Abstract
Osteocytes regulate bone turnover and mineralization in chronic kidney disease (CKD). Since osteocytes are derived from osteoblasts, alterations in osteoblast function may regulate osteoblast maturation, osteocytic transition, bone turnover and skeletal mineralization. Thus, primary osteoblasts-like cells were cultured from bone chips obtained from 24 pediatric ESKD patients. RNA expression in cultured cells was compared to RNA expression in cells from healthy individuals, to RNA expression in the bone core itself, and to parameters of bone histomorphometry. Proliferation and mineralization rates of patient cells were compared to rates in healthy control cells. Associations were observed between bone osteoid accumulation, as assessed by bone histomorphometry, and bone core RNA expression of osterix, matrix gla protein, PTH receptor 1, and RANKL. Gene expression of osteoblast markers was increased in cells from ESKD patients and signaling genes including Cyp24A1, Cyp27B1, VDR, and NHERF1 correlated between cells and bone cores. Cells from patients with high turnover renal osteodystrophy proliferated more rapidly and mineralized more slowly than did cells from healthy controls. Thus, primary osteoblasts obtained from patients with ESKD retain changes in gene expression ex vivo that are also observed in bone cores specimens. Evaluation of these cells in vitro may provide further insights into the abnormal bone biology that persists, despite current therapies, in patients with ESKD.
Keywords: osteoblasts, bone histomorphometry, RNA, CKD
INTRODUCTION
Recent discoveries in the field of chronic kidney disease mineral and bone disorder (CKD-MBD) have led to a major shift in the way that bone metabolism and bone cell biology are viewed in the context of systemic disease (1–5). Indeed, osteocytes, the predominant cell type in mineralized bone, were long thought to be relatively quiescent cells, with a function limited to transduction of signals related to mechanical stress (6). These cells, which are derived from osteoblasts, are now known to secrete hormones which regulate mineral metabolism and control skeletal mineralization. CKD is associated with altered osteocytic protein expression (7) and abnormally elevated circulating levels of at least one osteocytic hormone—fibroblast growth factor 23 (FGF23)—have been linked to systemic disease, including cardiovascular disease(8), in CKD.
Much current interest is focused on the role of osteocytes in the control of CKD-MBD; however, the study of these human cells, both in healthy individuals and in the context of kidney disease, is limited by the fact that osteocytes are terminally differentiated cells that exist in a mineralized milieu, limiting their potential to be isolated and propagated in cell culture. However, osteocytes are differentiated from osteoblasts and primary human osteoblast-like cells have been isolated from humans with CKD and other bone disorders and have been evaluated in culture since the 1980s (9, 10). These cells may be induced to transition to an osteocytic phenotype in culture under mineralizing conditions (9, 10), thus allowing for the study of osteoblast maturation and osteocyte transition in both healthy individuals and in patients with CKD. Previous in vivo and in vitro studies have demonstrated that both circulating factors and those intrinsic to osteoblasts themselves contribute to differences in osteoblast characteristics between patients with different types of renal osteodystophy; ideed, primary osteoblasts obtained from patients with end-stage kidney disease have been shown to exhibit differences in proliferative characteristics ex vivo (11, 12) (13). However, the mechanisms underlying the changes in bone cell phenotype observed in CKD patients remain poorly understood. Therefore, we created and characterized a model system that may be used to study this abnormal phenotype, performing analysis of gene expression in whole bone biopsy samples and in cultures of osteoblast precursors derived from the same patients.
RESULTS
Study subjects
Twenty-four pediatric patients (14M, 10F) with end-stage kidney disease (ESKD) treated with maintenance dialysis were included in the study. Subjects were 17.1 ± 0.7 years of age, were primarily Hispanic (83%), and had varying types of renal osteodystrophy (Figure 1). Eight subjects were treated with active vitamin D sterols (doxercalciferol or calcitriol) at the time of biopsy; 16 subjects were not on active vitamin D sterols for at least 4 weeks prior to the biopsy. Biochemical parameters of the subjects at the time of the biopsy are displayed in Table 1 and bone histologic variables are displayed in Table 2.
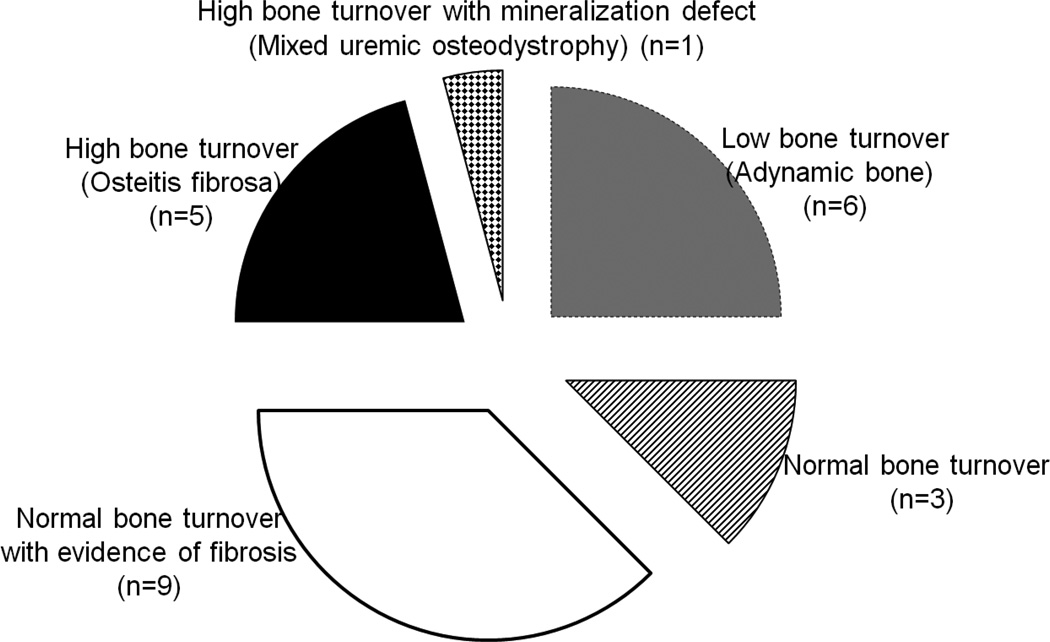
Figure 1.
Lesions of renal osteodystophy on bone biopsy represented in the cohort of ESKD patients evaluated for osteoblast-like cell and bone core RNA. Adynamic bone (low bone turnover) (grey): n=6; normal bone turnover (striped): n=3; normal bone turnover with fibrosis (white): n=9; osteitis fibrosa (high bone turnover) (black): n=5; mixed uremic osteodystrophy (high bone turnover with defective mineralization) (checkered): n=1.
Table 1.
Biochemical and growth parameters from 24 dialysis patients at the time of bone biopsy
| Parameter | Patients |
|---|---|
| Calcium (mg/dl) | 8.9 ± 0.2 |
| Phosphorus (mg/dl) | 6.5 ± 0.4 |
| 25(OH)vitamin D (ng/ml) | 20.4 ± 2.0 |
| Alkaline phosphatase (IU/l)* | 160 (95, 283) |
| PTH (pg/ml) * | 555 (357, 764) |
| FGF23 (RU/ml) * | 1826 (425, 4825) |
| Height Z score (SDS) * | −1.9 (−2.7, −0.2) |
| Weight Z score (SDS) * | −0.6 (−1.8, 0.8) |
Indicates that the value is displayed as the median (interquartile range) due to non-normal distribution. All other values are displayed as mean ± SEM.
Table 2.
Bone histomorphometry in 24 dialysis patients undergoing bone biopsy.
| Value | Normal range | |
|---|---|---|
| Turnover | ||
| Bone formation rate (BFR/BS) (µm3/um2/y) * | 25.6 (8.0, 63.9) | 8.0–73.4 |
| Eroded surface (ES/BS) (%) | 9.1 ± 1.0 | 0.5–4.3 |
| Mineralization | ||
| Osteoid volume (OV/BV) (%) | 4.5 ± 0.7 | 0.2–5.8 |
| Osteoid thickness (O.Th) (µm) | 8.9 ± 0.6 | 2.0–13.2 |
| Osteoid surface (OS/BS) (%) | 31.0 ± 3.2 | 4.3–37.0 |
| Osteoid maturation time (OMT) (d) | 12.1 (8.9, 14.9) | 1.2–11.5 |
| Mineralization lag time (MLT) (d) * | 32.8 (20.4, 56.2) | 2.3–63.8 |
| Volume | ||
| Bone volume (BV/TV) (%) | 32.0 ± 1.9 | 8.9–34.4 |
| Trabecular number (Tb.N) (#/mm2) | 2.1 ± 0.1 | 1.1–2.2 |
| Trabecular separation (Tb.Sp) (µm) | 347 ± 34 | 351–737 |
Indicates that the value is displayed as the median (interquartile range) due to non-normal distribution. All other values are displayed as mean ± SEM.
Bone core RNA, biochemical values, and bone histology
Consistent with the heterogeneous cellular composition of bone, both osteoblastic and osteocytic marker gene expression were readily detectable in bone cores from patients with ESKD, and, for the most part, the expression of these markers was increased compared to the healthy controls. These data suggest that the gene expression reflects abnormalities in osteoblast function that can be observed histologically. Differences in expression of osteoblast and osteocyte marker genes were not evident between patients with different types of bone histology (adynamic or normal bone turnover versus high bone turnover) or based on the presence or absence of active vitamin D sterol treatment and the tight range of ages precluded any assessment of the effect of age on gene expression. Therefore, data from the different patient groups were pooled for subsequent analysis.
When examining genes involved in vitamin D metabolism, expression of Cyp27B1 and VDR were found to be increased in cores from patients with ESKD with respect to controls, whereas expression of Cyp24A1 was decreased. Expression of FGFR1, the receptor for the classical actions of FGF23, was increased in patient biopsies compared with controls as were genes critical to PTH signaling, including the PTH receptor (PTHR1) and Na+/H+ Exchange Regulatory Co-Factor (NHERF1), a scaffold protein that interacts with the PTHR. Moreover, expression of osteocyte marker genes was significantly increased in patient samples (Table 3).
Table 3.
Gene expression, expressed as fold of expression in healthy controls, from bone cores and primary human osteoblasts from 24 dialysis patients undergoing bone biopsy. Data are expressed as medians (IQ ranges). A significant (p<0.05) difference in gene expression in CKD samples relative to healthy controls is denoted by an asterisk. Bold font denotes significant correlations between cores and cells.
| Core | Cells | Correlation between gene expression in cores and cells |
|
|---|---|---|---|
| Osteoblast markers | |||
| OSX | 8.84 (4.74, 13.84) * | 2.93 (1.01, 5.20) * | r=0.30, p=0.18 |
| Runx2 | 3.36 (2.53, 5.49) * | 2.61 (1.79, 3.40) * | r=0.30, p=0.18 |
| Col1A1 | 3.83 (3.28, 4.37) * | 1.37 (1.10, 1.70) * | r=0.34, p=0.12 |
| BGLAP | 7.91 (3.92, 10.44) * | 1.13 (0.56, 2.03) | r=0.19, p=0.39 |
| OPG | 4.23 (3.11, 7.08) * | 1.38 (0.64, 1.92) * | r=0.44, p=0.04 |
| ALPL | 5.15 (3.21, 6.41) * | 2.96 (1.79, 5.59) * | r=0.03, p=0.90 |
| MGP | 3.23 (1.69, 4.07) * | 2.35 (1.24, 3.65) * | r=0.18, p=0.43 |
| BMP2 | 2.93 (1.81, 5.14) * | 2.46 (1.74, 4.22) * | r= − 0.06, p=0.80 |
| Cell signaling | |||
| Cyp24A1 | 0.04 (0.01, 0.43) * | 1.28 (0.45, 3.69) | r=0.42, p=0.05 |
| Cyp27B1 | 2.45 (1.66, 4.13) * | 0.96 (0.77, 1.75) | r=0.73, p=0.0001 |
| VDR | 2.49 (2.07, 3.88) * | 2.79 (1.98, 3.97) * | r=0.45, p=0.04 |
| FGFR1 | 4.33 (2.52, 6.05) * | 1.68 (1.13, 2.09) * | r=0.49, p=0.02 |
| PHEX | 6.46 (4.42, 12.48) * | 0.99 (0.70, 1.60) | r=0.36, p=0.10 |
| RANKL | 6.50 (4.46, 12.08) * | 1.63 (0.24, 2.98) * | r= − 0.07, p=0.75 |
| NHERF1 | 1.04 (0.82, 1.65) | 2.42 (1.83, 3.19) * | r=0.64, p=0.001 |
| PTHR1 | 8.25 (4.91, 11.08) * | 4.77 (1.83, 6.73) * | r= − 0.18, p=0.43 |
| IGF1 | 1.60 (1.19, 2.51) * | 4.14 (1.96, 9.15) * | r=0.47, p=0.03 |
| Osteocyte markers | |||
| DMP 1 | 10.06 (5.21, 16.32) * | 0.96 (0.53, 1.04) | r=0.17, p=0.46 |
| FGF23 | 3.88 (1.55, 8.95) * | 1.10 (0.89, 1.22) | r= − 0.06, p=0.80 |
| MEPE | 6.29 (4.38, 11.62) * | 3.82 (2.22, 9.86) * | r=0.04, p=0.85 |
| SOST | 4.15 (2.77, 9.02) * | Not detected | NA |
Consistent with the previously observed correlation between plasma concentrations of FGF23, bone protein FGF23, and bone mineralization (14), bone core FGF23 RNA was inversely related to circulating alkaline phosphatase values (r= − 0.53, p=0.01), osteoid thickness (O.Th) (r= − 0.55, p=0.01, Figure 2), osteoid volume (OV/BV) (r= − 0.53, p=0.01), and mineralization lag time (MLT) (r= − 0.47, p=0.04). Osteocyte marker gene expression was markedly increased in bone cores of patients as compared to control bone. Measurements of osteoblast surface (Ob.S/BS) also correlated directly with multiple osteoblastic markers (osterix: r=0.53, p=0.01; matrix gla protein r=0.46, p=0.03; and type 1 collagen: r=0.55, p=0.01) as well as with signaling markers (PTH receptor 1 (PTHR1): r=0.51, p=0.02 and receptor activator of nuclear factor kappa-B ligand (RANKL): r=0.46, p=0.03). Vitamin D sterol therapy did not modify the correlations between bone core RNA and bone histomorphometry. No correlations were observed between bone formation rate and osteoblastic or osteocytic marker gene expression, either before or after normalizing bone core gene expression by osteoblast surface.
Figure 2.
Relationship between FGF23 RNA expression in bone cores and osteoid thickness (O.Th), as determined by bone histomorphometry.
Bone cell RNA, biochemical values, and bone histology
To determine whether primary cultures of ESKD patient-derived osteoblastic cells could serve as a model system for studying changes in variations in osteoblast and osteocyte biology associated with this disease, RNA was isolated from osteoblastic cells derived from bone biopsy samples and gene expression of osteoblast and osteocyte markers was assessed relative to expression in normal control cells. For the most part, expression of osteoblast marker genes was up-regulated in confluent cultures of primary osteoblasts obtained from patients as compared to healthy control cells (Figure 3). However, expression of osteocalcin (bone gamma-carboxyglutamic acid-containing protein, BGLAP) was not increased in these confluent cultures, demonstrating their early stage of osteoblastic differentiation (Figure 4). Interestingly, whereas expression of Cyp24a1 was decreased compared to controls in the bone cores, expression of this gene tended to increase in the cultured cells. Conversely, expression of Cyp27B1, which was increased in bone cores, was unchanged in the isolated cells. Consistent with the source of cells—i.e. of an osteoblast phenotype—PTHR1 expression from primary osteoblasts correlated with osteoblast surface on bone histomorphometry (r= −0.53, p<0.01). As expected, osteocytic gene expression was low in confluent cultures from both healthy controls and from ESKD patients and no correlations were observed between expression of these genes and parameters of bone histomorphometry. Unlike the bone cores, correlations between cell RNA and circulating biochemical values were not observed. Neither therapy with active vitamin D sterols nor subject age altered the relationship between cellular gene expression and bone histomorphometry.
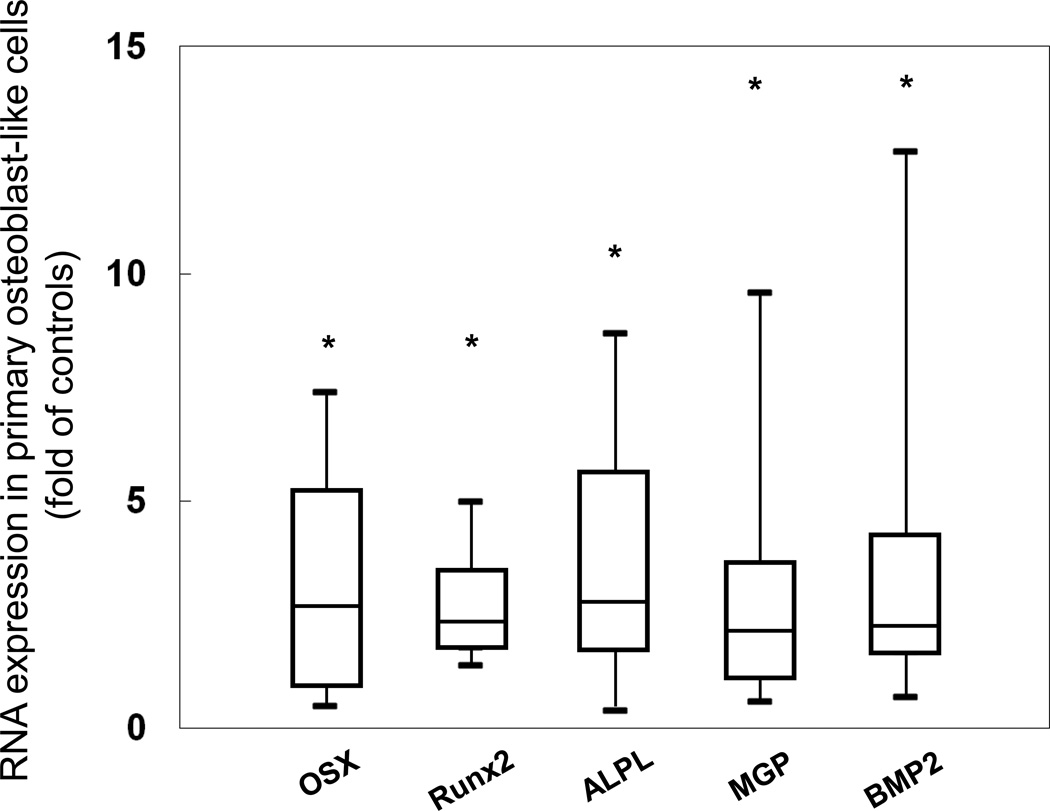
Figure 3.
Gene expression of osteoblast markers in primary osteoblasts from ESKD patients. Values are expressed as a multiple of (fold of) the expression found in healthy control cells. Box plots indicate the medians (interquartile ranges); the 5th and 95th percentiles are denoted by the lower and upper whiskers, respectively. The asterisks denote significant (p<0.001) differences from healthy controls.
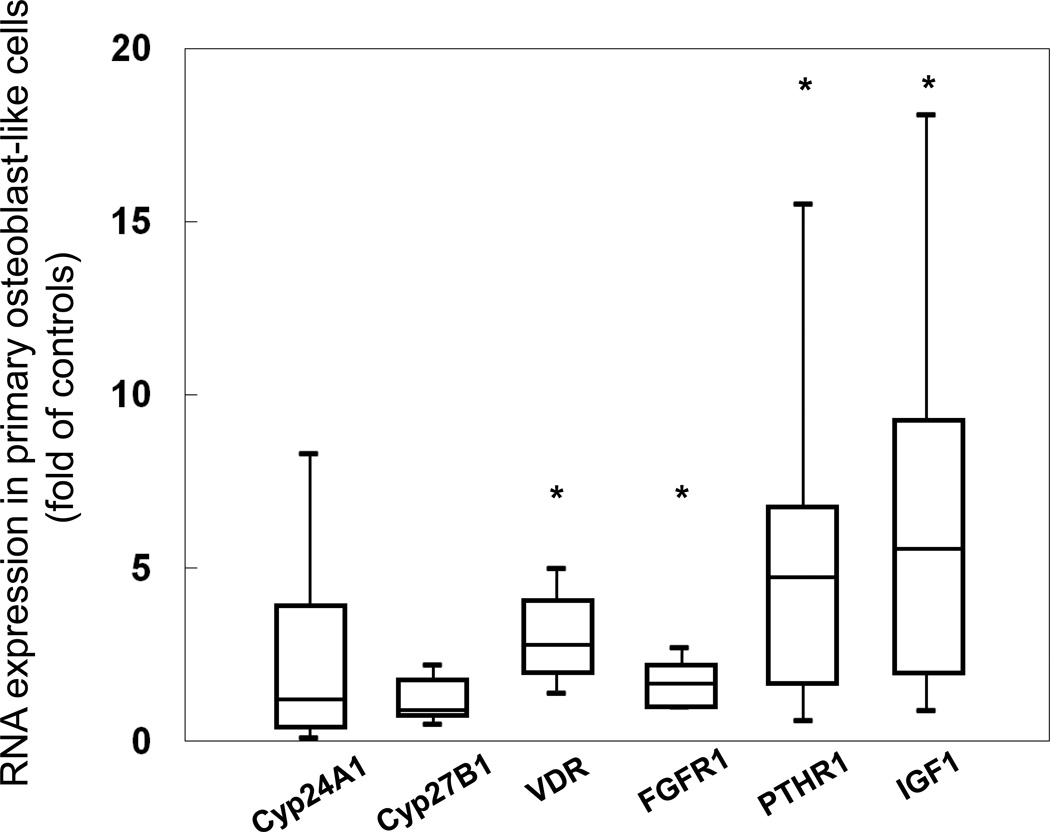
Figure 4.
Gene expression of cell signaling genes in primary osteoblasts from CKD patients. Values are expressed as a multiple of (fold of) the expression found in normal control cells. Box plots indicate the medians (interquartile ranges); the 5th and 95th percentiles are denoted by the lower and upper whiskers, respectively. The asterisks denote significant (p<0.001) differences from healthy controls.
In vitro osteoblast differentiation, mineralization, and proliferation
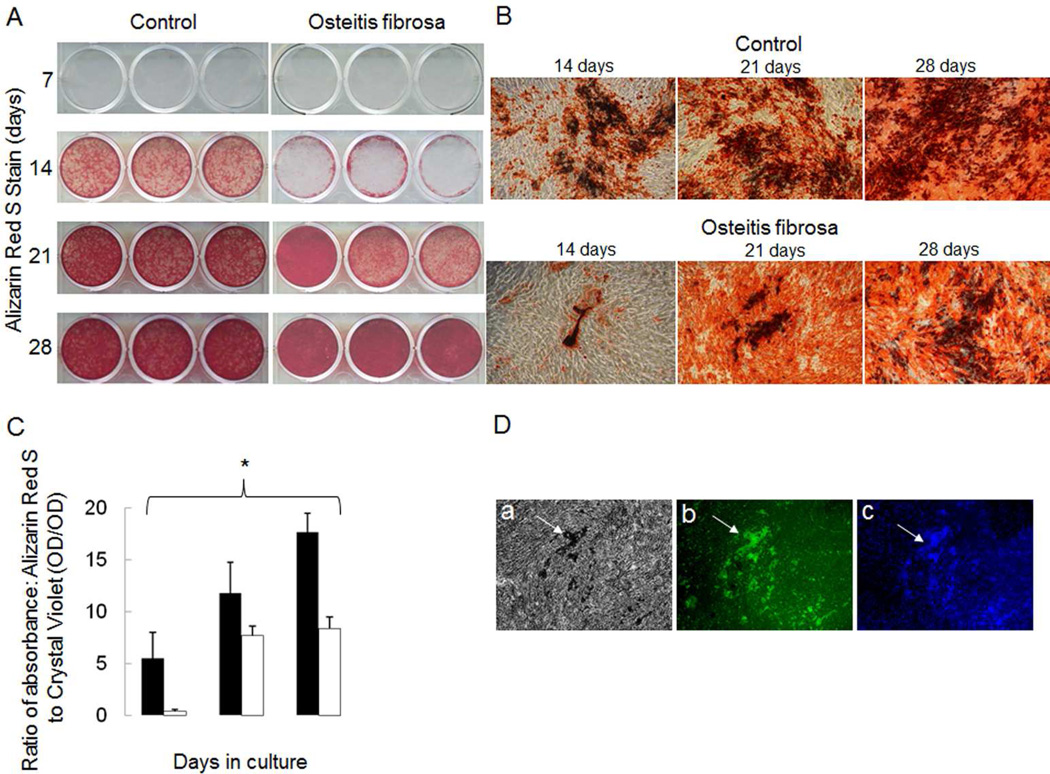
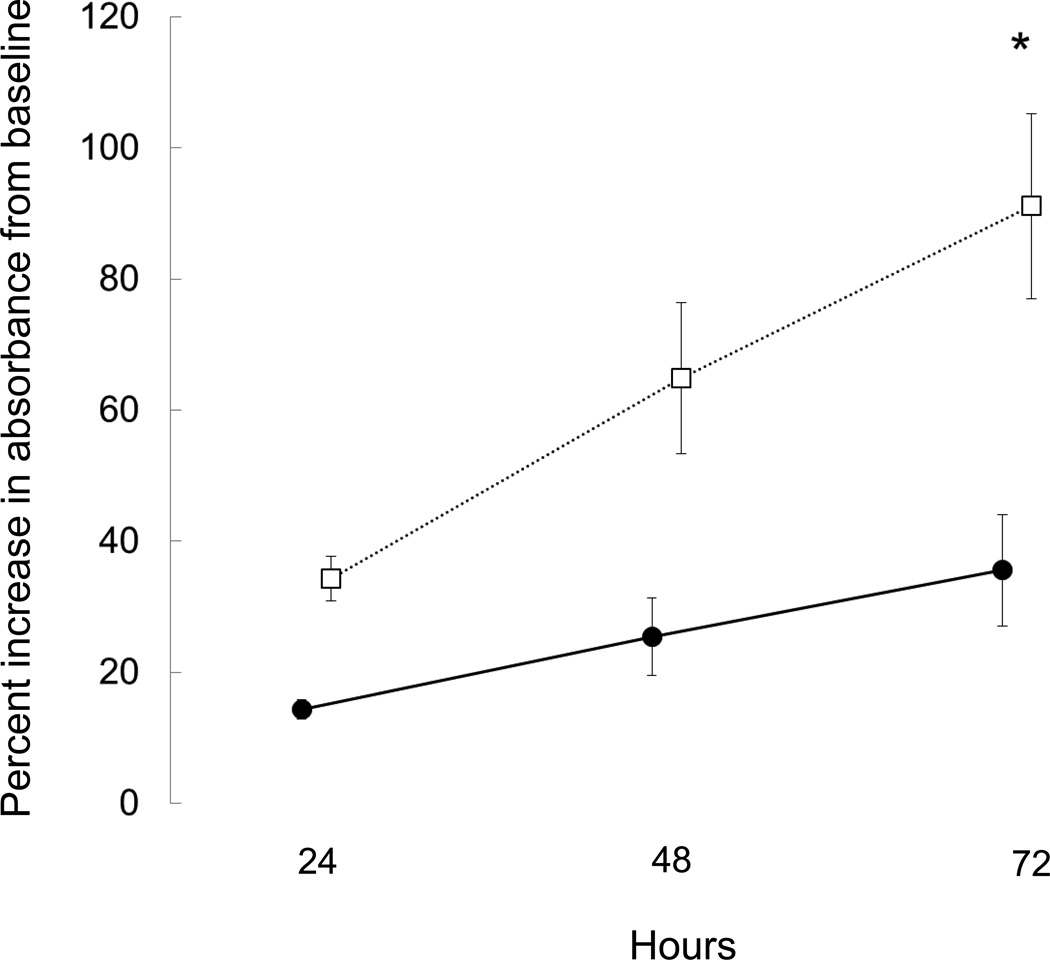
To confirm that the confluent cultures of osteoblastic cells possessed the ability to grow, differentiate and deposit mineralized matrix and to assess whether changes in gene expression observed in cells from ESKD patients were related to an altered phenotype, proliferation and mineralization assays were performed using osteoprogenitors from 3 healthy controls and from 3 patients. Patient cells were chosen from one representative subgroup of renal bone disease, namely high turnover renal osteodystrophy. The time from when the biopsies were performed until the cells had reached confluence was affected by the number and size of the bone chips obtained from each biopsy specimen; thus, progeny of the cells were plated at equal density and proliferation and mineralization characteristics evaluated in these progeny. The results demonstrated that the number of proliferating cells from healthy controls increased 35.6 ± 8.5% while cells from ESKD patients with high bone turnover plated at the same density grew by 91.1 ± 14.1% (p<0.05 from controls) by 72 hours (Figure 5). Cells from healthy controls maintained in an osteoblast differentiation cocktail containing ascorbic acid, β-glycerolphosphate, and dexamethasone began to mineralize by week 2 in culture while mineralization was delayed in cells from the ESKD patients (Figure 6). Immunocytochemistry for osteocalcin was positive in mineralizing cells, further confirming that the cultured cells were of an osteoblast lineage (Figure 5).
Figure 5.
Mineralization rates in cells from dialysis patients with high turnover renal osteodystrophy as compared to those from healthy controls. A) Alizarin Red S staining of primary human osteoblast-like cells from healthy controls and dialysis patients with high turnover renal osteodystrophy grown under mineralizing conditions (10 mM β-glycerolphosphate and 100 µg/ml ascorbic acid) for 7, 14, 21 and 28 days. B) Alizarin red stained mineralized nodules under light microscopy (40x) at 14, 21, and 28 days. C) Mineralization of primary osteoblast-like cells, as quantified by the ratio of absorbance of Alizarin Red S to Crystal violet (i.e. ratio of mineral content to cells), occurs more rapidly in cells from healthy controls (black bars) than in dialysis patients with high turnover renal osteodystrophy (open bars). The asterisk denotes significant (p<0.01) difference in Alizarin Red S to Crystal Violet absorbance ratios between dialysis patients with high turnover renal osteodystrophy and healthy controls. D) Primary osteoblast-like cells from healthy controls after 28 days in mineralizing conditions a) display mineralized nodules under light microscopy and b) express osteocalcin (green color) as detected by immunofluorescence. DAPI staining (c) confirms that osteocalcin expression localizes to living cells at the site of the mineralized nodule formation.
Figure 6.
Proliferation of primary human osteoblast-like cells as assessed by MTS incorporation. Data are expressed as percentage increase in absorbance from baseline. A higher proliferation rate was observed in dialysis patients with high turnover renal osteodystrophy (open symbols) than in controls (closed symbols) (p<0.01). The asterisk denotes a significant (p<0.01) difference in proliferation rates between groups.
Bone core RNA and bone cell RNA
Correlations between gene expression in cores and cells are displayed in Table 3. Most notably, genes involved in vitamin D signaling correlated well between RNA obtained from bone itself and RNA obtained from cultured primary osteoblasts (Cyp24A1: r=0.42, p=0.05; Cyp27B1: r=0.73, p=0.0001; and VDR: r=0.45, p=0.04). Similarly, selected elements of PTH signaling (NHERF1: r=0.64, p=0.001) and growth hormone signaling pathways (IGF1: r=0.47, p=0.03) correlated between whole bone and cultured primary osteoblast RNA. As expected, genes specific to an osteocytic phenotype did not correlate between whole bone cores, whose cellular make-up consists of a high percentage of osteocytes, and cultured primary osteoblasts, reflecting the early differentiation status of the confluent cultures. In this small sample, ongoing in vivo therapy with active vitamin D sterols at the time of bone biopsy did not affect the relationship between bone core and primary osteoblast cell RNA expression.
DISCUSSION
The current study demonstrates that primary human osteoblast-like cells can be cultured from bone fragments obtained from pediatric ESKD and healthy control subjects at the time of bone biopsy. These cells express osteocalcin and can be induced to mineralize, thus confirming an osteoblastic phenotype. Cells from these adolescent patients with ESKD have increased expression of genes crucial to the mineralization process and these changes in gene expression persist in culture as the cells divide and grow to confluence. Expression of osteoblastic genes in vitro correlate with osteoblastic gene expression from bone cores themselves and this expression is related to histomorphometric indices.
Currently, much interest and research are focused on the role of different osteocytic proteins in CKD and its complications. We have previously demonstrated that osteocytes in pediatric patients with all stages of CKD express excessive amounts of FGF23, a protein which appears to play in important role in the development of secondary hyperparathyroidism (7). These individuals display defects in skeletal mineralization, even in very early CKD when circulating concentrations of calcium, phosphorus, PTH and 1,25(OH)2vitamin D are within the normal range (14), suggesting that altered bone cell biology may contribute to these abnormalities. The current results demonstrate that abnormalities in RNA expression persist in primary human osteoblast-like cells from pediatric ESKD patients removed from the uremic milieu, as do abnormalities in proliferation and mineralization characteristics. Since abnormal expression of osteocytic proteins such as FGF23 has been linked to systemic consequences, such as cardiovascular disease, progressive renal dysfunction, and mortality in patients with CKD (8, 15), and since osteocytes are derived from osteoblasts, further characterization of the maturation and mineralization characteristics of these primary osteoblast-like cells may yield valuable insights into the pathogenesis of both bone and cardiovascular disease in CKD.
Previous in vivo and in vitro data have demonstrated that circulating factors contribute to differences in osteoblast proliferation; indeed, IGF1 and PTH both stimulate bone formation rates while circulating inhibitors of osteoblast mitogenesis have also been identified in uremic serum (16). It has also been suggested that factors intrinsic to osteoblasts themselves may contribute to differences in osteoblast characteristics between patients (11–13). The current study expands these findings, demonstrating that cellular gene expression of both early and later osteoblast markers from primary human osteoblast-like cells are increased in cells obtained from adolescent dialysis patients when compared to control cells from healthy adolescents. Osteocytic gene expression, however, appears to be very low in these cell cultures, demonstrating that the predominant cell type in ossified bone—namely osteocytes—do not migrate and/or survive in these cell culture conditions. However, osteocytes are derived from mature osteoblasts and since abnormal expression of osteocytic proteins such as FGF23 have been linked to systemic consequences such as cardiovascular disease, progressive renal dysfunction, and mortality in patients with CKD (8, 15, 17), the ability to use these primary osteoblast-like cells, which are able to mineralize, to study potential abnormalities in osteoblast to osteocyte transition may greatly improve understanding of the origins of not only bone, but also cardiovascular disease in CKD.
Although some osteoblastic markers in cells from adolescent CKD patients appear upregulated—including factors required for osteoblast maturation—expression of signaling genes were not increased. Induction—such as by vitamin D sterol therapy—might change gene expression in bone cores that is not carried over into subsequent generations of primary osteoblasts and circulating factors—including 1,25(OH)2vitamin D, 25(OH)vitamin D, and IGF-1—may similarly affect the bone cores in a way that is not reflected in primary osteoblasts and their progeny once removed from the uremic milieu. This may explain why many genes evaluated in the current study did not correlate directly between cells and bone cores. However, it is interesting to note that expression of factors involved in vitamin D (Cyp24 and Cyp27), PTH (PTHR1), and FGF23 (FGFR1) signaling did correlate directly between cells and their respective bone cores, suggesting that expression of these genes were not primarily influenced by circulating factors in the uremic milieu.
In the current study, expression of osteocytic genes, as well as of osteoblastic genes, was markedly upregulated in bone core tissue itself—an expected finding given that osteocytes comprise the majority of cells in bone. The increased amount of PTHR1 mRNA found in bone cores of dialysis patients appears to differ from previous data by Picton and colleagues (18) in which in situ hybridization noted a decrease in PTHR1 mRNA overlying osteoblasts in bone from patients with ESKD. However, the current data reflect RNA from whole bone, including bone marrow and osteocytes, in addition to the osteoblasts evaluated by Picton et al. Since PTHR1 is expressed on osteocytes, the most abundant cell-type in mineralized bone, the current findings may reflect increased expression of the PTH receptor on these cells specifically (19). The inverse correlations between bone core FGF23 RNA and osteoid accumulation are consistent with previous reports documenting the same relationship between osteocytic FGF23 protein expression, circulating FGF23 levels and osteoid accumulation in dialysis patients (3, 20). The reason behind this inverse relationship is not clear and warrants further investigation since it is not possible to tell from cross-sectional studies whether this represents a positive effect of FGF23 expression on skeletal mineralization or whether FGF23 expression is a reflection of the quality of mineral content in the surrounding bone, as suggested by recent studies by David et al (21). The mechanism by which FGF23 mRNA is increased in bone in CKD remains poorly understood. However, Martin et al suggest that stimulation of FGF23 may occur via FGFR1 (22), findings which are consistent with the current data in which FGF23 and FGFR1 expression are simultaneously increased in CKD bone.
Since the primary cells obtained from the current method were polyclonal, their RNA expression reflects that of osteoblasts from multiple precursors. This observation is important as osteocytes, the mature form of osteoblasts, have been found to be a heterogeneous cell type in patients with CKD, with osteocytes in different areas of both trabecular and cortical bone expressing different quantities of proteins such as FGF23 and DMP1 (20). Whether differences in osteocytic phenotype is related to cell maturity or is related to more fundamental differences in cell origin or environment is unknown, but warrants further investigation.
In summary, primary osteoblast-like cells obtained from patients with ESKD retain changes in gene expression, as well as in proliferation and mineralization characteristics, ex-vivo. These cells provide a novel in vitro model by which critical aspects of CKD-MBD, including changes in osteoblast proliferation and maturation and the effects of different circulating hormones on cells of this lineage, may be evaluated. In addition to advancing current understanding as to the pathogenesis of renal osteodystrophy, this model may also provide insights into the pathogenesis of systemic complications, including progressive renal dysfunction and cardiovascular disease, both of which have been linked to increased bone protein expression in the context of CKD.
METHODS
Patients
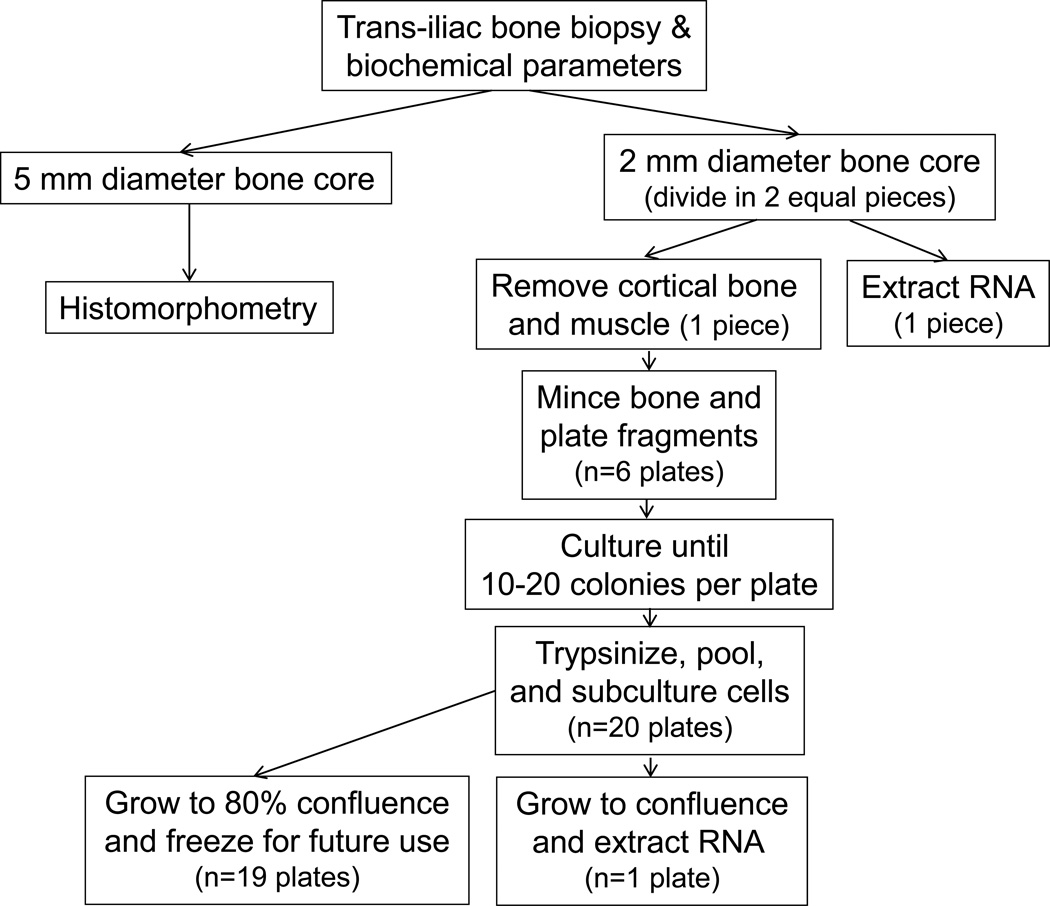
Sequential pediatric patients with ESKD who were treated with chronic cycling peritoneal dialysis (CCPD) and who underwent trans-iliac bone biopsy at UCLA between January 2008 and March 2011 were included in the study (Figure 7). Patients with metabolic bone disease other than renal osteodystrophy, those who had undergone parathyroidectomy within the preceding year, and those who had been treated with immunosuppressive therapy or recombinant human growth hormone within the previous year were excluded from study participation. All subjects underwent double tetracycline labeling prior to the bone biopsy. At the time of the bone biopsy, 2 bone cores were obtained. The first core, measuring 5 mm in diameter, was placed in 70% ethanol and then prepared, sectioned, and evaluated for bone histomorphometric parameters. The second core, which measured 2 mm in diameter, was collected in Dulbecco’s Modified Eagle’s Medium (DMEM). In the laboratory, this second core was divided into two equal pieces. The first piece was placed directly in TRIzol (Invitrogen, Carlsband, CA, USA) and frozen at −80°C. Muscle and cortical bone were removed from the second piece which was then minced and placed in culture (see below). Biochemical determinations of blood concentrations of calcium, phosphorus, PTH, alkaline phosphatase, 25(OH)vitamin D, and FGF23 were obtained at the time of bone biopsy. Serum calcium, phosphorus, albumin, creatinine, and alkaline phosphatase values were assessed using an Olympus AU5400 analyzer (Olympus America Incorporated, Center Valley, PA). PTH concentrations in EDTA plasma were measured by the 1st generation immunometric assay (ImmutopicsR, San Clemente, California, normal range: 10–65 pg/ml). FGF23 levels were determined in EDTA plasma by a 2nd generation C-terminal assay (ImmutopicsR, San Clemente, California). The normal range (<100 RU/mL) was defined by Isakova et al. (23) and confirmed in a cohort of 42 healthy children age 12 ± 4 years (24). 25(OH)vitamin D and 1,25(OH)2vitamin D levels were measured by radioimmunoassay (25).
Figure 7.
Flowchart of the study.
Control samples for the PCR data were obtained from 4 subjects, ages 14, 15, 16 and 38 years, undergoing surgery for idiopathic scoliosis or for maxillofacial surgery requiring grafting. All control subjects were of normal statural height with normal body mass index (height Z score SDS: −0.1 (−0.3, 0.8); weight Z score SDS: 0.7 (0.2, 1.1)), normal kidney function, and normal acid/base parameters. All samples were of trabecular bone obtained from either the hip or spine. All of the control fragments were obtained in the operating room and were immediately placed in preservative solutions by the operating surgeons. In the laboratory, the cortical bone was carefully removed from all control samples, similar to its removal from patient samples. Two of the controls were Hispanic and 3 were female. Due to their similarities in age to the study population, the three controls aged 14 to 16 years were selected for the proliferation and mineralization studies. The study was approved by the UCLA Institutional Review Board and consent/assent was obtained from all patients and their parents.
Isolation of RNA from bone cores
Total RNA was extracted from bone cores using TRIzol according to the manufacturer’s instructions. The pellet was then resuspended in RNase free water and measurement of RNA yield was performed using a NanoDrop 1000A Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Reverse transcription reaction was performed for cDNA synthesis using High Capacity cDNA RT kit (Applied Biosystems, Foster City, CA 94404) according to the manufacturer’s instructions. Samples were then stored at −80°C. Quantitative real-time PCR (qPCR) amplification was performed using QuaniTect® Probe PCR kit (QIAGEN Inc, Helden Germany). Taqman assays were used to quantify expression of genes listed in Table 4. Sequence-specific probes are listed in Table 4. The housekeeping gene GAPDH was analyzed in every sample and used as an endogenous control for data normalization. Samples were assayed in triplicate. Relative quantification studies of threshold cycle (Ct) were performed with Sequence Detector software (Applied Biosystems, Foster City, CA). Relative gene expression in patient samples was expressed relative to the healthy controls.
Table 4.
Osteoblast, osteocyte, and cell-signaling genes assessed in cells and cores.
| Protein name | Number | Gene name |
|---|---|---|
| Osteoblast markers | ||
| Osterix | Hs01866874_s1 | OSX |
| Cbfa1/Runx2 | Hs00231692_m1 | RUNX2 |
| Collagen Type 1 | Hs00164004_m1 | Col1A1 |
| Bone γ-Carboxyglutamate (gla) Protein | Hs01587814_g1 | BGLAP |
| Alkaline phosphatase | Hs00758162_m1 | ALPL |
| Matrix gla protein | Hs00179899_m1 | MGP |
| Bone morphogenic protein 2 | Hs00154192_m1 | BMP2 |
| Cell signaling markers | ||
| Cyp24 | H200167999_m1 | Cyp24A1 |
| Cyp27 | Hs00168017_m1 | Cyp27B1 |
| Vitamin D receptor | Hs01045840_m1 | VDR |
| FGF receptor 1 | Hs00241111_m1 | FGFR1 |
| Receptor activator of nuclear factor kappa-B ligand | Hs00243522_m1 | RANKL |
| Na(+)/H(+)-exchanger regulatory factor1 | Hs00188594_m1 | NHERF1 |
| Parathyroid hormone receptor 1 | Hs00174895_m1 | PTHR1 |
| Osteoprotegerin | Hs00900358_m1 | OPG |
| Insulin growth factor 1 | Hs01547656_m1 | IGF1 |
| Osteocyte markers | ||
| Dentin matrix protein 1 | Hs00189368_m1 | DMP1 |
| Fibroblast growth factor 23 | Hs00221003_m1 | FGF23 |
| Matrix extracellular phosphoglycoprotein | Hs00220237_m1 | MEPE |
| PHEX | Hs01011692_m1 | PHEX |
| Sclerostin | Hs00228830_m1 | SOST |
Isolation and propagation of human osteoblast-like cells
Trabecular bone fragments were obtained from the second bone biopsy core, as described above, and were prepared according to the procedure adapted from Dillon et al (26). Briefly, bone fragments were washed with fresh media containing antibiotic and antimycotic (Anti-Anti) (Gibco, Carlsbad, CA) upon arrival in the laboratory. The fragments were minced into very small pieces, divided into 6 tissue culture plates, and allowed to adhere for 2 hours in 5% CO2 at 37°C (Figure 7). DMEM supplemented with 20% FBS and ascorbic acid (100 ug/ml) was then added and the plates were incubated at 37°C. Once outgrowth of cells and their attachment to the plates were observed, the media was changed twice weekly until the formation of 10–20 cell colonies per plate was observed (1 to 4 weeks). The colonies of cell were trypsinized, pooled, and subcultured. When cells reached ~80% confluence, the contents of 19 plates were frozen in 20 ml of freezing media containing 10% DMSO and 30% FBS and placed in liquid nitrogen for future use. The remaining plate was allowed to grow to confluence prior to RNA extraction, as described above.
Mineralization matrix formation
In order to confirm the osteoblastic potential of the cells isolated from bone chips, an aliquot of frozen cells from the 3 controls and 3 patients with high turnover renal osteodystrophy were thawed and seeded into 12 well plates at a density of 1 × 104 cells per well. Cells were cultured in DMEM with 10% FBS. At confluence the media was changed to an osteoblast differentiation cocktail containing 10−8 M dexamethasone, 10 mM β-glycerol phosphate, and 100 µg/ml ascorbic acid for 1–4 weeks. The media was replaced twice weekly and the presence of mineralized nodules was assessed at weekly intervals using a 1% (w/v) solution of Alizarin Red S (pH 6.4) (Sigma-Aldrich, St. Louis MO); staining was quantified by extracting the dye from the cell layer using 10% acetic acid at 85°C for 30 minutes; the supernatant was analyzed at 405 nm. Cell number was estimated by staining parallel wells with 0.05% Crystal Violet; staining was quantified by extracting the dye from the cell layer by incubation in methanol for 30 min. The supernatant was analyzed at 570 nm. The quantity of Alizarin Red S was then normalized to that of Crystal Violet.
Immunocytochemistry
Cells were cultured on 8 chamber slides at a density of 1 × 104 cells per well. After 21 days of culture in osteoblast differentiation cocktail, cells were fixed with a 95% ethanol and 5% acetic acid for 10 minutes at −20°C. After washing with PBS, cells were permeabilized with 0.1% (v/v) Triton X-100 (Sigma) for 45 minutes and washed again with PBS. Nonspecific binding was blocked by incubating in 1:20 horse serum (Sigma) for 1 hour. Cells were then incubated with mouse anti-human osteocalcin (R&D Systems, Minneapolis MN) at 4°C overnight. After washing with PBS, cells were incubated with secondary antibody (donkey Alexa-Fluor-488) (Invitrogen, Carlsbad CA) for 1 hour at room temperature. Nuclear counterstaining was performed with mounting medium containing DAPI (Vector, Burlingame CA).
Proliferation
Proliferation was assessed by tetrazolium incorportation [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H–tetrazolium, MTS] assay, using the CellTiter96® AQueous One Cell Proliferation Assay (Promega, Fitchburg, WI) according to manufacturer’s instructions. In brief, a frozen aliquot of primary osteoblast-like cells from 3 healthy control subjects and 3 patients with high turnover renal osteodystrophy were thawed, grown to subconfluence and passaged 3 times. Cells were plated at 8,000 cells per well in 96 well plates. Viable cells were quantified by the addition of 20µl of CellTiter96® AQueous One Solution Reagent per well and analyzed by spectrophotometry at 490 nm.
Bone histomorphometry
Five mm diameter specimens of iliac crest were dehydrated in alcohol, cleared with xylene, and embedded in methylmethacrylate. Static histomorphometric parameters were evaluated in undecalcified 5 µm sections treated with Toluidine blue stain; tetracycline labeling was assessed in unstained 10 µm sections. Primary bone histomorphometric parameters were assessed in trabecular bone under 200× magnification using the OsteoMetricsR system (OsteoMetrics, Decatur, GA) by a histomorphometrist (RP). Mineralized bone was defined by dark blue staining areas; pale-blue seams at least 1.5 µm in width were included in measurements of osteoid. Derived indices were calculated as described by Parfitt et al (27). Normal parameters of bone histomorphometry were defined from bone biopsies obtained from a previously described control group of 31 pediatric patients with normal kidney function (mean age 12.4 ± 1.5 years, 71% male, 48% Caucasian and 26% Hispanic) who were undergoing elective orthopedic surgery (28).
Statistical analysis
Variables are reported as mean ± standard error or median (interquartile range). Skewed values were log-transformed prior to statistical analysis. Spearman correlation coefficients were used to express the relationship between bone histomorphometry and RNA expression for bone cores and cells. In select cases where distribution of both variables (or transformed values of those variables) was normal, Pearson correlation coefficients were also performed and the relationship between variables was displayed in a scatter plot. The Wilcoxon Signed Rank test was used to evaluate differences from healthy controls. Differences in proliferation rates between cells from healthy controls and CKD patients were assessed using a mixed model. All statistical analyses were performed using SAS software (SAS Institute Inc., Cary, NC) and all tests were two-sided. A probability of type I error less than 5% was considered statistically significant and ordinary p values are reported.
ACKNOWLEDGEMENTS
Sources of support: This work was supported in part by USPHS grants DK-67563, DK-35423, DK-080984 and CTSI grant UL1 TR-000124, funds from the UCLA Children’s Discovery and Innovation Institute, from the American Society of Nephrology Foundation’s Normal Siegel Research Scholar Award, and from the Casey Lee Ball Foundation.
Footnotes
DISCLOSURE
None of the authors of this paper have any financial interest in the information contained in this manuscript.
References
- 1.Shimada T, Hasegawa H, Yamazaki Y, Muto T, Hino R, Takeuchi Y, Fujita T, Nakahara K, Fukumoto S, Yamashita T. FGF-23 is a potent regulator of vitamin D metabolism and phosphate homeostasis. J.Bone Miner.Res. 2004;19:429–435. doi: 10.1359/JBMR.0301264. [DOI] [PubMed] [Google Scholar]
- 2.Sitara D, Razzaque MS, Hesse M, Yoganathan S, Taguchi T, Erben RG, Juppner H, Lanske B. Homozygous ablation of fibroblast growth factor-23 results in hyperphosphatemia and impaired skeletogenesis, and reverses hypophosphatemia in Phex-deficient mice. Matrix Biol. 2004;23:421–432. doi: 10.1016/j.matbio.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wesseling-Perry K, Pereira RC, Wang H, Elashoff RM, Sahney S, Gales B, Juppner H, Salusky IB. Relationship between plasma fibroblast growth factor-23 concentration and bone mineralization in children with renal failure on peritoneal dialysis. J Clin Endocrinol Metab. 2009;94:511–517. doi: 10.1210/jc.2008-0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sabbagh Y, Graciolli FG, O'Brien S, Tang W, Dos Reis LM, Ryan S, Phillips L, Boulanger J, Song W, Bracken C, Liu S, Ledbetter S, Dechow P, Canziani ME, Carvalho AB, Jorgetti V, Moyses RM, Schiavi SC. Repression of osteocyte Wnt/beta-catenin signaling is an early event in the progression of renal osteodystrophy. J Bone Miner Res. 2012;27:1757–1772. doi: 10.1002/jbmr.1630. [DOI] [PubMed] [Google Scholar]
- 5.Faul C, Amaral AP, Oskouei B, Hu MC, Sloan A, Isakova T, Gutierrez OM, Aguillon-Prada R, Lincoln J, Hare JM, Mundel P, Morales A, Scialla J, Fischer M, Soliman EZ, Chen J, Go AS, Rosas SE, Nessel L, Townsend RR, Feldman HI, St John Sutton M, Ojo A, Gadegbeku C, Di Marco GS, Reuter S, Kentrup D, Tiemann K, Brand M, Hill JA, Moe OW, Kuro OM, Kusek JW, Keane MG, Wolf M. FGF23 induces left ventricular hypertrophy. J Clin Invest. 2011;121:4393–4408. doi: 10.1172/JCI46122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonewald L. Osteocytes as multifunctional cells. J Musculoskelet Neuronal Interact. 2006;6:331–333. [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira RC, Juppner H, Azucena-Serrano CE, Yadin O, Salusky IB, Wesseling-Perry K. Patterns of FGF-23, DMP1, and MEPE expression in patients with chronic kidney disease. Bone. 2009;45:1161–1168. doi: 10.1016/j.bone.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Faul C, Amaral AP, Oskouei B, Hu MC, Sloan A, Isakova T, Gutierrez OM, Aguillon-Prada R, Lincoln J, Hare JM, Mundel P, Morales A, Scialla J, Fischer M, Soliman EZ, Chen J, Go AS, Rosas SE, Nessel L, Townsend RR, Feldman HI, St John Sutton M, Ojo A, Gadegbeku C, Di Marco GS, Reuter S, Kentrup D, Tiemann K, Brand M, Hill JA, Moe OW, Kuro OM, Kusek JW, Keane MG, Wolf M. FGF23 induces left ventricular hypertrophy. J Clin Invest. 2011;121:4393–4408. doi: 10.1172/JCI46122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gallagher JA, Beresford JN, McGuire MK, Ebsworth NM, Meats JE, Gowen M, Elford P, Wright D, Poser J, Coulton LA, et al. Effects of glucocorticoids and anabolic steroids on cells derived from human skeletal and articular tissues in vitro. Adv Exp Med Biol. 1984;171:279–291. [PubMed] [Google Scholar]
- 10.Beresford JN, Gallagher JA, Gowen M, Couch M, Poser J, Wood DD, Russell RG. The effects of monocyte-conditioned medium and interleukin 1 on the synthesis of collagenous and non-collagenous proteins by mouse bone and human bone cells in vitro. Biochim Biophys Acta. 1984;801:58–65. doi: 10.1016/0304-4165(84)90212-5. [DOI] [PubMed] [Google Scholar]
- 11.Marie PJ, Lomri A, de Vernejoul MC, Morieux C, Graulet AM, Gueris J, Llach F. Relationships between histomorphometric features of bone formation and bone cell characteristics in vitro in renal osteodystrophy. J Clin Endocrinol Metab. 1989;69:1166–1173. doi: 10.1210/jcem-69-6-1166. [DOI] [PubMed] [Google Scholar]
- 12.Marie PJ, de Vernejoul MC. Proliferation of bone surface-derived osteoblastic cells and control of bone formation. Bone. 1993;14:463–468. doi: 10.1016/8756-3282(93)90180-i. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez MC, Bajo MA, Selgas R, Mate A, Sanchez-Cabezudo MJ, Lopez-Barea F, Esbrit P, Martinez ME. Cultures of human osteoblastic cells from dialysis patients: influence of bone turnover rate on in vitro selection of interleukin-6 and osteoblastic cell makers. Am J Kidney Dis. 2001;37:30–37. doi: 10.1053/ajkd.2001.20574. [DOI] [PubMed] [Google Scholar]
- 14.Wesseling-Perry K, Pereira RC, Tseng CH, Elashoff R, Zaritsky JJ, Yadin O, Sahney S, Gales B, Juppner H, Salusky IB. Early skeletal and biochemical alterations in pediatric chronic kidney disease. Clin J Am Soc Nephrol. 2012;7:146–152. doi: 10.2215/CJN.05940611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fliser D, Kollerits B, Neyer U, Ankerst DP, Lhotta K, Lingenhel A, Ritz E, Kronenberg F, Kuen E, Konig P, Kraatz G, Mann JF, Muller GA, Kohler H, Riegler P. Fibroblast growth factor 23 (FGF23) predicts progression of chronic kidney disease: the Mild to Moderate Kidney Disease (MMKD) Study. J.Am.Soc.Nephrol. 2007;18:2600–2608. doi: 10.1681/ASN.2006080936. [DOI] [PubMed] [Google Scholar]
- 16.Andress DL, Howard GA, Birnbaum RS. Identification of a low molecular weight inhibitor of osteoblast mitogenesis in uremic plasma. Kidney Int. 1991;39:942–945. doi: 10.1038/ki.1991.118. [DOI] [PubMed] [Google Scholar]
- 17.Gutierrez OM, Mannstadt M, Isakova T, Rauh-Hain JA, Tamez H, Shah A, Smith K, Lee H, Thadhani R, Juppner H, Wolf M. Fibroblast growth factor 23 and mortality among patients undergoing hemodialysis. N.Engl.J.Med. 2008;359:584–592. doi: 10.1056/NEJMoa0706130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picton ML, Moore PR, Mawer EB, Houghton D, Freemont AJ, Hutchison AJ, Gokal R, Hoyland JA. Down-regulation of human osteoblast PTH/PTHrP receptor mRNA in end-stage renal failure. Kidney Int. 2000;58:1440–1449. doi: 10.1046/j.1523-1755.2000.00306.x. [DOI] [PubMed] [Google Scholar]
- 19.Lupp A, Klenk C, Rocken C, Evert M, Mawrin C, Schulz S. Immunohistochemical identification of the PTHR1 parathyroid hormone receptor in normal and neoplastic human tissues. Eur J Endocrinol. 2010;162:979–986. doi: 10.1530/EJE-09-0821. [DOI] [PubMed] [Google Scholar]
- 20.Pereira RC, Jüppner H, Azucena-Serrano CE, Yadin O, Salusky IB, Wesseling-Perry K. Patterns of FGF-23, DMP1, and MEPE expression in patients with chronic kidney disease. Bone. 2009;45:1161–1168. doi: 10.1016/j.bone.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.David V, Dai B, Martin A, Huang J, Han X, Quarles LD. Calcium regulates FGF-23 expression in bone. Endocrinology. 2013;154:4469–4482. doi: 10.1210/en.2013-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin A, Liu S, David V, Li H, Karydis A, Feng JQ, Quarles LD. Bone proteins PHEX and DMP1 regulate fibroblastic growth factor Fgf23 expression in osteocytes through a common pathway involving FGF receptor (FGFR) signaling. FASEB J. 2011;25:2551–2562. doi: 10.1096/fj.10-177816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isakova T, Wahl P, Vargas GS, Gutierrez OM, Scialla J, Xie H, Appleby D, Nessel L, Bellovich K, Chen J, Hamm L, Gadegbeku C, Horwitz E, Townsend RR, Anderson CA, Lash JP, Hsu CY, Leonard MB, Wolf M. Fibroblast growth factor 23 is elevated before parathyroid hormone and phosphate in chronic kidney disease. Kidney Int. 2011;79:1370–1378. doi: 10.1038/ki.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Portale A, Wolf M, Juppner H, Messinger S, Kumar J, Wesseling-Perry K, Schwartz G, Furth S, Warady B, Salusky I. Disordered FGF23 and Mineral Metabolism in the Chronic Kidney Disease in Children (CKiD) Cohort. Clin J Am Soc Nephrol. 2013 doi: 10.2215/CJN.05840513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hollis BW, Kamerud JQ, Kurkowski A, Beaulieu J, Napoli JL. Quantification of circulating 1,25-dihydroxyvitamin D by radioimmunoassay with 125I–labeled tracer. Clin.Chem. 1996;42:586–592. [PubMed] [Google Scholar]
- 26.Dillon JP. Primary Human Osteoblast Cultures. In: Helfrich MH, Ralston, Stuart H, editors. Bone Research Protocols. Vol. 816. 2012. pp. 3–18. [DOI] [PubMed] [Google Scholar]
- 27.Parfitt AM, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J.Bone Miner.Res. 2010;2:595–610. doi: 10.1002/jbmr.5650020617. [DOI] [PubMed] [Google Scholar]
- 28.Bakkaloglu SA, Wesseling-Perry K, Pereira RC, Gales B, Wang HJ, Elashoff RM, Salusky IB. Value of the new bone classification system in pediatric renal osteodystrophy. Clin.J.Am.Soc.Nephrol. 2010;5:1860–1866. doi: 10.2215/CJN.01330210. [DOI] [PMC free article] [PubMed] [Google Scholar]