Abstract
Coenzyme Q10 (CoQ10) deficiency has been associated with an increasing number of clinical phenotypes that respond to CoQ10 supplementation. In two siblings with encephalomyopathy, nephropathy and severe CoQ10 deficiency, a homozygous mutation was identified in the CoQ10 biosynthesis gene COQ2, encoding polyprenyl-pHB transferase. To confirm the pathogenicity of this mutation, we have demonstrated that human wild-type, but not mutant COQ2, functionally complements COQ2 defective yeast. In addition, an equivalent mutation introduced in the yeast COQ2 gene also decreases both CoQ6 concentration and growth in respiratory-chain dependent medium. Polyprenyl-pHB transferase activity was 33–45% of controls in COQ2 mutant fibroblasts. CoQ-dependent mitochondrial complexes activities were restored in deficient fibroblasts by CoQ10 supplementation, and growth rate was restored in these cells by either CoQ10 or uridine supplementation. This work is the first direct demonstration of the pathogenicity of a COQ2 mutation involved in human disease, and establishes yeast as a useful model to study human CoQ10 deficiency. Moreover, we demonstrate that CoQ10 deficiency in addition to the bioenergetics defect also impairs de novo pyrimidine synthesis, which may contribute to the pathogenesis of the disease.
INTRODUCTION
Coenzyme Q10 (CoQ10) is a vital molecule that transports electrons from mitochondrial respiratory chain complexes I and II to complex III (1). In addition, it functions as a cofactor for uncoupling proteins (2), as an antioxidant stabilizing plasma membrane and a regulator of the extracellularly-induced ceramide-dependent apoptotic pathway (1,3). CoQ10 also enhances survival of chemotherapy-treated cells (4) and is required for the stabilization of complex III in mitochondria (5). At least nine nuclear gene (COQ) products are involved in the CoQ biosynthesis complex pathway in yeast, which have homologous genes in all the species currently studied (6). This pathway and its regulation are still incompletely understood (1). CoQ10 deficiency (MIM 607426) has been associated with clinically heterogeneous diseases, which have been delineated into five major phenotypes: (i) an encephalomyopathic form (7,8), (ii) a predominantly ataxic form with cerebellar atrophy (9,10), (iii) a multisystem infantile variant with brain and renal disease (11,12), (iv) Leigh syndrome (13) and (v) isolated myopathy (14,15). It is anticipated that mutations in COQ genes or other components of the CoQ10 biosynthetic pathway are likely to cause these diverse syndromes (1).
We have recently demonstrated that two siblings with CoQ10 deficiency harbor a homozygous c.890 A>G mutation in the COQ2 gene (MIM 609825) (16), which encodes polyprenyl-p-hydroxybenzoate transferase (17). This mutation changes amino acid 297 from tyrosine into cysteine, in a conserved trans-membrane domain (16). To demonstrate pathogenicity of this mutation, we performed functional complementation in Saccharomyces cerevisiae using the BY4741Δcoq2 strain, which is unable to grow in nonfermentable carbon source. We have studied the role of CoQ10 supplementation on both respiratory chain complexes I + III and II + III and growth in deficient fibroblasts. Also, we have shown the requirement of uridine for maintaining growth of deficient fibroblasts demonstrating the dependence of these cells on pyrimidine biosynthesis.
RESULTS
Complementation of ΔCOQ2 yeast
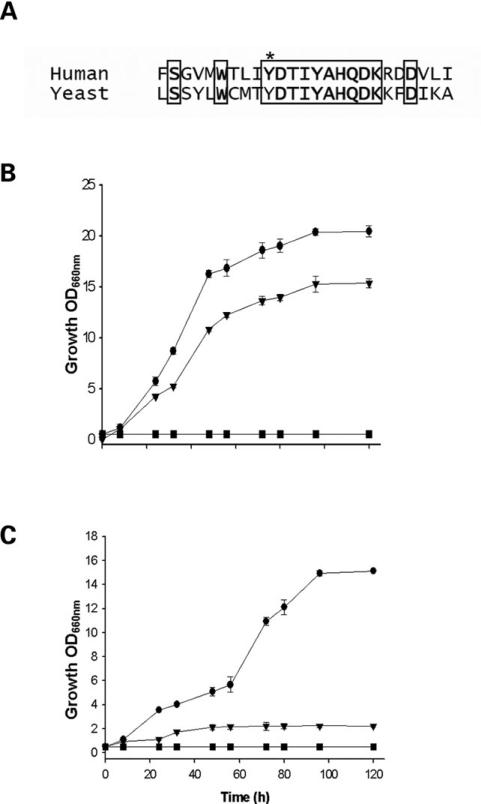
Figure 1A compares the 5′-nucleotide sequences of the cloned cDNA and shows that the human gene contains four ATG initiation codons. The third ATG produces a transcript of similar length to the yeast gene. Wild-type human COQ2 (hCOQ2) and yeast COQ2 (yCOQ2) cDNA sequences were sub-cloned into the yeast expression vector pYES2.1-TOPO TA/V5-His with a GAL promoter, and transformed into BY4741Δcoq2 strain, a null mutant strain for this gene. Increased growth in glycerol was observed when either homologous yCOQ2 or heterologous third ATG-initiated hCOQ2 was expressed in BY4741Δcoq2 (Fig. 1B). Lower growth was observed with cDNA initiated at the first or second ATG, and no growth was detected with hCOQ2 starting at the fourth ATG (Fig. 1B). The reported full-length hCOQ2 has been cloned as a hybrid molecule: the 3′-sequence was obtained from mRNA while the 5′-sequence was amplified from genomic DNA due to difficulties in amplifying the 5′-region of the mRNA (17). Complementation of COQ2 null yeast with this cDNA (initiating at the first ATG) induced ~50% growth at 120 h compared with yCOQ2 complementation (18), a similar growth rate as those we have obtained with first ATG hCOQ2 (Fig. 1B).
Figure 1.
Human COQ2 starting at the 3rd ATG can replace yeast COQ2. (A) Comparison of 5′ end of human and yeast COQ2 gene showing the four initiation codons (ATG). The initiation ATG in yeast gene corresponds to the 3rd ATG in human gene. (B) Culture densities of coq2 yeast strain transformed with clones of homologous yeast wild-type cDNA, and the heterologous human cDNAs starting from the four initiation codons. These cells were grown for 120 h in non-fermentable carbon source media (YPG). (C) Immunolocalization of human Coq2 peptide in mitochondria isolated from both wild-type (lane 1) and coq2 (lane 2) yeast strains. Lane 3 corresponds to mitochondria isolated from BY4741 coq2:hCOQ2 (3rd ATG), and lane 4 corresponds to mitochondria isolated from HeLa cells. Localization was performed using a polyclonal antibody against the human protein (Agrisera, Sweden). The inferior panel shows the same membrane probed with an antibody against yCox1p (molecular probes), a protein with known mitochondrial localization.
Localization of hCoq2p in yeast mitochondria
To determine the localization of human Coq2p protein in yeast mitochondria, an immunoblot was developed with an antibody raised against the human Coq2 peptide (Fig. 1C). This protein was not detected in either wild-type yeast or coq2 mutant strains, but it was detected in the mitochondria of both δcoq2:hCOQ2 and HeLa cells. The apparent molecular weight of Coq2p in HeLa cells is higher probably because of a different processing of the precursor polypeptide in yeast.
Complementation efficiency of mutated COQ2
To determine whether the mutation in hCOQ2 gene (16) caused a loss of function of the encoded protein affecting both CoQ biosynthesis and respiration, we carried out complementation experiments transforming the yeast yCOQ2 null mutant strain with cDNA cloned from patients, and the yeast gene engineered to harbor the c.783 A>G mutation (equivalent to hCOQ2 c.890 A>G mutation). Cells expressing the mutant yeast gene displayed a lower growth rate than those expressing the wild-type gene (Fig. 2B), demonstrating that functional complementation is sufficiently sensitive to study the effect of a missense mutation in a CoQ biosynthetic gene. Also, transformation of BY4741 Δcoq2 yeast strain with mutated hCOQ2 showed a considerably lower respiration-dependent growth than with wild-type hCOQ2, which was, however, still significantly increased compared with the deleted strain (Fig. 2C).
Figure 2.
The c.890 A>G mutation decreases the rate of growth in transformed yeast strains. (A) Comparison of the sequences of human and yeast Coq2 proteins. The asterisk indicates the mutated Y>C residue. Conserved amino acids inside a frame. (B) Growth curves of the Δcoq2 yeast strain (squares), and of this strain transformed with: wild-type yCOQ2 (circles) or c.783 A>G mutated yCOQ2 (triangles). (C) Growth of the Δcoq2 yeast strain (squares) transformed with wild-type (circles) or mutant (triangles) hCOQ2.
Effect on CoQ biosynthesis
Growth results correlated with the content of Coenzyme Q6 (CoQ6) in the different transformed strains (Table 1). The expression of yCOQ2 caused an accumulation of CoQ6 in transformed yeasts in the range of the wild-type strain, and strains harboring yCOQ2 (c.783 A>G) attained CoQ levels ~59% of wild-type. These results demonstrate the deleterious effect of the mutation on the Coq2 protein, which, however, apparently retains some residual enzymatic activity. The expression of wild-type hCOQ2 gene increased CoQ6 levels to ~64% of control, while expression of the mutated sequence produced CoQ6 levels that were only 11% of transformed cells with the human wild-type allele.
Table 1.
Content of CoQ6 in yeast strains after 120 h growing in non-fermentable media (YPG)
| Strain | Coenzyme Q6 (ng/mg protein ± SD) | Demethoxy-Q6 (ng/mg protein ± SD) | CoQ6/DMQ6 Ratio |
|---|---|---|---|
| BY4741 | 1448 ± 20 | 122 ± 10 | 11.86 |
| BY4741 Δcoq2 | ND | ND | — |
| BY4741 Δcoq2:hCOQ2 | 842 ± 12 | 73 ± 4 | 11.52 |
| BY4741 Δcoq2:hCOQ2 (c.890 A>G) | 92 ± 3 | 42 ± 9 | 2.19 |
| BY4741 Δcoq2:yCOQ2 | 1309 ± 45 | 110 ± 18 | 11.90 |
| BY4741 Δcoq2:yCOQ2 (c.783 A>G) | 774 ± 70 | 230 ± 25 | 3.36 |
ND, non-detectable.
Analysis of demetoxy-Q6 (DMQ) levels, an intermediate of coenzyme Q biosynthesis, showed an increase of DMQ relative to CoQ6, in strains transformed with the mutated genes (Table 1), indicating an inhibition of the biosynthetic process also downstream of CoQ2p.
COQ2 expression in patient's fibroblasts
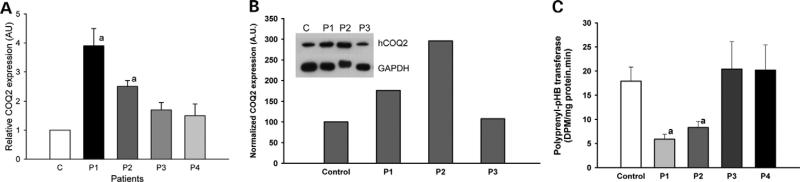
We then analyzed the expression of COQ2 in patient's fibro-blasts by real-time-PCR (Fig. 3A). mRNA levels of this gene were significantly increased in patients harboring COQ2 mutations (16) compared with controls and other CoQ10-deficient fibroblasts isolated from ataxic patients, which do not harbor mutations in this gene (10). Furthermore, immunoblot analysis of Coq2p in fibroblasts demonstrated that cells from both P1 and P2 patients contained higher amount of protein than both control and P3 patient fibroblasts (Fig. 3B). We also measured polyprenyl-pHB transferase activity using a sensitive radioactive method (19). This activity was significantly lower in fibroblasts of patients with hCOQ2 (c.890 A>G) gene (Fig. 3C) relative to controls and CoQ10-deficient fibroblasts from ataxic patients (10). Increased expression of both mRNA and protein appears to be a compensatory mechanism for the enzymatic deficiency in the patient's fibroblasts.
Figure 3.
COQ2 is over-expressed in fibroblasts of patients with mutation and Coq2p shows lower activity. (A) Expression of COQ2 gene measured by real-time PCR in patients 1 (P1) and 2 (P2) harboring the c.890 A>G mutation, compared with fibroblasts from control and patients 3 and 4, who have CoQ10 deficiency but no COQ2 mutation (10). a: Significant versus control (P < 0.01) and both P3 and P4 (P < 0.05). (B) Content of Coq2p in human fibroblasts. a: Significant versus both control and P3 (P ≤ 0.01). Significant versus both control and P3 (P < 0.05). (C) Biochemical analysis of the polyprenyl-pHB transferase activity in mitochondria isolated from fibroblasts of the patients described earlier. a: Significant versus both control and P3 (P < 0.01).
Effect of CoQ10 on mitochondrial complexes activities
It has been shown that CoQ added to human cells reaches the inner mitochondrial membrane and acts on the respiratory chain (20). To demonstrate whether CoQ10 deficiency was responsible for mitochondria defects observed in CoQ10− deficient patients (12), fibroblasts were incubated for 24 h with CoQ10, and both complexes I + III and II + III activities were determined (Table 2). Both activities were significantly increased in deficient fibroblasts, which were not affected in control fibroblasts.
Table 2.
Effect of supplementation of CoQ10 on the CoQ-dependent mitochondrial complexes activities in human fibroblasts
| Samples | Complex I + III (nmol/min/mg protein) | Complex II + III (nmol/min/mg protein) | ||
|---|---|---|---|---|
| No addition | 10 μm CoQ10 | No addition | 10 μm CoQ10 | |
| Control | 410 ± 36 | 445 ± 27 | 3.5 ± 0.23 | 3.8 ± 0.41 |
| P1 | 248 ± 25 | 386 ± 44a | 1.7 ± 0.02 | 3.6 ± 0.12a |
| P2 | 301 ± 37 | 480 ± 56a | 1.9 ± 0.07 | 3.1 ± 0.15a |
Significant versus no addition.
Effects of CoQ10 and uridine on fibroblasts growth rates
CoQ10 deficient fibroblasts showed slow rates of growth. To demonstrate that this phenotype was due to CoQ10 deficiency, cells (100 ± 20 cells/cm2) were seeded and grown in a medium supplemented with serum-solubilized CoQ10. Growth was significantly increased in deficient fibroblasts incubated with a supplement of CoQ10 (Fig. 4A). CoQ10 is required for the biosynthesis of pyrimidine nucleotides because it is an essential co-factor for dihydro-orotate dehydrogenase, an enzyme located in the inner mitochondrial membrane (21). To demonstrate that the deficiency of CoQ10 affects pyrimidine supply, and consequently cell growth, we incubated CoQ10-deficient fibroblasts with uridine and observed that growth was also increased compared with both control and respiratory chain-deficient fibroblasts from a patient with mitochondrial encephalomyopathy without CoQ10 deficiency (Fig. 4B). No cumulative effect of uridine and CoQ10 was noted (data not shown).
Figure 4.
Growth of CoQ10 deficient fibroblasts can be restored by exogenous CoQ10 or uridine. (A) Addition of CoQ10 to culture media containing 20% FCS induced significant increases of growth of fibroblasts of patients harboring the COQ2 mutation compared with control fibroblasts. Black bars: no CoQ10. Grey bars: plus 10 μm CoQ10. a: Significant versus no addition of CoQ10 (P ≤ 0.01). (B) Growth of fibroblasts in media containing 10% FCS supplemented with 10 μm uridine. Black bars: control fibroblasts; grey bars: fibroblasts from a patient with mitochondrial respiratory chain deficiency but normal concentration of CoQ10; white bars: fibroblasts harboring the COQ2 mutation (patient 2). a: Significant versus no addition of uridine (P < 0.05).
DISCUSSION
Despite the fact that CoQ10 deficiency has been described more than 15 years ago, its genetic bases have remained elusive until this year, when mutations were identified in two genes involved in CoQ10 biosynthesis, COQ2 and PDSS2 (16,22). The product of COQ2 catalyzes the transfer of para-hydroxybenzoate to the polyprenyl chain, one of the initial steps of CoQ biosynthesis. The aim of this work was to demonstrate the pathogenicity of the missense COQ2 mutation found in our patients using a functional complementation approach in S. cerevisiae, and to analyze its consequences on the mitochondrial electron transport chain and cell proliferation.
Human COQ2 contains four possible ATG codons on exon 1. The reported hCOQ2 sequence starts from ATG number 1. We observed complementation with constructs initiating from each of the first three ATG, but not with ATG number 4 possibly because the sequence between the third to fourth ATGs contains a critical signal for mitochondrial importation in yeast (18).
The highest complementation efficiency was achieved with constructs initiating at the third ATG, this sequence is the most similar to the yeast mitochondrial importation pre-sequence. However, analysis of COQ2 genes in other mammalian species such as dog and rat revealed that they contain only one possible ATG initiation codon, which corresponds to the human fourth ATG. We are currently investigating the functional significance of the four ATG codons in human cells.
Both the mutated human COQ2 gene and its yeast homolog engineered to harbor the corresponding mutation, failed to fully complement COQ2 deficient yeast strain. Nevertheless, complementation studies clearly indicate that mutated proteins still retain some enzymatic activity because yeast can grow on non-fermentable substrates and accumulate some amount of CoQ6, although at a significantly lower rate than the wild-type strains. Direct biochemical assays on patients P1 and P2 fibro-blasts confirm the presence of residual enzymatic activity, which correlates with the amount of Coq2p detected by western blot. The residual Coq2 function is probably essential for embryogenesis, and may explain why the patients did not present symptoms of the disease until 1 year of age. In both patients P1 and P2, COQ2 mRNA and Coq2 peptides are also hyper-expressed, suggesting that there is a feedback control mechanism regulating COQ2 expression in cells.
Interestingly, yeast cells transformed with the mutated genes show accumulation of DMQ, an intermediate of coenzyme Q biosynthesis that is synthesized by reactions downstream of Coq2p. This apparent paradox can be explained by the fact that enzymes that catalyze coenzyme Q biosynthesis in yeast are thought to function in a multienzyme complex (20). In this case, the mutation in Coq2 proteins not only impairs enzymatic function of Coq2p, but probably affects the whole complex, interfering also with some of the downstream enzymatic steps.
In a second set of experiments, we analyzed the effect of COQ2 deficiency in fibroblasts. We have previously shown that a CoQ10 analogue, decyl-ubiquinone, can rescue the complex II + III enzymatic defect in vitro (12). We have now checked the effect of ubiquinone supplementation in vivo on cell proliferation and mitochondrial complexes activities. Supplementation of human cells with different isoforms of CoQ shows that a small amount can reach inner mitochondrial membrane (23). Using this approach, we have shown that long-term incubation of deficient fibroblasts with CoQ10 increases significantly the activity of both complexes I + III and II + III. These results support the cause of mitochondrial dysfunction on the deficiency, and would explain the positive results observed after the treatment of deficient patients with CoQ10 (10,12).
We then analyzed if we could modulate the growth deficient phenotype in these cells. Surprisingly, the effect of uridine addition was even more pronounced than that of CoQ10 suggesting that this phenotype is largely due to an insufficient supply of nucleotides rather than to an impairment of ATP production. We believe that the relative lower efficiency of CoQ10 in rescuing the defect in proliferation is probably due to a low efficiency of its incorporation in cells (1,23).
Taken together, our results demonstrate the pathogenicity of the COQ2 mutation and that yeast is an optimal model to study the effects of mutations in COQ genes, because it is sensitive enough to unveil defects due to missense mutations. The high homology of yeast and human COQ genes may allow complementation studies to identify other human genes involved in CoQ10 deficiency. Moreover, we demonstrate that the pathogenesis of CoQ10 deficiency is related not only to a defect in bioenergetics, but also to an impairment of pyrimidine metabolism.
MATERIALS AND METHODS
Fibroblast cultures
Fibroblasts from CoQ10 deficient patients (P1 and P2) (12,16), from patients with cerebellar ataxia and CoQ10 deficiency (10) and controls were plated in separate six-well plates (40 000 cells/well) and cultured using DMEM with 20% fetal calf serum (FCS). Also, HeLa cells were grown in a similar procedure. Supplemental CoQ10 pre-diluted in FCS was added to the plates at a final concentration of 10 μm.
Cloning of human and yeast COQ2
yCOQ2 was cloned into pGEM-T Easy (Promega) and in pYES2.1V5HisTOPO (Invitrogen) vectors using primers listed in Table 3. Initially, hCOQ2 was cloned in a two-step protocol similarly to what has been described (17). pEGFPN1 was used as intermediate vector because of convenient cloning sites. The 5′ portion corresponding to each of the four ATG was amplified from genomic DNA and sub-cloned into pCRIITOPO (Invitrogen). The 3 portion was amplified from cDNA (both wild-type and patient's) and sub-cloned into the same vector. The fragments were then sequentially cloned into the PstI site of pEGFPN1. The full length cDNA was then cloned into pYES2.1V5HisTOPO.
Table 3.
Oligonucletides used in PCR amplifications
| Primer name | Sequence 5′ → 3′ |
|---|---|
| hCOQ2-Fext(ATG1) | 5′-ATGACCCCAATTTCACAA-3′ |
| hCOQ2-Rext(ATG1) | 5′-ACTCAAGGCCATCTCAAA-3′ |
| hCOQ2-Fext(ATG2) | 5′-AAGGATGAGGAAAGGTTCTG-3′ |
| hCOQ2-Rext(ATG2) | 5′-TTGTATCAGATTTTGTATTCAAATC-3′ |
| hCOQ2-Fext(ATG3) | 5′-GAATGACGTCAATCCGA-3′ |
| hCOQ2-Rext(ATG3) | 5′-TGTAAAAAATGTATTAAAAATTC-3′ |
| hCOQ2-Fext(ATG4) | 5′-AGCTCGTCCTGGCCTCAC-3′ |
| hCOQ2-Rext(ATG4) | 5′-TGGGCAGAAGAATGTATCAACTA-3′ |
| hCOQ2-Fint | 5′-TTACAAGAACAGCCAATCGTC-3′ |
| hCOQ2-Rint | 5′-TGTCCTGATGGGCATAAATAGTG-3′ |
| yCOQ2-Fext | 5′-CAACTAATGTTTATTTGGCAGAG-3′ |
| yCOQ2-Rext | 5′-CTACAAGAATCCAAACAGTCTCA-3′ |
| yMUTAG-F | 5′-GTATGTTGGGTATTTTCGGTG-3′ |
| yMUTAG-R | 5′-CGTATATAGTATCGCAAGTCATACA-3′ |
| hCOQ-RQ-F | 5′-AGAACAGCCAATCGTTCCAATAGC-3′ |
| hCOQ-RQ-R | 5′-CCCAATTAAATGTCAAGCCCAAGG-3′ |
Bold-face character is the mismatched nucleotide used in site-directed mutagenesis.
Finally, the ATG3-hCOQ2 was directly cloned from reverse transcriptase (RT)-PCR amplified from total fibroblast RNA in a single reaction in order to analyze patient samples. Site-directed mutagenesis of wild-type yCOQ2 was carried out by PCR using primers yMUTAG-F and yMUTAG-R. All inserts were directly sequenced using standard protocols and the ABI-Prism 310 automated sequencer (Perkin Elmer).
Yeast complementation analysis
Yeast coq2 mutants were transformed with the pYES2 vector containing the different versions of hCOQ2, and both wild-type and site-directed mutant yeast COQ2 and the empty vector for complementation experiments. Growth rate in nonfermentable carbon source (YPG medium) was determined as a marker of functional complementation.
CoQ determination
CoQ10 was extracted after addition of 4 ml hexane-ethanol (5/ 2 v/v) and vortexed for 2 min. After centrifugation at 1000 g at room temperature for 5 min, the upper phase was carefully transferred into a 20-ml glass scintillation vial (performed twice for each sample). The combined extract was evaporated under a gentle stream of N2 gas and the residue was dissolved in 0.1 ml of 1-propanol. An aliquot of 50 μl of the extract was directly injected into an HPLC system with a C18 reversed-phase column and an electrochemical detector.
Immunoblotting and real-time pcr
Western blotting was performed using standard methods, and a primary polyclonal antibody developed in hens against human Coq2 peptide, manufactured by Agrisera (Sweden), was used. The expression of hCOQ2 in fibroblasts was analyzed by SYBR Green quantitative PCR using mRNA extracts and primers hCOQ-RQ-F and hCOQ-RQ-R. Actin was used as housekeeping control gene.
Polyprenyl-pHB transferase activity
The activity was assayed by measuring the incorporation of radioactive 4-hydroxybenzoate (pHB) into nonaprenyl-4-hydroxybenzoate. Isolated mitochondria (0.1–1 mg protein) were mixed with assay buffer (50 mm phosphate buffer, pH 7.5, 10 mm MgCl2, 5 mm EGTA) containing 1 mm PMSF, 20 μg/ml each of the protease inhibitors chymostatin, leupeptin, antipain and pepstatin A, 5 μm solanesyl pyrophosphate solubilized in detergent solution (1% in water) and 105 DPM of [U-14C]pHB. Sufficient volume of a 10% detergent stock solution was also added to the reaction medium to achieve a final detergent concentration of up to 1%. The following detergents were tested: Triton X-100, Chaps, sodium cholate, sodium deoxycholate, lysophosphatidyl choline and octylglucoside. After incubation for 30 min at 37°C with gentle stirring, the reaction was stopped by chilling samples to 4°C. Prenylated [U-14C]pHB was separated by organic extraction with hexane and then measured using a liquid scintillation counter. Specific activity was expressed as DPM min−1mg protein−1.
Mitochondrial enzyme activities
Mitochondria-enriched fractions were obtained according to Fernández-Ayala et al. (23). and used to determine complex I + III and II + III activities. In both cases, mitochondria were incubated in 40 mm sodium phosphate buffer pH 7.5 plus 0.25 mm KCN and containing either 0.2 mm NADH or 5 mm succinate, and reduction of beef-heart cytochrome c (0.5 mm) was monitored spectrophotometrically at 550 nm for 5 min. To discriminate the rotenone-sensitive NADH-cytochrome c reductase activity (Complex I + III) from the rotenone-insensitive NADH-cytochrome c reductase activity due to the NADH-cytochrome b5 reductase located in the outer mitochondrial membrane, 5 μm rotenone was added for the last 2 min of incubation.
Statistical analysis
All results are expressed as mean ± SEM. Serial measurements were analyzed by using two-way ANOVA with Tukey's post-hoc test using SigmaStat software from SPSS Science (Chicago, IL). The level of significance was set at P < 0.05.
ACKNOWLEDGEMENTS
This research was supported by European Union contract LSHB-CT-2004-005151. J.M.L. and M.D.C. are supported by the EU contract and A.R.H. is a fellow from the Spanish Instituto de Salud Carlos III.
Footnotes
Conflict of Interest statement. None declared.
REFERENCES
- 1.Turunen M, Olsson J, Dallner G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta. 2004;1660:171–199. doi: 10.1016/j.bbamem.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Echtay KS, Winkler E, Klingenberg M. Coenzyme Q is an obligatory cofactor for uncoupling protein function. Nature. 2000;408:609–613. doi: 10.1038/35046114. [DOI] [PubMed] [Google Scholar]
- 3.Navas P, Manuel Villalba J. Regulation of ceramide signaling by plasma membrane coenzyme Q reductases. Methods Enzymol. 2004;378:200–206. doi: 10.1016/S0076-6879(04)78016-7. [DOI] [PubMed] [Google Scholar]
- 4.Brea-Calvo G, Rodriguez-Hernandez A, Fernández-Ayala DJM, Navas P, Sanchez-Alcazar JA. Chemotherapy induces an increase in coenzyme Q10 levels in cancer cell lines. Free Radic. Biol. Med. 2006;40:1293–1302. doi: 10.1016/j.freeradbiomed.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 5.Santos-Ocana C, Do TQ, Padilla S, Navas P, Clarke CF. Uptake of exogenous coenzyme Q and transport to mitochondria is required for bc1 complex stability in yeast coq mutants. J. Biol. Chem. 2002;277:10973–10981. doi: 10.1074/jbc.M112222200. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez-Aguilera JC, Gavilan A, Asencio C, Navas P. The role of ubiquinone in Caenorhabditis elegans longevity. Ageing Res. Rev. 2005;4:41–53. doi: 10.1016/j.arr.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Ogasahara S, Engel AG, Frens D, Mack D. Muscle coenzyme Q deficiency in familial mitochondrial encephalomyopathy. Proc. Natl. Acad. Sci. 1989;86:2379–2382. doi: 10.1073/pnas.86.7.2379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sobreira C, Hirano M, Shanske S, Keller RK, Haller RG, Davidson E, Santorelli FM, Miranda AF, Bonilla E, Mojon DS, et al. Mitochondrial encephalomyopathy with coenzyme Q10 deficiency. Neurology. 1997;48:1238–1243. doi: 10.1212/wnl.48.5.1238. [DOI] [PubMed] [Google Scholar]
- 9.Lamperti C, Naini A, Hirano M, De Vivo DC, Bertini E, Servidei S, Valeriani M, Lynch D, Banwell B, Berg M, et al. Cerebellar ataxia and coenzyme Q10 deficiency. Neurology. 2003;60:1206–1208. doi: 10.1212/01.wnl.0000055089.39373.fc. [DOI] [PubMed] [Google Scholar]
- 10.Artuch R, Brea-Calvo G, Briones P, Aracil A, Galvan M, Espinos C, Corral J, Volpini V, Ribes A, Andreu AL, et al. Cerebellar ataxia with coenzyme Q10 deficiency: Diagnosis and follow-up after coenzyme Q10 supplementation. J. Neurol. Sci. 2006;246:153–158. doi: 10.1016/j.jns.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 11.Rotig A, Appelkvist EL, Geromel V, Chretien D, Kadhom N, Edery P, Lebideau M, Dallner G, Munnich A, Ernster L, Rustin P. Quinone-responsive multiple respiratory-chain dysfunction due to widespread coenzyme Q10 deficiency. Lancet. 2000;356:391–395. doi: 10.1016/S0140-6736(00)02531-9. [DOI] [PubMed] [Google Scholar]
- 12.Salviati L, Sacconi S, Murer L, Zacchello G, Franceschini L, Laverda AM, Basso G, Quinzii C, Angelini C, Hirano M, et al. Infantile encephalomyopathy and nephropathy with CoQ10 deficiency: a CoQ10-responsive condition. Neurology. 2005;65:606–608. doi: 10.1212/01.wnl.0000172859.55579.a7. [DOI] [PubMed] [Google Scholar]
- 13.Van Maldergem L, Trijbels F, DiMauro S, Sindelar PJ, Musumeci O, Janssen A, Delberghe X, Martin JJ, Gillerot Y. Coenzyme Q-responsive Leigh's encephalopathy in two sisters. Ann. Neurol. 2002;52:750–754. doi: 10.1002/ana.10371. [DOI] [PubMed] [Google Scholar]
- 14.Lalani SR, Vladutiu GD, Plunkett K, Lotze TE, Adesina AM, Scaglia F. Isolated mitochondrial myopathy associated with muscle coenzyme Q10 deficiency. Arch. Neurol. 2005;62:317–320. doi: 10.1001/archneur.62.2.317. [DOI] [PubMed] [Google Scholar]
- 15.Horvath R, Schneiderat P, Schoser BG, Gempel K, Neuen-Jacob E, Ploger H, Muller-Hocker J, Pongratz DE, Naini A, DiMauro S, Lochmuller H. Coenzyme Q10 deficiency and isolated myopathy. Neurology. 2006;66:253–255. doi: 10.1212/01.wnl.0000194241.35115.7c. [DOI] [PubMed] [Google Scholar]
- 16.Quinzii C, Naini A, Salviati L, Trevisson E, Navas P, Dimauro S, Hirano M. A mutation in para-hydroxybenzoate-polyprenyl transferase (COQ2) causes primary coenzyme Q10 deficiency. Am. J. Hum. Genet. 2006;78:345–349. doi: 10.1086/500092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forsgren M, Attersand A, Lake S, Grunler J, Swiezewska E, Dallner G, Climent I. Isolation and functional expression of human COQ2, a gene encoding a polyprenyl transferase involved in the synthesis of CoQ. Biochem. J. 2004;382:519–526. doi: 10.1042/BJ20040261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uchida N, Suzuki K, Saiki R, Kainou T, Tanaka K, Matsuda H, Kawamukai M. Phenotypes of fission yeast defective in ubiquinone production due to disruption of the gene for p-hydroxybenzoate polyprenyl diphosphate transferase. J. Bacteriol. 2000;182:6933–6939. doi: 10.1128/jb.182.24.6933-6939.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burón MI, Herman MD, Alcain FJ, Villalba JM. Stimulation of polyprenyl 4-hydroxybenzoate transferase activity by sodium cholate and 3-[(cholamidopropyl)dimethylammonio]-1-propanesulfonate. Anal. Biochem. 2006;353:15–21. doi: 10.1016/j.ab.2006.03.029. [DOI] [PubMed] [Google Scholar]
- 20.Marbois B, Gin P, Faull KF, Poon WW, Lee PT, Strahan J, Shepherd JN, Clarke CF. Coq3 and Coq4 define a polypeptide complex in yeast mitochondria for the biosynthesis of coenzyme Q. J. Biol. Chem. 2005;280:20231–20238. doi: 10.1074/jbc.M501315200. [DOI] [PubMed] [Google Scholar]
- 21.Jones ME. Pyrimidine nucleotide biosynthesis in animals: genes, enzymes, and regulation of UMP biosynthesis. Annu. Rev. Biochem. 1980;49:253–279. doi: 10.1146/annurev.bi.49.070180.001345. [DOI] [PubMed] [Google Scholar]
- 22.Lopez LC, Schuelke M, Quinzii CM, Kanki T, Rodenburg RJ, Naini A, Dimauro S, Hirano M. Leigh syndrome with nephropathy and CoQ10 deficiency due to decaprenyl diphosphate synthase subunit 2 (PDSS2) mutations. Am. J. Hum. Genet. 2006;79:1125–1129. doi: 10.1086/510023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fernández-Ayala DJM, López-Lluch G, García-Valdés M, Arroyo A, Navas P. Specificity of coenzyme Q10 for a balanced function of respiratory chain and endogenous ubiquinone biosynthesis in human cells. Biochim. Biophys. Acta. 2005;1706:174–183. doi: 10.1016/j.bbabio.2004.10.009. [DOI] [PubMed] [Google Scholar]