Abstract
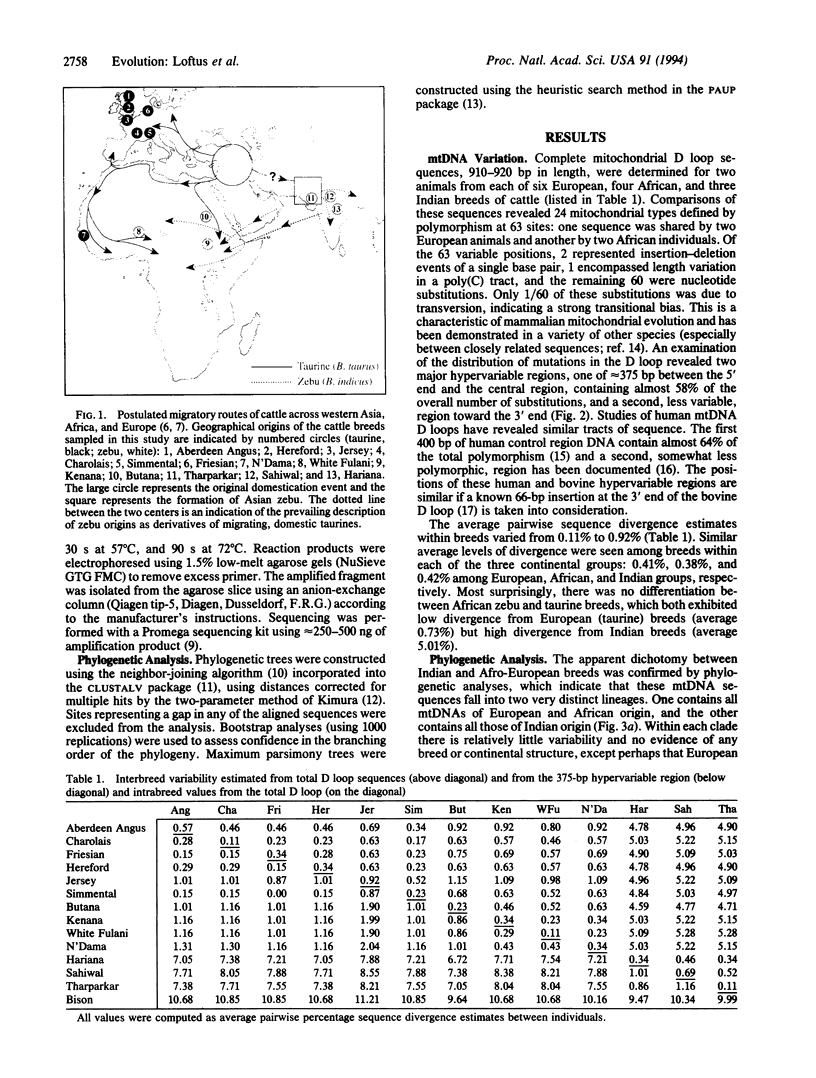
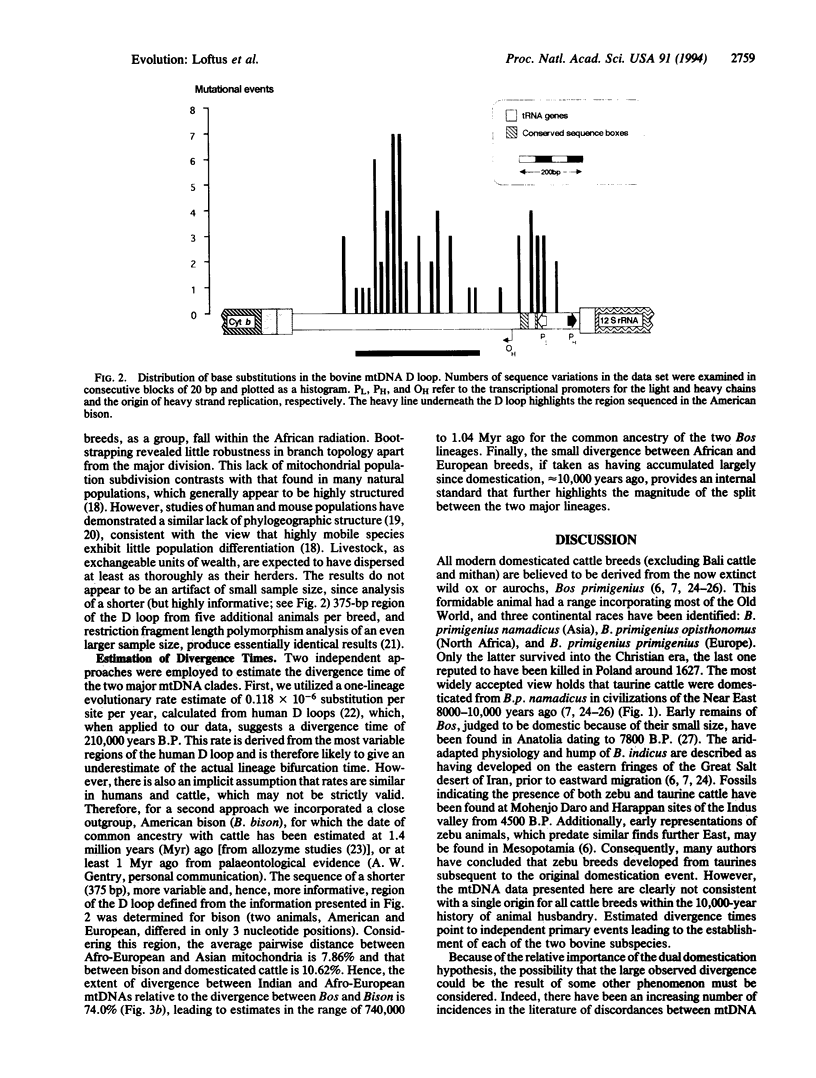
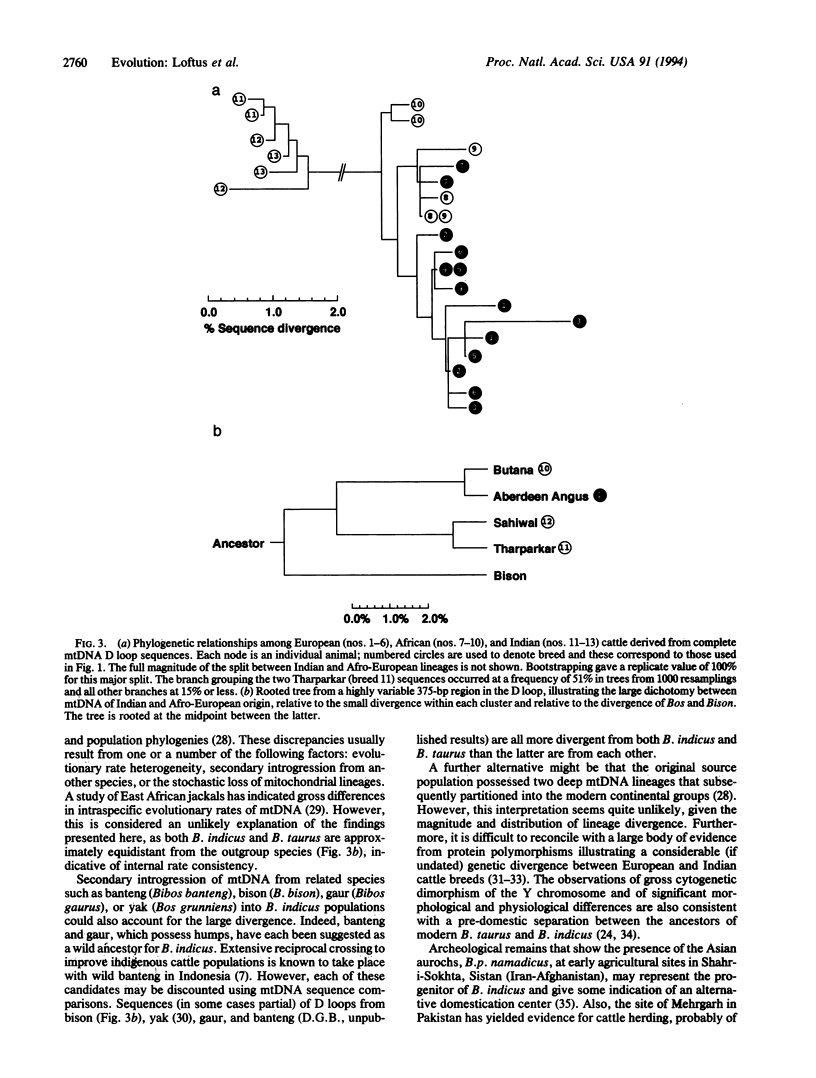
The origin and taxonomic status of domesticated cattle are controversial. Zebu and taurine breeds are differentiated primarily by the presence or absence of a hump and have been recognized as separate species (Bos indicus and Bos taurus). However, the most widely held view is that both types of cattle derive from a single domestication event 8000-10,000 years ago. We have examined mtDNA sequences from representatives of six European (taurine) breeds, three Indian (zebu) breeds, and four African (three zebu, one taurine) breeds. Similar levels of average sequence divergence were observed among animals within each of the major continental groups: 0.41% (European), 0.38% (African), and 0.42% (Indian). However, the sequences fell into two very distinct geographic lineages that do not correspond with the taurine-zebu dichotomy: all European and African breeds are in one lineage, and all Indian breeds are in the other. There was little indication of breed clustering within either lineage. Application of a molecular clock suggests that the two major mtDNA clades diverged at least 200,000, and possibly as much as 1 million, years ago. This relatively large divergence is interpreted most simply as evidence for two separate domestication events, presumably of different subspecies of the aurochs, Bos primigenius. The clustering of all African zebu mtDNA sequences within the taurine lineage is attributed to ancestral crossbreeding with the earlier B. taurus inhabitants of the continent.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bradley D. G., MacHugh D. E., Loftus R. T., Sow R. S., Hoste C. H., Cunningham E. P. Zebu-taurine variation in Y chromosomal DNA: a sensitive assay for genetic introgression in west African trypanotolerant cattle populations. Anim Genet. 1994 Feb;25(1):7–12. [PubMed] [Google Scholar]
- Di Rienzo A., Wilson A. C. Branching pattern in the evolutionary tree for human mitochondrial DNA. Proc Natl Acad Sci U S A. 1991 Mar 1;88(5):1597–1601. doi: 10.1073/pnas.88.5.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferris S. D., Sage R. D., Prager E. M., Ritte U., Wilson A. C. Mitochondrial DNA evolution in mice. Genetics. 1983 Nov;105(3):681–721. doi: 10.1093/genetics/105.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graml R., Ohmayer G., Pirchner F., Erhard L., Buchberger J., Mostageer A. Biochemical polymorphism in Egyptian Baladi cattle and their relationship with other breeds. Anim Genet. 1986;17(1):61–76. doi: 10.1111/j.1365-2052.1986.tb03188.x. [DOI] [PubMed] [Google Scholar]
- Higgins D. G., Bleasby A. J., Fuchs R. CLUSTAL V: improved software for multiple sequence alignment. Comput Appl Biosci. 1992 Apr;8(2):189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- Kidd K. K., Osterhoff D., Erhard L., Stone W. H. The use of genetic relationships among cattle breeds in the formulation of rational breeding policies: an example with South Devon (South Africa) and Gelbvieh (Germany). Anim Blood Groups Biochem Genet. 1974;5(1):21–28. doi: 10.1111/j.1365-2052.1974.tb01309.x. [DOI] [PubMed] [Google Scholar]
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980 Dec;16(2):111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- Lindberg G. L., Koehler C. M., Mayfield J. E., Myers A. M., Beitz D. C. Recovery of mitochondrial DNA from blood leukocytes using detergent lysis. Biochem Genet. 1992 Feb;30(1-2):27–33. doi: 10.1007/BF00554425. [DOI] [PubMed] [Google Scholar]
- Manwell C., Baker C. M. Chemical classification of cattle. 2. Phylogenetic tree and specific status of the Zebu. Anim Blood Groups Biochem Genet. 1980;11(3):151–162. doi: 10.1111/j.1365-2052.1980.tb01504.x. [DOI] [PubMed] [Google Scholar]
- Murphy G., Kavanagh T. Speeding-up the sequencing of double-stranded DNA. Nucleic Acids Res. 1988 Jun 10;16(11):5198–5198. doi: 10.1093/nar/16.11.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkins D., Jr Fauna of Catal Hüyük: evidence for early cattle domestication in Anatolia. Science. 1969 Apr 11;164(3876):177–179. doi: 10.1126/science.164.3876.177. [DOI] [PubMed] [Google Scholar]
- Saccone C., Pesole G., Sbisá E. The main regulatory region of mammalian mitochondrial DNA: structure-function model and evolutionary pattern. J Mol Evol. 1991 Jul;33(1):83–91. doi: 10.1007/BF02100199. [DOI] [PubMed] [Google Scholar]
- Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987 Jul;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Stoneking M., Sherry S. T., Redd A. J., Vigilant L. New approaches to dating suggest a recent age for the human mtDNA ancestor. Philos Trans R Soc Lond B Biol Sci. 1992 Aug 29;337(1280):167–175. doi: 10.1098/rstb.1992.0094. [DOI] [PubMed] [Google Scholar]
- Vigilant L., Pennington R., Harpending H., Kocher T. D., Wilson A. C. Mitochondrial DNA sequences in single hairs from a southern African population. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9350–9354. doi: 10.1073/pnas.86.23.9350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L., Stoneking M., Harpending H., Hawkes K., Wilson A. C. African populations and the evolution of human mitochondrial DNA. Science. 1991 Sep 27;253(5027):1503–1507. doi: 10.1126/science.1840702. [DOI] [PubMed] [Google Scholar]
- Wayne R. K., Meyer A., Lehman N., Van Valkenburgh B., Kat P. W., Fuller T. K., Girman D., O'Brien S. J. Large sequence divergence among mitochondrial DNA genotypes within populations of eastern African black-backed jackals. Proc Natl Acad Sci U S A. 1990 Mar;87(5):1772–1776. doi: 10.1073/pnas.87.5.1772. [DOI] [PMC free article] [PubMed] [Google Scholar]





