SUMMARY
The filoviruses, including Marburg and Ebola, express a single glycoprotein on their surface, termed GP, which is responsible for attachment and entry of target cells. Filovirus GPs differ by up to 70% in protein sequence, and no antibodies are yet described that cross-react among them. Here, we present the 3.6 Å crystal structure of Marburg virus GP in complex with a cross-reactive antibody from a human survivor, and a lower resolution structure of the antibody bound to Ebola virus GP. The antibody, MR78, recognizes a GP1 epitope conserved across the filovirus family, which likely represents the binding site of their NPC1 receptor. Indeed, MR78 blocks binding of the essential NPC1 domain C. These structures and additional small-angle X-ray scattering of mucin-containing MARV and EBOV GPs suggest why such antibodies were not previously elicited in studies of Ebola virus, and provide critical templates for development of immunotherapeutics and inhibitors of entry.
INTRODUCTION
The filovirus family includes Marburg virus and five ebolaviruses (Ebola-, Sudan-, Reston-, Bundibugyo- and Taï Forest viruses), most of which cause highly lethal hemorrhagic fever and multiple outbreaks among humans. Among the filoviruses, Marburg virus was the first to be identified when it sickened laboratory workers in Europe in 1967 (Malherbe and Strickland-Cholmley, 1968; Siegert et al., 1968). Marburg virus has since re-emerged multiple times, with modern strains conferring greater lethality (~90%) (Geisbert et al., 2007; Towner et al., 2006). Sudan virus has caused at least six outbreaks between 1976 and 2013 (Albarino et al., 2013; Bowen et al., 1977; Sanchez and Rollin, 2005; Shoemaker et al., 2012), Bundibugyo virus emerged in 2007 (Towner et al., 2008; Wamala et al., 2010) and again in 2012 (Albarino et al., 2013), and Reston virus was found to infect ranches of swine being raised for human consumption in Asia in 2009 and 2011 (Barrette et al., 2009; Pan et al., 2012; Sayama et al., 2012). Ebola virus is typically found in Central Africa, but re-emerged in Western Africa in 2014 to cause an outbreak unprecedented in magnitude and geographic spread (WHO, 2014). An experimental Ebola virus-specific monoclonal antibody (mAb) cocktail (Qiu et al., 2014) was used compassionately in several patients. No such treatment yet exists that could be used against Marburg virus or the other four ebolaviruses.
Filoviruses express a single protein on their envelope surface, a glycoprotein termed GP, which is responsible for attachment to, and entry of, host cells (Sanchez et al., 1996). GP forms a trimer on the viral surface. In the trimer, each monomer is comprised of GP1 and GP2 subunits that are anchored together by a GP1-GP2 disulfide bond (Volchkov et al., 1998). GP1 contains a receptor-binding core topped by a glycan cap and a heavily glycosylated mucin-like domain (Lee et al., 2008), while GP2 contains two heptad repeats and a transmembrane domain. Filoviruses initially enter cells via macropinocytosis (Aleksandrowicz et al., 2011; Nanbo et al., 2010; Saeed et al., 2010). Once in the endosome, the viral surface GP is cleaved by host cathepsins. Cleavage removes the mucin-like domains and glycan cap and renders GP competent to bind the Niemann Pick C1 (NPC1) receptor (Brecher et al., 2012; Carette et al., 2011; Chandran et al., 2005; Cote et al., 2011; Hood et al., 2010; Marzi et al., 2012a; Sanchez, 2007; Schornberg et al., 2006). Interestingly, Ebola virus entry requires cleavage by cathepsin B (Chandran et al., 2005; Martinez et al., 2010; Schornberg et al., 2006), while Marburg virus entry is independent of cathepsin B (Gnirss et al., 2012; Misasi et al., 2012). The reasons underlying these differences are unknown. After enzymatic cleavage and receptor binding, the GP2 subunit unwinds from its GP1 clamp and rearranges irreversibly into a six-helix bundle (Malashkevich et al., 1999; Weissenhorn et al., 1998a; Weissenhorn et al., 1998b) to drive fusion of virus and host membranes.
Antibody therapies recently have demonstrated effective post-exposure protection against filoviruses in animal models (Dye et al., 2012; Marzi et al., 2012b; Olinger et al., 2012; Pettitt et al., 2013; Qiu et al., 2012; Qiu et al., 2014). MAbs can be produced on large scale and offer more reproducible effects than polyclonal sera from survivors. However, most mAbs available only recognize Ebola virus. Very few are yet described against Marburg virus, and no antibodies are yet described that cross-react among the filoviruses. Indeed, Marburg and Ebola GP are 72% different in protein sequence, and the filoviruses are thought to be antigenically distinct. Further, there is no structure available for the unique Marburg virus GP, by which we may interpret differences in requirements for viral entry, or develop immunotherapeutics or inhibitors of entry.
Here, we report the crystal structure of the trimeric, receptor-competent form of Marburg virus GP in complex with a neutralizing antibody, termed MR78, that was identified in a recent human survivor of Marburg virus infection (Flyak et al., 2014) (co-submitted manuscript). Atypically, MR78 cross-reacts to cleaved Ebola virus GP, and an additional structure of MR78 in complex with Ebola virus GP illustrates the basis of the cross-reactivity. The antibody binds a hydrophobic “trough” at the top of GP1, the sequence and structure of which are conserved across the filoviruses. We propose that this trough is the binding site of the critical domain C of the NPC1 receptor, and indeed, MR78 blocks binding of domain C to Marburg GP. Further, the extended third complementarity-determining region of the heavy chain (CDR H3) of MR78 mimics the glycan cap that shields this site on Ebola virus prior to entry, and may mimic the receptor itself. These crystal structures plus additional biophysical analysis of complete, mucin-containing Ebola and Marburg GP ectodomains reveal that the receptor-binding site is masked on the surface of Ebola virus but more exposed on the surface of Marburg virus. These findings may explain why a cross-reactive antibody such as MR78 has not been identified in studies of Ebola virus.
RESULTS
Structure Determination
Trimeric GP ectodomains for Marburg virus (MARV; strain Ravn) or Ebola virus (EBOV, also known as Ebola Zaire; strain Mayinga) were expressed in Drosophila S2 cells, with or without their mucin-like domains (GP and GPΔmuc, respectively). MARV and EBOV GPΔmuc were further proteolyzed by trypsin or thermolysin, respectively, to produce cleaved GP (GPcl) resembling the version of GP competent for receptor binding in the endosome (Figure S1A). 300 versions of MARV GP were engineered and complexed with 22 different mAbs in order to find a crystallizable combination. Hundreds of crystals of the final MARV GPcl-MR78 combination were grown and screened for X-ray diffraction: just one crystal yielded suitable diffraction.
Diffraction to 3.6 Å resolution was obtained from a single crystal of the MARV GPcl-Fab MR78 complex. The structure was determined by molecular replacement using EBOV GP and Fab KZ52 (Lee et al., 2008) as search models and was refined to Rwork of 24.73 % and Rfree of 27.92 % (Table S1). Four GP-Fab complexes are contained in the asymmetric unit: one complete trimer and one other monomer, which forms its biologically relevant trimer around a crystallographic 3-fold axis.
Differences in GP Structure between EBOV and MARV
Although the overall organization is similar between Marburg and Ebola GPs (1.8 Å r.m.s.d. among 212 Cα atoms) (Figure 1A and 1B), several structural differences exist that may explain their differing requirements for cellular entry. The first difference is that the intra-GP1 disulfide bond formed by C121 and C147 in ebolavirus GP structures (Ebola (Lee et al., 2008) and Sudan (Bale et al., 2012; Dias et al., 2011)) does not exist in MARV. In MARV, the two Cysteines are replaced instead with L105 and H131 (Figure 1C and Figure S1B). As a result, the equivalent polypeptides, which form the crest of the receptor-binding subunit, differ in structure and flexibility. In the ebolaviruses, the polypeptide bearing C147 (residues 145 to 150) turns inward, toward the trimer center to disulfide bond to C121. In MARV, the equivalent polypeptide (residues 129 to 134) turns outward into solvent, away from the trimer center.
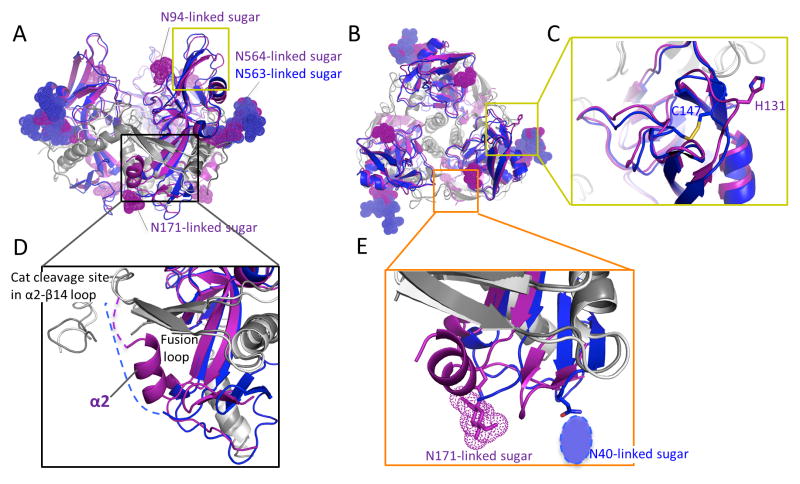
Figure 1. Structure of Marburg virus GP.
(A) Crystal structure of MARV GPcl (GP1; purple and GP2; dark gray) superimposed with the equivalent structure of EBOV (PDB ID; 3CSY, GP1; blue and GP2; light gray). The glycan cap of EBOV GP is deleted for clarity. The yellow box outlines the MR78 epitope and putative receptor-binding site. The black box outlines the interaction site of the MARV-specific helix α2 of GP1 (purple) with the fusion loop of GP2 (dark grey). The visible N-linked sugars on MARV and EBOV GPcl crystal structures are shown as dot models. MARV GPcl bears glycans at positions N94 and N171, which are not glycosylated in EBOV. See also Figure S1. (B) Top view of GP. (C) MARV GP lacks the intra-GP1 disulfide bond of EBOV. C147 of EBOV (blue) is replaced by H131 in MARV (purple), and the corresponding polypeptide traces outward from the trimer center. The orange box outlines the glycan attachment sites at the base of each GP. (D) Residues 172-180 of MARV form an α helix (α2) that packs against both N- and C-terminal arms of the fusion loop. In ebolaviruses, the equivalent residues are predict to form a loop rather than a helix and are disordered in crystal structures. (E) At the base of GP, MARV bears a glycan attached to N171 while EBOV bears a glycan attached to N40 (drawn as an oval as it was not included in the EBOV crystal structure).
A second difference between MARV and the ebolaviruses lies at the base of the cathepsin cleavage loop. In MARV, these residues (172-180) form a clear alpha helix (α2), which packs against the outside of the GP2 fusion loop, interacting with both the N- and C-terminal strands of the fusion loop (Figure 1D). In ebolaviruses, the equivalent residues predict to form a loop rather than a helix and are disordered (Bale et al., 2012; Dias et al., 2011; Lee et al., 2008). In MARV GP, the peptide connecting this α2 helix to β14 in the glycan cap would necessarily and immediately cover the both N- and C-terminal arms of the GP2 fusion loop, and if uncleaved, would hinder the conformational changes of fusion. Structural differences in α2 of MARV may prevent effective processing by cathepsin B.
The third difference in the MARV GP structure lies at the N terminus, in the base of the β sheet that forms the GP1 spool, about which the metastable GP2 subunit is wound. In EBOV, the base of the spool connects to the anchoring GP1-GP2 disulfide bond by a short stretch of polypeptide that intimately interacts with GP2. This short connecting polypeptide contains an N-linked glycan at Asn40, and also contains residue Asp47, which renders EBOV dependent on cathepsin B for entry (Misasi et al., 2012). In EBOV entry, cathepsin B removes an additional and critical 1 kDa of mass from GP beyond that removed by cathepsin L, but the site and consequences of that extra cleavage event are not yet known. We propose that if cathepsin B cleaves this connecting loop, EBOV GP2 would be freed from the constraints of the disulfide bond and more free to undergo the conformational rearrangements of fusion. Our crystal structure reveals that MARV, which is cathepsin B-independent, is structured differently from EBOV at the same site. In MARV, the base of the GP1 spool is more mobile, and is shifted toward the center of the trimer, inside of the fusion loop. Further, unlike in ebolaviruses (Dias et al., 2011; Bale et al., 2012), the polypeptide connection to the MARV GP1-GP2 disulfide could not be visualized and the N-linked glycan is absent (Figure 1E). The nearest glycan is instead attached to residue 171 on the MARV GP1 β sheet itself. These differences in sequence, glycosylation, mobility and conformation likely allow MARV to be cleaved by other enzymes and render MARV cathepsin B-independent.
Overall Organization of the MARV or EBOV GPcl bound to Fab MR78
The crystal structure of MARV GPcl in complex with the Fab fragment of MR78 indicates that MR78 binds the membrane-distal head of GP1 (Figure 2A). We determined an additional, low-resolution structure of EBOV GPcl bound to both MR78 and KZ52. The ternary EBOV complex, determined by molecular replacement, demonstrates that the MR78 antibody recognizes a similar site on both MARV and EBOV (Figure 2B, S2 and Table S1). MR78 binds into a highly conserved hydrophobic trough revealed at the top of the EBOV GP1 core, after removal of the glycan cap by proteolytic cleavage in the endosome. Although MARV and EBOV diverge significantly in sequence overall, residues contained in this site, the MR78 epitope, are 85% similar between the viruses (Figure 3A and S1B).
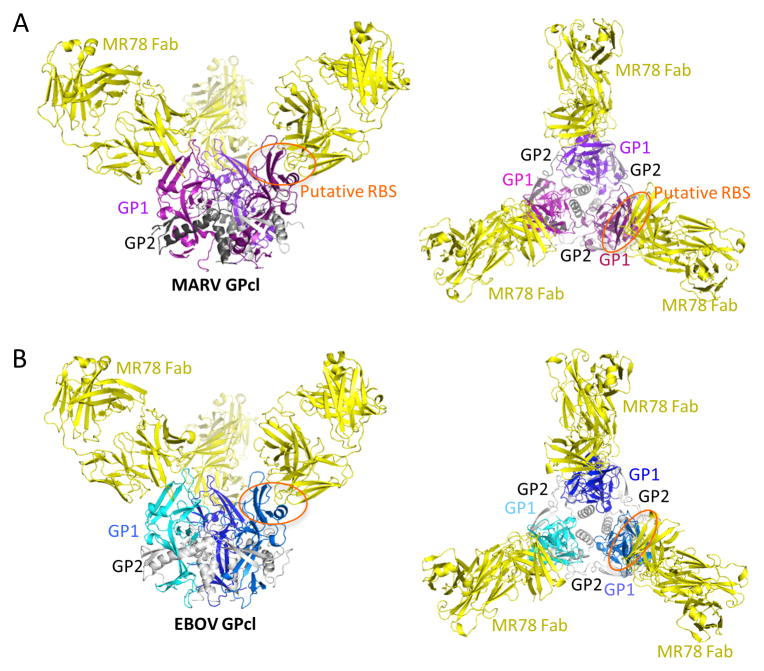
Figure 2. MR78 binds both MARV and EBOV GPcl at the apex of GP1.
(A) 3.6 Å crystal structure of MARV GPcl in complex with Fab MR78. Each GP1 is colored a different shade of purple, GP2 is gray, and the MR78 Fab is in yellow. (B) 8 Å structure of EBOV GPcl in complex with Fab MR78, determined by molecular replacement and rigid body refinement. Each EBOV GP1 is colored a different shade of blue and GP2 is gray. See also Figure S2. Fab MR78 (yellow) binds the apex of GP1 of both viruses.
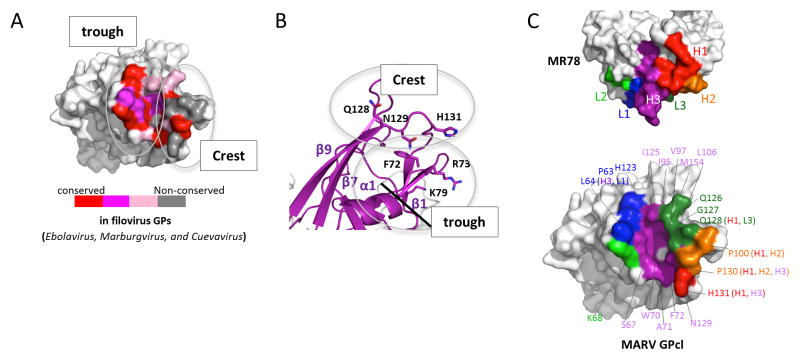
Figure 3. MR78 recognizes a conserved epitope at the apex of cleaved GP1.
(A) Conservation of the MR78 epitope among filovirus GPs, mapped onto one monomer of MARV GPcl. Sequence alignment was performed in ebolavirus (Ebola, Sudan, Reston, Taï Forest, Bundibugyo), marburgvirus (Musoke, Angola, Popp, Ci67, DRC1999, Ravn), and cuevavirus (Lloviu) genuses. Residues identical across the filoviruses are colored red; residues that possess strong similarity, magenta; weak similarity, pink; no similarity, gray. (B) The apex of cleaved MARV GP1, where Fab MR78 binds, forms a wave crest-and-trough morphology (magenta). The hydrophilic crest and the hydrophobic trough each contain residues previously shown to be critical for virus entry (Dube et al., 2009; Manicassamy et al., 2005; Manicassamy et al., 2007; Mpanju et al., 2006). The diagonal black line indicates the base of the trough. See also Figure S4. (C) Surface representation of the interface between one monomer of MARV GPcl (bottom) and Fab MR78 (top). CDR H1 is colored red; CDR H2, orange; CDR H3, purple; CDR L1, blue; CDR L2, green; CDR L3, forest green. The footprint on MARV GPcl is colored according to the CDR that mediates the contact. GP residues contacted by MR78 are indicated, and colored according to the CDR that mediates the contact (CDR names in parentheses).
Likely Receptor-Binding Site
The location and structural conservation of this site suggest that it could be the binding site of the NPC1 receptor, used by all known filoviruses (Carette et al., 2011; Cote et al., 2011; Miller et al., 2012; Ng et al., 2014). Indeed, in ELISA, MR78 inhibits binding of NPC1 domain C to MARV GP (Figure S3A). This site, at the apex of cleaved GP1, resembles an ocean wave morphology, with a lower trough beneath a rising crest. The trough is hydrophobic and is formed by α1, β4 and connecting loops (residues 63-74 in MARV). It is 22 Å wide and 8 Å deep at F72. The crest is hydrophilic, includes charged residues previously identified as essential for virus entry (Dube et al., 2009; Manicassamy et al., 2005; Manicassamy et al., 2007), and is formed by strands β7, β9 and their connecting loops (residues 92-106 and 120-134 in MARV). The 120-134 loop contains H131, which replaces the Cysteine and the intra-GP1 disulfide bond of EBOV (Figure 3B).
Here, we show by ELISA that a Q128S and N129S double mutant in MARV GP abrogate binding to NPC1 domain C (Figure S4A). Q128 and N129 are at the tip of the crest and could make direct hydrophilic interaction with NPC1. The trough itself is formed by hydrophobic side chains, such as F72 (equivalent to F88 in EBOV). Also forming the trough are the main chains of hydrophilic residues; these polar side chains reach away from the trough into the trimer to make key stabilizing contacts to GP2. Two examples are R73 and K79, previously shown to be essential for MARV infectivity (Manicassamy et al., 2007). In the crystal structure, R73 makes multiple hydrogen bonds to the fusion loop of the neighboring protomer in the trimer (Figure S4B), and likely plays a key role in maintaining the prefusion structure, or transmitting a conformational change to the fusion loop after receptor binding. K79 interacts with the main chain of residues 574-577 of GP2 (Figure S4C), residues that connect the separated helical segments of the first heptad repeat. We propose that binding of NPC1 domain C involves contact with the hydrophilic crest and hydrophobic trough, and that binding in the trough may transmit conformational changes to GP2 via R73 and K79 (equivalent to R89 and K95 in EBOV). Although MR78 binds both MARV and EBOV GPcl, it only outcompetes NPC1 domain C for binding of MARV GPcl (Figure S3B). MR78 may have lower affinity for EBOV GPcl than MARV GPcl or domain C may bind the GPs slightly differently.
GP - MR78 interactions
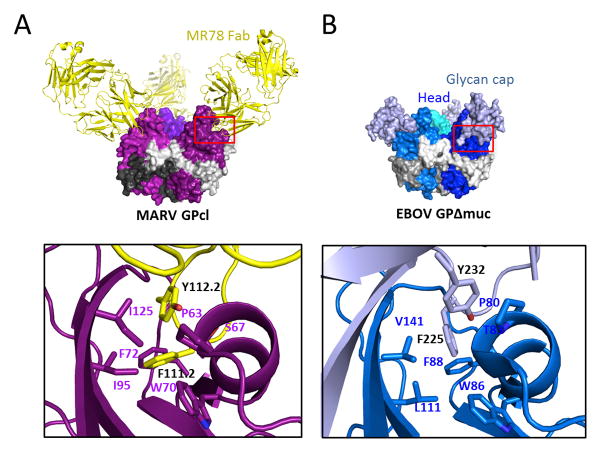
The interaction surface between the MR78 antibody and MARV GP buries 976 Å2 of molecular surface and is primarily hydrophobic. Contact is mediated by both the heavy and light chains, but the primary region of interaction is the 17-residue CDR H3 (Figure 3C and S3CD), which penetrates the hydrophobic trough in MARV GP1. In this interaction, F111.2 and Y112.2 of the CDR H3 interact with P63, S67, W70, F72, I95 and I125 of MARV GP (IMGT numbering, Figure 4A).
Figure 4. Similarity in recognition of the putative receptor-binding site by MR78 and the Ebola virus glycan cap.
(A) The CDR H3 of MR78 (yellow) reaches into the hydrophobic trough of GP1 (purple). F111.2 and Y112.2 of CDR H3 interact with P63, S67, W70, F72, I95 and I125 of MARV GP. (B) Similar residues of the EBOV glycan cap (white blue) bind into this trough on the surface of EBOV GP (blue), prior to enzymatic cleavage. Here, F225 and Y232 of the glycan cap interact with P80, T83, W86, F88, L111 and V141 in the trough (PDB ID; 3CSY).
Notably, these interactions are similar to those made by the Ebola virus glycan cap, which occupies this site prior to enzymatic cleavage in the endosome. In Ebola virus, the equivalent interactions are made by F225 and Y232 of the EBOV glycan cap interacting with P80, T83, W86, F88, L111 and V141 on EBOV GP (Figure 4B). Similarity may even extend to the key domain C of the NPC1 receptor itself, as domain C contains similar Phenylalanine and Tyrosine residues that are essential for binding GP. Further, F72 in MARV, which is equivalent to F88 in EBOV, interacts with CDR H3 in the bottom of the trough. Both F72 of MARV and F88 of EBOV are critical for attachment and entry (Martinez et al., 2013; Mpanju et al., 2006) and may interact directly with essential hydrophobic residues of domain C. The binding mode of MR78 is reminiscent of anti-influenza virus human mAbs in which long CDR H3s similarly reach into the conserved receptor-binding site (Barbey-Martin et al., 2002; Bizebard et al., 1995; Hong et al., 2013; Lee et al., 2014; Schmidt et al., 2013; Whittle et al., 2011; Xu et al., 2013). In many cases those influenza mAbs also use Phe or Tyr aromatic residues to interact with an aromatic residue in the viral receptor binding domain, suggesting that the favorable energetics and intermolecular interactions of common aromatic molecules may constitute a canonical mode of binding of antiviral antibodies to recessed receptor-binding sites.
Although the MR78 epitope is largely conserved in sequence and structure between MARV and EBOV, it differs in its exposure at different stages of virus entry. MR78 binds MARV GP equally well whether MARV GP is in its uncleaved, viral-surface form or its cleaved, endosomal form. In contrast, MR78 does not bind uncleaved EBOV GP. It only binds the endosomal, cleaved form from which the glycan cap has been removed. Together, these results suggest that in EBOV, the glycan cap effectively blocks the MR78 epitope and putative receptor-binding site on the (uncleaved) viral surface, but that in MARV, the epitope and at least part of the receptor-binding site is fully exposed on the viral surface. Better exposure of this site may explain why antibodies against the putative receptor-binding site were elicited by MARV infection (see companion paper by Flyak, et al. (Flyak et al., 2014)), but seem to be more rarely elicited and have not yet been described against EBOV.
Differences in mucin-like domains between MARV and EBOV, and possible effect on antibody reactivity
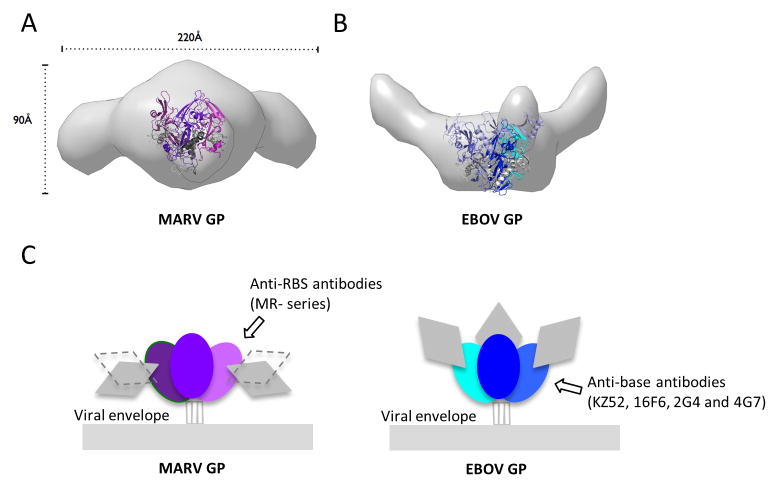
In addition to a glycan cap, the GP spike on the viral surface includes three heavily glycosylated mucin-like domains that are ~75 kDa each in mass and are predicted to have little secondary structure. All mucin-containing GPs thus far have been refractory to crystallization. In order to visualize the native glycoprotein ectodomain and position of the mucin-like domain relative to the receptor-binding core, we turned to Small-Angle X-ray Scattering (SAXS) in solution. SAXS data collected for mucin-containing EBOV or MARV GP trimers indicate that the mucin-like domains of both viruses are large and extending outward from the GP core. The radius of gyration, RG, for mucin-deleted and mucin-containing MARV GPs are 50 and 72 Å, respectively, and maximum dimension, Dmax, for mucin-deleted and mucin-containing GPs are 160 and 250Å, respectively, indicating that the mucin-like domain of MARV widens the molecule up to 90Å (Figure 5A and S5). The mucin-like domains of MARV are a bit larger than those of EBOV (67 Å RG and 225 Å Dmax for mucin-containing EBOV GP), consistent with their greater volume determined by SAXS (Figure S5C) and mass noted by SDS-PAGE (Figure S5D). The mucin-like domains of EBOV appear to project more upward (consistent with EM tomography (Tran et al., 2014)), while those of of MARV appear to project less upward, more equatorially, and to cover the sides of the GP trimer (Figure 5AB). Although the mucin-like domains are likely flexible (see Porod-Debey coefficient P in Figure S5C), an equatorial, rather than upward projection is consistent with attachment points of the mucin-like domain to both GP1 and GP2 in MARV. In EBOV, a different position of the furin cleavage site results in all of mucin-like domain being attached to GP1. The MARV GP2 portion of the mucin-like domain, residues 436-509, is attached to residue 510 on the side of the MARV GP trimer, but is flexible and disordered.
Figure 5. MARV and EBOV present different surfaces for antibody recognition.
(A and B) Molecular envelopes of mucin-containing MARV and EBOV GP ectodomains determined by SAXS. Rendered Gaussian distributions of molecular envelopes are illustrated in light gray, with ribbon models of the crystallized MARV GPcl and EBOV GPΔmuc trimers to scale and overlaid for comparison. The trimers are illustrated as ribbons. Note that the glycan cap was removed from MARV GP used in crystallization in order to improve diffraction, but was contained in the complete MARV GP used for SAXS. The glycan cap did not inhibit diffraction of EBOV GP and is included in the EBOV GP crystal structure. MARV GPcl is colored in purple (GP1) and gray (GP2). EBOV GPΔmuc is colored blue (GP1), white blue (GP1 glycan cap) and gray (GP2). MARV GP is drawn in two possible orientations because definitive placement of polypeptide is challenging at this resolution. In either orientation however, the mucin-like domains of MARV project sideways, equatorially or downwards from the core of GP. In MARV, the mucin-like domain is attached to both GP1 and GP2. By contrast, in EBOV, the mucin-like domain is attached solely to GP1, there is no anchor at the base. Both these SAXS experiments and previous electron tomography (Tran et al., 2014) agree on the upward projection of the mucin-like domains in EBOV. See also Figure S5. (C) Differing positions of the mucin-like domains between MARV and EBOV may lead to elicitation of different types of antibodies. The lower position and GP2 anchor of the mucin-like domain of MARV may better mask the base of GP, but expose its upper surfaces, allowing antibodies like mAb MR78 to be elicited. The upwards projection of the EBOV mucin-like domain and absence of any GP2 anchor, appear to better mask upper surfaces, but expose the base, allowing antibodies such as KZ52 (Lee et al., 2008), 2G4, 4G7 (Murin et al., 2014) and 16F6 (directed against Sudan ebolavirus (Dias et al., 2011; Bale et al., 2012)) to be elicited.
A differing position of the mucin-like domains between MARV and EBOV would leave different surfaces exposed for immune recognition. The equatorial projection of the MARV mucin-like domain, for example, would leave the expected receptor-binding site at the top more accessible on MARV than EBOV, and further supports the notion that antibodies against the expected receptor-binding site would be more likely to be elicited using marburgvirus antigens than ebolavirus antigens. The accompanying paper (Flyak et al.) and other immunization studies (Qiu et al., 2011; Wilson et al., 2000) support this notion.
In contrast, on EBOV, the upward projection of the mucin-like domains and the absence of mucin attached to EBOV GP2 would leave the EBOV base more exposed for antibody surveillance, compared to that of MARV. Indeed, in the accompanying paper by Flyak et al., none of the 18 neutralizing antibodies raised against MARV appear to bind the base of the MARV GP, while multiple neutralizing antibodies elicited by ebolaviruses are known to bind, or thought to bind, to the base of ebolavirus GP (Dias et al., 2011; Lee et al., 2008; Qiu et al., 2012; Murin et al., 2014) (Figure 5C).
DISCUSSION
In summary, the crystal structures and accompanying experiments indicate that MR78 binds a conserved site on the apex of GP1, that is available on the surface of MARV GP, but masked on EBOV GP prior to enzymatic cleavage. The epitope of MR78 likely overlaps with the receptor-binding site, and hydrophobic contacts made by CDR H3 to the hydrophobic trough may mimic those of the as-yet-unvisualized NPC1 domain C. MR78 does not neutralize authentic EBOV, likely because its epitope is masked on the EBOV surface by the mucin-like domain and glycan cap on the virus surface. MR78 does, however, neutralize authentic MARV (Flyak et al., 2014) and could be a valuable monoclonal antibody therapeutic against this extremely lethal virus. Importantly, no mAb therapeutic yet exists against MARV, and few mAbs are yet known against MARV from which such a therapeutic could be developed. The crystal structure of MARV GP presented here, and the highly conserved MR78 epitope, provide strategies for immunotherapy and templates for development of potentially broad-spectrum inhibitors of filovirus entry.
EXPERIMENTAL PROCEDURES
Construction, expression, and purification of MARV/EBOV GP
DNA encoding the MARV GPΔmuc ectodomain (residues 1-636 with a mucin deletion of residues 257-425), point mutants of MARV GPΔmuc and the EBOV GPΔmuc ectodomain (residues 1-637 with a mucin deletion of residues 314-462) were amplified by PCR using codon-optimized and whole-gene synthesized MARV or EBOV GPs as templates. Four point mutations in MARV GPΔmuc, F438L, W439A, F445G, and F447N, on GP2, located around the furin cleavage site were found to improve the efficiency of furin cleavage. GP constructs were cloned into a derivative of the expression vector pMT. This derivative vector contains the puromycin resistant gene and a C-terminal double-strep tag sequence. Expression plasmids were transfected using Effectin (Qiagen) into 80 % confluent Drosophila Schneider S2 cells. The cells were first cultured in complete Schneider’s medium supplemented with 10 % (v/v) FCS (LONZA), and were adapted to Insect Xpress medium by progressively modifying the Schneider/Insect Xpress medium ratio with 6.0 μg/ml puromycin. Large-scale expression of the MARV/EBOV GPΔmuc was performed using stable S2 cell lines in 2 L Erlenmeyer flask at 27.0 °C, induced with 0.5 mM CuSO4. Supernatants containing the expressed proteins were harvested 4 days after induction, and mixed with the Strept-Tactin affinity column binding buffer (100 mM Tris-HCl, 150 mM NaCl, 1 mM EDTA, 15 μg/ml Avidin, pH8.0). The proteins were purified via Strept-Tactin affinity, followed by Superdex 200GL 10/300 (GE Healthcare Life Sciences) size-exclusion chromatography (S200 SEC).
Preparation and crystallization of GP-antibody complexes
To mimic endosomal protease cleavage and produce MARV GPcl, MARVΔGP muc was incubated with 0.01 mg trypsin at 37 °C for 1 hour in 20 mM TBS pH 8.0, 100 mM NaCl. The reaction was stopped using 0.5 mM 4-(2-Aminoethyl) benzenesulfonyl fluoride hydrochloride (AEBSF), and the protein was purified by S200 SEC. EBOV GPcl was produced by incubating EBOV GPΔmuc with 0.02 mg thermolysin overnight at room temperature in 20 mM TBS pH 7.5, 100 mM NaCl, 1 mM CaCl2, and purified by S200 SEC. Hybridoma cells expressing the human MR78 antibody were generated from peripheral blood mononuclear cells (PBMCs) from a donor, who contracted MARV infection in the Python Cave in Queen Elizabeth National Park, Uganda in 2008 [companion paper, Flyak, et al.]. MR78 was expressed in serum-free medium (Hybridoma-SFM, Gibco), and culture supernatants were centrifuged, sterile-filtered, and purified over HiTrap Protein G columns (GE Healthcare Life Sciences). Fab fragments were generated by standard papain digestion, with released Fc and undigested IgG removed by Protein A chromatography, and remaining Fab fragments further purified by MonoQ ion-exchange chromatography. For crystallization, purified MARV GPcl was mixed with excess Fab MR78 for two days at 4°C. Complexes were purified from extra Fab via S200 SEC. Crystals were grown by hanging-drop vapor diffusion at 20°C using 0.8 μl protein (13.0 mg ml−1, in 20 mM Tris-HCl pH8.0, 100 mM NaCl) and 0.8 μl of mother liquor (100 mM NaCl, 50 mM MES pH6.5, 13 % PEG4000, 0.5 % ethyl acetate). These crystals were cryoprotected with 25% glycerol plus mother liquor before flash cooling in liquid nitrogen. One crystal diffracted to a resolution of 3.6 Å. EBOV GPcl was complexed with Fabs KZ52 and MR78 and crystallized using hanging-drop vapor diffusion at 20°C with 1.0 μl of protein (6 mg/mL, 150mM NaCl, 10 mM Tris pH7.5) and 1.0 μl of mother liquor (100mM NaAcetate pH 4.6, 200mM NH4SO4, 10% PEG 3350, 2% PEG 400). The crystals were then cryoprotected by washing in 100mM NaAcetate pH 4.6, 200mM NH4SO4, 12% PEG 3350, 10% PEG 400, 10% Ethylene Glycol. Only diffraction to 8 Å was obtained, but this data permitted molecular replacement using Phaser (McCoy et al., 2007), and EBOV GP and KZ52 (Lee et al., 2008) as search models.
Supplementary Material
Acknowledgments
We would like to thank Dr. Adrianna P.P. Zhang (TSRI) for help with data collection, Dr. Sebastien Iognet (TSRI and Calixar) for assistance with S2 cell expression, Dr. Kartik Chandran (Albert Einstein College of Medicine), C. Daniel Murin and Dr. Andrew Ward (TSRI) for valuable discussions, and the beamline staff of Photon Factory (Tsukuba, Japan) for technical help during data collection. We thank NIAID CETR U19 AI109762 (EOS, ABW), R01 AI089498 and R21AI069347 (EOS), Defense Threat Reduction Agency grant HDTRA1-13-1-0034, NIAID grant U19 AI109711 (JEC), MEXT KAKENHI Grant Number 26713018 (TH), 24115005 (YY), MEXT Platform for Drug Discovery Informatics and Structural Life Science (TH), JSPS Postdoctoral Fellowships (DC2 (RM), Research Abroad (TH)), a Research Fellowship of The Uehara Memorial Foundation (TH), an NSF predoctoral fellowship (CDM), an Investigators in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund (EOS) and an NIH grant MINOS GM105404 (MH) for support. SIBYLS beamline efforts to combine SAXS and crystallography at the Advanced Light Source of Lawrence Berkeley National Laboratory are supported in part by United States Department of Energy program Integrated Diffraction Analysis Technologies (IDAT). This is manuscript # 28060 from The Scripps Research Institute.
Footnotes
ACCESSION NUMBERS
Coordinate and structure factors have been deposited into the Protein Data Bank under the accession code 3X2D.
Supplemental Information, which includes Extended Experimental Procedures, five figures and one table, can be found with this article online at http://.
AUTHOR CONTRIBUTIONS
Experiments were conceived by E.O.S. with T.H., M.L.F., J.E.L. and Z.A.B. All structural and biochemical work was performed by T.H., M.L.F., Z.A.B., J.E.L., A.I.F., and analyzed by T.H., M.L.F., Z.A.B., J.E.L., A.I.F., R.M., D.K., Y.Y., M.H., J.E.C. and E.O.S. The manuscript was written by E.O.S. and T.H. All authors contributed to editing the manuscript and support the conclusions.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Albarino CG, Shoemaker T, Khristova ML, Wamala JF, Muyembe JJ, Balinandi S, Tumusiime A, Campbell S, Cannon D, Gibbons A, et al. Genomic analysis of filoviruses associated with four viral hemorrhagic fever outbreaks in Uganda and the Democratic Republic of the Congo in 2012. Virology. 2013;442:97–100. doi: 10.1016/j.virol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrowicz P, Marzi A, Biedenkopf N, Beimforde N, Becker S, Hoenen T, Feldmann H, Schnittler HJ. Ebola virus enters host cells by macropinocytosis and clathrin-mediated endocytosis. J Infect Dis. 2011;204(Suppl 3):S957–967. doi: 10.1093/infdis/jir326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale S, Dias JM, Fusco ML, Hashiguchi T, Wong AC, Liu T, Keuhne AI, Li S, Woods VL, Jr, Chandran K, et al. Structural basis for differential neutralization of ebolaviruses. Viruses. 2012;4:447–470. doi: 10.3390/v4040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology. 2002;294:70–74. doi: 10.1006/viro.2001.1320. [DOI] [PubMed] [Google Scholar]
- Barrette RW, Metwally SA, Rowland JM, Xu L, Zaki SR, Nichol ST, Rollin PE, Towner JS, Shieh WJ, Batten B, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- Bizebard T, Gigant B, Rigolet P, Rasmussen B, Diat O, Bosecke P, Wharton SA, Skehel JJ, Knossow M. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. doi: 10.1038/376092a0. [DOI] [PubMed] [Google Scholar]
- Bowen ET, Lloyd G, Harris WJ, Platt GS, Baskerville A, Vella EE. Viral haemorrhagic fever in southern Sudan and northern Zaire. Preliminary studies on the aetiological agent. Lancet. 1977;1:571–573. doi: 10.1016/s0140-6736(77)92001-3. [DOI] [PubMed] [Google Scholar]
- Brecher M, Schornberg KL, Delos SE, Fusco ML, Saphire EO, White JM. Cathepsin cleavage potentiates the Ebola virus glycoprotein to undergo a subsequent fusion-relevant conformational change. J Virol. 2012;86:364–372. doi: 10.1128/JVI.05708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carette JE, Raaben M, Wong AC, Herbert AS, Obernosterer G, Mulherkar N, Kuehne AI, Kranzusch PJ, Griffin AM, Ruthel G, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandran K, Sullivan NJ, Felbor U, Whelan SP, Cunningham JM. Endosomal Proteolysis of the Ebola Virus Glycoprotein Is Necessary for Infection. Science. 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cote M, Misasi J, Ren T, Bruchez A, Lee K, Filone CM, Hensley L, Li Q, Ory D, Chandran K, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for Ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias JM, Kuehne AI, Abelson DM, Bale S, Wong AC, Halfmann P, Muhammad MA, Fusco ML, Zak SE, Kang E, et al. A shared structural solution for neutralizing ebolaviruses. Nat Struct Mol Biol. 2011;18:1424–1427. doi: 10.1038/nsmb.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dube D, Brecher MB, Delos SE, Rose SC, Park EW, Schornberg KL, Kuhn JH, White JM. The primed Ebolavirus glycoprotein (‘19kDa’ GP1,2): Sequence and residues critical for host cellbinding. J Virol. 2009;83:2883–2891. doi: 10.1128/JVI.01956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dye JM, Herbert AS, Kuehne AI, Barth JF, Muhammad MA, Zak SE, Ortiz RA, Prugar LI, Pratt WD. Postexposure antibody prophylaxis protects nonhuman primates from filovirus disease. Proc Natl Acad Sci USA. 2012;109:5034–5039. doi: 10.1073/pnas.1200409109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flyak AI, Ilinykh PA, Murin CD, Garron T, Shen X, Fusco ML, Hashiguchi T, Bornholdt ZA, Slaughter JC, Sapparapu G, et al. Mechanism of Human Antibody-Mediated Neutralization of Marburg virus. 2015 doi: 10.1016/j.cell.2015.01.031. co-submitted manuscript. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Daddario-DiCaprio KM, Geisbert JB, Young HA, Formenty P, Fritz EA, Larsen T, Hensley LE. Marburg virus Angola infection of rhesus macaques: pathogenesis and treatment with recombinant nematode anticoagulant protein c2. J Infect Dis. 2007;196(Suppl 2):S372–381. doi: 10.1086/520608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnirss K, Kuhl A, Karsten C, Glowacka I, Bertram S, Kaup F, Hofmann H, Pohlmann S. Cathepsins B and L activate Ebola but not Marburg virus glycoproteins for efficient entry into cell lines and macrophages independent of TMPRSS2 expression. Virology. 2012;424:3–10. doi: 10.1016/j.virol.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE, Jr, et al. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol. 2013;87:12471–12480. doi: 10.1128/JVI.01388-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood CL, Abraham J, Boyington JC, Leung K, Kwong PD, Nabel GJ. Biochemical and structural characterization of Cathepsin L-processed Ebola virus glycoprotein: implications for viral entry and immunogenicity. J Virol. 2010;84:2972–2982. doi: 10.1128/JVI.02151-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JE, Fusco ML, Hessell AJ, Oswald WB, Burton DR, Saphire EO. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nature communications. 2014;5:3614. doi: 10.1038/ncomms4614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malashkevich VN, Schneider BJ, McNally ML, Milhollen MA, Pang JX, Kim PS. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-A resolution. Proc Natl Acad Sci USA. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malherbe H, Strickland-Cholmley M. Human disease from monkeys (Marburg virus) Lancet. 1968;1:1434. doi: 10.1016/s0140-6736(68)92023-0. [DOI] [PubMed] [Google Scholar]
- Manicassamy B, Wang J, Jiang H, Rong L. Comprehensive analysis of ebola virus GP1 in viral entry. J Virol. 2005;79:4793–4805. doi: 10.1128/JVI.79.8.4793-4805.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B, Wang J, Rumschlag E, Tymen S, Volchkova V, Volchkov V, Rong L. Characterization of Marburg virus glycoprotein in viral entry. Virology. 2007;358:79–88. doi: 10.1016/j.virol.2006.06.041. [DOI] [PubMed] [Google Scholar]
- Martinez O, Johnson J, Manicassamy B, Rong L, Olinger GG, Hensley LE, Basler CF. Zaire Ebola virus entry into human dendritic cells is insensitive to cathepsin L inhibition. Cellular Microbiology. 2010;12:148–157. doi: 10.1111/j.1462-5822.2009.01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez O, Ndungo E, Tantral L, Miller EH, Leung LW, Chandran K, Basler CF. A mutation in the Ebola virus envelope glycoprotein restricts viral entry in a host species and cell-type specific manner. J Virol. 2013;87:3324–3334. doi: 10.1128/JVI.01598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Reinheckel T, Feldmann H. Cathepsin B & L are not required for ebola virus replication. PLoS Nagl Trop Dis. 2012a;6:e1923. doi: 10.1371/journal.pntd.0001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzi A, Yoshida R, Miyamoto H, Ishijima M, Suzuki Y, Higuchi M, Matsuyama Y, Igarashi M, Nakayama E, Kuroda M, et al. Protective efficacy of neutralizing monoclonal antibodies in a nonhuman primate model of Ebola hemorrhagic fever. PLoS ONE. 2012b;7:e36192. doi: 10.1371/journal.pone.0036192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EH, Obernosterer G, Raaben M, Herbert AS, Deffieu MS, Krishnan A, Ndungo E, Sandesara RG, Carette JE, Kuehne AI, et al. Ebola virus entry requires the host-programmed recognition of an intracellular receptor. EMBO J. 2012;31:1947–1960. doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misasi J, Chandran K, Yang JY, Considine B, Filone CM, Cote M, Sullivan N, Fabozzi G, Hensley L, Cunningham J. Filoviruses require endosomal cysteine proteases for entry but exhibit distinct protease preferences. J Virol. 2012;86:3284–3292. doi: 10.1128/JVI.06346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mpanju OM, Towner JS, Dover JE, Nichol ST, Wilson CA. Identification of two amino acid residues on Ebola virus glycoprotein 1 critical for cell entry. Virus Res. 2006;121:205–214. doi: 10.1016/j.virusres.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Murin CD, Fusco ML, Bornholdt ZA, Qiu X, Olinger GG, Zeitlin L, Kobinger GP, Ward AB, Saphire EO. Structures of protective antibodies reveal sites of volunerability of Ebola virus. Proc Natl Acad Sci USA. 2014;111:17182–7. doi: 10.1073/pnas.1414164111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanbo A, Imai M, Watanabe S, Noda T, Takahashi K, Neumann G, Halfmann P, Kawaoka Y. Ebolavirus is internalized into host cells via macropinocytosis in a viral glycoprotein-dependent manner. PLoS Pathog. 2010;6:e1001121. doi: 10.1371/journal.ppat.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Ndungo E, Jangra RK, Cai Y, Postnikova E, Radoshitzky SR, Dye JM, Ramirez de Arellano E, Negredo A, Palacios G, et al. Cell entry by a novel European filovirus requires host endosomal cysteine proteases and Niemann Pick C1. Virology. 2014 doi: 10.1016/j.virol.2014.08.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinger GG, Jr, Pettitt J, Kim D, Working C, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, Morton J, et al. Delayed treatment of Ebola virus infection with plant-derived monoclonal antibodies provides protection in rhesus macaques. Proc Natl Acad Sci USA. 2012;109:18030–18035. doi: 10.1073/pnas.1213709109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhang W, Cui L, Hua X, Wang M, Zeng Q. Reston virus in domestic pigs in China. Arch Virol. 2014;159:1129–1132. doi: 10.1007/s00705-012-1477-6. [DOI] [PubMed] [Google Scholar]
- Pettitt J, Zeitlin L, Kim do H, Working C, Johnson JC, Bohorov O, Bratcher B, Hiatt E, Hume SD, Johnson AK, et al. Therapeutic intervention of Ebola virus infection in rhesus macaques with the MB-003 monoclonal antibody cocktail. Science Translational Medicine. 2013;5:199ra113. doi: 10.1126/scitranslmed.3006608. [DOI] [PubMed] [Google Scholar]
- Qiu X, Alimonti JB, Melito PL, Fernando L, Stroher U, Jones SM. Characterization of Zaire ebolavirus glycoprotein-specific monoclonal antibodies. Clin Immunol. 2011;141:218–227. doi: 10.1016/j.clim.2011.08.008. [DOI] [PubMed] [Google Scholar]
- Qiu X, Audet J, Wong G, Pillet S, Bello A, Cabral T, Strong JE, Plummer F, Corbett CR, Alimonti JB, et al. Successful treatment of ebola virus-infected cynomolgus macaques with monoclonal antibodies. Science Translational Medicine. 2012;4:138ra181. doi: 10.1126/scitranslmed.3003876. [DOI] [PubMed] [Google Scholar]
- Qiu X, Wong G, Audet J, Bello A, Fernando L, Alimonti JB, Fausther-Bovendo H, Wei H, Aviles J, Hiatt E, et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature. 2014;514:47–53. doi: 10.1038/nature13777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed MF, Kolokoltsov AA, Albrecht T, Davey RA. Cellular entry of ebola virus involves uptake by a macropinocytosis-like mechanism and subsequent trafficking through early and late endosomes. PLoS Pathog. 2010;6:e1001110. doi: 10.1371/journal.ppat.1001110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. Analysis of filovirus entry into vero e6 cells, using inhibitors of endocytosis, endosomal acidification, structural integrity, and cathepsin (B and L) activity. J Infect Dis. 2007;196(Suppl 2):S251–258. doi: 10.1086/520597. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Rollin PE. Complete genome sequence of an Ebola virus (Sudan species) responsible for a 2000 outbreak of human disease in Uganda. Virus Res. 2005;113:16–25. doi: 10.1016/j.virusres.2005.03.028. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Trappier SG, Mahy BW, Peters CJ, Nichol ST. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayama Y, Demetria C, Saito M, Azul RR, Taniguchi S, Fukushi S, Yoshikawa T, Iizuka I, Mizutani T, Kurane I, et al. A seroepidemiologic study of Reston ebolavirus in swine in the Philippines. BMC Vet Res. 2012;8:82. doi: 10.1186/1746-6148-8-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O’Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci USA. 2013;110:264–269. doi: 10.1073/pnas.1218256109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schornberg K, Matsuyama S, Kabsch K, Delos S, Bouton A, White J. Role of endosomal cathepsins in entry mediated by the Ebola virus glycoprotein. J Virol. 2006;80:4174–4178. doi: 10.1128/JVI.80.8.4174-4178.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker T, Macneil A, Balinandi S, Campbell S, Wamala JF, McMullan LK, Downing R, Lutwama J, Mbidde E, Stroher U, et al. Reemerging Sudan ebola virus disease in Uganda, 2011. Emerg Infect Dis. 2012;18:1480–1483. doi: 10.3201/eid1809.111536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert R, Shu HL, Slenczka W. Isolation and identification of the “Marburg virus”. Dtsch Med Wochenschr. 1968;93:604–612. doi: 10.1055/s-0028-1105103. [DOI] [PubMed] [Google Scholar]
- Towner JS, Khristova ML, Sealy TK, Vincent MJ, Erickson BR, Bawiec DA, Hartman AL, Comer JA, Zaki SR, Stroher U, et al. Marburgvirus genomics and association with a large hemorrhagic fever outbreak in Angola. J Virol. 2006;80:6497–6516. doi: 10.1128/JVI.00069-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towner JS, Sealy TK, Khristova ML, Albarino CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran EE, Simmons JA, Bartesaghi A, Shoemaker CJ, Nelson E, White JM, Subramaniam S. Spatial localization of the Ebola virus glycoprotein mucin-like domain determined by cryo-electron tomography. J Virol. 2014;88:10958–10962. doi: 10.1128/JVI.00870-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volchkov VE, Feldmann H, Volchkova VA, Klenk HD. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamala JF, Lukwago L, Malimbo M, Nguku P, Yoti Z, Musenero M, Amone J, Mbabazi W, Nanyunja M, Zaramba S, et al. Ebola hemorrhagic fever associated with novel virus strain, Uganda, 2007–2008. Emerg Infect Dis. 2010;16:1087–1092. doi: 10.3201/eid1607.091525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Calder LJ, Wharton SA, Skehel JJ, Wiley DC. The central structural feature of the membrane fusion protein subunit from the Ebola virus glycoprotein is a long triple-stranded coiled coil. Proc Natl Acad Sci USA. 1998a;95:6032–6036. doi: 10.1073/pnas.95.11.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissenhorn W, Carfi A, Lee KH, Skehel JJ, Wiley DC. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol Cell. 1998b;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci USA. 2011;108:14216–14221. doi: 10.1073/pnas.1111497108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO Ebola Response Team. Ebola Virus Disease in West Africa - The First 9 Months of the Epidemic and Forward Projections. New England Journal of Medicine. 2014 doi: 10.1056/NEJMoa1411100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JA, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn AL, Hart MK. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE, Jr, Wilson IA. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol. 2013;20:363–370. doi: 10.1038/nsmb.2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.