Abstract
Membrane association with mother centriole (M-centriole) distal appendages is critical for ciliogenesis initiation. How the Rab GTPase Rab11-Rab8 cascade functions in early ciliary membrane assembly is unknown. Here, we show that the membrane shaping proteins EHD1 and EHD3, in association with the Rab11-Rab8 cascade, function in early ciliogenesis. EHD1 and EHD3 localize to pre-ciliary membranes and the ciliary pocket. EHD-dependent membrane tubulation is essential for ciliary vesicle (CV) formation from smaller distal appendage vesicles (DAV). Importantly, this step functions in M-centriole to basal body transformation and recruitment of transition zone proteins and IFT20. SNAP29, a SNARE membrane fusion regulator and EHD1-binding protein, is also required for DAV-mediated CV assembly. Interestingly, only after CV assembly is Rab8 activated for ciliary growth. Our studies uncover molecular mechanisms informing a previously uncharacterized ciliogenesis step whereby EHD1 and EHD3 reorganize the M-centriole and associated DAV prior to coordinated ciliary membrane and axoneme growth.
INTRODUCTION
Primary cilia play essential roles in signal transduction and defects in cilia formation or function cause ciliopathies1, 2. Cilia form at the distal end of the mother centriole (M-centriole) via recruitment of pre-ciliary membranes, intraflagellar transport (IFT) machinery and transition zone components to enable microtubule-based axonemal assembly. Association of pre-ciliary membranes with M-centriole distal appendages is needed for basal body formation and ciliogenesis progression3-11. The membrane trafficking regulator Rab small GTPases and in particular, the Rab11-Rab8 cascade are required for ciliary membrane formation during ciliogenesis12-17. In this cascade, Rabin8, the guanine nucleotide exchange factor for Rab8, binds to Rab11 and is delivered to the centrosome on vesicles to activate Rab8 to promote ciliary membrane assembly15.
Over 50 years ago, Sergei Sorokin proposed a model whereby intracellular membranes organize at the distal end of the M-centriole prior to axoneme formation. A large ciliary vesicle (CV) assembles, reorganizes to form a sheath around the extending axoneme and later fuses with the plasma membrane10, 18. The requirement for the Rab11-Rab8 cascade in ciliogenesis provides a molecular explanation for these early ciliary assembly steps but is poorly understood. Furthermore, how ciliary membrane assembly is coordinated with other early ciliogenesis processes including establishment of the basal body, IFT recruitment, transition zone assembly and axoneme growth is largely unknown.
The Eps15 homology domain (EHD)-family of proteins, composed of EHD1-4, is associated with Rab11 and Rab8 membranes and regulates endosomal membrane trafficking19. EHDs are characterized by an ATP-binding G-domain, a central coiled-coil domain, and a COOH-terminal EH domain, which interacts with asparagine-proline-phenylalanine (NPF) motif containing proteins20. EHD1 and EHD3 exhibit 87% amino acid identity, whereas EHD2 and EHD4 are <74% identical to EHD1. EHD1 and EHD3 regulate Rab11-endosome recycling compartment (ERC) trafficking and bind to Rab11-FIP2, a Rab11 effector21. Additionally, EHD1 and EHD3 bind to the Rab8 effector MICAL-L1 and affect membrane tubulo-vesicle formation and scission19, 22-25. EHD1 also associates with the membrane fusion regulator SNAP2926. Here, we investigated EHD protein involvement in ciliary membrane biogenesis. Using advanced microscopy imaging approaches, we dynamically observed the recruitment of proteins essential for early ciliogenesis processes. Moreover, depletion of EHD proteins shed light on a previously uncharacterized but required step in ciliogenesis. Our data suggest a model in which EHD1 and EHD3 coordinate critical steps at the onset of ciliogenesis.
RESULTS
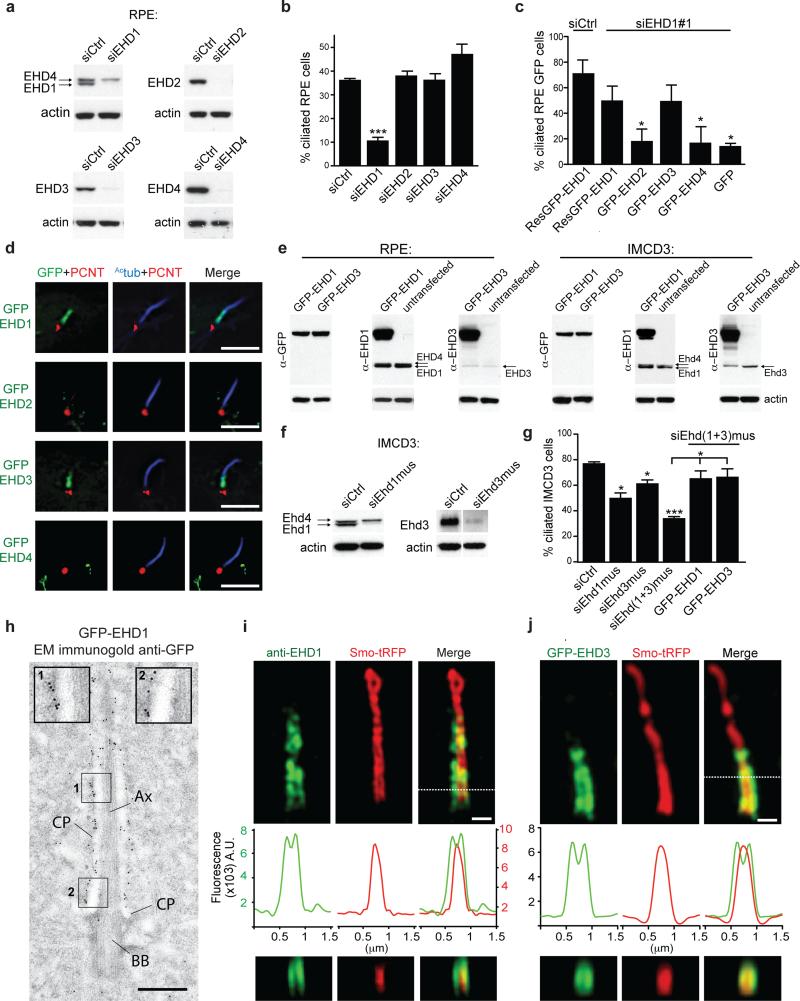
EHD1 and EHD3 function in ciliogenesis and localize to the ciliary pocket membrane
Because EHD1 and EHD3 have been linked to both Rab11 and Rab8 membrane compartments19 we tested their role in ciliogenesis. SiRNA-mediated knockdown of EHD1, but not EHD2-4, impaired ciliation in hTERT-RPE (RPE) cells (Fig. 1a,b, Supplementary Fig. 1a-c). Importantly, an siRNA resistant form of GFP-EHD1 or GFP-EHD3 but not GFP, GFP-EHD2 or GFP-EHD4 rescued ciliation (Fig. 1c, Supplemental Fig. 1d-f) suggesting that EHD1 and EHD3 function in ciliogenesis. Interestingly, only GFP-EHD1 and GFP-EHD3 were detected at the proximal ciliary region (Fig. 1d, Supplementary Fig. 1d). Endogenous EHD1 was also detected in the proximal ciliary region in 30 ± 5% (± SD) of cells (n=162, pooled from 3 experiments) and did not completely overlap with ciliary Smo-tRFP or GFP-Rab8a (Supplementary Fig. 1g,h). Overexpressed GFP-EHD3 colocalizes with EHD1 at this region (Supplementary Fig. 1g). Immunoblotting of RPE cell lysates revealed that EHD1 levels were >5 times higher than EHD3 (Fig. 1e) indicating that EHD3 may be dispensable for RPE cell ciliogenesis. In IMCD3 cells, EHD1 and EHD3 had similar expression levels (Fig. 1e) and were both required for ciliogenesis (Fig. 1f,g). GFP-fused EHD1 and EHD3 were detected in the proximal ciliary region in 8 ± 2% (± SD, n=164, pooled from 2 experiments) and 8 ± 4 % (± SD, n= 152, pooled from 2 experiments) respectively (Supplementary Fig. 1i).
Figure 1. EHD1 and EHD3 function in ciliogenesis and localize to the ciliary pocket membrane.
a. Western analysis of EHD proteins from 72h siRNA treated RPE cells (siRNA #1 from Supplementary Table 1).
b. Ciliation quantification in 72h siRNA treated RPE cells (siEHD1#2, siEHD2#1, siEHD3#1, siEHD4#1), with 24h serum starvation, followed by staining with Actub and pericentrin (PCNT) antibodies. Mean ± SEM from n=3 independent experiments is shown (>100 cells per treatment). Two-tailed t-test analysis compared with siCtrl.
c. Quantification of ciliation in RPE cells siRNA treated for 6h, followed by transfection with siRNA resistant (Res)GFP-EHD proteins or GFP. At 48h post-transfection, cells were serum starved and stained as in (b). Mean ± SEM from n=3 independent experiments is shown (>50 cells per condition). Two-tailed t-test analysis compared with siEHD1 ResGFP-EHD1.
d. Localization of transiently expressing GFP-EHD proteins in RPE cells after 24h serum starvation and staining as in (b); Deconvolved single xy-plane from epifluorescence stack is shown. Scale bar: 5 μm.
e. Western analysis comparing EHD1 and EHD3 expression in cell lines transiently expressing GFP-EHD1 or GFP-EHD3.
f. Western analysis of EHD protein depletion in IMCD3 cells treated with siRNA for 72h.
g. Quantification of ciliation in IMCD3 cells treated with siRNAs for 72h, and where indicated, rescued with human GFP-EHD proteins and stained as in (a-c). Mean ± SEM from n=3 independent experiments is shown (>300 cells per condition). Two-tailed t-test analysis compared with siCtrl or siEhd(1+3)mus,
h. Immuno-electron micrograph of RPE cells expressing GFP-EHD1 stained with anti-GFP antibody. 10 nm gold particles accumulate on the ciliary pocket (CP) membrane. Magnification of region 1 and 2 (inset). Ax: axoneme; BB: basal body. Scale bar: 500 nm.
i,j. SIM imaging of 24h serum starved RPE cells transiently expressing Smo-tRFP stained with anti-EHD1 antibody (i) or transiently expressing GFP-EHD3 (j). Middle panels show fluorescence profile plots and lower panels show orthogonal views of dotted line region. Representative image of > 10 cilia. Scale bar: 500 nm.
*** P<0.0001, * P<0.05. Un-cropped blots are shown in Supplementary Fig 6. Fig 1 b,c,g statistics source data can be found in Supplementary Table 2.
We investigated EHD protein ciliary localization using correlative light and electron microscopy (CLEM), immuno-electron microscopy and super-resolution structured illumination microscopy (SIM) and demonstrated that EHD1 and EHD3 are primarily localized to the ciliary pocket membrane unlike Rab8 and Smoothened (Smo), largely present in the ciliary membrane (Fig. 1h,i,j, Supplementary Fig. 1j,k). Additionally, EHD1 localization did not overlap with Inversin (Supplementary Fig. 1l), a proximal intra-ciliary marker27. The localization of EHD1 and EHD3 in IMCD3 cells is consistent with lower levels of ciliary pockets reported in these cells compared to RPE cells, which could point to different ciliogenesis mechanisms28. Together our results indicate that EHD1 and EHD3 are important for ciliogenesis and localize to the ciliary pocket membrane.
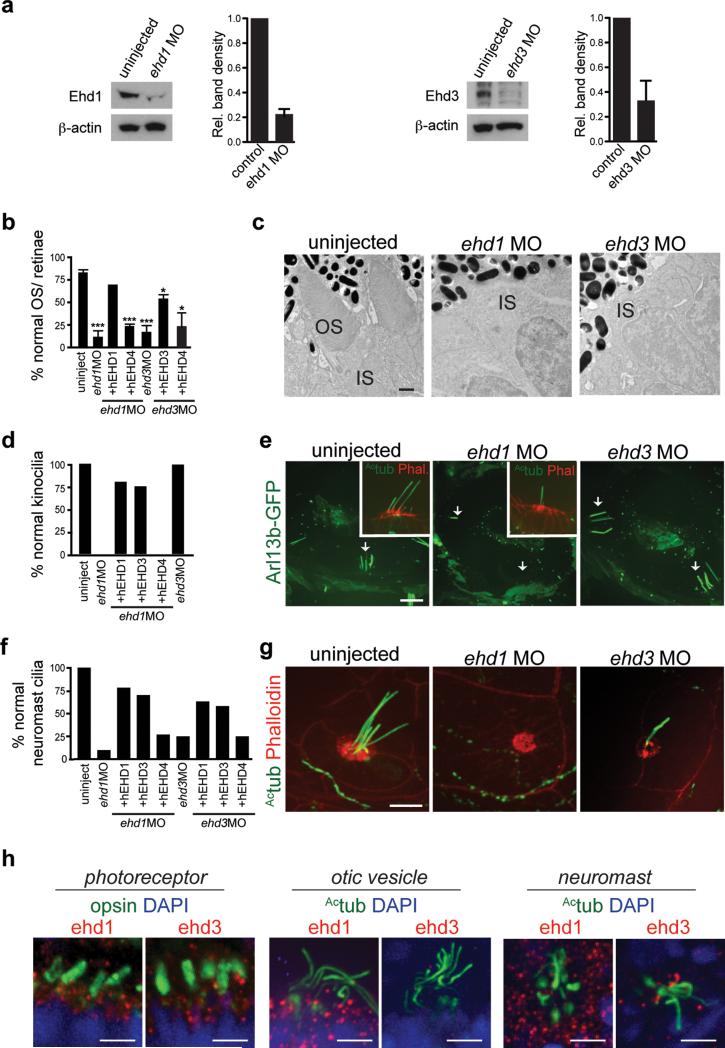
Requirements for Ehd1 and Ehd3 in ciliogenesis during development in zebrafish
We investigated the ciliogenesis function of EHD proteins in zebrafish embryos. Zebrafish have ehd homologs: ehd1a, ehd1b, ehd2a, ehd2b and ehd3. ehd1a and 1b (89% identity) are closely related to human EHD1 (85% and 87% respectively) and ehd3 is 93% identical to human EHD3. Combination of ehd1(a+b) MO gave synergistic effects while the triple knockdown with ehd3 MO had 70 ± 6% lethality (± SD, n>150, from 3 experiments). As was observed in mammalian cells, hEHD1 and hEHD3 rescued phenotypes observed in ehd3 and ehd1 morphants unlike hEHD4 (Fig. 2b,d,f, Supplementary Fig. 2a). Histological and electron microscopy (EM) analysis in ehd1 and ehd3 morphants revealed a failure to form photoreceptor cilia-outer segments (OS, Fig. 2b,c) suggesting that both proteins are important for photoreceptor ciliogenesis, similar to IMCD3 cell requirements. However, in other tissues, requirements for Ehd proteins varied. Ehd1, but not Ehd3, was required for kinocilia formation in otic vesicles (OV, Fig. 2d,e). Similarly, Ehd1 was essential for neuromast ciliation whereas ehd3 morphants showed a partial reduction in kinocilia (Fig. 2f,g, Supplementary Fig. 2b). Consistent with these results, Ehd1 and Ehd3 were expressed in photoreceptors and neuromasts whereas only Ehd1 but not Ehd3 was expressed in the OV during embryonic development (Fig. 2h, Supplementary Fig. 2c,d,e). In these tissues, Ehd1 and Ehd3 localized to puncta throughout the cell, with some protein detected at or close to the ciliary base. Together these results indicate that Ehd1 and Ehd3 have overlapping, albeit tissue specific, functions in ciliogenesis in zebrafish embryos.
Figure 2. ehd1 and ehd3 regulate ciliogenesis in zebrafish.
a. Western analysis and quantification of Ehd1 and Ehd3 protein expression in 3 dpf MO injected embryos. Mean ± SEM from n=3 independent experiments (lysates with 50 embryos per condition) is shown.
b. Quantification of photoreceptor OS number from histological sections of 3 dpf retinae stained with toluidin blue. Mean ± SEM from n=3 independent experiments is shown (>100 photoreceptors per condition). ehd1 MO+EHD1 from n=2 independent experiments (>100 photoreceptors), Two-tailed t-test analysis compared with uninjected.
c. Representative electron micrographs from n=3 independent experiments (1-2 fish per condition) showing two photoreceptor cells in control, ehd1 and ehd3 morphants at 3 dpf. OS: outer segment, IS: inner segment. Scale bar: 500nm.
d. Quantification of otic vesicles with kinocilia in 2 dpf Tg(arl13b-GFP) embryos uninjected or injected with ehd1 or ehd3 MO. uninjected, n=18; ehd1MO, n=13; ehd1MO+ hEHD1, n=6; ehd1MO+ hEHD3, n=4; ehd1MO+ hEHD4, n=5; ehd3MO, n=10.
e. Representative images of otic vesicles described in (d) with insets showing sensory patches fixed and stained with anti-Actub antibody and phalloidin. Note that ehd1 morphants lack kinocilia. Scale bar: 10 μm.
f. Quantification as in (d) of neuromasts with normal cilia in 2 dpf embryos stained with Actub and phalloidin antibodies. Uninjected, n=20; ehd1MO, n=10; ehd1MO+ hEHD1, n=23; ehd1MO+ hEHD3, n=27; ehd1MO+ hEHD4, n=22; ehd3MO, n=16; ehd3MO+ hEHD3, n=41; ehd3MO+ hEHD1, n=12; ehd3MO+ hEHD4, n=16. Note that ehd3 morphants present a partial phenotype with reduced number of kinocilia (quantification in Supplemental Fig 2b). Pooled data across 3 independent experiments is shown in (d) and (f).
g. Representative images of neuromasts as described in (f). Scale bar: 10 μm.
h. Representative images of photoreceptors in retinae of 3 dpf embryos, anterior cristae of otic vesicles and neuromasts stained with anti-Ehd1 or anti-Ehd3 and anti-rhodopsin or anti-Actub antibodies showing punctate accumulation of proteins in the cytosol and around the base of cilia in these organelles. Organs were imaged with the same fluorescence microscopy settings, which show that Ehd3 levels were low in the otic vesicles. Scale bar: 10 μm.
Un-cropped images of blots are shown in Supplementary Fig 6. *P<0.05, *** P<0.0001.
Statistics source data for Fig 2 a,b found in Supplementary Table 2.
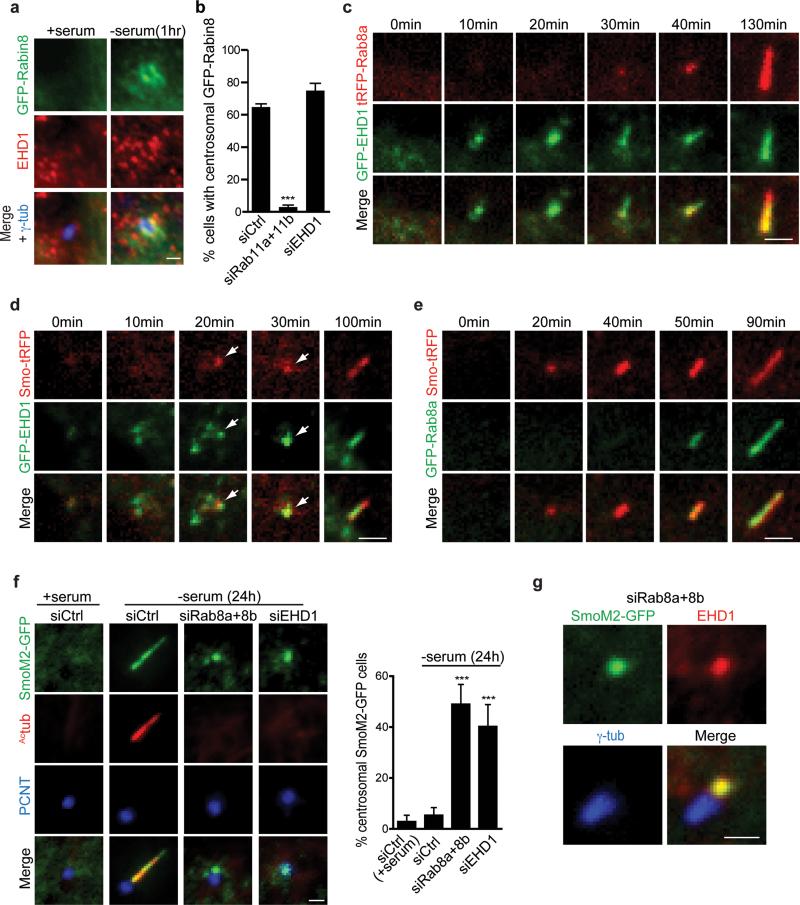
EHD1 localizes to pre-ciliary membranes but is dispensable for their centrosomal trafficking
Next, we investigated EHD1 association with Rab11-dependent pre-ciliary vesicle transport of Rabin8. EHD1 co-localizes with GFP-Rabin8 vesicles following serum starvation, but not in the presence of serum (Fig. 3a). Moreover, in contrast to Rab11 requirements15, depletion of EHD1 did not affect GFP-Rabin8 centrosomal accumulation (Fig. 3b) suggesting that EHD1 is associated with pre-ciliary vesicles but is not required for their centrosomal trafficking. We performed live imaging on GFP-EHD1 and tRFP-Rab8a expressing RPE cells and determined that EHD1 and Rab8 associate with developing cilia (Fig. 3c, Supplementary Fig. 3, Movie 1). Remarkably, GFP-EHD1 was detected at a site in the cell, which later accumulated tRFP-Rab8a and extended ciliary structures suggesting that EHD1 accumulates at M-centriole-associated membranes before Rab8. Additionally, Smo-tRFP, a ciliary receptor marking early ciliary membrane structures6, localized to developing cilia about the same time as GFP-EHD1 (Fig. 3d, Supplementary Movie 2) and prior to GFP-Rab8 (Fig. 3e, Supplementary Movie 3) suggesting EHD1 and Smo occupy the same pre-ciliary vesicles. Similarly, the constitutive active SmoM2 mutant localized to the M-centriole following serum starvation and its trafficking was not affected by EHD1 or Rab8 depletion (Fig. 3f), indicating that SmoM2 M-centriole transport is independent of these proteins. Furthermore, SmoM2-GFP and EHD1 co-localize at the centrosome in the absence of Rab8 (Fig. 3g). Together these results indicate that EHD1 functions upstream of Rab8 in ciliogenesis.
Figure 3. EHD1 localize to pre-ciliary membranes and the developing cilia.
a. Representative images of GFP-Rabin8 stably expressed in RPE cells grown in the presence or absence (1h) of serum and stained with antibodies to EHD1 and centrosomal marker γ-tubulin. Scale bar: 1 μm.
b. Quantification of RPE GFP-Rabin8 cells treated with siRNA for 72h and imaged live by epifluorescence microscopy for GFP-Rabin8 centrosomal accumulation 1h after serum starvation. Mean ± SEM from n=3 independent experiments is shown (total number of cells counted in all experiments: siControl, 198 cells; siRab11a+b, 318 cells; siEHD1, 139 cells). Two-tailed t-test analysis compared with siCtrl.
c. RPE GFP-EHD1 cells transiently expressing tRFP-Rab8a were serum starved for 1h and imaged live by spinning disk confocal microscopy. Images are maximum intensity projections of z-stacks.
d. RPE GFP-EHD1 cells transiently expressing Smo-tRFP were imaged as in (c). Arrows show accumulation of EHD1 and Smo over time in developing cilia.
e. RPE GFP-Rab8a cells transiently expressing Smo-tRFP were imaged as in (c). Images are single xy planes.
Note that panels c-e are representative image series of > 10 cilia assembly events observed. Scale bars c-e: 2 μm
f. RNAi of EHD1 or Rab8a+8b in stably expressing RPE SmoM2-GFP cell line. Cells were stained with cilia marker (Actub) and centrosome marker (PCNT) (left panel).
Quantification of SmoM2-GFP centrosomal accumulation (right panel). Means ± SD are pooled data from n= 9 areas imaged in 3 independent experiments (total number of cells counted in all experiments: siCtrl (+ serum), 123; siCtrl (-serum), 179; siRab8a+8b, 172; siEHD1#1, 241). Two-tailed t-test analysis compared with siCtrl (-serum), Scale bar: 1 μm.
g. RPE cells stably expressing SmoM2-GFP were transfected with Rab8a+8b siRNAs for 48h, serum starved for 24h and stained with anti-EHD1 and anti-γ-tubulin antibodies. Imaging was performed by epifluorescence microscopy. Scale bar: 1 μm.
*** P<0.0001. Statistics source data for Fig3 b,f can be found in Supplementary Table 2.
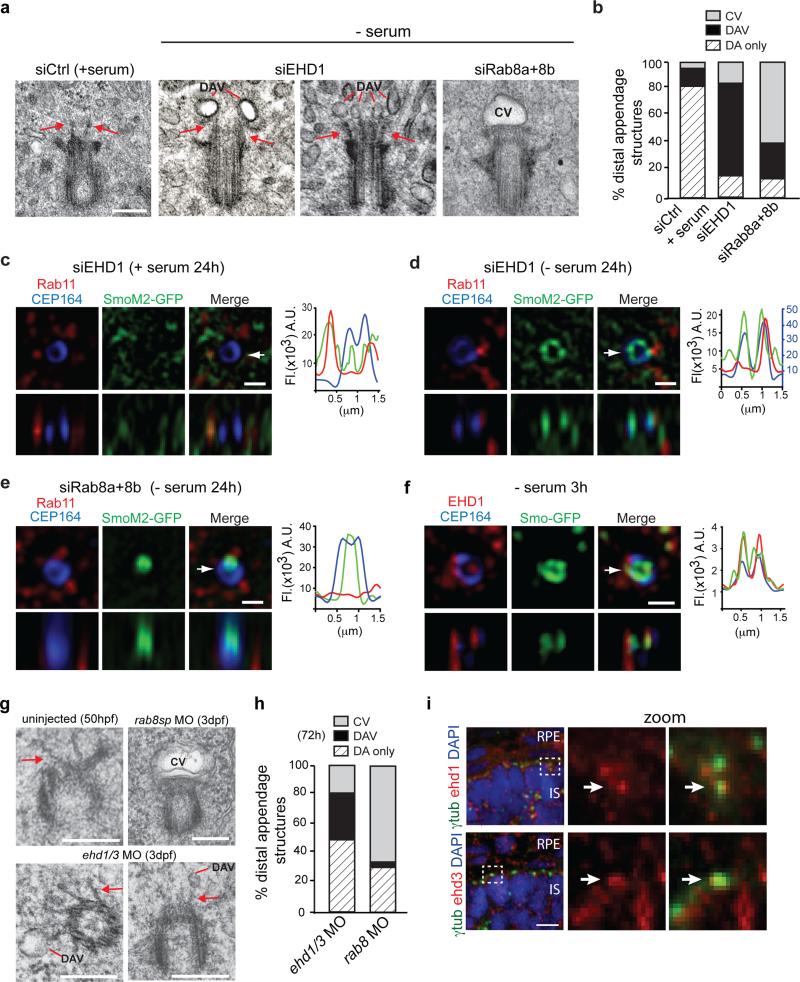
EHD1 is required for small distal appendage vesicles assembly into the CV, while Rab8 functions in CV extension
RPE cells use an intracellular ciliogenesis pathway with formation of the CV as a critical step5. To investigate EHD1 function in this pathway, we performed EM studies on serum starved EHD1-depleted cells (Fig 4a). Strikingly, the majority (68%) of the non-ciliated cells had small ~40-60nm diameter vesicles associated with the M-centriole distal appendages (Fig. 4a,b, Supplementary Fig 4a), that we called distal appendage vesicles (DAVs). CVs were detected in 16% of EHD1 knockdown cells. By comparison, DAVs were detected in 13% of siControl treated unstarved cells and 81% had no detectable membranes at the distal appendages suggesting that DAVs are the precursor membranes for the CV. In contrast to EHD1 depleted cells, 63% of Rab8 siRNA treated unciliated cells had CV structures while only 28% had DAVs (Fig. 4a,b, Supplementary Fig 4a). Combined with our live imaging studies, these results suggest that EHD1 and Rab8 regulate different steps in ciliogenesis, with EHD1 important for CV formation and Rab8 functioning in CV extension.
Figure 4. EHD1 functions in ciliary vesicle formation upstream of Rab8.
a. Representative electron micrographs of RPE cell M-centrioles treated with siRNA for 72h, with serum starvation (- serum) the last 24h. Scale bar: 200 nm.
b. Quantification of M-centriole non-cilia distal appendages structures from (a). Pooled data from 2-4 independent experiments (total number of cells counted in all experiments: siControl+serum, 48 cells; siEHD1, 44 cells; siRab8a+8b, 51 cells).
c,d,e. SIM images of RPE SmoM2-GFP cells treated with siRNA as in (a) and stained with Rab11 and CEP164 antibodies. Arrow marks orthogonal view in lower panels and corresponds to fluorescence profile plots. (c) Representative images from 5 random serum fed cells showing no centriolar SmoM2-GFP accumulation. (d) 5 out of 8 cells (63%) showed SmoM2-GFP positive DAV like structures. (e) 5 out of 9 cells (56%) showed CV-like structures. Note that 37.5% of siEHD1 treated cells showed CV-like structures, while 22% of siRab8 depleted cells had DAV-like structures. Profile plots, CEP164 values were normalized by a factor 2 (c) and SmoM2-GFP values were normalized by factor 2 (d). Scale bars: 500 nm.
f. 3h serum starved RPE Smo-GFP cells stained with anti-CEP164 and anti-EHD1 antibodies and imaged by SIM. 7 out of 17 cells (41%) showed distal appendage Smo-GFP co-localization with EHD1. Arrow marks orthogonal view in lower panels and corresponds to fluorescence profile plots. Scale bar: 500 nm.
g. Representative electron micrographs of M-centrioles (MC) from uninjected zebrafish photoreceptor at 50 hpf and 3 dpf photoreceptors injected with ehd1 and ehd3 or rab8sp MO. ehd1 and ehd3 MO were co-injected to maximize depletion of both proteins. Because of high lethality resulting in ehd1 and ehd3 MO, only viable embryos with small eyes were analyzed. Scale bar: 250 nm.
h. Quantification of M-centriole distal appendages structures observed in electron micrographs described in (g). Averages from pooled data across 3 independent experiments (>25 photoreceptors per treatment in total across the experiments).
i. Representative images of M-centrioles in zebrafish photoreceptors at 50hpf stained with anti-Ehd1 or Ehd3 and anti-γ-tubulin antibodies and imaged by spinning disk confocal microscopy. Scale bar: 10 μm.
DAV: Distal appendages vesicles, CV: ciliary vesicle or DA: non-membrane associated distal appendages.
SIM imaging has been used to resolve the structure of CEP164, a distal appendage protein forming a ring ~300nm in diameter at the distal end of the M-centriole29. We performed SIM on SmoM2-GFP expressing RPE cells to attempt to resolve pre-ciliary membrane assembly steps at the M-centriole distal end (Fig. 4c-e). In serum fed cells depleted of EHD1 SmoM2-GFP did not co-localize with the CEP164 ring, although SmoM2-GFP and Rab11 partially co-localized on pericentriolar vesicles (Fig. 4c). However, in serum starved EHD1 depleted cells, SmoM2-GFP vesicles were detected at the CEP164 ring and partially co-localized with Rab11 both at the distal appendages and in the pericentriolar region (Fig. 4d). From our EM studies, the SmoM2-GFP vesicles are likely DAVs. In contrast, in Rab8 depleted cells SIM showed a single large structure at the M-centriole distal end, which is likely a CV (Fig. 4e). Based on our previous work15 examining ciliogenesis initiation following serum withdrawal, we looked for DAVs similar to those observed in EHD1 depleted cells using SIM (Fig. 4 d,f). Following 3h starvation, we observed Smo-GFP at the pericentriolar region and on DAVs that co-localized with EHD1 supporting an association of EHD1 with pre-ciliary vesicles and DAVs (Fig. 4f).
Because intracellular ciliogenesis has been observed in mouse photoreceptors30, we tested Ehd1 and Ehd3 ciliogenesis function in zebrafish photoreceptors. Consistent with our human cell data, in ehd1 and ehd3 double morphants DAVs were frequently observed on the M-centriole distal appendages whereas in rab8a morphants, the majority of non-ciliated M-centrioles had CVs (Fig. 4g,h, Supplementary Fig. 4b). Interestingly, Ehd proteins were detected near the centrioles prior to outer segment formation at 50hpf (Fig. 4i). Together our results in human RPE cells and zebrafish photoreceptors support a model wherein EHD1 is recruited on pre-ciliary vesicles to the M-centriole to function in CV assembly from DAVs, whereas Rab8 is associated with post-CV ciliogenesis processes.
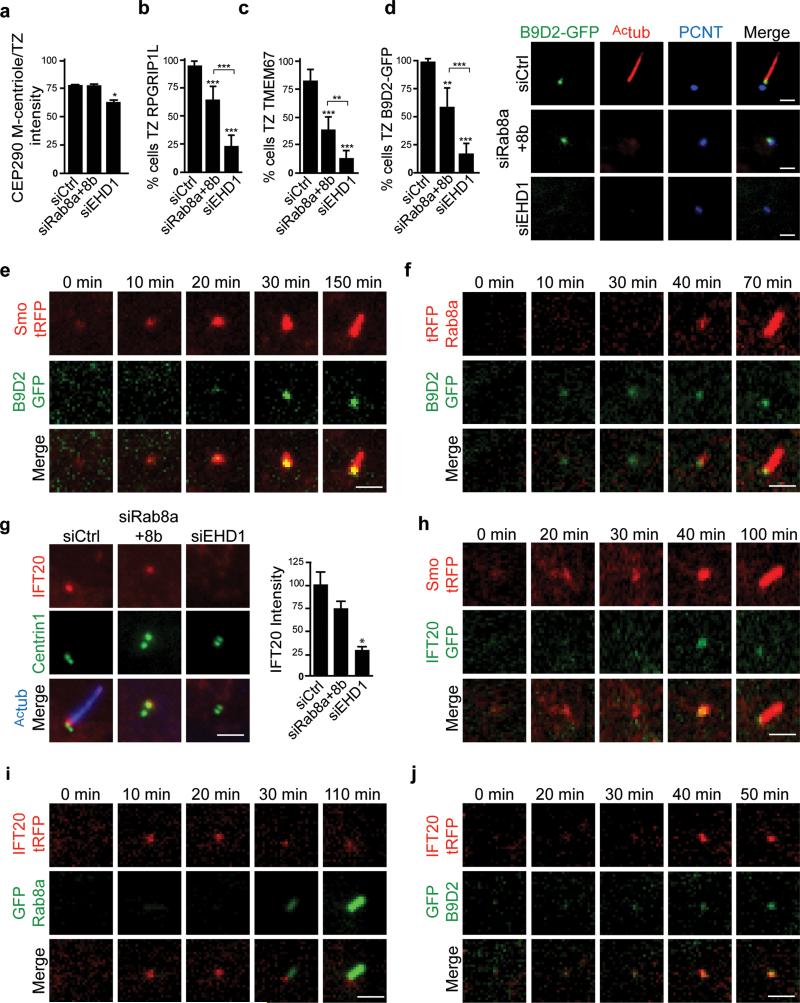
EHD1-dependent CV formation is required for recruitment of transition zone proteins and IFT20
To study the relationship between DAV docking, CV assembly steps and other ciliogenesis processes we examined recruitment of transition zone (TZ) proteins and intraflagellar transport proteins (IFT). We discovered that EHD1 was required for the recruitment of TZ proteins CEP290, RPGRIP1L, TMEM67 and B9D2-GFP to the M-centriole distal region, whereas Rab8 appeared to be dispensable (Fig. 5a-d). We further confirmed that pre-ciliary membranes accumulated at the M-centriole before TZ protein recruitment by live cell imaging B9D2-GFP and Smo-tRFP (Fig. 5e), and another ciliary receptor 5-HT6-tRFP (Supplementary Fig. 5). B9D2-GFP also localized to the developing cilium prior to Rab8a (Fig. 5f). IFT20, a protein known to localize to the Golgi, centrosome and cilia, is thought to be important for CV formation6, 31. Similarly to TZ proteins, IFT20 accumulation at the M-centriole was strongly blocked in EHD1 but not Rab8 depleted cells (Fig. 5g). Furthermore, IFT20-GFP accumulated at the M-centriole after Smo-tRFP recruitment (Fig. 5h, Supplementary Movie 4), but before Rab8 localization (Fig. 5i, Supplementary Movie 5). IFT20 basal body enrichment coincided with B9D2 recruitment (Fig. 5j). Therefore, our studies suggest that EHD1-dependent CV assembly function is required prior to IFT20 and TZ proteins recruitment, while Rab8 targets to the CV following these steps.
Figure 5. Transition zone proteins and IFT20 are recruited after distal appendage vesicle reorganization and before Rab8-dependent ciliary membrane extension.
a. Quantification of CEP290 centrosome levels as previously described48 in RPE cells treated with siRNA for 72h, and serum starved for the final 24h, followed by staining with CEP164 and CEP290 antibodies. Mean ± SEM from n=3 independent experiments is shown (total number of cells from all 3 experiments: siCtrl, 127 cells; siRab8a+8b, 110 cells; siEHD1#1, 143
b, c. Quantification of RPGRIP1L (b) and TMEM67 (c) -positive M-centrioles-basal bodies in RPE cells treated as in (a), stained with γ-tubulin and RPGRIP1L or TMEM67 antibodies. Mean ± SD are pooled data from 3 independent experiments with (b) n=15 and (c) n=6 areas imaged (total number of cells counted in all experiments: b: siCtrl, 288; siRab8a+8b, 323; siEHD1#1, 218, c: siCtrl, 131; siRab8a+8b, 138; siEHD1#1, 117).
d. RPE B9D2-GFP cells were treated with siRNA as described (a), serum starved for 24h, and stained with Actub and PCNT antibodies (right panel). Scale bars: 2 μm. Quantification cells with GFP-B9D2-positive M-centrioles-basal bodies (left panel). Means ± SD are pooled data from 3 independent experiments with n=7 areas imaged (total number of cells counted in all experiments: siCtrl, 134; siRab8a+8b, 171; siEHD1#1, 149). TZ: transition zone. Two tailed t-test analyses compared with siCtrl or between siRab8a+b and siEHD1 in a-d.
e, f. RPE B9D2-GFP cells transiently expressing tRFP-Smo or tRFP-Rab8a (f) were serum starved for 1h and imaged live by spinning disk confocal microscopy. Images show single xy-planes from z-stacks.
g. RPE GFP-Centrin1 cells treated with siRNA as described in (a) and stained for IFT20 and Actub. Quantification of IFT20 fluorescence intensity at the M-centriole-basal bodies in cells from left panel. Mean ± SEM from n=3 independent experiments is shown (total number of cells counted in all experiments: siCtrl, 183; siRab8a+8b, 142; siEHD1#1, 127). Two tailed t-test analysis compared with siCtrl.
h,i,j. RPE cells transiently expressing IFT20-GFP and Smo-tRFP (h), RPE GFP-Rab8a cells transiently expressing IFT20-tRFP (i), RPE GFP-B9D2 cells transiently expressing IFT20-tRFP (j) were imaged and analyzed as described in (e).
Scale bars: 2 μm. Note that panels e,f,h-j are representative image series of > 5 cilia assembly events observed. * P<0.05, ** P<0.001, *** P<0.001. Statistics source data for Fig5 a-d,g, can be found in Supplementary Table 2.
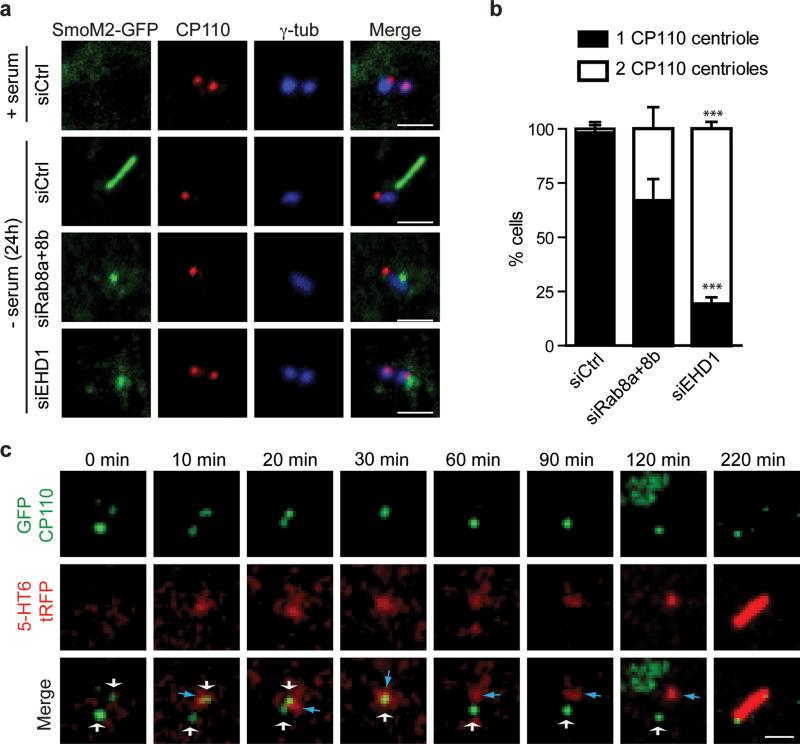
EHD1 is required for CP110 loss from the distal end of the M-centriole
Because CP110 loss from the M-centriole is required for basal body formation and precedes axoneme growth32, we examined its relationship with EHD1-dependent CV assembly. Rab8 appears to be dispensable for CP110 loss with ~70% of cells having only daughter centriolar CP110. In contrast, in EHD1-depleted cells 80% of cells had CP110 localized to both centrioles (Fig. 6a,b). To further demonstrate requirements for DAV membranes in CP110 loss, during ciliogenesis, we investigated this process in live cells using pre-ciliary membrane markers. We observed 5-HT6-tRFP membranes accumulating at the M-centriole followed by the loss of CP110-GFP a few minutes later (Fig. 6c). Thus, these results indicate that CP110 loss from the M-centriole occurs after DAV docking to the M-centriole and prior to CV assembly.
Figure 6. EHD1-dependent ciliary vesicle formation stimulates CP110 loss from the distal end of the mother-centriole.
a. RPE SmoM2-GFP cells treated with siRNA for 72h, grown in serum or serum starved for the last 24h, and stained with anti-CP110 and anti-γ-tubulin antibodies. Scale bar: 2 μm.
b. Quantification of serum starved RPE cells described in (a) showing CP110 localization on the mother and daughter centriole (2 dots) or only the daughter centriole (1 dot). Note that we disregarded cells with more than 2 dots in our quantification. Means ± SD are pooled data from 3 independent experiments with n=6 areas imaged (total number of cells counted in all experiments: siCtrl, 184; siRab8a+8b, 161; siEHD1#1, 157). Two tailed t-test analysis compared with siCtrl, *** P<0.0001. Statistics source data can be found in Supplementary Table 2.
c. RPE cells transiently co-expressing GFP-CP110 and 5-HT6-tRFP, serum starved for 1h and imaged live over time by spinning disk confocal microscopy as in Fig 5c. Representative images of maximum intensity projections of z-stack imaging series from 5 cilia assembly events observed. White and blue arrows indicate CP110-positive centrioles and 5-HT6 vesicles respectively. For presentation purposes images were smoothed with a Gaussian blur filter. Scale bar: 2 μm.
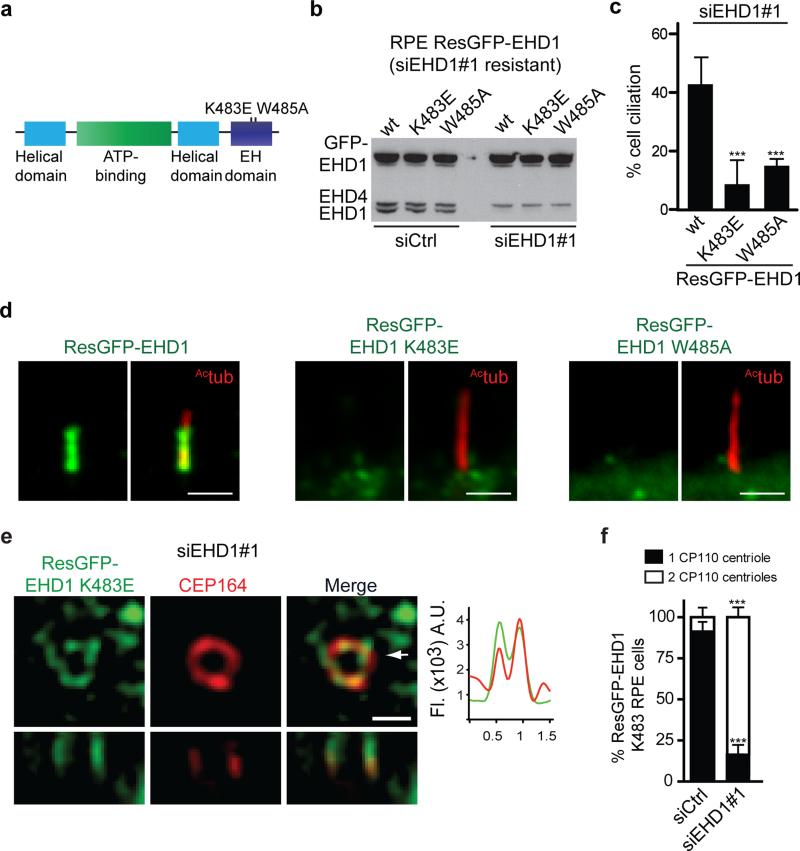
EHD1 membrane tubulation function is required for CV formation
Because EHD1-dependent CV assembly is critical for ciliogenesis initiation, we wanted to determine how EHD1 regulates DAV reorganization. We examined the ability of specific EHD1 loss-of-function mutations to rescue ciliation in RPE cells depleted of endogenous EHD1. EH domain mutants specifically affecting either NPF-substrate binding and membrane recruitment (W485A) or tubulo-vesicular (K483E) membrane functions failed to rescue compared to the wildtype siRNA resistant protein 33, 34 (Fig. 7a-c). Importantly, neither mutant protein accumulated significantly in the CP (Fig. 7d). Remarkably, using SIM we found that the GFP-K483E mutant localized to structures near the distal appendages when endogenous EHD1 was depleted following starvation (Fig. 7e). Furthermore, CP110 removal failed in K483E expressing cells (Fig. 7f). Together, these results indicate that the EHD1 K483 membrane tubulation function is critical for DAV assembly into the CV and M-centriole reorganization
Figure 7. EHD1 tubulation function is required for distal appendages vesicles assembly into the ciliary vesicle.
a. Domain structure of EHD1 and loss of function mutations.
b. Immunoblot analysis of siRNA resistant (Res) GFP-EHD1 wildtype, -K483E and -W485A proteins stably expressed in RPE cells 72h after transfection with siControl or siEHD1#1. Endogenous and GFP-EHD proteins expression levels were detected using anti-EHD1 antibody. Note that the EHD1 antibody also recognizes endogenous EHD4 as indicated. Un-cropped images of blots are shown in Supplementary Fig 6.
c. Quantification of cilia in cells treated with siEHD1#1 as described in (b) and serum starved the last 24h. Means ± SD are pooled data from 3 independent experiments with n=8 areas imaged (total number of cells from all experiments: ResGFP-EHD1, 293; ResGFP-EHD1 K483E, 269; ResGFP-EHD1 W485A, 263).Two tailed t-test analysis compared with WT.
d. Cell lines described in (b) were serum starved for 24h and stained with Actub antibody to mark the cilia. Scale bar: 2 μm.
e. Representative SIM image of RPE cells expressing the siRNA resistant GFP-K483E mutant transfected with EHD1 siRNA as in (c) and stained with CEP164 antibody. 9 out of 15 cells (60%) imaged expressing the K483E showed DAV-like structures. Arrow marks orthogonal view (bottom panels) and corresponds to fluorescence profile plots. Scale bar: 500nm.
f. Quantification of CP110 localization on the mother and daughter centrioles in RPE cells treated as in (c) and stained as described in Fig 6b. Means ± SD are pooled data from 3 independent experiments with n=6 areas imaged (total number of cells from all experiments: siCtrl, 148; siEHD1#1, 114). Two tailed t-tests compared with siCtrl.
*** P<0.0001. Statistics source data for Fig7 c,f can be found in Supplementary Table 2.
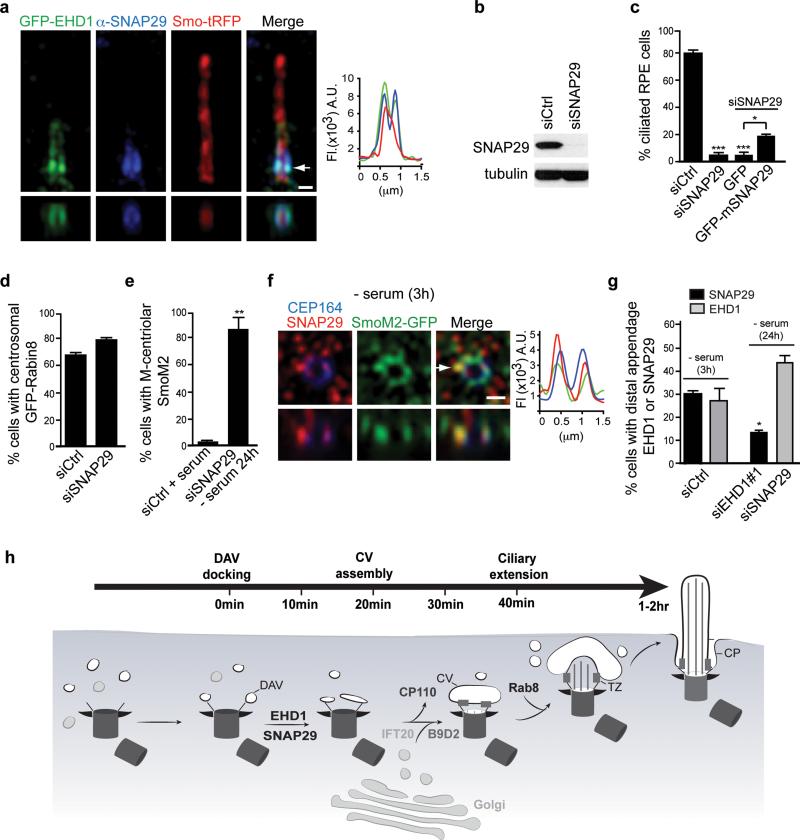
EHD1 is required for the fusion of DAVs into the CV
Based on our findings for EHD1 and EHD3 functioning in CV assembly from DAVs, we predicted that the SNARE membrane fusion machinery would play a critical role in early ciliogenesis. Because the SNARE protein SNAP29 directly interacts with EHD1 and EHD326, we tested its function in ciliogenesis. Consistent with the EHD-SNAP29 interaction, SNAP29 co-localized with EHD1 in the ciliary pocket and was required for ciliogenesis in RPE cells (Fig. 8a,b,c). Depletion of SNAP29 also did not affect GFP-Rabin8 pre-ciliary trafficking, nor SmoM2-GFP recruitment to the M-centriole (Fig. 8 d,e). As was observed with EHD1, SNAP29 co-localized with SmoM2-GFP on DAV-like membranes in 3h starved cells (Fig. 8f,g). Interestingly, SNAP29 failed to co-localize with SmoM2-GFP at the M-centriole distal end following EHD1 depletion (24 h starved) compared to 3h starved siControl treated cells, whereas SNAP29 was dispensable for EHD1 recruitment to early ciliary membranes (Fig. 8g). Given these findings and the requirement for EHD1 and EHD3 tubulation function in CV assembly, we theorized that EHD proteins recruit SNAP29 to DAVs where the SNARE functions in fusion of tubulated DAVs.
Figure 8. SNAP29, a SNARE and EHD1 and EHD3 binding protein, functions in CV assembly.
a. SIM image of RPE cells expressing Smo-tRFP and GFP-EHD1 stained with anti-SNAP29 antibody. Arrow corresponds to orthogonal view (bottom panels) and fluorescence intensity plot. Representative image of > 10 cells. Scale bar: 500 nm.
b. Western analysis of SNAP29 depletion in RPE cells treated for 72h. Un-cropped blots are shown in Supplementary Fig 6.
c. Quantification of ciliogenesis in RPE cells treated with siRNA for 72h (- serum the last 24h) with and without expression of GFP or GFP-mSnap29, stained with Actub and PCNT antibodies. Not included in the plot are short cilia (<1 μm) detected in 44 ± 2.4% in mSNAP29-positive cells. Mean ± SEM from n=3 independent experiments (>150 cells per treatment).
d. Quantification of RPE GFP-Rabin8 cells transfected with siRNAs for 72h and imaged live for centrosomal GFP-Rabin8 accumulation after 1hr starvation. Mean ± SEM from n=3 independent experiments (>150 cells per treatment).
e. Quantification of SmoM2-GFP at the M-centriole in RPE cells treated as described in (c) and stained with CEP164 antibodies. Mean ± SEM from n=3 independent experiments (>150 cells per treatment).
f. Representative SIM image of RPE SmoM2-GFP cells serum starved for 3h and stained with SNAP29 and CEP164 antibodies. 6 out of 14 cells (42%) showed SNAP29 co-localization with Smo-GFP at the distal appendages. Arrow corresponds to orthogonal view (bottom panels) and fluorescence intensity plot. Scale bars: 500nm.
g. Quantification of EHD1 and SNAP29 M-centriolar accumulation in RPE cells treated with siRNAs for 48h followed by 24h or 3h (siControl) serum starvation, and stained with CEP164 and EHD1 or SNAP29 antibodies. Mean ± SEM from n=3 independent experiments is shown (total number of cells counted in all experiments: siCtrl SNAP29, 234; siCtrl EHD1, 215: siEHD1#1, 116, siSNAP29, 149). Two tailed t-test analysis compared with siCtrl.
h. Model of intracellular ciliogenesis and time-line for the accumulation of ciliogenesis proteins at the M-centriole and developing cilia.
Two tailed t-test analyses compared with siCtrl or GFP Figure 8c,d,e.* P<0.05, ** P<0.001. Statistics source data for Fig 8c-e,g can be found in Supplementary Table 2.
DISCUSSION
In the intracellular ciliogenesis pathway, the ciliary membrane is thought to arise from a CV that is reshaped into a membrane sheath around the developing axoneme. However, the molecular details governing this process remained unresolved. Based on Rab11-Rabin8 pre-ciliary vesicle transport observed following cues to ciliate15, we reasoned that the recruitment of smaller pre-ciliary vesicles to the M-centriole would be followed by fusion into a larger CV. Here, we describe a role for EHD1 and EHD3, regulators of Rab compartments, in early primary cilium assembly. We find that EHD1 and EHD3 are indispensable for CV formation from DAVs and orchestrate other ciliogenesis events, whereas Rab8 functions downstream in CV extension. In addition, we show that the EHD interacting protein SNAP29, a SNARE membrane fusion protein, regulates CV assembly.
A role for EHD1 and EHD3 in CV formation
EHD1−/− mice were reported to have ocular defects which could be associated with defective photoreceptor cilia formation35, as we have observed in zebrafish. These mice also had higher rates of embryonic lethality suggesting an essential role of EHD1. Unlike EHD1-null mice, EHD3 knockout mice had no discernable pathology36. Our studies in human cells and zebrafish suggest that EHD1 and EHD3 display functional overlap in cilia formation likely due to their ability to heterodimerize37. Importantly, EHD1 and EHD3 share the same binding partners through interaction via NPF motif-EH domain binding21,26,38. Our results are also consistent with prior studies showing that EHD proteins have tissue-specific and redundant roles39.
The connection between EHD1 and EHD3 and the Rab11-Rab8 cascade is supported by our findings that these proteins co-localize with both Rab proteins in ciliary-associated membrane compartments. Although our data do not exclude the possibility that endocytic recycling dysfunction might affect ciliogenesis indirectly, EHD1 co-localized with Rab11, Rabin8, and Smo on pre-ciliary vesicles and DAVs, and with Rab8 on the growing ciliary membrane. EHD1 is presumably transported to the centrosome via pre-ciliary vesicles, likely originating from the ERC15, which subsequently associate with distal appendages and fuse to form the CV. In non-ciliating cells, an interaction between Rab11-ERC at the M-centriole was previously described40. EHD1 and EHD3 function in tubular membrane regulation of the ERC, although in HeLa cells EHD1 appears to function as a vesiculator of tubular recycling endosomes24. Our finding that GFP-EHD1 K483E fails to rescue ciliation and yet still localized to DAVs suggests that tubulo-vesicular functioning of EHD proteins is important for CV formation. Thus we hypothesize that EHD1- and EHD3-membrane remodeling function is responsible for bringing DAV membranes in close proximity to promote SNAP29-dependent fusion during CV assembly. Our discovery that SNAP29 localizes to the ciliary pocket and functions in ciliogenesis in RPE cells is also interesting as SNAP29 mutations are responsible for CEDNIK syndrome and SNAP29 knockout mice are embryonic lethal41, 42. CEDNIK patients do not appear to have classic ciliopathy, but this is something that cannot be ruled out at this time. In addition to SNAP29 other SNAREs would be expected to function in membrane fusion during the CV assembly stage. Given the observed importance of intracellular membrane assembly during ciliogenesis, investigation of SNARE protein function in CV assembly and ciliary progression merits further investigation.
Pre-ciliary membrane reorganization and ciliogenesis initiation
Failure to dock membranes to the distal appendages prevents CP110 loss from the M-centriole needed for basal body formation and ciliogenesis progression5,6,8,11,43. Our work further clarifies the molecular mechanism of this critical ciliogenesis initiation step by demonstrating that M-centriole modifications are initiated following docking of DAVs to the M-centriole. To our knowledge, EHD1 and EHD3 are the first direct membrane-associated proteins whose function affects M-centriole reorganization. How EHD1-dependent DAV assembly into the CV affects CP110 loss is not clear, but there are provocative possible mechanisms. CP110 loss from the M-centriole is associated with ubiquitination44. One possibility is that pre-ciliary membranes and/or EHD proteins recruit components of the ubiquitination machinery to the M-centriole. Alternatively, changes in the membrane accompanying DAV to CV formation could help establish the basal body.
The orchestration of DAV reorganization by EHD proteins is also a prerequisite step for TZ proteins and IFT20 recruitment, which is consistent with other reports showing that without membrane association at M-centriole, TZ and IFT components fail to accumulate5,6,45,46. Likewise, Rab8 recruitment to the developing cilia appears to be different from IFT20 and TZ. Importantly, our work suggests that a mechanism may be present to control the timing of Rab8 localization to the CV. Could this be a checkpoint to ensure proper membrane association with the growing axoneme? Hence, only following IFT20 and TZ proteins recruitment does coordinated axoneme and ciliary membrane growth occur (Fig. 8h).
EHD1 membrane shaping and the ciliary pocket
A surprising finding was the localization of EHD1 and SNAP29 to the ciliary pocket membrane. The ciliary pocket is a site for TGF-β signaling47 and its proximity to the ciliary membrane suggests it is important for ciliary trafficking28. The ciliary pocket membrane resembles a tubular structure surrounding the proximal part of the cilium that much like a vesicle or tubule has a positive curvature membrane facing the cytosol, a preferential site for EHD proteins binding. In contrast, the intracellular face of the ciliary membrane has a negative curvature. Our work suggests that EHD1 and EHD3 are excluded from the ciliary membrane, possibly by the TZ established prior to CV reshaping and axoneme growth. We can speculate that TZ gating of EHD proteins is important for establishment of negative curvature after the CV stage to allow for close association with the axoneme and the developing ciliary membrane. In mature cilia, EHD proteins could also be important for ciliary pocket maintenance. Finally, whether EHD1 and EHD3 are important for signaling associated with the ciliary pocket or the cilium remains an open question.
METHODS
Antibodies and Reagents
Commercial antibodies used were mouse anti-γ-tubulin (clone GTU-88, 1/5000, Sigma), anti-acetylated α-tubulin (Actub) (clone 6-11B-1, 1/10000, Sigma), anti-rhodopsin (clone 4D2, 1/100), anti-β–actin (clone AC-15, 1/30000, Sigma) and anti-β-tubulin (clone 2-28-33, 1/2000, Sigma), anti-EHD3 (1/1000, Abnova for IF and 1/100, Abcam, 1/500, Proteintech for WB), rabbit anti-pericentrin (1/5000, Novus Biologicals, 1/5000), anti-EHD2 (1/1000, Abcam), anti-EHD4 (1/1000, Abcam), anti-GFP (1/50, Abcam), anti-HA HRP (1:1000, Roche), anti-CEP290 (1/200, Bethyl), anti-RPGRIP1L (1/200, Proteintech), anti-IFT20 (1/200, Proteintech), anti-TMEM67 (1/200, Proteintech), anti-SNAP29 (1/1000 for WB Proteintech and 1/200 for IF from Dr. Andrew Peden) and goat anti-CEP164 (1/500, Santa Cruz). Rabbit anti-Rab11a (1/300) was a gift from Dr. Jim Goldenring and rabbit anti-CP110 (1/500) was a gift from Dr. Monica Diaz. Polyclonal rabbit anti-EHD1 (1/1000) has been previously described33. Fluorescent secondary antibodies and phalloidin (1/50) were from Life Technologies (Molecular probes, Eugene, OR). Nuclei were labeled with Hoechst (Molecular probes, Eugene, OR).
Cell lines, Plasmids, and RNAi
EHD1 (MGC 8143828; Invitrogen) and EHD3 (RC214168: Origene) cDNAs were purchased from Invitrogen. EHD4 (BC006287) was purchased from Open Biosystems. EHD1, -3, -4 was PCR amplified and cloned into pDONR221 plasmid using BP clonase (Invitrogen). pENTR-EHD2 (IOH13420) was purchased from Invitrogen. The siRNA resistant (Res) EHD1 and EHD1 K483E, W485A mutants were made by QuikChange mutagenesis kit (Agilent Technologies). EHD1-4, ResEHD1 constructs and CP110 (BC036654, purchased from DNASU) cDNAs were cloned into pGLAP1 destination vectors15 (LAP referring to the GFP-Tev-Stag49) using the Invitrogen Gateway cloning system. Mouse Snap29 (BC030066) constructs were purchased from DNASU and cloned into pGLAP1. EHD1-4 were also subcloned into pCS2+ vectors. EHD1, ResEHD1 and EHD1 mutants were Gateway cloned into pHUSH-LAP, alentivirus expression vector50 reengineered by PCR to have the gLAP1 vector CMV promoter, TetO2 and LAP-tag upstream of the Gateway forward acceptor site for in-frame Tet-inducible LAP-fusion construct expression.
GFP-Rab8a, tRPF-Rab8a and tRPF-Rabin8 constructs were previously described15. Mouse IFT20 (NM_018854) was synthesized with flanking Gateway compatible attB sites (DNA2.0). Human Smo (BC009989) and the Smo-W535L (SmoM2) cDNAs51, B9D2 (BC004157, purchased from DNASU), and mIFT20 were cloned into pDONR221 and subcloned into pGLAP752. Smo, 5HT-615 and mIFT20 were cloned into pDEST-tRFP7, a pGLAP7 vector reengineered to replace the LAP tag with Tag-RFP (tRFP, Evrogen). The mouse DsRed-Inversin construct was a gift from Dr Tim Stearns.
GFP-Rab8 and GFP-Rabin8 RPE cell lines have been previously described15. FuGENE 6 (Promega, Madison,WI) was used for plasmid DNA transfections into cells. RPE-FRT cell lines stably expressing SmoWT, SmoM2 and B9D2 were made using the FLP-IN system (Invitrogen). GFP-EHD1, siRNA resistant GFP-EHD1, GFP-EHD1-K483E and GFP-EHD1-W485A RPE cell lines were made using the pHUSH-LAP lentivirus expression system. Lentiviral infections were performed as described53. RPE cells infected with lentivirus were selected with 4ug/ml of puromycin. siRNA duplexes were purchased from Dharmacon (Table Supplementary 1). Rab11a and Rab11b siRNA have been described15. Cells were transfected with siRNA duplexes (50 nM) using RNAiMAX (Invitrogen).
Immunofluorescence and time-lapse microscopy
For RPE and IMCD3 ciliation quantification, cells were serum-starved for 24h, unless otherwise stated, fixed and processed for immuno-staining and indirect immunofluorescence as described15. Briefly, cells were fixed in 4% paraformaldehyde for 10 min or cold methanol for 5 min and blocked with 1% BSA in PBS 0.1% TX100 or BSA solution alone, respectively for 10 min followed by incubation with primary antibody in blocking solution for 1h at RT or overnight at 4°C. Fluorophore conjugated secondary antibodies were incubated for 1h at RT. Pre-ciliary membranes, cilia, transition zone proteins, IFT20 and CP110 were imaged in > six fields or as a montage of 4-6 images from 2 or more areas using a 40× 1.4 NA or 63× 1.3 NA objective, unless otherwise indicated. Imaging was performed using a Zeiss Axio Scan.Z1 inverted epifluorescence microscope equipped with a Coolsnap HQ2 camera. Images were analyzed using the Slidebook software. Immuno-cytochemistry on cryosections of 3dpf zebrafish retinae was performed as previously described54. For whole-mount zebrafish cilia studies, embryos were fixed in Dent's fixative overnight at 4°C and processed for immuno-staining as described previously55. Briefly, immunostainings were performed using blocking solution (PBS with 0.1% TX100 and 5% goat serum) for 30 min at room temperature (RT) followed by overnight incubation in blocking solution with anti-acetylated α-tubulin antibody and phalloidin for wholemounts or anti-Ehd1, anti-Ehd3 and anti-rhodopsin or anti-γ-tubulin 2h at RT. Fluorophore-conjugated antibodies were incubated in blocking solution for 2h at RT for wholemounts and 30 min at RT for cyrosections. Note that in our zebrafish ciliation assays, morphants were selected for small eye defects whereas embryos from rescue experiments were chosen randomly. Organs with absent or strongly reduced cilia numbers were counted and this data was used to calculate the percentage of normal versus total number of organs observed. Imaging was performed using a 40× 1.4 NA or 63× 1.6 NA oil objective and a Marianas spinning disk confocal (SDC) microscope (3I, Denver)
Time-lapse imaging of RFP- and GFP-fusions was performed using the Marianas SDC. The environmental chamber containing the XYZ automatic stage (ASI) was set at 37°C and 5% CO2. For SDC time-lapse experiments, image acquisition of z-stacks (10×00B5;m z-stack with 500nm step size) was performed every 5-20 minutes using an Evolve 512 EMCCD camera and 40× 1.4 NA (325 nm/pixel) or 63× 1.3 NA (206 nm/pixel) oil objective and Slidebook software. For all zebrafish studies, image acquisition (35 μm z-stacks with 1 μm step size) was performed using a 40×1.4 NA or 63× 1.3 NA oil objective lens. Processing of images and fluorescence intensity measurements were carried out using either ImageJ or Slidebook software. GFP-Rabin8 vesicle centrosome localization was detected by time-lapse epifluorescence microscopy and analyzed as described15 using a HQ2 digital camera (Photometrics).
Structural illumination microscopy (SIM)
Cells were grown on #1.5 cover glasses (Zeiss). After immunofluorescence processing, cover glasses were mounted onto glass slides with Vectashield (Vector) and sealed with nail polish. 5 phases and 3 rotations of 3D SIM images were captured using a Zeiss or Nikon N-SIM microscopes. Channels were aligned using parameters obtained from calibration measurements with 100 nm TetraSpeck beads. Zeiss SIM images were taken with a 63× 1.4 NA oil objective and a PCO edge sCMOS camera. Nikon N-SIM images were taken with an Apo TIRF 100X/1.49 oil objective and an EMCCD camera (Andor DU-897). The pixel size of both cameras is 16 μm. The raw data pixel size is 80 nm with the Zeiss SIM using a 63X objective and 60 nm with Nikon SIM using 100X objective. Raw images captured by the Nikon SIM microscope in Fig. 1i, 1j, 4f, 7e, 8a and 8f are 14 bit and raw images captured by the Zeiss SIM microscope in Fig. 4c-e, Supplementary Fig. 1k,l are 16 bit. Laser power and exposure time were optimized to use a large portion of the camera's dynamic range while minimizing bleaching. To avoid reconstruction artifacts we have done the following: 1) during the sample preparation antibody dilutions, coverslip thickness and mounting media conditions were optimized to give the best signal to noise ratio, 2) samples were checked by wide-field or confocal microscopy prior to SIM imaging to check integrity of sample, 3) SIM images were acquired using the largest possible portion of the dynamic range provided by the camera, 4) images were reconstructed using the Zeiss Zen software and Nikon NIS-Elements software, with Wiener filter settings to reduce image noise. The estimated resolutions after reconstruction were: Zeiss channel 488: ~110 nm lateral and 350-400 nm axial; Zeiss channel 561: ~130 nm lateral and 350-400 nm axial; Zeiss channel 640: ~200 nm lateral and 350-400 nm axial; Nikon channel 488: ~130 nm lateral and ~400 nm axial; Nikon channel 561: ~140 nm lateral and ~400 nm axial; Nikon channel 640: ~160 nm lateral and ~400 nm axial. Histograms of reconstructed images were adjusted in the Nikon NIS-Elements or Zeiss Zen software and tiffs cropped in ImageJ. Intensity profile plots were created in the NIS-elements or the Zen software by drawing lines across the structure on the reconstructed images (non-histogram adjusted) and by exporting the values into Excel and GraphPad Prism 6 for Macintosh OS.
Transmission Electron Microscopy and Histology
Electron microscopy processing in RPE cells and zebrafish embryos were carried out as previously described54, 56. Briefly, cells or zebrafish embryos were fixed in a solution of 2% glutaraldehyde with or without 4% paraformaldehyde in 0.1 M sodium cacodylate buffer followed by post-fixation with 1% Osmium tetroxide and 1% Uranyl acetate. After dehydration in graded ethanol, cells were embedded in EMbed 812 (Electron Microscopy Sciences) and 80 nm sections were cut using a Leica EM UC7 microtome. Electron micrographs were acquired using a transmission electron microscope (Hitachi 7650 TEM). For photoreceptor cilia counting from histological analysis, semi-thin sections (250nm) cut from samples embedded for TEM analysis were stained with toluidine blue. Photoreceptor outer segments with intact inner segments were counted (>100). For immuno-EM, RPE cell line expressing GFP-EHD1 were fixed with 4% paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium cacodylate buffer. After dehydration in graded ethanol, cells were embedded in LR White resin. Ultra-thin sections were immuno-stained with an anti-GFP antibody (1/100) for 2h at RT in blocking solution (AURION blocking solution Electron Microscopy sciences) and a secondary antibody conjugated with 10 nm gold particles for 1h RT. For correlative light and electron microscopy (CLEM), cells were plated on gridded-glass bottom dishes (MatTek). Fluorescence and DIC images of fixed cells were taken with a Zeiss fluorescence microscope. The position of cells was recorded using grid numbers on cover glasses. After imaging, cells were processed for electron microscopy. Serial ultra-thin sections of the recorded region were collected on formvar coated slot grids and low magnification TEM images were matched with fluorescence images to identify cells of interest.
Morpholino knockdown
Fish care and husbandry were performed in compliance with NIH guideline for animal care. Zebrafish used in this study were TAB-5 and Tg(arl13b-GFP) line (a gift from Dr. Zhaoxia Sun). Knockdowns for each gene were performed using the following translation blocking morpholinos obtained from GENETOOLS, Philomath, OR: ehd1a CTGAACATGGTGGACGTTACACGAC, ehd1b ATCTTTGTTAGACCAACTGAACATT and ehd3 CATCGGTACCCAACCAGCTGAACAT.
The splice-blocking morpholino oligonucleotide rab8- SP1 was previously described57 Injections were done using a microinjector PLI-90 (Harvard Apparatus, Cambridge, MA). For rescue experiments, the full-length sequences of human EHD1, EHD3 and EHD4 were cloned into PCS2+ vectors and mRNAs were transcribed using the mMESSAGE mMACHINE kit (Ambion) according to manufacturer's instructions. Embryos were co-injected with 250 μM of morpholinos and 300 pg/nl of capped mRNAs at the one-cell stage.
Expression analysis
Taqman gene expression probes for EHD1-4 were purchased from Applied Biosystems and qRT-PCR was performed as previously described15.
Western blots on cell lysates were performed as previously described15. Briefly, RPE cells were homogenized in lysis buffer (20 mM Tris pH8, 137 mM NaCl, 10% glycerol, 1% TX100) containing protease inhibitor cocktail (PIC, Roche) and zebrafish embryos were lysed in RIPA buffer with PIC. Lysates were centrifuged for 10min at 13000rpm. Sample buffer was added to the supernatants and samples were boiled.
Standard whole mount in situ hybridizations were performed as previously described58 at 48hpf and 72 hpf with both ehd1a+b and ehd3 probes. Due to the high sequence similarity between these genes, we designed probes in the 3'UTR regions to maximize the specificity. However, the ehd1 probe was predicted to recognize both ehd1a and ehd1b transcripts. Primers: ehd1 (F) primer: 5’GTTTTAACCCAAGCGCCACC 3’, ehd1 (R) primer: 5’ATCTGGAGGAATTGCGCAGC 3’ (amplified product: 394pb, specific to ehd1a+b 3’UTR), ehd3 (F) primer: 5’ ATGCTTCACTAGGAGGAATGG 3’. ehd3 (R) primer: 5’GTTGACTGTTGGGTTTTACGG 3’ (amplified product: 411bp. Specific to ehd3 3’UTR).
Statistics and Reproducibility
Statistical analyses were performed as specified with GraphPad Prism 6 for Macintosh OS. All data are presented as mean ± S.E.M or S.D as specified in the figure legends, and two group comparisons were done with an unpaired two-tailed Student's t-test. A value of P<0.05 was considered as statistically significant. All P values were indicated on graphs in figures as followed: * P<0.05, **P<0.001. ***P<0.0001. Exact P values can be found in the Statistics Source Data Sheet (Supplementary Table 2).
Experiments were reproducibly performed as follows: one experiment (Fig 1h, Supplementary Fig 2d), two independent experiments (Fig 1j, 2h, 3a, 4i, 5e, 5f, 7e, Supplementary Fig 1i, 1l, 2a, 2e, 5), 3 independent experiments (Fig 1a, 1d-f, 2a, 2c-e, 2g, 3f-g, 4c-g, 5d, 5g, 6a, 6c, 7b, 8a-b, 8f, Supplementary Figure 1d, 1g, 1k, 2c, 4b), two-four independent experiments (Fig 4a, Supplementary Fig 4a) and more than 3 independent experiments (Fig 1i, 3c-e, 5h-j, 7d, Supplementary Fig 1h, 1j, 3, Movie 1-5).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Kunio Nagashima and Anne Kamata for help with IEM and EM, Christopher Lamont for data analysis, Stephen Lockett for help with SIM imaging, Suzanne Specht for help with cell culture, Dr. Andrew Peden for SNAP29 antibodies, Dr. Jim Goldenring for Rab11 antibodies, Dr. Zhaoxia Sun for the transgenic line and Dr. Shawn Burgess for assistance raising zebrafish embryos. We are grateful to Dr. Julie Donaldson for critical reading of this manuscript.
Footnotes
CONTRIBUTIONS
Q.L. and C.I. performed most experiments with crucial help from: C.W. (RNAi, fluorescence imaging, and RT-PCR); C.O. (SIM imaging); P.P., S.L., I.O.D. and Y.H. (in situ expression in zebrafish); U.B. (electon microscopy and CLEM); V.W. (immunoblotting); J.S. (zebrafish MO experiments); A.C. (cell line generation and RNAi rescue experiments). S.C. and J.R. provided critical reagents. S.C., C.O., J.L-S, and P.K.J. discussed the results and commented on the manuscript; C.W. and C.I. wrote the paper with suggestions from Q.L. and C.O. C.W, Q.L and C.I conceived and designed the research.
The authors declare no competing financial interests.
REFERENCES
- 1.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat Rev Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hildebrandt F, Benzing T, Katsanis N. Ciliopathies. N Engl J Med. 2011;364:1533–1543. doi: 10.1056/NEJMra1010172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt KN, et al. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol. 2012;199:1083–1101. doi: 10.1083/jcb.201202126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Joo K, et al. CCDC41 is required for ciliary vesicle docking to the mother centriole. Proc Natl Acad Sci U S A. 2013;110:5987–5992. doi: 10.1073/pnas.1220927110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sillibourne JE, et al. Primary ciliogenesis requires the distal appendage component Cep123. Biology open. 2013;2:535–545. doi: 10.1242/bio.20134457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanos BE, et al. Centriole distal appendages promote membrane docking, leading to cilia initiation. Genes Dev. 2013;27:163–168. doi: 10.1101/gad.207043.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kobayashi T, Kim S, Lin YC, Inoue T, Dynlacht BD. The CP110-interacting proteins Talpid3 and Cep290 play overlapping and distinct roles in cilia assembly. J Cell Biol. 2014;204:215–229. doi: 10.1083/jcb.201304153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorokin S. Centrioles and the formation of rudimentary cilia by fibroblasts and smooth muscle cells. J Cell Biol. 1962;15:363–377. doi: 10.1083/jcb.15.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye X, Zeng H, Ning G, Reiter JF, Liu A. C2cd3 is critical for centriolar distal appendage assembly and ciliary vesicle docking in mammals. Proc Natl Acad Sci U S A. 2014;111:2164–2169. doi: 10.1073/pnas.1318737111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129:1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 13.Yoshimura S, Egerer J, Fuchs E, Haas AK, Barr FA. Functional dissection of Rab GTPases involved in primary cilium formation. J Cell Biol. 2007;178:363–369. doi: 10.1083/jcb.200703047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knodler A, et al. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci U S A. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Westlake CJ, et al. Primary cilia membrane assembly is initiated by Rab11 and transport protein particle II (TRAPPII) complex-dependent trafficking of Rabin8 to the centrosome. Proc Natl Acad Sci U S A. 2011;108:2759–2764. doi: 10.1073/pnas.1018823108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato T, et al. Rab8a and Rab8b are essential for multiple apical transport pathways but insufficient for ciliogenesis. J Cell Sci. 2013 doi: 10.1242/jcs.136903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bryant DM, et al. A molecular network for de novo generation of the apical surface and lumen. Nat Cell Biol. 2010;12:1035–1045. doi: 10.1038/ncb2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 19.Zhang J, Naslavsky N, Caplan S. Rabs and EHDs: alternate modes for traffic control. Biosci Rep. 2012;32:17–23. doi: 10.1042/BSR20110009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naslavsky N, Caplan S. EHD proteins: key conductors of endocytic transport. Trends Cell Biol. 2011;21:122–131. doi: 10.1016/j.tcb.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naslavsky N, Rahajeng J, Sharma M, Jovic M, Caplan S. Interactions between EHD proteins and Rab11-FIP2: a role for EHD3 in early endosomal transport. Mol Biol Cell. 2006;17:163–177. doi: 10.1091/mbc.E05-05-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roland JT, Kenworthy AK, Peranen J, Caplan S, Goldenring JR. Myosin Vb interacts with Rab8a on a tubular network containing EHD1 and EHD3. Mol Biol Cell. 2007;18:2828–2837. doi: 10.1091/mbc.E07-02-0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sharma M, Giridharan SS, Rahajeng J, Naslavsky N, Caplan S. MICAL-L1 links EHD1 to tubular recycling endosomes and regulates receptor recycling. Mol Biol Cell. 2009;20:5181–5194. doi: 10.1091/mbc.E09-06-0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cai B, et al. Differential roles of C-terminal Eps15 homology domain proteins as vesiculators and tubulators of recycling endosomes. J Biol Chem. 2013;288:30172–30180. doi: 10.1074/jbc.M113.488627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giridharan SS, Cai B, Vitale N, Naslavsky N, Caplan S. Cooperation of MICAL-L1, syndapin2, and phosphatidic acid in tubular recycling endosome biogenesis. Mol Biol Cell. 2013;24:1776–1790. S1771–1715. doi: 10.1091/mbc.E13-01-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rotem-Yehudar R, Galperin E, Horowitz M. Association of insulin-like growth factor 1 receptor with EHD1 and SNAP29. J Biol Chem. 2001;276:33054–33060. doi: 10.1074/jbc.M009913200. [DOI] [PubMed] [Google Scholar]
- 27.Shiba D, et al. Localization of Inv in a distinctive intraciliary compartment requires the C-terminal ninein-homolog-containing region. J Cell Sci. 2009;122:44–54. doi: 10.1242/jcs.037408. [DOI] [PubMed] [Google Scholar]
- 28.Molla-Herman A, et al. The ciliary pocket: an endocytic membrane domain at the base of primary and motile cilia. J Cell Sci. 2010;123:1785–1795. doi: 10.1242/jcs.059519. [DOI] [PubMed] [Google Scholar]
- 29.Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biology open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sedmak T, Wolfrum U. Intraflagellar transport proteins in ciliogenesis of photoreceptor cells. Biol Cell. 2011;103:449–466. doi: 10.1042/BC20110034. [DOI] [PubMed] [Google Scholar]
- 31.Follit JA, Tuft RA, Fogarty KE, Pazour GJ. The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol Biol Cell. 2006;17:3781–3792. doi: 10.1091/mbc.E06-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 33.Naslavsky N, Boehm M, Backlund PS, Jr., Caplan S. Rabenosyn-5 and EHD1 interact and sequentially regulate protein recycling to the plasma membrane. Mol Biol Cell. 2004;15:2410–2422. doi: 10.1091/mbc.E03-10-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Naslavsky N, Rahajeng J, Chenavas S, Sorgen PL, Caplan S. EHD1 and Eps15 interact with phosphatidylinositols via their Eps15 homology domains. J Biol Chem. 2007;282:16612–16622. doi: 10.1074/jbc.M609493200. [DOI] [PubMed] [Google Scholar]
- 35.Rainey MA, et al. The endocytic recycling regulator EHD1 is essential for spermatogenesis and male fertility in mice. BMC Dev Biol. 2010;10:37. doi: 10.1186/1471-213X-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.George M, et al. Renal thrombotic microangiopathy in mice with combined deletion of endocytic recycling regulators EHD3 and EHD4. PLoS One. 2011;6:e17838. doi: 10.1371/journal.pone.0017838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galperin E, et al. EHD3: a protein that resides in recycling tubular and vesicular membrane structures and interacts with EHD1. Traffic. 2002;3:575–589. doi: 10.1034/j.1600-0854.2002.30807.x. [DOI] [PubMed] [Google Scholar]
- 38.de Beer T, et al. Molecular mechanism of NPF recognition by EH domains. Nat Struct Biol. 2000;7:1018–1022. doi: 10.1038/80924. [DOI] [PubMed] [Google Scholar]
- 39.George M, et al. Shared as well as distinct roles of EHD proteins revealed by biochemical and functional comparisons in mammalian cells and C. elegans. BMC Cell Biol. 2007;8:3. doi: 10.1186/1471-2121-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hehnly H, Chen CT, Powers CM, Liu HL, Doxsey S. The centrosome regulates the Rab11- dependent recycling endosome pathway at appendages of the mother centriole. Current biology : CB. 2012;22:1944–1950. doi: 10.1016/j.cub.2012.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprecher E, et al. A mutation in SNAP29, coding for a SNARE protein involved in intracellular trafficking, causes a novel neurocutaneous syndrome characterized by cerebral dysgenesis, neuropathy, ichthyosis, and palmoplantar keratoderma. Am J Hum Genet. 2005;77:242–251. doi: 10.1086/432556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuchs-Telem D, et al. CEDNIK syndrome results from loss-of-function mutations in SNAP29. The British journal of dermatology. 2011;164:610–616. doi: 10.1111/j.1365-2133.2010.10133.x. [DOI] [PubMed] [Google Scholar]
- 43.Tsang WY, et al. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li J, et al. USP33 regulates centrosome biogenesis via deubiquitination of the centriolar protein CP110. Nature. 2013;495:255–259. doi: 10.1038/nature11941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cajanek L, Nigg EA. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci U S A. 2014;111:E2841–2850. doi: 10.1073/pnas.1401777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Goetz SC, Liem KF, Jr., Anderson KV. The spinocerebellar ataxia-associated gene Tau tubulin kinase 2 controls the initiation of ciliogenesis. Cell. 2012;151:847–858. doi: 10.1016/j.cell.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clement CA, et al. TGF-beta signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 2013;3:1806–1814. doi: 10.1016/j.celrep.2013.05.020. [DOI] [PubMed] [Google Scholar]
- 48.Klinger M, et al. The novel centriolar satellite protein SSX2IP targets Cep290 to the ciliary transition zone. Mol Biol Cell. 2014;25:495–507. doi: 10.1091/mbc.E13-09-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheeseman IM, Desai A. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Science's STKE : signal transduction knowledge environment 2005. 2005:l1. doi: 10.1126/stke.2662005pl1. [DOI] [PubMed] [Google Scholar]
- 50.Gray DC, et al. pHUSH: a single vector system for conditional gene expression. BMC Biotechnol. 2007;7:61. doi: 10.1186/1472-6750-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murone M, Rosenthal A, de Sauvage FJ. Sonic hedgehog signaling by the patched-smoothened receptor complex. Current biology : CB. 1999;9:76–84. doi: 10.1016/s0960-9822(99)80018-9. [DOI] [PubMed] [Google Scholar]
- 52.Wright KJ, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jaiswal BS, et al. Combined targeting of BRAF and CRAF or BRAF and PI3K effector pathways is required for efficacy in NRAS mutant tumors. PLoS One. 2009;4:e5717. doi: 10.1371/journal.pone.0005717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Insinna C, Pathak N, Perkins B, Drummond I, Besharse JC. The homodimeric kinesin, Kif17, is essential for vertebrate photoreceptor sensory outer segment development. Dev Biol. 2008;316:160–170. doi: 10.1016/j.ydbio.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Malicki J, Avanesov A, Li J, Yuan S, Sun Z. Analysis of cilia structure and function in zebrafish. Methods Cell Biol. 2011;101:39–74. doi: 10.1016/B978-0-12-387036-0.00003-7. [DOI] [PubMed] [Google Scholar]
- 56.Lu Q, et al. Chromatin-bound NLS proteins recruit membrane vesicles and nucleoporins for nuclear envelope assembly via importin-alpha/beta. Cell research. 2012;22:1562–1575. doi: 10.1038/cr.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Omori Y, et al. Elipsa is an early determinant of ciliogenesis that links the IFT particle to membrane-associated small GTPase Rab8. Nat Cell Biol. 2008;10:437–444. doi: 10.1038/ncb1706. [DOI] [PubMed] [Google Scholar]
- 58.Thisse C, Thisse B. High-resolution in situ hybridization to whole-mount zebrafish embryos. Nature protocols. 2008;3:59–69. doi: 10.1038/nprot.2007.514. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.