund JN, Jarry BP. The rudimentary gene of Drosophila melanogaster encodes four enzymic functions. J Mol Biol. 1987;193:1–13. doi: 10.1016/0022-2836(87)90621-8. [DOI] [PubMed] [Google Scholar]
- Faure M, Camonis JH, Jacquet M. Molecular characterization of a Dictyostelium discoideum gene encoding a multifunctional enzyme of the pyrimidine pathway. Eur J Biochem. 1989;179:345–358. doi: 10.1111/j.1432-1033.1989.tb14560.x. [DOI] [PubMed] [Google Scholar]
- Simmer JP, Kelly RE, Rinker A. G., Jr., Zimmermann BH, Scully JL, Kim H, Evans DR. Mammalian dihydroorotase: nucleotide sequence, peptide sequences, and evolution of the dihydroorotase domain of the multifunctional protein CAD. Proc Natl Acad Sci U S A. 1990;87:174–178. doi: 10.1073/pnas.87.1.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy HI, Evans DR. Function of the major synthetase subdomains of carbamyl-phosphate synthetase. J Biol Chem. 1996;271:13762–13769. doi: 10.1074/jbc.271.16.9567. [DOI] [PubMed] [Google Scholar]
- Guy HI, Schmidt B, Herve G, Evans DR. Pressure-induced dissociation of carbamoyl-phosphate synthetase domains. The catalytically active form is dimeric. J Biol Chem. 1998;273:14172–14178. doi: 10.1074/jbc.273.23.14172. [DOI] [PubMed] [Google Scholar]
- Serre V, Guy H, Penverne B, Lux M, Rotgeri A, Evans D, Herve G. Half of Saccharomyces cerevisiae carbamoyl phosphate synthetase produces and channels carbamoyl phosphate to the fused aspartate transcarbamoylase domain. J Biol Chem. 1999;274:23794–23801. doi: 10.1074/jbc.274.34.23794. [DOI] [PubMed] [Google Scholar]
- Jaquet L, Lollier M, Souciet JL, Potier S. Genetic analysis of yeast strains lacking negative feedback control: a one-step method for positive selection and cloning of carbamoylphosphate synthetase-aspartate transcarbamoylase mutants unable to respond to UTP. Mol Gen Genet. 1993;241:81–88. doi: 10.1007/BF00280204. [DOI] [PubMed] [Google Scholar]
- Jaquet L, Serre V, Lollier M, Penverne B, Herve G, Souciet JL, Potier S. Allosteric regulation of carbamoylphosphate synthetase-aspartate transcarbamylase multifunctional protein of Saccharomyces cerevisiae: selection, mapping and identification of missense mutations define three regions involved in feedback inhibition by UTP. J Mol Biol. 1995;248:639–652. doi: 10.1006/jmbi.1995.0248. [DOI] [PubMed] [Google Scholar]
- Penverne B, Herve G. In situ behavior of the pyrimidine pathway enzymes in Saccharomyces cerevisiae. I. Catalytic and regulatory properties of aspartate transcarbamylase. Arch Biochem Biophys. 1983;225:562–575. doi: 10.1016/0003-9861(83)90068-1. [DOI] [PubMed] [Google Scholar]
- Rubio V, Cervera J, Lusty CJ, Bendala E, Britton HG. Domain structure of the large subunit of Escherichia coli carbamoyl phosphate synthetase. Location of the binding site for the allosteric inhibitor UMP in the COOH-terminal domain. Biochemistry. 1991;30:1068–1075. doi: 10.1021/bi00218a027. [DOI] [PubMed] [Google Scholar]
- Liu X, Guy HI, Evans DR. Identification of the regulatory domain of the mammalian multifunctional protein CAD by the construction of an Escherichia coli hamster hybrid carbamyl-phosphate synthetase. J Biol Chem. 1994;269:27747–27755. [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Villoutreix BO, Spassov VZ, Atanasov BP, Herve G, Ladjimi MM. Structural modeling and electrostatic properties of aspartate transcarbamylase from Saccharomyces cerevisiae. Proteins. 1994;19:230–243. doi: 10.1002/prot.340190307. [DOI] [PubMed] [Google Scholar]
- Nagy M, Le Gouar M, Potier S, Souciet JL, Herve G. The primary structure of the aspartate transcarbamylase region of the URA2 gene product in Saccharomyces cerevisiae. Features involved in activity and nuclear localization. J Biol Chem. 1989;264:8366–8374. [PubMed] [Google Scholar]
- Thoden JB, Raushel FM, Benning MM, Rayment I, Holden HM. The structure of carbamoyl phosphate synthetase determined to 2.1 A resolution. Acta Crystallogr D Biol Crystallogr. 1999;55:8–24. doi: 10.1107/S0907444998006234. [DOI] [PubMed] [Google Scholar]
- Ke HM, Lipscomb WN, Cho YJ, Honzatko RB. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988;204:725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Braxton BL, Mullins LS, Raushel FM, Reinhart GD. Quantifying the allosteric properties of Escherichia coli carbamyl phosphate synthetase: determination of thermodynamic linked-function parameters in an ordered kinetic mechanism. Biochemistry. 1992;31:2309–2316. doi: 10.1021/bi00123a015. [DOI] [PubMed] [Google Scholar]
- Delannay S, Charlier D, Tricot C, Villeret V, Pierard A, Stalon V. Serine 948 and threonine 1042 are crucial residues for allosteric regulation of Escherichia coli carbamoylphosphate synthetase and illustrate coupling effects of activation and inhibition pathways. J Mol Biol. 1999;286:1217–1228. doi: 10.1006/jmbi.1999.2561. [DOI] [PubMed] [Google Scholar]
- Thoden JB, Raushel FM, Wesenberg G, Holden HM. The binding of inosine monophosphate to Escherichia coli carbamoyl phosphate synthetase. J Biol Chem. 1999;274:22502–22507. doi: 10.1074/jbc.274.32.22502. [DOI] [PubMed] [Google Scholar]
- Denis-Duphil M, Mathien-Shire Y, Herve G. Proteolytically induced changes in the molecular form of the carbamyl phosphate synthetase-uracil-aspartate transcarbamylase complex coded for by the URA2 locus in Saccharomyces cerevisiae. J Bacteriol. 1981;148:659–669. doi: 10.1128/jb.148.2.659-669.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serre V, Guy H, Liu X, Penverne B, Herve G, Evans D. Allosteric regulation and substrate channeling in multifunctional pyrimidine biosynthetic complexes: analysis of isolated domains and yeast-mammalian chimeric proteins. J Mol Biol. 1998;281:363–377. doi: 10.1006/jmbi.1998.1856. [DOI] [PubMed] [Google Scholar]
- Guillou F, Rubino SD, Markovitz RS, Kinney DM, Lusty CJ. Escherichia coli carbamoyl-phosphate synthetase: domains of glutaminase and synthetase subunit interaction. Proc Natl Acad Sci U S A. 1989;86:8304–8308. doi: 10.1073/pnas.86.21.8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowlan SF, Kantrowitz ER. Superproduction and rapid purification of Escherichia coli aspartate transcarbamylase and its catalytic subunit under extreme derepression of the pyrimidine pathway. J Biol Chem. 1985;260:14712–14716. [PubMed] [Google Scholar]
- Lowry OH, Rosenbrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- Penverne B, Belkaid M, Herve G. In situ behavior of the pyrimidine pathway enzymes in Saccharomyces cerevisiae. 4. The channeling of carbamylphosphate to aspartate transcarbamylase and its partition in the pyrimidine and arginine pathways. Arch Biochem Biophys. 1994;309:85–93. doi: 10.1006/abbi.1994.1089. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]