Abstract
Objective
To identify single nucleotide polymorphisms (SNPs) and pathways associated with bronchopulmonary dysplasia (BPD) because O2 requirement at 36 weeks’ post-menstrual age risk is strongly influenced by heritable factors.
Study design
A genome-wide scan was conducted on 1.2 million genotyped SNPs, and an additional 7 million imputed SNPs, using a DNA repository of extremely low birth weight infants. Genome-wide association and gene set analysis was performed for BPD or death, severe BPD or death, and severe BPD in survivors. Specific targets were validated using gene expression in BPD lung tissue and in mouse models.
Results
Of 751 infants analyzed, 428 developed BPD or died. No SNPs achieved genome-wide significance (p<10−8) although multiple SNPs in adenosine deaminase (ADARB2), CD44, and other genes were just below p<10−6. Of approximately 8000 pathways, 75 were significant at False Discovery Rate (FDR) <0.1 and p<0.001 for BPD/death, 95 for severe BPD/death, and 90 for severe BPD in survivors. The pathway with lowest FDR was miR-219 targets (p=1.41E-08, FDR 9.5E-05) for BPD/death and Phosphorous Oxygen Lyase Activity (includes adenylate and guanylate cyclases) for both severe BPD/death (p=5.68E-08, FDR 0.00019) and severe BPD in survivors (p=3.91E-08, FDR 0.00013). Gene expression analysis confirmed significantly increased miR-219 and CD44 in BPD.
Conclusions
Pathway analyses confirmed involvement of known pathways of lung development and repair (CD44, Phosphorus Oxygen Lyase Activity) and indicated novel molecules and pathways (ADARB2, Targets of miR-219) involved in genetic predisposition to BPD.
Keywords: Bronchopulmonary dysplasia, Infant, premature, Infant mortality, Single nucleotide polymorphisms
Bronchopulmonary dysplasia (BPD) is common in extremely preterm infants, and genetic factors may account for much of the variance in risk for BPD.1 Targeted candidate gene analyses suggest single nucleotide polymorphisms (SNPs) in certain cytokines, surfactant proteins, and related molecules2, 3 but not others4 are associated with BPD. Hadchouel et al5 identified the SPOCK2 gene as associated with BPD in a genome-wide association study (GWAS) that evaluated the entire genome in an unbiased manner. However, Wang et al6 did not find SNPs associated with BPD in a GWAS.
Most complex diseases (such as BPD) involve gene-environment interactions and interactions among different loci. However, conventional single marker analysis does not explicitly look for interactions among different genes in the same biological pathway that have a multiplicative or a threshold effect.7 Most GWAS that focus on analysis of single markers lack the power to identify the small contribution of most genetic variants.8 Pathway-based approaches, which consider multiple contributing factors in the same biological pathway, complement the single marker approach and provide understanding of GWAS data in many diseases.9
In this study, we utilized a GWAS combined with pathway-based approaches to increase our understanding of the role of genetics in BPD susceptibility, and integrated these results with gene expression comparing BPD with controls, and a newborn mouse model of hyperoxia exposure simulating BPD. We hypothesized that SNPs in biological pathways involved in lung development and injury will be enriched in infants who develop BPD or die. The combined outcome of BPD or death was used because death is a competing outcome for BPD i.e., infants who die early cannot develop BPD even though they may be at the highest risk of BPD.
METHODS
Patients included were a subset of infants enrolled in the Eunice Kennedy Shriver NICHD Neonatal Research Network’s Cytokines study that enrolled infants 401–1000 g at birth, < 72 h age, and free of major congenital anomalies.10 The study was approved by institutional review boards (IRBs) at participating centers, and written informed consent was obtained from parent(s). Additional IRB review allowed the GWA genotyping results with a limited phenotype data to be included in the NHGRI Database of Genotypes and Phenotypes (DbGaP).
DNA was extracted from the earliest age blood spot collected on filter paper. Whole genome amplification was used for samples that did not provide adequate genomic DNA. Genotyping was done on the Illumina HumanOmni1-Quad_v1-0_B BeadChip.
BPD was defined by supplemental O2 at 36 weeks’ postmenstrual age. Severe BPD was defined as therapy with O2>21% for at least 28 days plus use of ≥30% O2 and/or positive pressure (ventilation or nasal continuous positive airway pressure) at 36 weeks’ post-menstrual age.11 Death was defined as in-hospital death prior to discharge.
Ancestry was classified as Black (African-American), White (non-Hispanic Caucasians), Hispanic (Hispanic Caucasian), and others including Asian and multi-racial using GWASTools12 to generate eigenvalues for the entire dataset.
Imputation was run using beagle 3.3.1. 769,757 SNPs were used for imputation with 7,500,443 SNPs being imputed.13
Analysis of SNPs was done using two complementary methods: a standard GWA analysis followed by a pathway analysis.
SNPs were analyzed using PLINK14 using logistic regression under an additive model. Three models were run: BPD or death vs. survival without BPD, Severe BPD or death vs. survival without severe BPD, Severe BPD in survivors vs. survivors without severe BPD. The regression model included covariates for GA, small for GA, sex, Apgar at 5 min < 5, antenatal steroids, and the race ethnicity Eigenvalues 1–4. The top 10 SNPs (by lowest p-value) for each of the 3 models were mapped to genes.
We assigned genes to pathways (gene sets) using the Molecular Signatures Database (MSigDB) (http://www.broadinstitute.org/gsea/msigdb/collections.jsp). SNPs were assigned to gene(s) based on being exonic, intronic, untranslated region, or within 20 kb of the ends of the gene model. Pathways were analyzed using Gene Set Enrichment Analysis (GSEA).15
Gene expression values for individual members of pathways considered most important were extracted from an existing dataset describing genome-wide expression in lung tissue obtained from BPD cases or controls and assessed for differential expression.16 Two selected molecules (miR-219 and CD44) were further evaluated by TaqMan Gene Expression assays (Life Technologies, Grand Island, NY) from RNA isolated using Qiagen RNeasy FFPE kit (Qiagen, Valencia, CA) from paraffin-embedded formalin-fixed samples of lungs collected at autopsy from extremely preterm infants (24–28 weeks gestation) who died soon after birth, term stillborn infants, and preterm infants who died due to BPD at term corrected age (36–44 weeks post-menstrual age) (n=4/group).
Three molecules (miR-219, ADARB2, and CD44) were selected for further evaluation in a mouse model. Gene expression was evaluated at different points during alveolar septation and hyperoxia exposure, using samples from studies approved by the UAB Institutional Animal Care and Use Committee.17,18 RNA was isolated from lung homogenates for real-time RT-PCR using specific primers.19
RESULTS
The GWAS cohort included 834 infants whose DNA samples were successfully genotyped. 172 (20%) samples required whole genome amplification. 751 infants met inclusion criteria with adequate information on BPD phenotype and genotyping (> 97% call rate). Characteristics of the study cohort are listed in Table I (available at www.jpeds.com). As expected, infants who developed outcomes of interest (BPD/death; severe BPD/death; Severe BPD in survivors) were more immature, of lower birth weight, more likely to be male, mechanically ventilated, and ventilated for a longer duration as compared with those who did not develop these outcomes.
Table 1.
Characteristics of enrolled infants
| Variable | Entire population |
BPD or Death vs. Survival without BPD |
Severe BPD or Death vs. Survival without severe BPD |
Severe BPD in survivors vs. Survivors without severe BPD |
|||
|---|---|---|---|---|---|---|---|
| BPD or Death |
Survival without BPD |
Severe BPD or Death |
Survival without severe BPD |
Severe BPD in survivors |
Survival without severe BPD |
||
| Sample size | 751 | 428 | 322 | 243 | 469 | 102 | 469 |
| Birth weight in grams (mean, SD) |
758 (140) |
723 (135) |
804 (133) |
688 (129) |
787 (132) |
707 (121) |
787 (132) |
| Gestational age in weeks (mean, SD, Range) |
25.8 (1.96) (20–33) |
25.3 (1.9) (20–33) |
26.5 (1.9) (20–32) |
24.9 (1.8) (21–33) |
26.1 (1.9) (22–32) |
25.4 (1.8) (21–33) |
26.1 (1.9) (22–32) |
| Multiple gestation (%) |
137 (18.2) |
84 (19.6) |
53 (16.5) |
43 (17.7) |
86 (18.3) |
11 (10.8) |
86 (18.3) |
| Antenatal steroids (%) |
545 (73) | 301 (71) | 244 (76) | 167 (69) | 348 (74) | 78 (77) | 348 (74) |
| SGA (%) | 105 (14.0) |
46 (19.6) |
59 (18.3) |
26 (10.7) |
70 (14.9) |
15 (14.7) |
70 (14.9) |
| Race: White | 339 (45.1) |
186 (43.5) |
153 (47.5) |
98 (40.3) |
219 (46.7) |
40 (39.2) |
219 (46.7) |
| Race: Black | 404 (53.8) |
236 (55.1) |
167 (51.9) |
142 (58.4) |
245 (52.2) |
61 (59.8) |
245 (52.2) |
| Race: Other | 8 (1) |
6 (1.4) |
2 (0.6) |
3 (1.2) |
5 (1.1) |
1 (1) |
5 (1.1) |
| Ethnicity: Hispanic |
156 (20.8) |
82 (19.2) |
74 (23.0) |
49 (20.2) |
97 (20.7) |
20 (19.6) |
97 (20.7) |
| Male sex | 354 (47.1) |
227 (53.0) |
127 (39.4) |
128 (52.7) |
210 (44.8) |
53 (52) |
210 (44.8) |
| Apgar score at 5 minutes (mean, SD) |
6.6 (1.8) | 6.4 (1.9) | 6.8 (1.6) | 6.1 (1.9) | 6.7 (1.9) | 6.2 (2.1) | 6.7 (1.7) |
| Cesarean delivery Yes (%) |
430 (57.3) |
229 (53.5) |
201 (62.4) |
120 (49.4) |
284 (60.6) |
59 (57.8) |
284 (60.6) |
| Any Mechanical ventilation (%) |
697 (92.8) |
426 (99.5) |
270 (83.9) |
243 (100) |
422 (90) |
102 (100) |
422 (90) |
| Days of
assisted ventilation (Days, SD) |
26.9 (27.5) |
37.5 (29.8) |
12.8 (15.2) |
41.7 (35) |
20.7 (19.6) |
62.2 (31.6) |
20.7 (19.6) |
GWA analysis
None of the SNPs were significant at the genome-wide significance level (p<10−8). The analysis for top ten SNPs for BPD/death (Table II) identified 4 SNPs in adenosine deaminase (ADARB2), two SNPs in CD44, one in NSMC4A, one in WDR45L, and two associated with no known gene. Similarly, the top ten SNPs for severe BPD/death were 4 SNPs in ADARB2, one in CD44, one in NSMC4A, one in NUAK1, one in KCNH7, and two associated with no known gene (Table II). The analysis for severe BPD in survivors also found ADARB2, CD44, NUAK1, KCNH7, WDR45B, in addition to GRIP1 and GALNTL6 (Table II). Most of these SNPs had p-values of 10−6 to 10−7.
TABLE 2.
Important single nucleotide polymorphisms (SNPs) and associated gene (using the NCBI database of SNPs (dbSNP; http://www.ncbi.nlm.nih.gov/snp/) and from UCSC Genome browser at http://genome.ucsc.edu), in relation to p-value for outcomes
| Chromosome | SNP | Gene | p value |
|---|---|---|---|
| BPD or Death vs. Survival without BPD | |||
| 10 | rs59582957 | ADARB2 (adenosine deaminase) | 7.18E-07 |
| 10 | chr10:123726948 | NSMC4A (non-SMC element 4 homolog A (S. | 7.08E-07 |
| 10 | rs17119652 | No gene | 1.38E-06 |
| 10 | chr10:1488099 | ADARB2 | 6.69E-07 |
| 10 | chr10:1488186 | ADARB2 | 6.42E-07 |
| 10 | chr10:1488126 | ADARB2 | 4.71E-07 |
| 11 | chr11:35165510 | CD44 | 8.60E-07 |
| 11 | chr11:35167447 | CD44 | 1.72E-06 |
| 12 | rs1504316 | No gene | 6.30E-07 |
| 17 | rs8082435 | WDR45L (WD repeat domain 45B) | 0.008752 |
| Severe BPD or death vs. survival without severe BPD | |||
| 10 | chr10:1488126 | ADARB2 | 4.71E-07 |
| 10 | chr10:1488186 | ADARB2 | 6.42E-07 |
| 10 | chr10:1488099 | ADARB2 | 6.69E-07 |
| 10 | chr10:123726948 | NSMC4A | 7.08E-07 |
| 10 | rs59582957 | ADARB2 | 7.18E-07 |
| 11 | chr11:35165510 | CD44 | 8.60E-07 |
| 12 | rs1427793 | NUAK1 (NUAK family SNF1-like kinase 1) | 1.09E-06 |
| 10 | rs57481375 | No gene | 1.10E-06 |
| 10 | rs17119652 | No gene | 1.38E-06 |
| 4 | rs2653829 | KCNH7 (Potassium voltage-gated channel | 1.48E-06 |
| Severe BPD in survivors vs. Survivors without severe BPD | |||
| 11 | chr11:35165510 | CD44 | 4.64E-07 |
| 12 | rs1504316 | GRIP1 (glutamate receptor interacting protein 1) | 6.30E-07 |
| 10 | rs17119652 | No gene | 8.89E-07 |
| 11 | chr11:35167447 | CD44 | 9.04E-07 |
| 17 | rs8082435 | WDR45B (WD repeat domain 45B) | 9.97E-07 |
| 4 | rs2653829 | KCNH7 | 1.01E-06 |
| 10 | chr10:1488126 | ADARB2 | 1.72E-06 |
| 4 | rs2610201 | GALNTL6 (UDP-N-acetyl-alpha-D- | 1.91E-06 |
| 4 | rs2653824 | GALNTL6 | 2.04E-06 |
| 12 | rs1427793 | NUAK1 | 2.06E-06 |
Pathway /Gene Set Enrichment Analysis
Of the approximately 7650 gene sets evaluated, 75 were significant at a False Discovery Rate (FDR) of <0.1 (suggesting about 10% of the pathways are false positives) and p <0.001 for the BPD or death vs. no BPD comparison. 95 pathways were significant for severe BPD or death, and 90 for severe BPD in survivors.
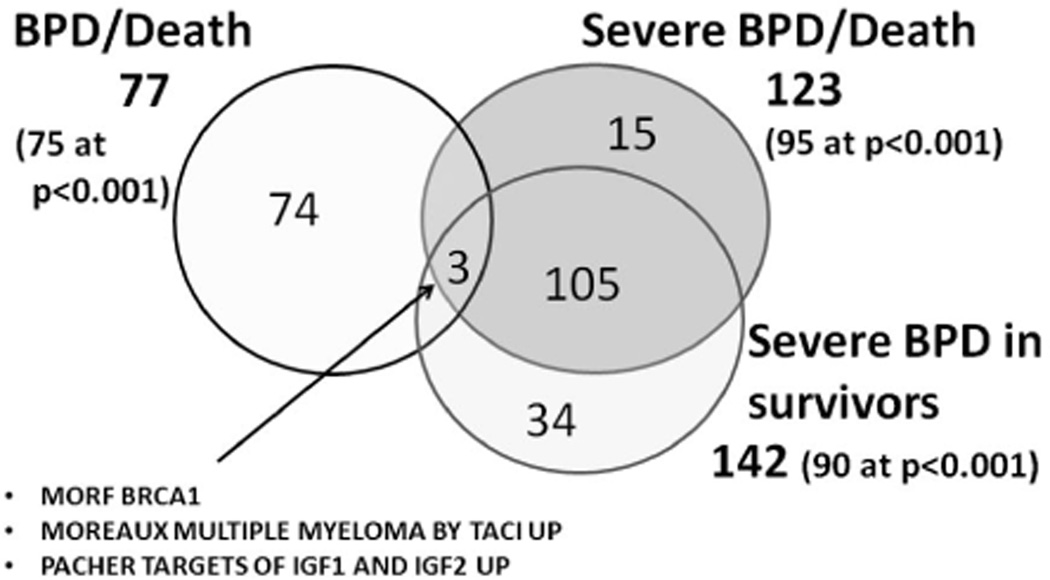
Pathways associated with BPD or death vs. survivors without BPD (Table III; available at www.jpeds.com): 77 pathways were identified with a FDR <0.1 (75 significant at p<0.001). Of these 77 pathways, only 3 were shared with Severe BPD or death, or Severe BPD in survivors (MORF_BRCA1, MOREAUX_MULTIPLE_MYELOMA_BY_TACI_UP, and PACHER_TARGETS_OF_IGF1_AND_IGF2_UP) (Figure 1).The top pathway was MIR-219 (http://www.broadinstitute.org/gsea/msigdb/cards/GACAATC,MIR-219.html), which includes 143 genes.
Pathways associated with severe BPD or death vs. survivors without severe BPD (Table IV; available at www.jpeds.com): 123 pathways were identified with a FDR <0.1, of which 95 were significant at p<0.001. Of these 123 pathways, 3 were shared with those involved in BPD or death. 108 of these pathways (which included the 3 shared with BPD or death) were shared with those involved with severe BPD in survivors, including the top 43 pathways, indicating significant overlap in the models for these outcomes. The top pathway associated with severe BPD or death (and survivors with severe BPD) was Phosphorus Oxygen Lyase Activity (http://www.broadinstitute.org/gsea/msigdb/cards/PHOSPHORUS_OXYGEN_LYASE_ACTIVITY.html), which includes ten genes consisting of adenylate cyclases and guanylate cyclases.
Pathways associated with Severe BPD in survivors (Table V; available at www.jpeds.com): 142 pathways were identified with a FDR <0.1, of which 90 were significant at p<0.001. 108 of these 142 pathways (including the top 43) were also associated with Severe BPD or death.
Pathways associated with BPD or death by race (Table VI; available at www.jpeds.com): Of the 77 pathways identified at FDR<0.1 in all infants, 20 were noted in Black infants, 13 in Hispanic infants, and 24 in White infants for the same FDR threshold. Importantly, there was little overlap in the major pathways between these racial/ethnic groups. For example, targets of miR-219, which was the top pathway for all infants (FDR 9.52E-05, p=1.41E-08), was ranked 415th (FDR 0.29, p=0.018) for Black infants, 2597th (FDR 0.34, p=0.13) for Hispanic infants (but with FDR 5.92 E-43, p=7.48E-44 for severe BPD in survivors for the same cohort of Hispanic infants), and 1477th (FDR 0.25, p=0.055) for White infants (but with FDR 2.68E-44, p=2.66E-45 for severe BPD in survivors for the same cohort of White infants).
Table 3.
Biological pathways associated with BPD or death, as compared to survivors without BPD. Pathways from the annotated gene sets of the molecular signatures database at the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp) are listed in order of increasing false discovery rate (FDR). Only pathways with FDR ≤0.1 are shown.
| Pathway Names | P values | FDR |
|---|---|---|
| GACAATC,MIR-219 | 1.41E-08 | 9.52E-05 |
| NUCLEAR_UBIQUITIN_LIGASE_COMPLEX | 2.16E-07 | 0.00073 |
| YRCCAKNNGNCGC_UNKNOWN | 7.11E-07 | 0.0016 |
| V$E2F_Q2 | 9.88E-07 | 0.001668 |
| NEURON_PROJECTION | 1.37E-05 | 0.015893 |
| V$GABP_B | 1.41E-05 | 0.015893 |
| TOMLINS_METASTASIS_DN | 1.66E-05 | 0.015965 |
| JAEGER_METASTASIS_UP | 3.34E-05 | 0.018801 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_KIN ASE_ACTIVITY |
2.74E-05 | 0.018801 |
| RUIZ_TNC_TARGETS_DN | 2.99E-05 | 0.018801 |
| SUGAR_BINDING | 2.71E-05 | 0.018801 |
| V$E2F_Q4 | 3.27E-05 | 0.018801 |
| CARBOHYDRATE_BINDING | 4.87E-05 | 0.024581 |
| UBIQUITIN_LIGASE_COMPLEX | 5.10E-05 | 0.024581 |
| V$E2F_Q6 | 5.64E-05 | 0.025396 |
| LIPID_KINASE_ACTIVITY | 6.62E-05 | 0.027946 |
| V$FREAC7_01 | 7.88E-05 | 0.031284 |
| CAGCTTT,MIR-320 | 0.000106 | 0.033841 |
| DAZARD_RESPONSE_TO_UV_NHEK_UP | 0.000101 | 0.033841 |
| HALMOS_CEBPA_TARGETS_DN | 9.54E-05 | 0.033841 |
| MOREAUX_MULTIPLE_MYELOMA_BY_TACI_ UP |
0.00011 | 0.033841 |
| TAGHAVI_NEOPLASTIC_TRANSFORMATION | 0.000107 | 0.033841 |
| COLLER_MYC_TARGETS_UP | 0.000116 | 0.034047 |
| V$AML_Q6 | 0.000138 | 0.038786 |
| REACTOME_CALCITONIN_LIKE_LIGAND_RE CEPTORS |
0.00017 | 0.045858 |
| AMIT_EGF_RESPONSE_480_HELA | 0.000182 | 0.04725 |
| BIOCARTA_CTCF_PATHWAY | 0.000253 | 0.047743 |
| BIOCARTA_RAC1_PATHWAY | 0.0002 | 0.047743 |
| CTCTATG,MIR-368 | 0.000226 | 0.047743 |
| GNF2_SPINK1 | 0.000219 | 0.047743 |
| IIZUKA_LIVER_CANCER_PROGRESSION_L0_L 1_UP |
0.000242 | 0.047743 |
| IVANOVA_HEMATOPOIESIS_LATE_PROGENIT OR |
0.000263 | 0.047743 |
| module_320 | 0.000223 | 0.047743 |
| MUELLER_COMMON_TARGETS_OF_AML_FU SIONS_DN |
0.000246 | 0.047743 |
| REACTOME_ACTIVATION_OF_NMDA_RECEP | 0.000263 | 0.047743 |
| TOR_UPON_GLUTAMATE_BINDING_AND_PO STSYNAPTIC_EVENTS |
||
| RESPONSE_TO_STEROID_HORMONE_STIMUL US |
0.000269 | 0.047743 |
| SCGGAAGY_V$ELK1_02 | 0.000241 | 0.047743 |
| V$E47_01 | 0.000207 | 0.047743 |
| CELLULAR_BIOSYNTHETIC_PROCESS | 0.000294 | 0.048217 |
| DAZARD_RESPONSE_TO_UV_SCC_DN | 0.000296 | 0.048217 |
| REGULATION_OF_CELLULAR_PROTEIN_MET ABOLIC_PROCESS |
0.0003 | 0.048217 |
| V$ZF5_01 | 0.000283 | 0.048217 |
| GAGACTG,MIR-452 | 0.000334 | 0.050215 |
| KAPOSI_LIVER_CANCER_POOR_SURVIVAL_U P |
0.000342 | 0.050215 |
| MORF_BRCA1 | 0.00034 | 0.050215 |
| PACHER_TARGETS_OF_IGF1_AND_IGF2_UP | 0.000341 | 0.050215 |
| chr4q26 | 0.000363 | 0.052097 |
| BIOCARTA_MCM_PATHWAY | 0.000395 | 0.054476 |
| GGCACAT,MIR-455 | 0.000391 | 0.054476 |
| BEGUM_TARGETS_OF_PAX3_FOXO1_FUSION _UP |
0.000423 | 0.056037 |
| SENESE_HDAC1_AND_HDAC2_TARGETS_DN | 0.000415 | 0.056037 |
| CTCNANGTGNY_UNKNOWN | 0.000445 | 0.056717 |
| REGULATION_OF_PROTEIN_METABOLIC_PR OCESS |
0.000437 | 0.056717 |
| BIOCARTA_RAS_PATHWAY | 0.000475 | 0.057752 |
| chr10q23 | 0.000468 | 0.057752 |
| module_321 | 0.00048 | 0.057752 |
| V$NRF2_01 | 0.000488 | 0.057752 |
| KENNY_CTNNB1_TARGETS_DN | 0.000518 | 0.060337 |
| chr2q13 | 0.000568 | 0.064501 |
| REGULATION_OF_CELL_CYCLE | 0.000573 | 0.064501 |
| GNF2_SERPINI2 | 0.000604 | 0.066831 |
| V$T3R_Q6 | 0.00066 | 0.071888 |
| ACCGAGC,MIR-423 | 0.000686 | 0.073179 |
| HELLER_HDAC_TARGETS_UP | 0.000694 | 0.073179 |
| LEE_DIFFERENTIATING_T_LYMPHOCYTE | 0.000712 | 0.073942 |
| CHESLER_BRAIN_HIGHEST_EXPRESSION | 0.00079 | 0.080012 |
| MATSUDA_NATURAL_KILLER_DIFFERENTIA TION |
0.000794 | 0.080012 |
| MICROTUBULE_MOTOR_ACTIVITY | 0.000807 | 0.080139 |
| AGGGCCA,MIR-328 | 0.000836 | 0.081055 |
| HOFMANN_CELL_LYMPHOMA_DN | 0.000841 | 0.081055 |
| CYTOSKELETAL_PART | 0.000873 | 0.082976 |
| CYTOSKELETON | 0.00092 | 0.085062 |
| OHM_EMBRYONIC_CARCINOMA_UP | 0.000909 | 0.085062 |
| CAGCCTC,MIR-485-5P | 0.000934 | 0.085164 |
| PROTEIN_UBIQUITINATION | 0.000992 | 0.089296 |
| chr9q22 | 0.001106 | 0.096946 |
| FERREIRA_EWINGS_SARCOMA_UNSTABLE_V S_STABLE_DN |
0.001105 | 0.096946 |
Figure 1.
Pathways at FDR<0.1. Venn diagram indicating number of pathways significant at FDR<0.1 and overlap for outcomes of BPD/death, severe BPD/death, and severe BPD in survivors.
Table 4.
Biological pathways associated with severe BPD or death, as compared to survivors without severe BPD. Pathways from the annotated gene sets of the molecular signatures database at the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp) are listed in order of increasing false discovery rate (FDR). Only pathways with FDR ≤0.1 are shown.
| Pathway Names | P values | FDR |
|---|---|---|
| PHOSPHORUS_OXYGEN_LYASE_ACTIVITY | 5.68E-08 | 0.000192 |
| CYCLASE_ACTIVITY | 4.57E-08 | 0.000192 |
| GNF2_PRDX2 | 2.16E-07 | 0.000487 |
| MORF_RAP1A | 6.18E-07 | 0.001043 |
| KUMAMOTO_RESPONSE_TO_NUTLIN_3A_UP | 1.87E-06 | 0.002526 |
| MITOCHONDRION | 2.73E-06 | 0.003076 |
| SHEDDEN_LUNG_CANCER_POOR_SURVIVAL _A6 |
3.48E-06 | 0.003357 |
| VALK_AML_CLUSTER_2 | 5.04E-06 | 0.003783 |
| GLUCOSE_CATABOLIC_PROCESS | 4.73E-06 | 0.003783 |
| REACTOME_TRANSMISSION_ACROSS_CHEMI CAL_SYNAPSES |
6.68E-06 | 0.00435 |
| BIOCARTA_CCR5_PATHWAY | 7.09E-06 | 0.00435 |
| GRAHAM_CML_DIVIDING_VS_NORMAL_QUI ESCENT_UP |
1.09E-05 | 0.006114 |
| MORF_BRCA1 | 1.35E-05 | 0.006995 |
| MAP_KINASE_ACTIVITY | 3.16E-05 | 0.009928 |
| REACTOME_NOREPINEPHRINE_NEUROTRAN SMITTER_RELEASE_CYCLE |
3.12E-05 | 0.009928 |
| KCCGNSWTTT_UNKNOWN | 3.24E-05 | 0.009928 |
| RESPONSE_TO_ENDOGENOUS_STIMULUS | 2.73E-05 | 0.009928 |
| DELAYED_RECTIFIER_POTASSIUM_CHANNE L_ACTIVITY |
2.79E-05 | 0.009928 |
| module_428 | 2.35E-05 | 0.009928 |
| MORF_ATRX | 2.86E-05 | 0.009928 |
| TAGCTTT,MIR-9 | 3.21E-05 | 0.009928 |
| REACTOME_CLASS_C3_METABOTROPIC_GLU TAMATE_PHEROMONE_RECEPTORS |
2.80E-05 | 0.009928 |
| RESPONSE_TO_DNA_DAMAGE_STIMULUS | 3.54E-05 | 0.010382 |
| TOMLINS_PROSTATE_CANCER_UP | 4.63E-05 | 0.012494 |
| MCCLUNG_DELTA_FOSB_TARGETS_2WK | 4.48E-05 | 0.012494 |
| TRANSCRIPTION_COACTIVATOR_ACTIVITY | 5.68E-05 | 0.014751 |
| RODRIGUES_NTN1_AND_DCC_TARGETS | 6.01E-05 | 0.015023 |
| V$CREB_02 | 6.67E-05 | 0.016079 |
| GCM_GSPT1 | 7.38E-05 | 0.017173 |
| FERRARI_RESPONSE_TO_FENRETINIDE_DN | 8.00E-05 | 0.017994 |
| ATAAGCT,MIR-21 | 8.47E-05 | 0.018436 |
| NAKAMURA_CANCER_MICROENVIRONMENT _DN |
9.69E-05 | 0.020443 |
| LEE_INTRATHYMIC_T_PROGENITOR | 0.00011 | 0.022509 |
| TCCCCAC,MIR-491 | 0.000128 | 0.025452 |
| CYTOSOLIC_PART | 0.000133 | 0.025637 |
| WINTER_HYPOXIA_UP | 0.000169 | 0.029972 |
| TRANSCRIPTION_ACTIVATOR_ACTIVITY | 0.000162 | 0.029972 |
| MOREAUX_MULTIPLE_MYELOMA_BY_TACI_ UP |
0.000165 | 0.029972 |
| NELSON_RESPONSE_TO_ANDROGEN_UP | 0.000179 | 0.030963 |
| PATTERSON_DOCETAXEL_RESISTANCE | 0.000187 | 0.031433 |
| MORF_PPP5C | 0.000191 | 0.031433 |
| DNA_REPAIR | 0.000198 | 0.031826 |
| V$NFY_Q6_01 | 0.000206 | 0.032314 |
| XU_HGF_TARGETS_INDUCED_BY_AKT1_48H R_UP |
0.000218 | 0.033408 |
| WATANABE_RECTAL_CANCER_RADIOTHER APY_RESPONSIVE_DN |
0.000245 | 0.035718 |
| MORF_CCNF | 0.000241 | 0.035718 |
| REGULATION_OF_LIPID_METABOLIC_PROCE SS |
0.000249 | 0.035718 |
| WHITE_NEUROBLASTOMA_WITH_1P36.3_DEL ETION |
0.000258 | 0.036278 |
| REGULATION_OF_SMALL_GTPASE_MEDIATE D_SIGNAL_TRANSDUCTION |
0.000338 | 0.043476 |
| LAMELLIPODIUM | 0.000344 | 0.043476 |
| REACTOME_GLUTAMATE_NEUROTRANSMIT TER_RELEASE_CYCLE |
0.000331 | 0.043476 |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSI ON_SUSTAINED_IN_GRANULOCYTE_UP |
0.000325 | 0.043476 |
| V$IRF_Q6 | 0.000318 | 0.043476 |
| V$CRX_Q4 | 0.000348 | 0.043476 |
| PENG_LEUCINE_DEPRIVATION_UP | 0.000363 | 0.044551 |
| chr6q24 | 0.000386 | 0.046488 |
| INDUCTION_OF_APOPTOSIS_BY_INTRACELL ULAR_SIGNALS |
0.000409 | 0.048445 |
| module_471 | 0.000425 | 0.049432 |
| PUJANA_CHEK2_PCC_NETWORK | 0.000442 | 0.050452 |
| V$CREB_Q4_01 | 0.00047 | 0.050452 |
| PERINUCLEAR_REGION_OF_CYTOPLASM | 0.000486 | 0.050452 |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_ER YTHROID_DN |
0.000471 | 0.050452 |
| MATTIOLI_MULTIPLE_MYELOMA_WITH_14Q 32_TRANSLOCATIONS |
0.000482 | 0.050452 |
| PROTEIN_HOMODIMERIZATION_ACTIVITY | 0.000464 | 0.050452 |
| GRAHAM_CML_DIVIDING_VS_NORMAL_DIVI DING_DN |
0.000465 | 0.050452 |
| LIU_TARGETS_OF_VMYB_VS_CMYB_UP | 0.000498 | 0.050942 |
| REACTOME_IONOTROPIC_ACTIVITY_OF_KAI NATE_RECEPTORS |
0.000527 | 0.053129 |
| PYEON_CANCER_HEAD_AND_NECK_VS_CER VICAL_DN |
0.000553 | 0.054085 |
| ZHANG_PROLIFERATING_VS_QUIESCENT | 0.000548 | 0.054085 |
| EXTERNAL_SIDE_OF_PLASMA_MEMBRANE | 0.000604 | 0.058227 |
| GNF2_APEX1 | 0.00062 | 0.058953 |
| ZUCCHI_METASTASIS_DN | 0.000638 | 0.059768 |
| SNIJDERS_AMPLIFIED_IN_HEAD_AND_NECK_ TUMORS |
0.000651 | 0.060177 |
| V$USF_C | 0.000708 | 0.062681 |
| RNTCANNRNNYNATTW_UNKNOWN | 0.000689 | 0.062681 |
| DEPHOSPHORYLATION | 0.000715 | 0.062681 |
| chr5q34 | 0.000713 | 0.062681 |
| RHEIN_ALL_GLUCOCORTICOID_THERAPY_D N |
0.000727 | 0.062882 |
| CELLULAR_CARBOHYDRATE_CATABOLIC_P ROCESS |
0.000743 | 0.063492 |
| REGULATION_OF_RAS_PROTEIN_SIGNAL_TR ANSDUCTION |
0.000757 | 0.06388 |
| RICKMAN_HEAD_AND_NECK_CANCER_D | 0.000767 | 0.06388 |
| module_441 | 0.00079 | 0.064239 |
| REGULATION_OF_RAS_GTPASE_ACTIVITY | 0.000815 | 0.064239 |
| CHIBA_RESPONSE_TO_TSA_UP | 0.000816 | 0.064239 |
| SEIDEN_ONCOGENESIS_BY_MET | 0.000811 | 0.064239 |
| MORI_MATURE_B_LYMPHOCYTE_DN | 0.000818 | 0.064239 |
| ION_TRANSMEMBRANE_TRANSPORTER_ACT IVITY |
0.000836 | 0.064863 |
| TIEN_INTESTINE_PROBIOTICS_24HR_UP | 0.000879 | 0.067459 |
| SENGUPTA_NASOPHARYNGEAL_CARCINOM A_WITH_LMP1_UP |
0.000902 | 0.068386 |
| REGULATION_OF_CATABOLIC_PROCESS | 0.000926 | 0.069418 |
| TSAI_DNAJB4_TARGETS_DN | 0.000946 | 0.069432 |
| DNA_DAMAGE_RESPONSESIGNAL_TRANSDU CTION_RESULTING_IN_INDUCTION_OF_APOP TOSIS |
0.000939 | 0.069432 |
| KEGG_DRUG_METABOLISM_OTHER_ENZYM ES |
0.00099 | 0.070374 |
| ENK_UV_RESPONSE_EPIDERMIS_UP | 0.000973 | 0.070374 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_AND_N UCLEIC_ACID_TRANSMEMBRANE_TRAN SPORTER_ACTIVITY |
0.000986 | 0.070374 |
| KEGG_LIMONENE_AND_PINENE_DEGRADATI ON |
0.001016 | 0.07146 |
| SAGIV_CD24_TARGETS_UP | 0.001083 | 0.075354 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_AN D_NUCLEIC_ACID_TRANSPORT |
0.001155 | 0.076817 |
| JISON_SICKLE_CELL_DISEASE_UP | 0.001119 | 0.076817 |
| module_528 | 0.001156 | 0.076817 |
| ONDER_CDH1_TARGETS_1_UP | 0.001145 | 0.076817 |
| TAKEDA_TARGETS_OF_NUP98_HOXA9_FUSI ON_16D_UP |
0.001161 | 0.076817 |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRAN SITION_UP |
0.001194 | 0.078262 |
| chr6p | 0.001286 | 0.083098 |
| ZIRN_TRETINOIN_RESPONSE_DN | 0.001293 | 0.083098 |
| PACHER_TARGETS_OF_IGF1_AND_IGF2_UP | 0.001389 | 0.087629 |
| PAPASPYRIDONOS_UNSTABLE_ATEROSCLER OTIC_PLAQUE_DN |
0.001384 | 0.087629 |
| REACTOME_GLYCOGEN_BREAKDOWN_GLY COGENOLYSIS |
0.001428 | 0.089249 |
| GGTAACC,MIR-409-5P | 0.001467 | 0.090596 |
| EXTRACELLULAR_SPACE | 0.001476 | 0.090596 |
| ZHAN_MULTIPLE_MYELOMA_CD2_DN | 0.001539 | 0.093601 |
| REGULATION_OF_GTPASE_ACTIVITY | 0.00161 | 0.096446 |
| REACTOME_ACTIVATION_OF_CHAPERONES_ BY_IRE1_ALPHA |
0.001615 | 0.096446 |
| TGAGATT,MIR-216 | 0.001668 | 0.096635 |
| OKUMURA_INFLAMMATORY_RESPONSE_LPS | 0.001654 | 0.096635 |
| V$IRF1_Q6 | 0.001668 | 0.096635 |
| MOOTHA_HUMAN_MITODB_6_2002 | 0.001675 | 0.096635 |
| TGAYRTCA_V$ATF3_Q6 | 0.001731 | 0.098992 |
| LOPES_METHYLATED_IN_COLON_CANCER_ UP |
0.0018 | 0.099652 |
| BASSO_HAIRY_CELL_LEUKEMIA_DN | 0.001801 | 0.099652 |
| HONRADO_BREAST_CANCER_BRCA1_VS_BR CA2 |
0.001772 | 0.099652 |
| MORF_PSMF1 | 0.001769 | 0.099652 |
Table 5.
Biological pathways associated with severe BPD in survivors, as compared to survivors without BPD. Pathways from the annotated gene sets of the molecular signatures database at the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp) are listed in order of increasing false discovery rate (FDR). Only pathways with FDR <0.1 are shown.
| Pathway Names | P values | FDR |
|---|---|---|
| PHOSPHORUS_OXYGEN_LYASE_ACTIVITY | 3.91E-08 | 0.000132037 |
| CYCLASE_ACTIVITY | 2.55E-08 | 0.000132037 |
| KUMAMOTO_RESPONSE_TO_NUTLIN_3A_UP | 1.22E-06 | 0.002374878 |
| GNF2_PRDX2 | 1.41E-06 | 0.002374878 |
| SHEDDEN_LUNG_CANCER_POOR_SURVIVAL_A6 | 2.69E-06 | 0.003636936 |
| MORF_RAP1A | 4.01E-06 | 0.004511149 |
| GRAHAM_CML_DIVIDING_VS_NORMAL_QUIESC ENT_UP |
5.99E-06 | 0.005773791 |
| TOMLINS_PROSTATE_CANCER_UP | 6.89E-06 | 0.005811578 |
| VALK_AML_CLUSTER_2 | 9.42E-06 | 0.006927232 |
| GLUCOSE_CATABOLIC_PROCESS | 1.03E-05 | 0.006927232 |
| MAP_KINASE_ACTIVITY | 1.57E-05 | 0.008805869 |
| MITOCHONDRION | 1.49E-05 | 0.008805869 |
| V$CREB_02 | 2.13E-05 | 0.011048338 |
| REACTOME_TRANSMISSION_ACROSS_CHEMICAL _SYNAPSES |
2.51E-05 | 0.012111145 |
| MORF_BRCA1 | 5.17E-05 | 0.018918766 |
| ATAAGCT,MIR-21 | 6.17E-05 | 0.018918766 |
| RODRIGUES_NTN1_AND_DCC_TARGETS | 5.41E-05 | 0.018918766 |
| TCCCCAC,MIR-491 | 5.87E-05 | 0.018918766 |
| REACTOME_NOREPINEPHRINE_NEUROTRANSMI TTER_RELEASE_CYCLE |
5.27E-05 | 0.018918766 |
| NAKAMURA_CANCER_MICROENVIRONMENT_DN | 6.01E-05 | 0.018918766 |
| KCCGNSWTTT_UNKNOWN | 5.54E-05 | 0.018918766 |
| BIOCARTA_CCR5_PATHWAY | 5.95E-05 | 0.018918766 |
| RESPONSE_TO_ENDOGENOUS_STIMULUS | 7.72E-05 | 0.020857418 |
| TRANSCRIPTION_COACTIVATOR_ACTIVITY | 7.32E-05 | 0.020857418 |
| XU_HGF_TARGETS_INDUCED_BY_AKT1_48HR_U P |
7.59E-05 | 0.020857418 |
| DELAYED_RECTIFIER_POTASSIUM_CHANNEL_A CTIVITY |
8.90E-05 | 0.022241771 |
| RESPONSE_TO_DNA_DAMAGE_STIMULUS | 8.76E-05 | 0.022241771 |
| FERRARI_RESPONSE_TO_FENRETINIDE_DN | 9.70E-05 | 0.02337315 |
| module_428 | 0.000103 | 0.023984694 |
| MORF_ATRX | 0.000113 | 0.025358723 |
| GCM_GSPT1 | 0.000122 | 0.026655373 |
| WINTER_HYPOXIA_UP | 0.000139 | 0.029219502 |
| TRANSCRIPTION_ACTIVATOR_ACTIVITY | 0.00015 | 0.030772585 |
| NELSON_RESPONSE_TO_ANDROGEN_UP | 0.000176 | 0.034427242 |
| WATANABE_RECTAL_CANCER_RADIOTHERAPY_ RESPONSIVE_DN |
0.000179 | 0.034427242 |
| TAGCTTT,MIR-9 | 0.000189 | 0.034900695 |
| EXTERNAL_SIDE_OF_PLASMA_MEMBRANE | 0.000196 | 0.034900695 |
| MCCLUNG_DELTA_FOSB_TARGETS_2WK | 0.000192 | 0.034900695 |
| REGULATION_OF_SMALL_GTPASE_MEDIATED_SI GNALTRANSDUCTION |
0.000216 | 0.036492426 |
| PUJANA_CHEK2_PCC_NETWORK | 0.000216 | 0.036492426 |
| WHITE_NEUROBLASTOMA_WITH_1P36.3_DELETI ON |
0.000228 | 0.037550752 |
| CYTOSOLIC_PART | 0.000242 | 0.038012621 |
| chr6q24 | 0.000241 | 0.038012621 |
| BENPORATH_ES_CORE_NINE_CORRELATED | 0.000249 | 0.038221556 |
| SENGUPTA_NASOPHARYNGEAL_CARCINOMA_W ITH_LMP1_UP |
0.000265 | 0.039765851 |
| SAGIV_CD24_TARGETS_UP | 0.000315 | 0.04177443 |
| KEGG_DRUG_METABOLISM_OTHER_ENZYMES | 0.00031 | 0.04177443 |
| RHEIN_ALL_GLUCOCORTICOID_THERAPY_DN | 0.000309 | 0.04177443 |
| REACTOME_CLASS_C3_METABOTROPIC_GLUTA MATE_PHEROMONE_RECEPTORS |
0.000306 | 0.04177443 |
| chr3p25 | 0.000316 | 0.04177443 |
| V$CREB_Q4_01 | 0.000306 | 0.04177443 |
| LAMELLIPODIUM | 0.000338 | 0.043890934 |
| ZUCCHI_METASTASIS_DN | 0.000372 | 0.047406175 |
| MOREAUX_MULTIPLE_MYELOMA_BY_TACI_UP | 0.000383 | 0.047933234 |
| REACTOME_STEROID_HORMONE_BIOSYNTHESIS | 0.000393 | 0.048235755 |
| chr3p14 | 0.00041 | 0.048235755 |
| LEE_INTRATHYMIC_T_PROGENITOR | 0.000404 | 0.048235755 |
| SARRIO_EPITHELIAL_MESENCHYMAL_TRANSITI ON_UP |
0.000414 | 0.048235755 |
| module_441 | 0.000431 | 0.049268618 |
| GNF2_APEX1 | 0.00044 | 0.049520971 |
| PERINUCLEAR_REGION_OF_CYTOPLASM | 0.000448 | 0.049520971 |
| V$USF_C | 0.000482 | 0.052423429 |
| MORF_CCNF | 0.000495 | 0.053037318 |
| V$NFY_Q6_01 | 0.000536 | 0.056482951 |
| REGULATION_OF_RAS_PROTEIN_SIGNAL_TRANS DUCTION |
0.000597 | 0.057783747 |
| LINDGREN_BLADDER_CANCER_CLUSTER_2B | 0.000599 | 0.057783747 |
| LOPES_METHYLATED_IN_COLON_CANCER_UP | 0.000594 | 0.057783747 |
| BASSO_HAIRY_CELL_LEUKEMIA_DN | 0.00057 | 0.057783747 |
| GAZDA_DIAMOND_BLACKFAN_ANEMIA_ERYTH ROID_DN |
0.000561 | 0.057783747 |
| REACTOME_GLUTAMATE_NEUROTRANSMITTER _RELEASE_CYCLE |
0.000584 | 0.057783747 |
| TSAI_DNAJB4_TARGETS_DN | 0.000646 | 0.058786732 |
| PYEON_CANCER_HEAD_AND_NECK_VS_CERVIC AL_DN |
0.000669 | 0.058786732 |
| TONKS_TARGETS_OF_RUNX1_RUNX1T1_FUSION_ SUSTAINED_IN_GRANULOCYTE_UP |
0.000661 | 0.058786732 |
| GGTAACC,MIR-409-5P | 0.000671 | 0.058786732 |
| REGULATION_OF_CATABOLIC_PROCESS | 0.000647 | 0.058786732 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_AND_N UCLEIC_ACID_TRANSPORT |
0.000645 | 0.058786732 |
| CELLULAR_CARBOHYDRATE_CATABOLIC_PROC ESS |
0.000663 | 0.058786732 |
| MCBRYAN_PUBERTAL_BREAST_5_6WK_UP | 0.000698 | 0.059610192 |
| RICKMAN_HEAD_AND_NECK_CANCER_D | 0.000696 | 0.059610192 |
| PATTERSON_DOCETAXEL_RESISTANCE | 0.000719 | 0.060660099 |
| DNA_REPAIR | 0.000744 | 0.061980193 |
| REGULATION_OF_RAS_GTPASE_ACTIVITY | 0.000769 | 0.063320275 |
| SNIJDERS_AMPLIFIED_IN_HEAD_AND_NECK_TU MORS |
0.000804 | 0.065381496 |
| MYLLYKANGAS_AMPLIFICATION_HOT_SPOT_25 | 0.000831 | 0.066760855 |
| REGULATION_OF_GTPASE_ACTIVITY | 0.000917 | 0.071987568 |
| CHIARETTI_T_ALL_REFRACTORY_TO_THERAPY | 0.000917 | 0.071987568 |
| module_471 | 0.000932 | 0.072274597 |
| MAYBURD_RESPONSE_TO_L663536_UP | 0.001005 | 0.074582373 |
| V$IRF_Q6 | 0.000987 | 0.074582373 |
| TGAGATT,MIR-216 | 0.000994 | 0.074582373 |
| MATTIOLI_MULTIPLE_MYELOMA_WITH_14Q 32_TRANSLOCATIONS |
0.001002 | 0.074582373 |
| REACTOME_IONOTROPIC_ACTIVITY_OF_KAINAT E_RECEPTORS |
0.001081 | 0.077418566 |
| REACTOME_GLYCOGEN_BREAKDOWN_GLYCOG ENOLYSIS |
0.00109 | 0.077418566 |
| CCCNNGGGAR_V$OLF1_01 | 0.001076 | 0.077418566 |
| RNTCANNRNNYNATTW_UNKNOWN | 0.001062 | 0.077418566 |
| HENDRICKS_SMARCA4_TARGETS_UP | 0.001129 | 0.079382915 |
| MORF_PPP5C | 0.001202 | 0.081971096 |
| CHIBA_RESPONSE_TO_TSA_UP | 0.001195 | 0.081971096 |
| GTGTGAG,MIR-342 | 0.001186 | 0.081971096 |
| PUJANA_BRCA1_PCC_NETWORK | 0.001284 | 0.086662282 |
| PENG_LEUCINE_DEPRIVATION_UP | 0.00132 | 0.087158417 |
| ZHANG_RESPONSE_TO_IKK_INHIBITOR_AND_TN F_UP |
0.001309 | 0.087158417 |
| VICENT_METASTASIS_UP | 0.00133 | 0.087158417 |
| GNF2_ANP32B | 0.001384 | 0.088728588 |
| DEPHOSPHORYLATION | 0.001389 | 0.088728588 |
| V$IRF2_01 | 0.001393 | 0.088728588 |
| HASINA_NOL7_TARGETS_DN | 0.001413 | 0.089142284 |
| INDUCTION_OF_APOPTOSIS_BY_INTRACELLULA R_SIGNALS |
0.001441 | 0.090064396 |
| OKUMURA_INFLAMMATORY_RESPONSE_LPS | 0.001493 | 0.092456662 |
| NUCLEAR_UBIQUITIN_LIGASE_COMPLEX | 0.001523 | 0.093434244 |
| TGAYRTCA_V$ATF3_Q6 | 0.001538 | 0.093522358 |
| FERREIRA_EWINGS_SARCOMA_UNSTABLE_VS_S TABLE_UP |
0.001631 | 0.09598751 |
| REGULATION_OF_LIPID_METABOLIC_PROCESS | 0.001594 | 0.09598751 |
| V$PAX3_01 | 0.001635 | 0.09598751 |
| BLUM_RESPONSE_TO_SALIRASIB_UP | 0.00165 | 0.09598751 |
| chr6p | 0.001628 | 0.09598751 |
| ENK_UV_RESPONSE_EPIDERMIS_UP | 0.001673 | 0.096498142 |
| BIOCARTA_LONGEVITY_PATHWAY | 0.001722 | 0.097653706 |
| chr5q34 | 0.001707 | 0.097653706 |
| NUCLEOLUS | 0.00174 | 0.097890086 |
| PACHER_TARGETS_OF_IGF1_AND_IGF2_UP | 0.001832 | 0.098235946 |
| AXON | 0.001769 | 0.098235946 |
| TIEN_INTESTINE_PROBIOTICS_24HR_UP | 0.001796 | 0.098235946 |
| JISON_SICKLE_CELL_DISEASE_UP | 0.00181 | 0.098235946 |
| CTGCAGY_UNKNOWN | 0.001848 | 0.098235946 |
| REACTOME_ACTIVATION_OF_CHAPERONES_BY_ IRE1_ALPHA |
0.001839 | 0.098235946 |
| KEGG_LIMONENE_AND_PINENE_DEGRADATION | 0.001808 | 0.098235946 |
| REACTOME_DOWN_STREAM_SIGNAL_TRANSDU CTION |
0.002053 | 0.099165696 |
| NUCLEOBASENUCLEOSIDENUCLEOTIDE_AND_N UCLEIC_ACID_TRANSMEMBRANE_TRANSPORTE R_ACTIVITY |
0.001938 | 0.099165696 |
| REACTOME_PYRUVATE_METABOLISM | 0.001953 | 0.099165696 |
| FUNG_IL2_SIGNALING_1 | 0.002071 | 0.099165696 |
| PAPASPYRIDONOS_UNSTABLE_ATEROSCLEROTI C_PLAQUE_DN |
0.002003 | 0.099165696 |
| ZHANG_PROLIFERATING_VS_QUIESCENT | 0.001967 | 0.099165696 |
| CAAGGAT,MIR-362 | 0.001995 | 0.099165696 |
| PENG_RAPAMYCIN_RESPONSE_UP | 0.002047 | 0.099165696 |
| V$CRX_Q4 | 0.002058 | 0.099165696 |
| EXTRACELLULAR_SPACE | 0.001993 | 0.099165696 |
| CARBOHYDRATE_CATABOLIC_PROCESS | 0.001922 | 0.099165696 |
| TTCCGTT,MIR-191 | 0.002022 | 0.099165696 |
| HONRADO_BREAST_CANCER_BRCA1_VS_BRCA2 | 0.001971 | 0.099165696 |
| SEIDEN_ONCOGENESIS_BY_MET | 0.001986 | 0.099165696 |
| module_318 | 0.002096 | 0.099652829 |
Table 6.
Biological pathways associated with BPD or death classified by race, as compared to survivors without BPD. Pathways from the annotated gene sets of the molecular signatures database at the Broad Institute (http://www.broadinstitute.org/gsea/msigdb/index.jsp) are listed in order of increasing false discovery rate (FDR). Only the top 12 pathways are shown for All infants, White infants, and Black infants.
| All infants | White infants | Black infants | ||||||
|---|---|---|---|---|---|---|---|---|
| Pathway | P value |
FDR | Pathway | P value |
FDR | Pathway | P value | FDR |
| GACAATC, MIR-219 |
1.41E- 08 |
9.52E -05 |
module_ 320 |
1.82E -49 |
1.23 E-45 |
RODRIGU ES_THYR OID_CAR CINOMA_ DN |
7.80E- 07 |
0.005262 |
| NUCLEAR_ UBIQUITIN _LIGASE_C OMPLEX |
2.16E- 07 |
0.000 73 |
GNF2_B UB1 |
2.29E -41 |
7.73 E-38 |
TOMLINS _METAST ASIS_DN |
4.91E- 06 |
0.007876 |
| YRCCAKN NGNCGC_U NKNOWN |
7.11E- 07 |
0.001 6 |
GNF2_T TK |
1.03E -31 |
2.32 E-28 |
ATAAGCT ,MIR-21 |
5.14E- 06 |
0.007876 |
| V$E2F_Q2 | 9.88E- 07 |
0.001 668 |
GNF2_S MC2L1 |
1.38E -31 |
2.33 E-28 |
KEGG_CE LL_ADHE SION_MO LECULES _CAMS |
2.39E- 06 |
0.007876 |
| NEURON_P ROJECTION |
1.37E- 05 |
0.015 893 |
GNF2_H MMR |
4.80E -24 |
6.48 E-21 |
module_34 9 |
5.83E- 06 |
0.007876 |
| V$GABP_B | 1.41E- 05 |
0.015 893 |
GNF2_E SPL1 |
8.12E -23 |
9.14 E-20 |
BEGUM_T ARGETS_ OF_PAX3_ FOXO1_F USION_UP |
8.48E- 06 |
0.008177 |
| TOMLINS_ METASTAS ISDN |
1.66E- 05 |
0.015 965 |
GNF2_C ENPE |
1.23E -22 |
CHEOK_R ESPONSE_ TO_MERC APTOPUR INE_AND_ HD_MTX_ UP |
8.15E- 06 |
0.008177 | |
| NUCLEOBA SENUCLEO SIDENUCL EOTIDE_KI NASE_ACTI VITY |
2.74E- 05 |
0.018 801 |
GNF2_C DC20 |
5.47E -22 |
AMIT_EG F_RESPO NSE_480_ HELA |
1.00E- 05 |
0.008437 | |
| V$E2F_Q4 | 3.27E- 05 |
0.018 801 |
module_ 244 |
1.14E -21 |
8.55 E-19 |
CELLULA R_MACRO MOLECUL E_METAB OLIC_PRO CESS |
1.98E- 05 |
0.014845 |
| RUIZ_TNC_ TARGETS_ DN |
2.99E- 05 |
0.018 801 |
GNF2_C KS1B | 3.48E -20 |
2.35 E-17 |
KENNY_C TNNB1_T ARGETS_ DN |
2.26E- 05 |
0.015232 |
| JAEGER_M ETASTASIS _UP |
3.34E- 05 |
0.018 801 |
MORI_L ARGE_P RE_BII_ LYMPH OCYTE_ UP |
2.86E -19 |
1.76 E-16 |
PROTEIN_ UBIQUITI NATION |
2.94E- 05 |
0.017542 |
| SUGAR_BI NDING |
2.71E- 05 |
0.018 801 |
GNF2_C KS2 |
7.66E -19 |
4.31 E-16 |
CELLULA R_PROTEI N_METAB OLIC_PRO CESS |
3.27E- 05 |
0.017542 |
Evaluation of individual SNPs and pathways/gene sets using gene expression dataset
Gene expression for six of the nine genes with the lowest single SNP p-values could be assessed by a total of 20 probe sets present in the data set.16 Two (NUAK1 and GRIP1) of these six genes were significantly dysregulated in BPD lung tissue, with lower expression in BPD when compared with controls. In addition to these significant genes, 2 probe sets for CD44 demonstrated a trend for increased expression in BPD lungs (p<0.01) (Table VII; available at www.jpeds.com).
Table 7.
Six of the nine genes (represented by 20 probesets) identified as having the top 10 SNPs are on HG-U133 plus array for gene expression. Two genes (NUAK1 and GRIP1) show significantly reduced expression in BPD lung tissue at p<0.05 and CD44 shows a trend towards increased expression in BPD, but none of the identified SNPs were found to have a more than two-fold change in expression level.
| Gene Symbol | Gene Description | Entrez Gene Id |
HU- 133plus2 ProbesetID |
Adj P- value |
Fold Change |
Log Fold Change |
|---|---|---|---|---|---|---|
| NUAK1 | “NUAK family, SNF1-like kinase, 1” |
9891 | 204589_at | 0.0061 | 0.60197 1 |
− 0.73224 |
| GRIP1 | glutamate receptor interacting protein 1 |
23426 | 235957_at | 0.0227 | 0.82473 | − 0.27801 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 204489_s_at | 0.0638 | 1.22524 1 |
0.29306 6 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 210916_s_at | 0.1006 | 1.34114 9 |
0.42346 9 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 209835_x_at | 0.1152 | 1.25162 9 |
0.32380 7 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 204490_s_at | 0.1160 | 1.25926 2 |
0.33257 9 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 212014_x_at | 0.1312 | 1.23871 3 |
0.30884 3 |
| CD44 | CD44 molecule | 960 | 1557905_s_a | 0.1570 | 1.29494 | 0.37288 |
| (Indian blood group) | t | 5 | ||||
| CD44 | CD44 molecule (Indian blood group) |
960 | 212063_at | 0.2456 | 1.08290 7 |
0.11491 |
| GALNTL6 | UDP-N-acetyl- alpha-D- galactosamine:polyp eptide N- acetylgalactosaminyl transferase-like 6 |
442117 | 1555273_at | 0.3811 | 1.04195 8 |
0.05929 8 |
| KCNH7 | “potassium voltage- gated channel, subfamily H (eagrelated), member 7” |
90134 | 224099_at | 0.4570 | 1.05710 3 |
0.08011 7 |
| ADARB2 | “adenosine deaminase, RNA- specific, B2” |
105 | 237437_s_at | 0.5247 | 1.02396 5 |
0.03416 6 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 229221_at | 0.5401 | 1.11443 1 |
0.15630 7 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 1565868_at | 0.6141 | 0.92827 6 |
− 0.10737 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 217523_at | 0.6336 | 1.05519 6 |
0.07751 1 |
| ADARB2 | “adenosine | 105 | 220648_at | 0.6381 | 1.04929 | 0.06941 |
| deaminase, RNA- specific, B2” |
4 | |||||
| GRIP1 | glutamate receptor interacting protein 1 |
23426 | 214018_at | 0.6900 | 1.01674 9 |
0.02396 3 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 234418_x_at | 0.7458 | 1.05579 9 |
0.07833 5 |
| KCNH7 | “potassium voltage- gated channel, subfamily H (eag- related), member 7” |
90134 | 1555316_a_a t |
0.7512 | 1.01244 6 |
0.01784 5 |
| CD44 | CD44 molecule (Indian blood group) |
960 | 234411_x_at | 0.8856 | 1.01904 | 0.02721 |
We selected four pathways for further evaluation using data from the lung tissue gene expression data set.16 These pathways were (1) miR-219 pathway, the top pathway for BPD/death, (2) PACHER_TARGETS_OF_IGF1_AND_IGF2_UP, one of the three pathways shared among all three outcomes, as IGF1 is important in lung development20 and is increased in BPD,21 (3) Phosphorus Oxygen Lyase Pathway, the top pathway associated with severe BPD/death as well as severe BPD in survivors, and (4) Cell Cycle: G2/M DNA Damage Checkpoint Regulation canonical pathway, previously appreciated as the top pathway in the BPD gene expression dataset16 but not specifically evaluated in this study (as it is not defined in MSigDB), but with overlap with MORF_BRCA1, a pathway shared among all three outcomes.
MiR-219 Pathway (Table VIII; available at www.jpeds.com): Gene expression for all 143 genes in this pathway was assessed. 32 of 143 (22%) of pathway genes were dysregulated in BPD lung tissue (vs. 7 expected at random, p <0.0001). Fourteen genes had increased expression in BPD lung and 19 genes had decreased expression. Interestingly, independent probe sets for MAPT had increased (1 probe set) or decreased (3 probe sets) expression. Likewise, THRB had increased (1 probe set) or decreased (1 probe set) expression. These observations might suggest alternative splicing.
Targets of IGF1 and IGF2 Pathway (Table IX; available at www.jpeds.com): Gene expression for 34 of the 36 genes in this pathway was assessed using 78 probe sets. Two of the 34 genes had significantly increased expression in BPD; IGF1 (fold change>2, p<0.01) and SFMBT2 (fold change >1.07, p<0.05). Four independent probe sets demonstrated significance for IGF1.
Phosphorus Oxygen Lyase Activity Pathway: Gene expression for all 10 genes was assessed. ADCY8 had significantly reduced expression (fold change=0.59, p=0.0041) in BPD.
Cell Cycle Pathway (Table X; available at www.jpeds.com): Gene expression for all 23 genes was assessed using 61 probe sets. 35% of all pathway genes (8 of 23) were dysregulated in BPD, with increased expression. Many of these observations were demonstrated by multiple probe sets (15 probe sets different). Brca1 was increased by 1.21 fold in one probe set, with a p= 0.07, and by 1.3 fold in another, with p=0.09.
Table 8.
Targets of miR-219. Table showing the 32 genes (represented by 42 probe sets) of the 143 unique genes (represented by 515 probe sets) in the miR-219 pathway that were significant by t-test in the gene expression data set.
| Gene Symbol | Gene Description | Entrez Gene Id |
HU- 133plus2 ProbesetID |
Adj P- value |
Fold Change |
Log Fold Change |
|---|---|---|---|---|---|---|
| AKAP13 | A kinase (PRKA) anchor protein 13 |
11214 | 243450_at | 0.00705 5 |
0.80614 6 |
− 0.31089 |
| BTBD7 | BTB (POZ) domain containing 7 |
55727 | 220297_at | 0.01765 4 |
0.76128 6 |
− 0.39349 |
| BTBD7 | BTB (POZ) domain containing 7 |
55727 | 1556000_s_a t |
0.04300 6 |
0.90249 3 |
− 0.14801 |
| CAMK2G | calcium/calmodulin- dependent prote… |
818 | 212669_at | 0.03227 8 |
0.87526 7 |
− 0.19221 |
| CBFA2T3 | core-binding factor, runt domain, … |
863 | 208056_s_at | 0.01536 5 |
0.71140 8 |
− 0.49125 |
| CC2D1A | coiled-coil and C2 domain containi… |
54862 | 221888_at | 0.02882 9 |
0.92234 7 |
− 0.11662 |
| CD164 | CD164 molecule, sialomucin |
8763 | 208654_s_at | 0.02680 3 |
1.14101 | 0.19031 2 |
| CELF2//CUG BP2 |
10659 | 1554569_a_a t |
0.02572 2 |
1.45132 1 |
0.53736 7 |
|
| CHD7 | chromodomain | 55636 | 218829_s_at | 0.03446 | 0.86567 | −0.2081 |
| helicase DNA binding … |
3 | 9 | ||||
| CHD7 | chromodomain helicase DNA binding … |
55636 | 226123_at | 0.03789 9 |
0.88107 8 |
− 0.18266 |
| CPEB3 | cytoplasmic polyadenylation elemen… |
22849 | 237508_at | 0.02606 9 |
0.90919 5 |
− 0.13734 |
| CXXC5 | CXXC finger 5 | 51523 | 236516_at | 0.01953 8 |
1.06521 3 |
0.09114 2 |
| ELK1 | ELK1, member of ETS oncogene family |
2002 | 203617_x_at | 0.02835 7 |
1.16351 9 |
0.21849 5 |
| ELMOD2 | ELMO/CED-12 domain containing 2 |
255520 | 1553928_at | 0.02606 6 |
1.26317 2 |
0.33705 1 |
| FMNL2 | formin-like 2 | 114793 | 230663_at | 0.04878 2 |
1.31819 7 |
0.39856 6 |
| GTPBP1 | GTP binding protein 1 |
9567 | 219357_at | 0.04833 7 |
1.12997 3 |
0.17628 8 |
| HAS3 | hyaluronan synthase 3 |
3038 | 228179_at | 0.02455 | 0.83261 4 |
− 0.26428 |
| ING3 | inhibitor of growth family, member 3 |
54556 | 205070_at | 0.03146 9 |
0.81135 4 |
−0.3016 |
| INPP5J//PIB5 PA |
27124 | 213651_at | 0.04666 6 |
0.88371 5 |
− 0.17835 |
|
| KCNH8 | potassium voltage- gated channel, s… |
131096 | 1552742_at | 0.02507 3 |
0.52641 4 |
− 0.92573 |
| MAPT | microtubule- associated protein tau |
4137 | 203929_s_at | 0.00363 2 |
0.69750 1 |
− 0.51973 |
| MAPT | microtubule- associated protein tau |
4137 | 206401_s_at | 0.02041 4 |
0.79484 3 |
− 0.33126 |
| MAPT | microtubule- associated protein tau |
4137 | 225379_at | 0.04582 2 |
0.76185 | − 0.39242 |
| MTAP | methylthioadenosine phosphorylase |
4507 | 204956_at | 0.01869 | 1.16245 8 |
0.21717 8 |
| NEK6 | NIMA (never in mitosis gene a)-rel… |
10783 | 237761_at | 0.04877 7 |
1.41764 7 |
0.50349 8 |
| PHACTR2 | phosphatase and actin regulator 2 |
9749 | 204047_s_at | 0.02978 4 |
0.75942 1 |
− 0.39703 |
| PHF19 | PHD finger protein 19 |
26147 | 227212_s_at | 0.02077 9 |
1.49380 4 |
0.57899 1 |
| PHF19 | PHD finger protein 19 |
26147 | 227211_at | 0.02539 3 |
1.46813 | 0.55398 |
| RECK | reversion-inducing- cysteine-rich p… |
8434 | 1558116_x_ at |
0.02616 8 |
1.26418 2 |
0.33820 4 |
| RECK | reversion-inducing- cysteine-rich p… |
8434 | 216156_at | 0.03733 5 |
1.16256 4 |
0.21731 |
| RNF6 | ring finger protein (C3H2C3 type) 6 |
6049 | 210931_at | 0.04062 3 |
1.49443 7 |
0.57960 2 |
| SDK1 | sidekick homolog 1 (chicken) |
221935 | 229407_at | 0.00500 4 |
0.69252 3 |
− 0.53007 |
| SH3D19 | - | 152503 | 243636_s_at | 0.01745 5 |
0.63818 3 |
− 0.64796 |
| SH3D19 | - | 152503 | 237157_at | 0.02170 8 |
0.61934 8 |
− 0.69118 |
| SLC31A1 | solute carrier family 31 (copper t… |
1317 | 203971_at | 0.04906 4 |
1.50404 1 |
0.58884 4 |
| SNRK | SNF related kinase | 54861 | 237942_at | 0.02227 5 |
0.76360 8 |
−0.3891 |
| SNRK | SNF related kinase | 54861 | 209481_at | 0.02994 6 |
0.86885 8 |
− 0.20281 |
| SOX6 | SRY (sex determining region Y)-box 6 |
55553 | 243255_at | 0.04205 9 |
0.89038 6 |
−0.1675 |
| TACC1 | transforming, acidic coiled-coil c… |
6867 | 234010_at | 0.00250 4 |
0.70495 9 |
− 0.50439 |
| THRB | thyroid hormone receptor, beta (er… |
7068 | 228716_at | 0.02175 3 |
0.80693 3 |
− 0.30948 |
| THRB | thyroid hormone receptor, beta (er… |
7068 | 235927_at | 0.02974 5 |
1.2889 | 0.36614 |
Table 9.
Targets of IGF-1 and IGF-2. Table showing the 2 genes (represented by 5 probe sets) of the 34 unique genes (represented by 78 probe sets) out of the 36 listed in the IGF-1 and IGF-2 pathway that were significant by t-test in the gene expression data set.
| Gene Symbol |
Gene Description |
Entrez Gene Id |
ProbesetID (HU-133+) |
Adj P- value |
Fold Change |
Log Fold Change |
|---|---|---|---|---|---|---|
| IGF1 | insulin-like growth factor 1 (somatomed… |
3479 | 209542_x_at | 0.0089 | 2.035383 | 1.0253 |
| IGF1 | insulin-like growth factor 1 (somatomed… |
3479 | 211577_s_at | 0.009588 | 1.990326 | 0.993005 |
| IGF1 | insulin-like growth factor 1 (somatomed… |
3479 | 209540_at | 0.013246 | 1.940347 | 0.956315 |
| IGF1 | insulin-like growth factor 1 (somatomed… |
3479 | 209541_at | 0.014723 | 1.875297 | 0.907119 |
| SFMBT2 | Scm-like with four mbt domains 2 |
57713 | 232938_at | 0.049014 | 1.075057 | 0.104413 |
Table 10.
Cell Cycle: G2/M DNA Damage Checkpoint Regulation canonical pathway. Table showing the 8 genes (represented by 15 probe sets) of the 23 unique genes (represented by 61 probe sets) of the cell cycle pathway that were significant by t-test in the gene expression data set.
| Molecule Name |
ProbesetID (HU-133+) |
Gene Symbol |
Gene Title | Entrez Gene |
Adj P- value |
Fold Change |
|---|---|---|---|---|---|---|
| Cdc2 | 231534_at | CDC2 | Cell division cycle 2, G1 to S and G2 to M |
983 | 0.00291 | 2.282306 |
| Cdc2 | 203214_x_at | CDC2 | cell division cycle 2, G1 to S and G2 to M |
983 | 0.003788 | 2.055242 |
| Cdc2 | 210559_s_at | CDC2 | cell division cycle 2, G1 to S and G2 to M |
983 | 0.004146 | 2.102061 |
| CKS2 | 204170_s_at | CKS2 | CDC28 protein kinase regulatory subunit 2 |
1164 | 0.004267 | 1.522256 |
| Chk1 | 205393_s_at | CHEK1 | CHK1 checkpoint homolog (S. pombe) |
1111 | 0.005303 | 1.723429 |
| Chk1 | 205394_at | CHEK1 | CHK1 checkpoint homolog (S. pombe) |
1111 | 0.009462 | 1.73522 |
| MDM2 | 237891_at | MDM2 | Mdm2, transformed 3T3 cell double minute 2, p53 binding protein (mouse) |
4193 | 0.010258 | 1.447509 |
| Cdc2 | 203213_at | CDC2 | cell division cycle 2, G1 to S and G2 to M |
983 | 0.013499 | 1.861818 |
| p19Arf | 209644_x_at | CDKN2A | cyclin- dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) |
1029 | 0.015326 | 1.424757 |
| Chk1 | 238075_at | CHEK1 | CHK1 checkpoint homolog (S. pombe) |
1111 | 0.025811 | 1.552763 |
| Plk1 | 202240_at | PLK1 | polo-like kinase 1 (Drosophila) |
5347 | 0.026476 | 1.455546 |
| 14-3-3σ | 33323_r_at | SFN | stratifin | 2810 | 0.029624 | 2.786214 |
| 14-3-3σ | 33322_i_at | SFN | stratifin | 2810 | 0.033602 | 2.215685 |
| ATR | 209903_s_at | ATR | ataxia telangiectasia and Rad3 related |
545 | 0.040315 | 1.171942 |
| Chk2 | 210416_s_at | CHEK2 | CHK2 checkpoint homolog (S. pombe) |
11200 | 0.048167 | 1.2259 |
Evaluation of miR-219 and CD44 in mouse models and in human lung
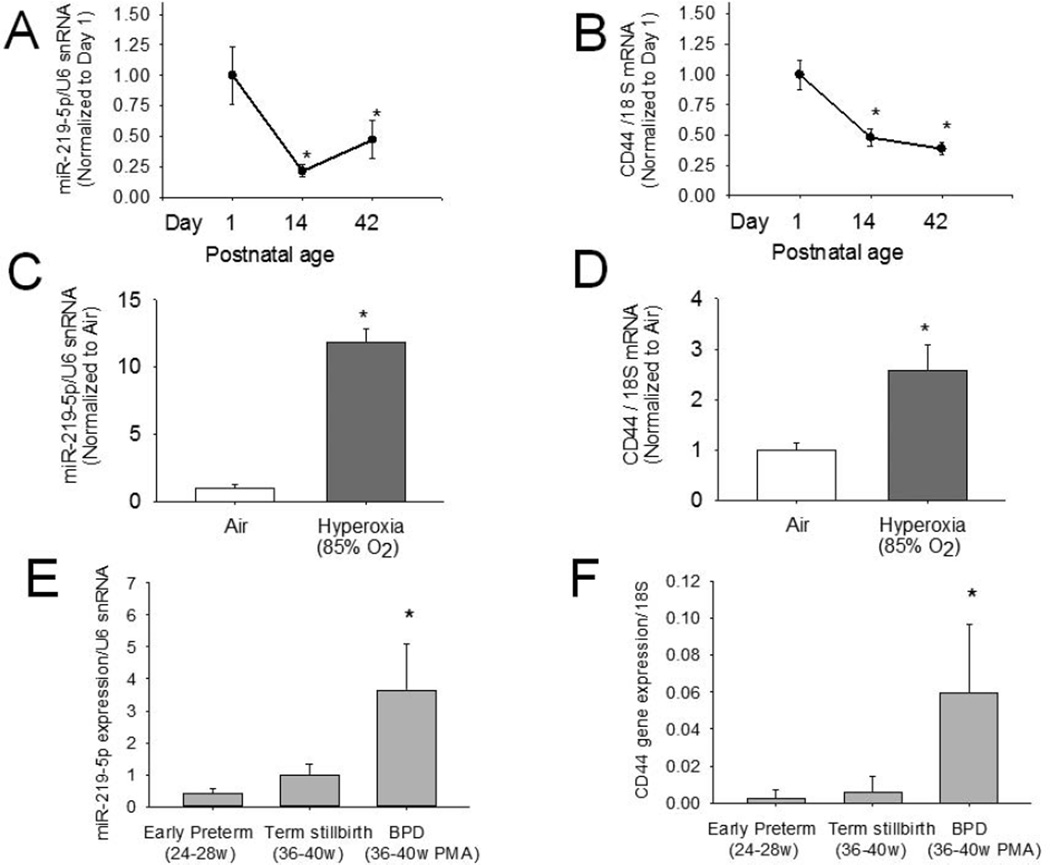
Expression of miR-219 and CD44 decreased over the course of alveolar septation as they were reduced on postnatal day 14 and 42 compared with day 1. Exposure to hyperoxia was associated with increased miR-219 and CD44 on day 14. ADARB2 transcripts were not detected in the lung in significant amounts (detected at more than 35 cycles of qPCR).
Expression of miR-219 and CD44 were both increased in human BPD lung compared with preterm and term lung (Figure 2).
Figure 2.
Evaluation of miR-219 and CD44 in a newborn mouse model (Panels A-D) and in human lung (Panels E-F). Lung miR-219 (Panel A) and CD44 mRNA (Panel B) decreased during alveolar septation, with expression on postnatal days 14 and 42 significantly less as compared with day 1; *p<0.05. Lung miR-219 (Panel C) and CD44 mRNA (Panel D) were also increased on postnatal day 14 during hyperoxia exposure (*p<0.05 compared with air). Lung miR-219 (Panel E) and CD44 mRNA (Panel F) were increased in human lungs with BPD as compared to early preterm or term stillbirth lungs (Mean±SEM; n=4/group)
DISCUSSION
BPD has a strong genetic component, but conventional single-marker approaches have not successfully explained more than a small fraction of the heritability of BPD. In this exploratory analysis, we identified biological pathways that contribute to the heritability of BPD using gene set analysis. Our analysis suggests involvement of known pathways (e.g. phosphorus oxygen lyase activity) and molecules (e.g. CD44) involved in lung development and repair. In addition, we identified novel pathways (e.g. targets of miR-219) and molecules (e.g. ADARB2, CD44) that may be involved in genetic predisposition to BPD or death. We validated this survey of gene sets associated with BPD in extremely preterm infants using a gene expression dataset from an independent population and evaluated selected molecules in a newborn mouse model and by gene expression in autopsy lung samples of BPD lung compared with normal preterm and term lung. Our results also indicate that severe BPD or death are associated with pathways distinct from mild/moderate BPD, suggesting that they have a different pathophysiologic basis, and that much variation is present in genetic predisposition to BPD by race/ethnicity.
To date, analysis of the pathways affected in BPD has relied on two GWAS5, 6 and a genome-wide transcriptional profiling study.16 The GWAS by Hadchouel et al.5 identified SPOCK2 gene as associated with BPD, but the GWAS by Wang et al6 did not identify any SNPs associated with BPD at a p<5 ×10−8 and pathway analyses were also not informative. Bhattacharya et al16 analyzed RNA from lung tissue obtained at autopsy from 11 BPD cases and 17 age-matched controls without BPD. 159 genes were differentially expressed in BPD, and pathway analysis confirmed previously known (e.g. DNA damage regulation of cell cycle) and novel (e.g. B-cell development) pathways.
In the present study, we identified multiple pathways associated with BPD/death, severe BPD/death, and severe BPD in survivors. Notably, the overlap in pathways between any BPD/death and severe BPD/death (or severe BPD in survivors) was limited to only 3 pathways, a small fraction of the total number of pathways associated with each outcome. This suggests that the pathways associated with any BPD/death but not with severe BPD/death are those associated with mild or moderate BPD. This suggests that the difference in clinical phenotype between mild and moderate BPD versus severe BPD is also manifest at the genomic level. Similarly, the 105 pathways in the large overlap between severe BPD/death and severe BPD in survivors, especially the top 43 pathways, are probably pathways associated with severe BPD. The 15 pathways in severe BPD/death that do not overlap with severe BPD in survivors may be those associated with death. These results suggest that distinct biologic pathways are involved in the pathogenesis of mild/moderate BPD as compared with severe BPD or death and indicate that they do not represent a continuum in lung disease severity. A detailed evaluation of the specific pathways involved may shed light on the possible differences in pathogenesis.
The pathway “Targets of MicroRNA GACAATC, MIR-219” was the top pathway for BPD/death. Many members of this pathway are transcription factors. Other members include the alpha-type platelet-derived growth factor receptor (PDGFRA) important in lung alveolar septation.22 miR-219 is involved in resolution of acute inflammation23 which may be relevant to BPD. Not all targets of miR-219 were dysregulated in BPD lung, perhaps because most genes are regulated by multiple miRNA as well as by other factors (transcription factors, lncRNA, DNA methylation etc). A preliminary evaluation of highly conserved targets of miR-219 in hyperoxia-vs. air-exposed mice using publicly-available datasets (e.g. GSE25293) found that all targets were reduced with hyperoxia (data not shown). Our findings that miR-219 in the murine newborn lung reduced over the course of alveolar septation and increased during hyperoxia, and was increased in the human BPD lung suggests that this miRNA may regulate normal lung development and injury response.
The more important clinical outcomes are probably those related to severe BPD or death, as most infants with mild/moderate BPD improve over time. The top pathway associated with severe BPD/death and in survivors with severe BPD was Phosphorus Oxygen Lyase Activity. The second pathway was Cyclase Activity, which shares considerable overlap (10 of 11 genes) with Phosphorus Oxygen Lyase Activity. Cyclic AMP produced by adenylate cyclase is important in lung development.24 Cyclic GMP produced by guanylate cyclase mediates nitric oxide signaling, and guanylate cyclase is involved in lung injury and development.25 These results suggest that modulation of the cGMP and cAMP pathways may be specifically relevant to severe BPD, and perhaps less important in mild/moderate BPD.
A major finding was that of the top ten SNPs in the model for BPD/death, four were SNPs associated with ADARB2 and two were SNPs associated with CD44. These genes were also highly represented in the models for severe BPD/death and severe BPD in survivors. ADARB2 is RNA-editing deaminase 2,26 a double-stranded RNA adenosine deaminase expressed mostly in the brain.27 It is unclear at the current time why there is a strong association of ADARB2 with BPD/death. CD44 is a hyaluronic acid cell surface receptor important in leukocyte trafficking and involved in lung injury. In mouse models, CD44 is protective during hyperoxia-induced lung injury28. However, severe lung fibrosis is promoted by CD44 in adult mice, indicating that CD44 may also have detrimental effects.29 We observed in the murine newborn lung that CD44 decreased over the course of alveolar septation and increased during hyperoxia, and was increased in human BPD lung, suggesting a role of this molecule in neonatal lung development and injury. The role of ADARB2 and CD44 in the pathophysiology of BPD requires further study.
Our study did not confirm findings of previous GWA studies5, 6 or all pathways of the gene expression study,16 perhaps due to different methods (pathways vs. single gene; genetic predisposition via SNPs vs. gene expression in established disease that may mask signals of early initiating events) or the populations being studied. For example, the population studied by Wang et al6 was mainly of Mexican Hispanic origin, and our study was about 54% Black and 45% White. We observed marked differences in pathways by race/ethnicity. The large differences in pathways by race/ethnicity suggest that although the clinical phenotype of BPD may be similar, the underlying genetic predisposition may differ significantly. This may be considered anticipated, as ancestry-specific associations contribute to chronic lung diseases such as asthma30 and emphysema.31 This also suggests that potential therapies may need to be specifically targeted at pathways that are found to be involved, and therefore suggests a role of “personalized genomics” in BPD.
The results of this study provide complementary information to conventional single-marker analysis, help fill in the ‘missing heritability”, and provide useful information to guide mechanistic studies based on pathway inhibition/augmentation. Future studies will need to validate the gene set analysis, perhaps by analysis of gene expression and epigenetic data to determine if similar pathways are involved. In addition, sequencing methods may help identify individuals who might be genetically predisposed to severe lung disease, such as those with mutations in SFTPB, ABCA3, FOXF1 or NKX2-1.Finally, translational studies are required to identify “druggable” mechanistic pathways and evaluate drug development strategies targeting these pathways.
Acknowledgments
Supported by the National Institutes of Health (General Clinical Research Center M01 RR30, M01 RR32, M01 RR39, M01 RR70, M01 RR80, M01 RR633, M01 RR750, M01 RR997, M01 RR6022, M01 RR7122, M01 RR8084, M01 RR16587, UL1 RR24979) and the Eunice Kennedy Shriver NICHD (U01 HD36790, U10 HD21364, U10 HD21373, U10 HD21385, U10 HD21397, U10 HD21415, U10 HD27851, U10 HD27853, U10 HD27856, U10 HD27871, U10 HD27880, U10 HD27881, U10 HD27904, U10 HD34216, U10 HD40461, U10 HD40492, U10 HD40498, U10 HD40689, U10 HD53109). J.M. received assistance for the GENEVA study from the National Human Genome Research Institute (U01 HG4423). Data collected at participating sites of the NICHD Neonatal Research Network were transmitted to RTI International, the data coordinating center for the network, which stored, managed, and analyzed the data for this study.
Abbreviations
- ADARB2
adenosine deaminase
- FDR
False Discovery Rate
- SNP
Single Nucleotide Polymorphism
Appendix
The following investigators, in addition to those listed as authors, are members of the Genomics and Cytokine Subcommittees of the Eunice Kennedy Shriver National Institute of Child Health and Human Development Neonatal Research Network:
Abhik Das (DCC PI) and Grier Page (DCC Statistician) had full access to all the data in the study and take responsibility for the integrity of the data and accuracy of the data analysis. NRN Steering Committee Chair: Alan H. Jobe, MD PhD (University of Cincinnati).
Genomics Subcommittee: C. Michael Cotten, MD MHS (chair); Jeffrey C. Murray, MD (vice chair); Edward F. Bell, MD; Ronald N. Goldberg, MD; Kurt Schibler, MD; Beena G. Sood, MD MS; David K. Stevenson, MD; Barbara J. Stoll, MD; Krisa P. Van Meurs, MD; Abhik Das, PhD; Rosemary D. Higgins, MD; Karen J. Johnson, RN; Kristin M. Zaterka-Baxter, RN BSN. Cytokines Subcommittee: Waldemar A. Carlo, MD (chair); Richard A. Ehrenkranz, MD; Ronald N. Goldberg, MD; Seetha Shankaran, MD; Barbara J. Stoll, MD; Jon E. Tyson, MD MPH; Abhik Das, PhD; Rosemary D. Higgins, MD.
Case Western Reserve University, Rainbow Babies & Children’s Hospital (U10 HD21364, M01 RR80) – Michele C. Walsh, MD MS; Avroy A. Fanaroff, MD; Nancy S. Newman, RN; Bonnie S. Siner, RN.
Cincinnati Children’s Hospital Medical Center, University Hospital, and Good Samaritan Hospital (U10 HD27853, M01 RR8084) – Kurt Schibler, MD; Edward F. Donovan, MD; Vivek Narendran, MD MRCP; Barbara Alexander, RN; Cathy Grisby, BSN CCRC; Jody Hessling, RN; Marcia Worley Mersmann, RN CCRC; Holly L. Mincey, RN BSN.
Duke University School of Medicine University Hospital, Alamance Regional Medical Center, and Durham Regional Hospital (M01 RR30, U10 HD40492) – Ronald N. Goldberg, MD; Kathy J. Auten, MSHS.
Emory University, Children’s Healthcare of Atlanta, Grady Memorial Hospital, and Emory Crawford Long Hospital (U10 HD27851, M01 RR39) – Barbara J. Stoll, MD; Ellen C. Hale, RN BS CCRC.
Eunice Kennedy Shriver National Institute of Child Health and Human Development – Linda L. Wright, MD; Sumner J. Yaffe, MD; Elizabeth M. McClure, Med; Stephanie Wilson Archer, MA.
RTI International (U10 HD36790) – Abhik Das, PhD; Scott A. McDonald, BS; Kristin M. Zaterka-Baxter, RN BSN; W. Kenneth Poole, PhD; Betty K. Hastings; Jeanette O’Donnell Auman, BS.
Stanford University, Lucile Packard Children’s Hospital (U10 HD27880, M01 RR70) – Krisa P. Van Meurs, MD; David K. Stevenson, MD; M. Bethany Ball, BS CCRC.
University of Alabama at Birmingham Health System and Children’s Hospital of Alabama (U10 HD34216, M01 RR32) – Namasivayam Ambalavanan, MD; Monica V. Collins, RN BSN MaEd; Shirley S. Cosby, RN BSN.
University of California – San Diego Medical Center and Sharp Mary Birch Hospital for Women (U10 HD40461) – Neil N. Finer, MD; Maynard R. Rasmussen, MD; David Kaegi, MD; Kathy Arnell, RNC; Clarence Demetrio, RN; Wade Rich, BSHS RRT.
University of Iowa Children’s Hospital and Mercy Medical Center (U10 HD53109, M01 RR59, UL1 TR442) – Edward F. Bell, MD; Karen J. Johnson, RN.
University of Miami Holtz Children’s Hospital (U10 HD21397, M01 RR16587) – Shahnaz Duara, MD; Charles R. Bauer, MD; Ruth Everett-Thomas, RN MSN.
University of Tennessee (U10 HD21415) – Sheldon B. Korones, MD; Henrietta S. Bada, MD; Tina Hudson, RN BSN.
University of Texas Southwestern Medical Center at Dallas, Parkland Health & Hospital System, and Children’s Medical Center Dallas (U10 HD40689, M01 RR633) – Pablo J. Sánchez, MD; Abbot R. Laptook, MD; Walid A. Salhab, MD; Susie Madison, RN; Nancy A. Miller, RN; Gaynelle Hensley, RN; Alicia Guzman.
University of Texas Health Science Center at Houston Medical School, Children’s Memorial Hermann Hospital, and Lyndon B. Johnson General Hospital (U10 HD21373) – Jon E. Tyson, MD MPH; Kathleen A. Kennedy, MD MPH; Esther G. Akpa, RN BSN; Patty A. Cluff, RN; Claudia I. Franco, RNC MSN; Anna E. Lis, RN BSN; Georgia E. McDavid, RN; Patti Pierce Tate, RCP.
Wayne State University, Hutzel Women’s Hospital, and Children’s Hospital of Michigan (U10 HD21385) – Seetha Shankaran, MD; Beena G. Sood, MD MS; G. Ganesh Konduri, MD; Rebecca Bara, RN BSN; Geraldine Muran, RN BSN.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Bhandari V, Bizzarro MJ, Shetty A, Zhong X, Page GP, Zhang H, et al. Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics. 2006;117:1901–1906. doi: 10.1542/peds.2005-1414. [DOI] [PubMed] [Google Scholar]
- 2.Weber B, Borkhardt A, Stoll-Becker S, Reiss I, Gortner L. Polymorphisms of surfactant protein A genes and the risk of bronchopulmonary dysplasia in preterm infants. Turk J Pediatr. 2000;42:181–185. [PubMed] [Google Scholar]
- 3.Rova M, Haataja R, Marttila R, Ollikainen V, Tammela O, Hallman M. Data mining and multiparameter analysis of lung surfactant protein genes in bronchopulmonary dysplasia. Hum Mol Genet. 2004;13:1095–1104. doi: 10.1093/hmg/ddh132. [DOI] [PubMed] [Google Scholar]
- 4.Lin HC, Su BH, Chang JS, Hsu CM, Tsai CH, Tsai FJ. Nonassociation of interleukin 4 intron 3 and 590 promoter polymorphisms with bronchopulmonary dysplasia for ventilated preterm infants. Biol Neonate. 2005;87:181–186. doi: 10.1159/000082937. [DOI] [PubMed] [Google Scholar]
- 5.Hadchouel A, Durrmeyer X, Bouzigon E, Incitti R, Huusko J, Jarreau PH, et al. Identification of SPOCK2 as a susceptibility gene for bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2011;184:1164–1170. doi: 10.1164/rccm.201103-0548OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang H, St Julien KR, Stevenson DK, Hoffmann TJ, Witte JS, Lazzeroni LC, et al. A genome-wide association study (GWAS) for bronchopulmonary dysplasia. Pediatrics. 2013;132:290–297. doi: 10.1542/peds.2013-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- 8.Wang K, Li M, Hakonarson H. Analysing biological pathways in genome-wide association studies. Nature reviews Genetics. 2010;11:843–854. doi: 10.1038/nrg2884. [DOI] [PubMed] [Google Scholar]
- 9.Wang K, Li M, Bucan M. Pathway-based approaches for analysis of genomewide association studies. American journal of human genetics. 2007;81:1278–1283. doi: 10.1086/522374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carlo WA, McDonald SA, Tyson JE, Stoll BJ, Ehrenkranz RA, Shankaran S, et al. Cytokines and neurodevelopmental outcomes in extremely low birth weight infants. J Pediatr. 2011;159:919–925. doi: 10.1016/j.jpeds.2011.05.042. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jobe AH, Bancalari E. Bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2001;163:1723–1729. doi: 10.1164/ajrccm.163.7.2011060. [DOI] [PubMed] [Google Scholar]
- 12.Gogarten SM, Bhangale T, Conomos MP, Laurie CA, McHugh CP, Painter I, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hancock DB, Levy JL, Gaddis NC, Bierut LJ, Saccone NL, Page GP, et al. Assessment of genotype imputation performance using 1000 Genomes in African American studies. PLoS One. 2012;7:e50610. doi: 10.1371/journal.pone.0050610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. American journal of human genetics. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharya S, Go D, Krenitsky DL, Huyck HL, Solleti SK, Lunger VA, et al. Genome-wide transcriptional profiling reveals connective tissue mast cell accumulation in bronchopulmonary dysplasia. Am J Respir Crit Care Med. 2012;186:349–358. doi: 10.1164/rccm.201203-0406OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nicola T, Hagood JS, James ML, Macewen MW, Williams TA, Hewitt MM, et al. Loss of Thy-1 inhibits alveolar development in the newborn mouse lung. Am J Physiol Lung Cell Mol Physiol. 2009;296:L738–L750. doi: 10.1152/ajplung.90603.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.James ML, Ross AC, Nicola T, Steele C, Ambalavanan N. VARA Attenuates Hyperoxia-Induced Impaired Alveolar Development and Lung Function in Newborn Mice. Am J Physiol Lung Cell Mol Physiol. 2013 doi: 10.1152/ajplung.00257.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wan G, Lim QE, Too HP. High-performance quantification of mature microRNAs by realtime RT-PCR using deoxyuridine-incorporated oligonucleotides and hemi-nested primers. Rna. 2010;16:1436–1445. doi: 10.1261/rna.2001610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li J, Masood A, Yi M, Lau M, Belcastro R, Ivanovska J, et al. The IGF-I/IGF-R1 pathway regulates postnatal lung growth and is a nonspecific regulator of alveologenesis in the neonatal rat. Am J Physiol Lung Cell Mol Physiol. 2013;304:L626–L637. doi: 10.1152/ajplung.00198.2012. [DOI] [PubMed] [Google Scholar]
- 21.Chetty A, Andersson S, Lassus P, Nielsen HC. Insulin-like growth factor-1 (IGF-1) and IGF-1 receptor (IGF-1R) expression in human lung in RDS and BPD. Pediatr Pulmonol. 2004;37:128–136. doi: 10.1002/ppul.10415. [DOI] [PubMed] [Google Scholar]
- 22.McGowan SE, Grossmann RE, Kimani PW, Holmes AJ. Platelet-derived growth factor receptor-alpha-expressing cells localize to the alveolar entry ring and have characteristics of myofibroblasts during pulmonary alveolar septal formation. Anat Rec (Hoboken) 2008;291:1649–1661. doi: 10.1002/ar.20764. [DOI] [PubMed] [Google Scholar]
- 23.Recchiuti A, Krishnamoorthy S, Fredman G, Chiang N, Serhan CN. MicroRNAs in resolution of acute inflammation: identification of novel resolvin D1-miRNA circuits. FASEB J. 2011;25:544–560. doi: 10.1096/fj.10-169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nijjar MS. Role of cyclic AMP and related enzymes in rat lung growth and development. Biochimica et biophysica acta. 1979;586:464–472. doi: 10.1016/0304-4165(79)90036-9. [DOI] [PubMed] [Google Scholar]
- 25.Moreno L, Gonzalez-Luis G, Cogolludo A, Lodi F, Lopez-Farre A, Tamargo J, et al. Soluble guanylyl cyclase during postnatal porcine pulmonary maturation. Am J Physiol Lung Cell Mol Physiol. 2005;288:L125–L130. doi: 10.1152/ajplung.00244.2004. [DOI] [PubMed] [Google Scholar]
- 26.Mittaz L, Antonarakis SE, Higuchi M, Scott HS. Localization of a novel human RNA-editing deaminase (hRED2 or ADARB2) to chromosome 10p15. Human genetics. 1997;100:398–400. doi: 10.1007/s004390050523. [DOI] [PubMed] [Google Scholar]
- 27.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg PH. RED2, a brain-specific member of the RNA-specific adenosine deaminase family. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 28.van der Windt GJ, Schouten M, Zeerleder S, Florquin S, van der Poll T. CD44 is protective during hyperoxia-induced lung injury. Am J Respir Cell Mol Biol. 2011;44:377–383. doi: 10.1165/rcmb.2010-0158OC. [DOI] [PubMed] [Google Scholar]
- 29.Li Y, Jiang D, Liang J, Meltzer EB, Gray A, Miura R, et al. Severe lung fibrosis requires an invasive fibroblast phenotype regulated by hyaluronan and CD44. The Journal of experimental medicine. 2011;208:1459–1471. doi: 10.1084/jem.20102510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pilger D, Rodrigues LC. Genetic variation and the risk of asthma: does it drive the differences in asthma prevalence among ethnic groups in North America? Ann Allergy Asthma Immunol. 2012;108:206–207. doi: 10.1016/j.anai.2011.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Manichaikul A, Hoffman EA, Smolonska J, Gao W, Cho MH, Baumhauer H, et al. Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. Am J Respir Crit Care Med. 2014;189:408–418. doi: 10.1164/rccm.201306-1061OC. [DOI] [PMC free article] [PubMed] [Google Scholar]