Abstract
Microorganisms associated with the roots of plants have an important function in plant growth and in soil carbon sequestration. Rice cultivation is the second largest anthropogenic source of atmospheric CH4, which is a significant greenhouse gas. Up to 60% of fixed carbon formed by photosynthesis in plants is transported below ground, much of it as root exudates that are consumed by microorganisms. A stable isotope probing (SIP) approach was used to identify microorganisms using plant carbon in association with the roots and rhizosphere of rice plants. Rice plants grown in Italian paddy soil were labeled with 13CO2 for 10 days. RNA was extracted from root material and rhizosphere soil and subjected to cesium gradient centrifugation followed by 16S rRNA amplicon pyrosequencing to identify microorganisms enriched with 13C. Thirty operational taxonomic units (OTUs) were labeled and mostly corresponded to Proteobacteria (13 OTUs) and Verrucomicrobia (8 OTUs). These OTUs were affiliated with the Alphaproteobacteria, Betaproteobacteria, and Deltaproteobacteria classes of Proteobacteria and the “Spartobacteria” and Opitutae classes of Verrucomicrobia. In general, different bacterial groups were labeled in the root and rhizosphere, reflecting different physicochemical characteristics of these locations. The labeled OTUs in the root compartment corresponded to a greater proportion of the 16S rRNA sequences (∼20%) than did those in the rhizosphere (∼4%), indicating that a proportion of the active microbial community on the roots greater than that in the rhizosphere incorporated plant-derived carbon within the time frame of the experiment.
INTRODUCTION
The interaction between plants and microorganisms within the root and in the rhizosphere is complex and poorly understood. Early studies focused on specific plant growth-promoting bacteria (1, 2) and more recently on characterizing the rhizosphere microbial community (3). For example, studies with Arabidopsis thaliana, a model plant species, have shown that the root endophytic microbial communities are highly specific (4, 5). In rice, the microbial communities associated with the rhizosphere and the phyllosphere have been characterized by a metaproteogenomic approach (6), and a metagenomic approach was used to characterize the endophytic root community in rice (7).
A variety of factors act to shape the microbial community within the roots and in the rhizosphere, including the volume and nature of carbon substrates transported by the plant to the roots (8) and highly evolved signaling and interaction mechanisms between plants and microbes and the plant immune system (9). Plants actively recruit and sustain microorganisms in the root environment in part by the translocation of organic compounds from the leaves to the roots and into the rhizosphere that serve as growth substrates. In annual plants, this has been shown to account for 30 to 60% of net fixed carbon, 40 to 90% of which is excreted by the root and ultimately sequestered or respired by the root-associated microorganisms (10).
Rice is a major world food source, with 2012 production estimated at 720 million tons (http://faostat3.fao.org/), which was exceeded only by production of sugarcane and maize crops. Rice is cultivated in flooded soil, which becomes predominantly anoxic, leading to CH4 formation by methanogenic archaea (11). The estimate for CH4 emission associated with rice cultivation from 1996 to 2001 was 112 Tg year−1 (12), making it the largest anthropogenic source of atmospheric CH4 after production associated with farming of ruminant livestock (13). CH4 is the most important greenhouse gas after CO2, contributing about 30% to the total net anthropogenic radiative forcing (13). Up to 60% of the CH4 released from rice field soil originates from fresh plant carbon in the form of root exudates or decomposing root material (14). In addition to supplying organic carbon to the soil that is respired to CH4, the rice vascular system acts as a conduit for CH4 from the soil to the atmosphere, which bypasses the oxic-anoxic interface near the soil surface where aerobic methanotrophic bacteria consume CH4 (15).
A variety of microorganisms can colonize the root (16, 17) and rhizosphere (18, 19) and can use plant-derived compounds as a growth substrate. The incorporation of plant carbon by bacteria (19) and archaea (18, 20, 21) in the rice rhizosphere has been studied. Also, the association between root longevity and fungal colonization has been studied (22), indicating the particular importance of these organisms in regulating nutrient cycles and metabolizing root-derived organic compounds.
Stable isotope probing (SIP) is an approach that can be used to identify the microorganisms consuming plant carbon. The principle is that plants are cultivated in chambers supplied with 13CO2 and the microorganisms incorporating this plant carbon into biomass become enriched with 13C. 13CO2 pulse-labeling experiments in grassland soils demonstrated that the incorporation of photosynthesized 13C into soil microbial biomass occurs in less than 24 h with an isotope half-life of 4.7 days and maximum incorporation into microbial RNA in 4 to 8 days (23, 24). Labeled and unlabeled microbial RNA can be separated by density gradient centrifugation in order to identify the microbial communities using plant carbon. This approach has been used to identify archaea (18, 21), bacteria (19, 25–27), and fungi (26) incorporating root carbon, as well as to identify bacterial endophytes of plant shoots (28). In particular, the combination of RNA-SIP with 454 pyrosequencing represents a powerful molecular tool for the sensitive detection of labeled microorganisms (29).
Previous studies have characterized the microbial diversity in the rice rhizosphere (6) as well as rice root endophytes (7). Other studies have used a SIP approach to identify the archaea (18) and bacteria (19) that incorporate rice plant carbon in the rhizosphere. A follow-up to the SIP experiments was worthwhile since 16S rRNA amplicon sequencing technology now enables a deeper and higher-resolution analysis than was previously available. Building on previous studies, our aim was to characterize and compare the root and rhizosphere microbial communities of rice and determine which microbial groups consume plant carbon in these compartments.
MATERIALS AND METHODS
Planted rice microcosms.
Air-dried soil was collected in spring 2009 from an experimental rice field at the Italian Rice Research Institute in Vercelli. Soil samples were crushed and sieved (<2 mm) prior to use. Soil characteristics have been previously reported (30). Rice seeds (Oryza sativa, Japonica group, cultivar Koral) were germinated for 10 days on moist filter paper at room temperature and then transplanted into handmade nylon root bags (25-μm mesh, 15 cm in width by 18 cm in height) containing 0.8 kg dry Vercelli paddy soil and inserted into pots containing a further 1.0 kg of soil. In total were 15 pots, corresponding to five treatments performed in triplicate as described below. Each pot contained a single rice plant. Pots were flooded with 940 ml of demineralized water and 45 ml of fertilizer solution (31). The plants were incubated in a Conviron PGV36 phytochamber (Winnipeg, Canada) with a 12-h photoperiod, a light intensity of 860 μmol m−2 s−1, 70% humidity, and a 28/22°C day/night temperature cycle. Water levels in the pots were continuously maintained at 4 to 5 cm above the soil surface during the entire growth of the plants. Starting on day 45 after transplantation, when rice plants were in the late vegetative growth stage, labeling chambers (18) were placed over the plants. Pulse-labeling was performed seven times per day (ca. every 60 min) for 10 days. During the first 5 days, plants were in 5-liter chambers and 15-ml CO2 pulses were administered; during the subsequent 5 days of labeling, the plants were in 7-liter chambers and received 25-ml pulses of CO2. In general, labeling conditions were similar to those used by Lu and Conrad (18).
A schema depicting the treatments and analyses performed in the study is shown in Fig. S1 in the supplemental material. Treatments were as follows: microcosms pulsed with 13CO2 (13C, 99%; Cambridge Isotope Laboratories, Inc., USA); microcosms pulsed with unlabeled CO2 (Messer Group GmbH, Germany); microcosms pulsed with 13CO2 during the dark phase (diffusion control); plants not in chambers and exposed to ambient CO2 only (planted control); and finally, soil not containing rice plants (unplanted control). The control diffusion (CD) treatment was established to determine whether microorganisms in the root or rhizosphere environments could be labeled by 13CO2 transported through the plant. CO2 uptake was low for this control, and therefore, it was pulsed only three times per day during the 10 days, in between which the chambers were opened to remove background unlabeled CO2 that had accumulated.
Destructive sampling was performed at the end of the incubation. Rhizosphere soil, defined here as the layer of soil adhered to the root surface, was gently removed by hand while wearing clean nitrile gloves. Aliquots of rhizosphere soil were immediately placed in 2-ml screw-cap tubes and frozen in liquid nitrogen. Roots, from which the rhizosphere soil had been manually removed, were placed in small plastic bags and frozen in liquid nitrogen. Frozen samples were subsequently stored at −80°C until RNA extraction.
The isotopic signature of the rhizosphere soil and dried plant material was determined in a NA 1110 CN elemental analyzer (CE Instruments, Rodano, Italy), interfaced to a Delta Plus isotope mass spectrometer with a ConFlo III interface (Finnigan MAT, Bremen, Germany) (32). Rhizosphere soil samples were incubated with 1 M HCl at 65°C overnight before the measurement. A similar analysis of the dried plant material was performed by pulverizing with a mortar and pestle, before measurement. These measurements were performed at the Institute for Soil Science and Forest Nutrition (IBW) at the University of Göttingen (Göttingen, Germany).
Gas and soil pore water analyses.
Gas samples (CO2 and CH4) were taken from the labeling chambers every hour during pulse-labeling using a Shimadzu GC-8A gas chromatograph equipped with a flame ionization detector and methanizer. Measurements of soil pore water were performed daily by collection into Venoject blood collection tubes, as described previously (31). Stable isotope 13C/12C ratios in gas samples and soil pore water were determined using gas chromatography combustion-isotope ratio mass spectrometry (GCC-IRMS). A GC-IsoLink with a ConFlo IV interface (Thermo Fisher Scientific, Bremen, Germany) was connected with the Delta V Advantage isotope ratio mass spectrometer (Thermo Fisher Scientific, Bremen, Germany).
RNA purification.
RNA extractions were performed as described by Ma et al. (33). The first bead-beating lysate with Tris buffer was discarded since it was heavily contaminated with humus-like substances and contained very little or no detectable RNA. The extracts from the two subsequent rounds of bead-beating with lysis buffer were pooled and processed as described previously (33). Frozen roots were pulverized with a mortar and pestle, and 0.5 g was extracted using the same protocol. Traces of DNA were removed, and its absence was verified by PCR as described previously (34). RNA integrity was checked by electrophoresis using Experion RNA High Sense chips (Experion; Bio-Rad) as shown in Fig. S2 in the supplemental material and by spectrophotometry (NanoDrop 1000 spectrophotometer; Thermo).
Isopycnic centrifugation.
The isopycnic centrifugation of the RNA in cesium trifluoroacetate (CsTFA; GE Healthcare) gradients and RNA purification were performed as described previously (34). RNA was stored at −80°C until cDNA synthesis.
Reverse transcription.
RNA was converted to single-stranded cDNA by reverse transcription using random hexamer primers as previously described (34). The cDNA was stored at −20°C for further analyses.
454 pyrosequencing.
Pyrosequencing was used to determine the microbial composition in the SIP fractions and from unfractionated RNA from the diffusion and planted controls. PCR primers F515 and R806 were selected since they offer a wide coverage of bacterial and archaeal taxa (35, 36). Individual PCRs were barcoded with 6-bp molecular barcodes integrated in the forward primer and were unique for each sample. The amplification mix contained 0.6 μM (each) primer, 1 μl of Taq AccuPrime (Invitrogen) with 5 μl of 10× AccuPrime PCR buffer II, and 1 μl of cDNA (diluted 10 times) in a final volume of 50 μl. Cycling conditions consisted of an initial denaturation at 94°C for 5 min, followed by 28 cycles of 94°C for 30 s, 50°C for 30 s, and 68°C for 30 s and a final extension at 68°C for 10 min. Amplicons were purified using a PCR cleanup kit (Sigma) and quantified using a Qubit 2.0 fluorometer (Invitrogen). Finally, samples were pooled in an equimolar concentration and analyzed by using standard procedures in a Roche GS-FLX 454 automated pyrosequencer. Sequencing was performed at the Max Planck Genome Centre (MPGC), Cologne, Germany. MPGC requires the use of a 16-bp adaptor for each forward primer and also for the reverse primer, i.e., forward primer, 5′-adaptor + barcode + F515; reverse primer, 5′-adaptor + R806. Table S1 in the supplemental material shows the sequences of barcodes, adaptors, and primers.
Bioinformatic analysis.
The taxonomic assignment of pyrosequencing reads was performed with the mothur (37) software platform. All raw sequences were first denoised to remove sequencing errors by flow gram clustering using the shhh.flows command, and chimeric sequences were removed using UCHIME (38). Classification was carried out using the naive Bayesian classifier in mothur using the SILVA 16S rRNA reference taxonomy. To assess significant differences in the phyla (in labeled and unlabeled root samples), one-way analysis of variance followed by a Tukey post hoc test was performed using the vegan package in R (http://www.r-project.org/).
Sequences were assigned to operational taxonomic units (OTUs) using a cutoff of 97% sequence identity with UPARSE (39). The author's recommended parameters were used for the preprocessing and OTU assignment, with the exception that singleton OTUs were retained. An OTU corresponding to Pseudomonas was identified as a probable contaminant since its relative abundance was always high in the heavy gradient fractions from controls with unlabeled CO2 (data not shown). Therefore, this Pseudomonas OTU was removed from the data set. The number of reads per replicate sample ranged between 1,053 and 4,383 (see Table S1 in the supplemental material). The OTU table was used for statistical analysis. Data transformation has been reported to be superior to subsampling to control for differences in sampling intensity (40), and therefore, the data set was normalized by a Hellinger transformation (41) using the decostand function (42). We also compared the results obtained by subsampling the data set to the sample with the lowest number of reads using the sub.sample command in mothur. The results of principal coordinate analyses (PCoAs) were similar for the two strategies, but the resolution was superior for the transformed compared with the subsampled data (results not shown). Bray-Curtis distances were calculated using the vegdist function (42), and PCoA was performed using the vegan package (version 2.0-10) in R (43) (http://www.r-project.org/). metastats (44) was also performed based on the OTU table using the mothur implementations of the calculations. For this analysis, the OTU table was first subsampled to the minimum number of sequences obtained for a sample, using the sub.sample function in mothur. A phylogenetic analysis of significantly (P ≤ 0.05) overrepresented OTUs identified using metastats was performed using the ARB phylogenetic program package (45). The representative trimmed 16S rRNA sequences were aligned using SINA (46) and added to the SILVA 115 reference tree (47) by parsimony using the bacterial positional variability filter.
Nucleotide sequence accession numbers.
The 454 pyrosequencing reads (raw data) were deposited under the study number SRP043264 in the NCBI Sequence Read Archive (SRA) with the following accession numbers: SRX620410 to SRX620424 for root samples and SRX620425 to SRX620439 for rhizosphere samples (see details in Table S1 in the supplemental material).
RESULTS
CO2 consumption and CH4 emission.
The CO2 concentration in the chambers decreased from ca. 3,000 ppm immediately after injection to ca. 200 ppm after 45 to 60 min. The 13C/12C ratio of pore water CO2 and CH4 increased during the 10 days of labeling with 13CO2 (Fig. 1). The 13C atom% of plant biomass increased to nearly 50% but rose only slightly (1.2 to 1.5 atom%) for the rhizosphere soil (see Fig. S3 in the supplemental material). Control diffusion samples (plants incubated with 13CO2 in the dark) did not show a substantial increase of the 13C/12C ratios during the labeling. The 13C contents of plant and soil carbon in microcosms incubated with unlabeled CO2 were always 1.07 atom% (data not shown). The pH in soil pore water was always between 6.8 and 7.3 and tended to be slightly higher in unplanted than planted pots.
FIG 1.

Time course of 13C atom% in CO2 (A) and CH4 (B) in the soil pore water. 13-1, 13-2, and 13-3, microcosms labeled with 13CO2; CD, control diffusion (plants incubated with 13CO2 in the dark).
16S rRNA sequencing.
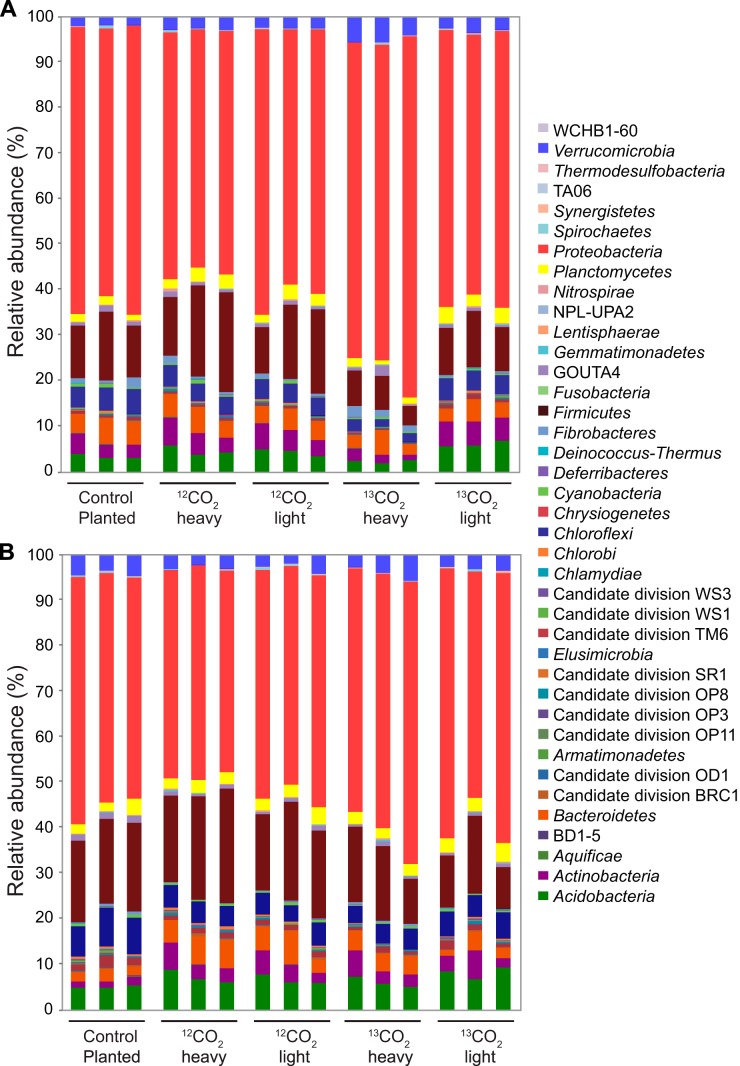
The microorganisms in the rhizosphere and on rice roots were determined by barcoded 16S rRNA amplicon pyrosequencing. Taxonomic assignments were made using the naive Bayesian classifier in mothur with the SILVA 16S rRNA reference taxonomy. For the root samples, 37 phyla were identified across all samples (Fig. 2A). Differences in the relative abundance of phyla between labeled and unlabeled fractions could be seen for Proteobacteria and Verrucomicrobia. Proteobacteria were 72.64% ± 5.66% in the 13CO2 heavy fraction, compared with 59.60% ± 2.20% in 13CO2 light. Verrucomicrobia were 5.18% ± 0.84% in the 13CO2 heavy fraction, compared with 3.03% ± 0.49% in 13CO2 light. These differences were significant (analysis of variance [ANOVA], P = 0.000 for Proteobacteria and Verrucomicrobia). The abundances of Acidobacteria, Actinobacteria, Chloroflexi, and Firmicutes were lower in the 13CO2 heavy fraction than in the light fraction (Fig. 2A), indicating that these phyla were not extensively labeled. For the rhizosphere samples, 38 phyla were observed, but no significant differences were found between heavy and light gradient fractions (Fig. 2B).
FIG 2.
Abundances of different bacterial phyla for root (A) and rhizosphere (B) samples from both heavy and light gradient fractions of labeled (13CO2) and unlabeled (12CO2) microcosms and for the ambient CO2 planted control. Taxonomic classification of 16S rRNA amplicons was performed in mothur using the SILVA 16S rRNA taxonomy.
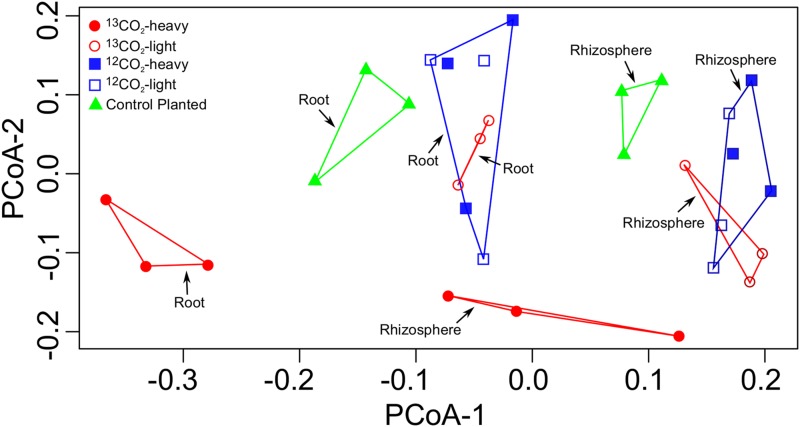
The assignment and analysis of operational taxonomic units (OTUs) enable a higher-resolution analysis of sequences associated with different samples and treatments than by taxonomic analysis described above. A total of 3,928 and 4,312 OTUs for root and rhizosphere samples, respectively, were obtained at a sequence similarity level of 97%. Principal coordinate analysis (PCoA) for root and rhizosphere samples as well as control planted samples indicated differences in community composition or structure between treatments and samples (Fig. 3). There was a clear difference between rhizosphere and root communities, as well as between the labeled (13C, heavy) and the unlabeled (13C, light) fractions of both roots and rhizosphere. There was no difference between light and heavy fractions for the controls incubated with unlabeled CO2. Also, the light fractions of the labeled treatments clustered with the unlabeled treatments. The control planted samples also clustered separately from the unlabeled control, indicating that factors associated with growth in a chamber or the elevated CO2 concentrations also influenced the microbial communities on the root and rhizosphere; however, these differences were less than that between labeled and unlabeled communities. A separate clustering of heavy gradient fractions of the control diffusion samples distinct from the unlabeled samples was not found (data not shown), suggesting that extensive labeling of autotrophic microorganisms on the root or in the rhizosphere via 13CO2 transport through the plant did not occur.
FIG 3.
Principal coordinate analysis (PCoA) based on abundances of 16S rRNA pyrosequencing OTUs (97% sequence similarity). The legend indicates the origin of the sample.
The statistical significance of OTU abundance differences between the microbial communities was determined by metastats analysis. Thirteen OTUs (representing 16.41% of sequences on the roots) were significantly higher in root than rhizosphere samples (where they represented 4.66%), as shown in Table 1. These included Gaiellales, myxobacteria, methanotrophs, Rhizobiaceae, and Opitutus bacteria (Table 1). On the other hand, 28 OTUs were significantly enriched in rhizosphere samples (where they represented 24.27% of sequences) compared with root samples (8.38%). These belonged mainly to Actinobacteria, Firmicutes, Sphingobacteriales, and various Proteobacteria (Table 2).
TABLE 1.
Differentially abundant taxa significantly enriched in root samples when compared with rhizosphere samples from unlabeled microcosms (metastats, P ≤ 0.05)
| Taxonomya | OTUb | Root (%) | Rhizosphere (%) |
|---|---|---|---|
| Bacteria | |||
| Actinobacteria: Gaiellales | 1 | 0.20 | 0.03 |
| Cyanobacteria | |||
| Subsection III | 1 | 0.26 | 0.10 |
| Anabaena | 1 | 0.52 | 0.23 |
| Fibrobacteres: Fibrobacterales | 1 | 0.13 | 0.00 |
| Proteobacteria | |||
| Alphaproteobacteria: Rhizobiaceae | 1 | 0.65 | 0.13 |
| Deltaproteobacteria | |||
| Anaeromyxobacter | 2 | 4.37 | 1.21 |
| Haliangium | 1 | 0.49 | 0.03 |
| Gammaproteobacteria | |||
| Methylomonas | 1 | 4.53 | 0.52 |
| Methylococcales | 1 | 4.24 | 2.25 |
| Betaproteobacteria: Denitratisoma | 1 | 0.13 | 0.00 |
| Verrucomicrobia: Opitutus | 2 | 0.88 | 0.16 |
| Sum | 13 | 16.41 | 4.66 |
Taxonomy was determined by the naive Bayesian classifier in mothur using the SILVA 16S rRNA taxonomy.
OTU indicates the number of OTUs assigned to the taxon.
TABLE 2.
Differentially abundant taxa significantly enriched in rhizosphere samples compared with root samples from unlabeled microcosms (metastats, P ≤ 0.05)
| Taxonomya | OTUb | Rhizosphere (%) | Root (%) |
|---|---|---|---|
| Archaea: Euryarchaeota: Methanosaeta | 1 | 0.29 | 0.03 |
| Bacteria | |||
| Actinobacteria | |||
| Modestobacter | 1 | 0.20 | 0.00 |
| Nocardioides | 1 | 0.16 | 0.03 |
| Gaiellales | 1 | 0.29 | 0.13 |
| Conexibacter | 1 | 0.49 | 0.03 |
| Bacteroidetes: Sphingobacteriales: vadinHA17 | 2 | 0.98 | 0.26 |
| Firmicutes | |||
| Bacillus | 3 | 0.85 | 0.13 |
| Planococcaceae | 2 | 5.15 | 2.32 |
| Clostridium | 2 | 1.50 | 0.46 |
| Heliobacteriaceae | 1 | 1.70 | 0.65 |
| Unclassified | 1 | 0.13 | 0.00 |
| Nitrospirae: Nitrospirales | 1 | 0.20 | 0.03 |
| Proteobacteria | |||
| Alphaproteobacteria | |||
| Beijerinckiaceae | 1 | 0.29 | 0.03 |
| Hyphomicrobium | 1 | 0.20 | 0.00 |
| Methylocystis | 1 | 1.57 | 0.62 |
| Rhodobium | 1 | 0.98 | 0.29 |
| Sphingomonas | 1 | 0.16 | 0.03 |
| Betaproteobacteria: Paucimonas | 1 | 0.16 | 0.03 |
| Deltaproteobacteria | |||
| Geobacter | 1 | 7.70 | 3.00 |
| Cystobacteraceae | 1 | 0.20 | 0.00 |
| Desulfobacca | 1 | 0.82 | 0.29 |
| Verrucomicrobia: OPB35 soil group | 2 | 0.26 | 0.00 |
| Sum | 28 | 24.27 | 8.38 |
Taxonomy was determined by the naive Bayesian classifier in mothur using the SILVA 16S rRNA taxonomy.
OTU indicates the number of OTUs assigned to the taxon.
We next focused on the OTUs that were identified by metastats as being labeled, which meant having significantly higher (P ≤ 0.05) relative abundance in heavy gradient fractions than in the light gradient fractions of the 13C incubations. For the root samples, we observed that 17 OTUs had abundances significantly higher in the heavy gradient fractions than in the light gradient fractions. Included in this category was rice chloroplast rRNA (OTU-22), which can be explained as a consequence of 13C incorporation into the plant biomass. Within these 17 OTUs, 7 belonged to the Opitutae and “Spartobacteria” classes of the Verrucomicrobia and 8 OTUs belonged to Proteobacteria, comprising mainly Comamonadaceae and Rhodocyclaceae. We added representative sequences of these OTUs to a phylogenetic tree (Fig. 4), which resulted in classification similar to that obtained by the Bayesian classifier. For the rhizosphere samples, 13 OTUs were significantly enriched in the heavy fractions compared with the light gradient fractions (Fig. 4). These included sequences most similar to Sphingomonas, Desulfobacterium, Geobacter, Paludibacter, Clostridium, Acidobacterium, and an OTU from the phylum Armatimonadetes. In general, the OTUs labeled on the roots and in the rhizosphere belonged to different phylogenetic taxa.
FIG 4.
Phylogenetic tree including representative 16S rRNA gene sequences of OTUs that were identified as being labeled (metastats, P ≤ 0.05) in the root compartment (green circles) and rhizosphere (red circles). The relative abundance of the OTUs in the heavy (labeled) and light (unlabeled) gradient fractions is indicated in parentheses next to the OTU name. The GenBank accession numbers of reference sequences are indicated. The scale bar corresponds to 5% 16S rRNA sequence divergence.
DISCUSSION
Rice plants were labeled with 13CO2 for 10 days during their vegetative growth stage, which would have resulted in all plant biomass synthesized during this period being labeled. At the end of the incubation, the total plant biomass was labeled to approximately 50% 13C (see Fig. S3 in the supplemental material). The 13C content of dissolved CO2 in the soil pore water increased during the incubation (Fig. 1A). This labeled CO2 in soil would have arisen from a number of processes, including plant respiration and microbial respiration of root exudates. In addition, some 13CO2 in the soil pore water could have resulted from the direct transport of labeled CO2 from the chamber through the plant since gases are continuously transported to the root to ensure oxygenation (48). As a control for this passive transport, a setup (referred to as “control diffusion” [CD]) was included whereby plants were incubated with 13CO2 in the dark, during which time gas transport would continue but photosynthesis would be absent. The atom% 13C of pore water CO2 showed only a slight increase above background level in this treatment, indicating that this direct transport of 13CO2 from the chamber was minimal.
The 13C atom% in CH4 increased during the incubation with 13CO2, indicating that the production of 13CH4 exceeded that of 12CH4 (Fig. 1B). This increase was not observed in plants incubated with labeled 13CO2 in the dark (CD) (Fig. 1B). CH4 is formed by methanogenic archaea primarily via the hydrogenotrophic or acetoclastic pathways, whereby either CO2 or the methyl group of acetate is reduced (49). Vercelli soil contains a wide diversity of methanogenic archaea, including acetoclastic Methanosarcinaceae and Methanosaetaceae as well as hydrogenotrophic Methanocellales, Methanomicrobiales, and Methanobacteriales (11, 20). CH4 is formed by both pathways in Vercelli soil (50), but on rice roots, CH4 is formed mostly by the reduction of CO2 (51). Furthermore, Lu and Conrad (18) showed that in particular Methanocellales incorporate rice plant carbon in the rhizosphere, but Methanosaetaceae and Methanomicrobiaceae are also active (52).
The pyrosequencing data set allowed us to evaluate differences in the relative abundance of rRNA from microorganisms in the roots and rhizosphere. Analyzing the relative abundance of rRNA has several advantages over comparing rRNA gene abundances, as discussed in the literature (53). One advantage is that rRNA abundance is in many cases a good reflection of cellular activity. On the other hand, rRNA abundance should not be confused as a proxy for cellular abundance. In general, the groups identified in the present study as having a higher relative rRNA abundance on roots and in the rhizosphere included both aerobic and anaerobic microorganisms (Tables 1 and 2); however, the root environment was represented by a greater proportion of strict aerobes and the rhizosphere included more organisms characterized by anaerobic metabolism. This is consistent with the root environment being more oxic and the rhizosphere becoming more anoxic with distance from the root (48). Gilbert and Frenzel (54) showed that O2 diffuses up to 0.62 mm from the root into the soil, meaning that the rhizosphere will include both oxic and anoxic sites. The OTUs that were identified as higher in the rhizosphere included strict anaerobes, such as Methanosaeta, which is an acetoclastic methanogenic archaeon. This is consistent with other studies that have shown a higher abundance of Methanosaeta in the rice rhizosphere than in the roots (55). Other strict anaerobes enriched in the rhizosphere included the following: Clostridium, a genus possessing strictly anaerobic fermentative metabolism (56); Heliobacteriaceae, anaerobes with either photosynthetic or fermentative metabolism (57); and Geobacter species, which are iron reducers (58).
In contrast to the rhizosphere, the root environment was represented by a greater proportion of known aerobes, for example, Gaiellales (59), Rhizobiaceae (60), Haliangium (61), and Methylomonas (62). The most abundant OTUs found to be higher in roots than rhizosphere included one belonging to Methylomonas, a member of the Methylococcaceae, and another OTU belonging to an unidentified species in the Methylococcales (Table 1). The Methylococcales are aerobic methanotrophs, which are a physiological group of organisms that consume CH4 and are highly active at oxic-anoxic interfaces (63, 64). An OTU corresponding to Methylocystis, a methanotroph belonging to the Methylocystaceae, was found to have higher relative abundance in the rhizosphere (Table 2). These patterns for methanotrophs are consistent with previous studies that have shown higher abundances of Methylococcaceae on roots and a more balanced distribution of Methylocystis between rice roots and the soil environment (65, 66). CH4 oxidation by methanotrophs is an important mitigating factor for CH4 emissions from rice field soils and is estimated to reduce emissions by approximately 20% (54). The activity of methanotrophs appears to be at least partly limited by O2 availability (54), which is exacerbated by competition for O2 with heterotrophic microorganisms (67). It is remarkable that methanotroph rRNA equates to more than 10% of the total on rice roots, suggesting that these organisms are relatively successful at establishing themselves on the root despite this competition.
RNA-SIP was used to identify microorganisms on the roots and in the rhizosphere by incubating rice plants with 13CO2 (68). We compared the 16S rRNA profiles in the labeled and unlabeled gradient fractions by pyrosequencing analysis and identified the OTUs that were overrepresented in the labeled fraction (Fig. 4). Some of the results matched expectations. For example, we expected rice plastid rRNA sequences to be labeled (OTU-22), as this is a reflection of plant biomass labeling. Second, a previous study using a similar labeling strategy identified Azospirillum-like and Burkholderiaceae-related microorganisms as major consumers of rice carbon (19). We also found Azospirillum sequences to be labeled (OTU-94), and although we identified no Burkholderiaceae, we identified several Betaproteobacteria OTUs. The study by Lu et al. (19) used terminal restriction fragment length polymorphism (T-RFLP) and small clone libraries to evaluate the labeling of rRNA, which has lower resolution and sensitivity than the deep sequencing that we have performed. The Azospirillum sequences detected were closely related to Azospirillum oryzae, which was isolated from rice (69). Field experiments with seeds inoculated with Azospirillum demonstrated that the bacterium provided between 19 and 47% of nitrogen consumed by the plant and increased rice yield by 22% (70). This relationship between Azospirillum and rice is a classic mutualistic symbiosis where the bacterium receives plant carbon and the plant receives fixed nitrogen. It is not possible to know the phenotype of the Azospirillum species detected in this study, but its high 16S rRNA sequence similarity to A. oryzae suggests that it is also an N2-fixing symbiont of rice. OTU-58, closely related to Paludibacter propionicigenes (Fig. 4), is another example of a labeled OTU previously shown to be associated with rice. P. propionicigenes is a strict anaerobe isolated from rice straw that ferments various sugars primarily to acetate and propionate (71), which is consistent with the labeling of OTU-58 in the rice rhizosphere.
In addition to labeling microorganisms that are closely related to known rice symbionts, microorganisms not previously shown to be associated with rice plants were also identified as using plant-derived carbon on the roots and in the rhizosphere of rice. The most abundant labeled taxon was OTU-3, corresponding to more than 10% of the 16S rRNA sequences in the heavy fraction of the root sample. OTU-3 corresponds to an uncultivated betaproteobacterium, distantly related to Comamonas. Another cluster of sequences, represented by OTU-1876, was more closely related to Comamonas. Comamonadaceae are common in soils and in the rhizosphere of terrestrial plants. In potato cultivars, Comamonadaceae were found to account for up to 25% of 16S rRNA genes, whereas they were not detected in the bulk soil (72). In the rhizosphere of wheat, they are believed to play a role in desulfonation reactions, which could provide an important source of sulfur for plant nutrition (73).
The indication that Spartobacteria (e.g., OTU-110) were using plant carbon in the root environment was unforeseen given that a role in plant microbes has not been previously reported. Four closely related OTUs belonging to Spartobacteria were identified as significantly enriched in the labeled fraction of the root samples. Spartobacteria are among the most abundant microorganisms in soil and believed to have been underestimated in ecological studies due to mismatches with common 16S rRNA PCR primers (74). “Chthoniobacter flavus” and related species are the only characterized isolates of this group (75, 76), and a representative genome has been sequenced (77). This organism was shown to be aerobic and to grow using many of the saccharide components of cell biomass, such as xylan, starch, cellulose, pectin, and alginate (75). The strain could not grow on amino acids or any organic acids besides pyruvate. Since Spartobacteria are abundant aerobes in soil that grow using plant saccharides, it is plausible that they have evolved in association with plants as indicated here. More studies are needed to identify the exact nature of their association with plants and rice in particular.
Also within the Verrucomicrobia, four OTUs belonging to the class Opitutae were identified in the labeled fractions (Fig. 4). Three of these were identified in the root samples, and one was identified in the rhizosphere. The closest cultivated representative was Opitutus terrae, an organism that was isolated from rice soil (78). It is a strict anaerobe that grows by fermentation or nitrate reduction using a variety of plant saccharides. As with Spartobacteria, the apparent adaptation for growth using plant polysaccharides by Opitutus makes it a good candidate for colonizing the roots and rhizosphere of rice plants. Unlike the Spartobacteria OTUs that were labeled exclusively in the root samples, OTU-122 was apparently labeled in the rhizosphere, which could be a reflection of its anaerobic metabolism.
As discussed above, some of the labeled OTUs are related to microorganisms that have been previously implicated in the degradation of plant material or isolated from plant roots or rhizosphere soil. This implies that these microbial groups are widespread and exist in diverse soil types and in association with various crops. There were additional examples of labeled OTUs that were most closely related to 16S rRNA recovered from rhizosphere soil. For example, OTU-84, classified as belonging to the Fibrobacteres, is closely related to a sequence (GenBank accession number JX489861, Fig. 4) from an unpublished study examining microbial diversity in the rhizosphere of cucumber plants. A second example is that OTU-1976, which could not be classified at the phylum level, is closely related to a sequence (GenBank accession number JN039008) recovered from the rhizosphere of Phragmites australis. Again, these examples suggest that some of the OTUs labeled in our rice experiment are related to microorganisms associated with other plant species.
If labeled microorganisms used plant carbon for growth, the corollary could be construed that unlabeled microorganisms do not use plant carbon for growth; however, it is not this straightforward. Indeed some of the unlabeled microorganisms are likely to specialize in using soil carbon and not fresh plant material, but there are other scenarios. For example, some of the unlabeled microorganisms might use plant carbon under conditions not tested in this study. Alternatively, they might be active at another growth stage of the rice plant or could be slow growing and have lacked sufficient time to become labeled in this experiment. Also, we used heavy and light gradient fractions differing in density by about 0.03 g ml−1, which would require a relatively high level of 13C incorporation into RNA to cause a shift to the heavy gradient fraction. This means that it would be unlikely to detect labeling of microorganisms that simultaneously used both labeled plant carbon and another unlabeled soil carbon. As an example, methanotrophs were not labeled although CH4 was enriched to 5 to 15% 13C by the end of the experiment (Fig. 1). Therefore, it should be noted that this study reveals more about the microorganisms that were labeled than those that were not.
The results of this study highlight how little is known about plant-microbe interactions, even for rice, which is an important food crop. Future work should determine the nature of the interaction between rice plants and the microbial community on the roots and in the rhizosphere. It will be important to determine which of these microbial groups have plant growth-promoting properties or enhance disease resistance of crops. Furthermore, understanding the degradation pathway of rice plant material is not only of academic interest, as it could aid in managing agriculture practice to mitigate greenhouse gas formation during cultivation.
Supplementary Material
ACKNOWLEDGMENTS
We thank P. Claus for technical assistance, J. Pump for providing Vercelli soil, and two anonymous reviewers for constructive comments.
M.H. was supported by Alexander von Humboldt and Max Planck Society postdoctoral fellowships. Q.Y. received a fellowship from the Max Planck Society, Germany. This work was supported by the LOEWE center for synthetic microbiology (SYNMIKRO), Germany.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03209-14.
REFERENCES
- 1.van Loon LC. 2007. Plant responses to plant growth-promoting rhizobacteria. Eur J Plant Pathol 119:243–254. doi: 10.1007/s10658-007-9165-1. [DOI] [Google Scholar]
- 2.Lugtenberg B, Kamilova F. 2009. Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63:541–556. doi: 10.1146/annurev.micro.62.081307.162918. [DOI] [PubMed] [Google Scholar]
- 3.Berendsen RL, Pieterse CM, Bakker PA. 2012. The rhizosphere microbiome and plant health. Trends Plant Sci 17:478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 4.Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 5.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, Tremblay J, Engelbrektson A, Kunin V, del Rio TG, Edgar RC, Eickhorst T, Ley RE, Hugenholtz P, Tringe SG, Dangl JL. 2012. Defining the core Arabidopsis thaliana root microbiome. Nature 488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knief C, Delmotte N, Chaffron S, Stark M, Innerebner G, Wassmann R, von Mering C, Vorholt JA. 2012. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J 6:1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sessitsch A, Hardoim P, Döring J, Weilharter A, Krause A, Woyke T, Mitter B, Hauberg-Lotte L, Friedrich F, Rahalkar M, Hurek T, Sarkar A, Bodrossy L, van Overbeek L, Brar D, van Elsas JD, Reinhold-Hurek B. 2012. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact 25:28–36. doi: 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 8.Dennis PG, Miller AJ, Hirsch PR. 2010. Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72:313–327. doi: 10.1111/j.1574-6941.2010.00860.x. [DOI] [PubMed] [Google Scholar]
- 9.Jones JDG, Dangl JL. 2006. The plant immune system. Nature 444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.Lynch JM, Whipps JM. 1990. Substrate flow in the rhizosphere. Plant Soil 129:1–10. doi: 10.1007/BF00011685. [DOI] [Google Scholar]
- 11.Chin KJ, Lueders T, Friedrich MW, Klose M, Conrad R. 2004. Archaeal community structure and pathway of methane formation on rice roots. Microb Ecol 47:59–67. doi: 10.1007/s00248-003-2014-7. [DOI] [PubMed] [Google Scholar]
- 12.Chen YH, Prinn RG. 2006. Estimation of atmospheric methane emissions between 1996 and 2001 using a three-dimensional global chemical transport model. J Geophys Res Atmos 111:1–25. doi: 10.1029/2005JD00605. [DOI] [Google Scholar]
- 13.Intergovernmental Panel on Climate Change. 2007. Fourth assessment report: climate change 2007. Intergovernmental Panel on Climate Change, Geneva, Switzerland: http://www.ipcc.ch/pdf/assessment-report/ar4/syr/ar4_syr_spm.pdf. [Google Scholar]
- 14.Watanabe A, Takeda T, Kimura M. 1999. Evaluation of origins of CH4 carbon emitted from rice paddies. J Geophys Res Atmos 104:23623–23629. doi: 10.1029/1999JD900467. [DOI] [Google Scholar]
- 15.Nouchi I, Mariko S, Aoki K. 1990. Mechanism of methane transport from the rhizosphere to the atmosphere through rice plants. Plant Physiol 94:59–66. doi: 10.1104/pp.94.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bowen GD, Rovira AD. 1976. Microbial colonization of plant roots. Annu Rev Phytopathol 14:121–144. doi: 10.1146/annurev.py.14.090176.001005. [DOI] [Google Scholar]
- 17.Haichar FZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. 2008. Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 18.Lu Y, Conrad R. 2005. In situ stable isotope probing of methanogenic archaea in the rice rhizosphere. Science 309:1088–1090. doi: 10.1126/science.1113435. [DOI] [PubMed] [Google Scholar]
- 19.Lu Y, Rosencrantz D, Liesack W, Conrad R. 2006. Structure and activity of bacterial community inhabiting rice roots and the rhizosphere. Environ Microbiol 8:1351–1360. doi: 10.1111/j.1462-2920.2006.01028.x. [DOI] [PubMed] [Google Scholar]
- 20.Conrad R, Klose M, Noll M, Kemnitz D, Bodelier PLE. 2008. Soil type links microbial colonization of rice roots to methane emission. Glob Chang Biol 14:657–669. doi: 10.1111/j.1365-2486.2007.01516.x. [DOI] [Google Scholar]
- 21.Zhu W, Lu H, Hill J, Guo X, Wang H, Wu W. 2014. 13C pulse-chase labeling comparative assessment of the active methanogenic archaeal community composition in the transgenic and nontransgenic parental rice rhizospheres. FEMS Microbiol Ecol 87:746–756. doi: 10.1111/1574-6941.12261. [DOI] [PubMed] [Google Scholar]
- 22.Eissenstat DM, Wells CE, Yanai RD, Whitbeck JL. 2000. Building roots in a changing environment: implications for root longevity. New Phytol 147:33–42. doi: 10.1046/j.1469-8137.2000.00686.x. [DOI] [Google Scholar]
- 23.Ostle N, Whiteley AS, Bailey MJ, Sleep D, Ineson P, Manefield M. 2003. Active microbial RNA turnover in a grassland soil estimated using a 13CO2 spike. Soil Biol Biochem 35:877–885. doi: 10.1016/S0038-0717(03)00117-2. [DOI] [Google Scholar]
- 24.Rangel-Castro JI, Prosser JI, Ostle N, Scrimgeour CM, Killham K, Meharg AA. 2005. Flux and turnover of fixed carbon in soil microbial biomass of limed and unlimed plots of an upland grassland ecosystem. Environ Microbiol 7:544–552. doi: 10.1111/j.1462-2920.2005.00722.x. [DOI] [PubMed] [Google Scholar]
- 25.Rangel-Castro JI, Killham K, Ostle N, Nicol GW, Anderson IC, Scrimgeour CM, Ineson P, Meharg A, Prosser JI. 2005. Stable isotope probing analysis of the influence of liming on root exudate utilization by soil microorganisms. Environ Microbiol 7:828–838. doi: 10.1111/j.1462-2920.2005.00756.x. [DOI] [PubMed] [Google Scholar]
- 26.Vandenkoornhuyse P, Mahé S, Ineson P, Staddon P, Ostle N, Cliquet JB, Francez AJ, Fitter AH, Young JP. 2007. Active root-inhabiting microbes identified by rapid incorporation of plant-derived carbon into RNA. Proc Natl Acad Sci U S A 104:16970–16975. doi: 10.1073/pnas.0705902104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Haichar FZ, Roncato MA, Achouak W. 2012. Stable isotope probing of bacterial community structure and gene expression in the rhizosphere of Arabidopsis thaliana. FEMS Microbiol Ecol 81:291–302. doi: 10.1111/j.1574-6941.2012.01345.x. [DOI] [PubMed] [Google Scholar]
- 28.Rasche F, Lueders T, Schloter M, Schaefer S, Buegger F, Gattinger A, Hood-Nowotny RC, Sessitsch A. 2009. DNA-based stable isotope probing enables the identification of active bacterial endophytes in potatoes. New Phytol 181:802–807. doi: 10.1111/j.1469-8137.2008.02744.x. [DOI] [PubMed] [Google Scholar]
- 29.Pilloni G, von Netzer F, Engel M, Lueders T. 2011. Electron acceptor-dependent identification of key anaerobic toluene degraders at a tar-oil-contaminated aquifer by Pyro-SIP. FEMS Microbiol Ecol 78:165–175. doi: 10.1111/j.1574-6941.2011.01083.x. [DOI] [PubMed] [Google Scholar]
- 30.Holzapfel-Pschorn A, Seiler W. 1986. Methane emission during a cultivation period from an Italian rice paddy. J Geophys Res Atmos 91:11803–11814. doi: 10.1029/JD091iD11p11803. [DOI] [Google Scholar]
- 31.Yuan Q, Pump J, Conrad R. 2012. Partitioning of CH4 and CO2 production originating from rice straw, soil and root organic carbon in rice microcosms. PLoS One 7:e49073. doi: 10.1371/journal.pone.0049073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werner RA, Bruch BA, Brand WA. 1999. ConFlo III—an interface for high precision δ13C and δ15N analysis with an extended dynamic range. Rapid Commun Mass Spectrom 13:1237–1241. doi:. [DOI] [PubMed] [Google Scholar]
- 33.Ma K, Conrad R, Lu Y. 2012. Responses of methanogen mcrA genes and their transcripts to an alternate dry/wet cycle of paddy field soil. Appl Environ Microbiol 78:445–454. doi: 10.1128/AEM.06934-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dumont MG, Pommerenke B, Casper P, Conrad R. 2011. DNA-, rRNA- and mRNA-based stable isotope probing of aerobic methanotrophs in lake sediment. Environ Microbiol 13:1153–1167. doi: 10.1111/j.1462-2920.2010.02415.x. [DOI] [PubMed] [Google Scholar]
- 35.Bates ST, Berg-Lyons D, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. Examining the global distribution of dominant archaeal populations in soil. ISME J 5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF. 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Edgar RC. 2013. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 40.McMurdie PJ, Holmes S. 2014. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol 10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Legendre P, Gallagher ED. 2001. Ecologically meaningful transformations for ordination of species data. Oecologia 129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 42.Oksanen J. 2013. Multivariate analysis of ecological communities in R: vegan tutorial. University of Oulu, Oulu, Finland: http://cc.oulu.fi/∼jarioksa/opetus/metodi/vegantutor.pdf. [Google Scholar]
- 43.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Vegan: community ecology package. R package version 2.0-10 The R Project for Statistical Computing, Vienna, Austria: http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 44.White JR, Nagarajan N, Pop M. 2009. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput Biol 5:e1000352. doi: 10.1371/journal.pcbi.1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer KH. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Colmer PD, Pedersen O. 2008. Oxygen dynamics in submerged rice (Oryza sativa). New Phytol 178:326–334. doi: 10.1111/j.1469-8137.2007.02364.x. [DOI] [PubMed] [Google Scholar]
- 49.Conrad R. 2007. Microbial ecology of methanogens and methanotrophs. Adv Agron 96:1–63. doi: 10.1016/S0065-2113(07)96005-8. [DOI] [Google Scholar]
- 50.Penning H, Conrad R. 2006. Effect of inhibition of acetoclastic methanogenesis on growth of archaeal populations in an anoxic model environment. Appl Environ Microbiol 72:178–184. doi: 10.1128/AEM.72.1.178-184.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Conrad R, Klose M. 1999. Anaerobic conversion of carbon dioxide to methane, acetate and propionate on washed rice roots. FEMS Microbiol Ecol 30:147–155. doi: 10.1111/j.1574-6941.1999.tb00643.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhu W, Lu H, Hill J, Guo X, Wang H, Wu W. 2014. 13C pulse-chase labeling comparative assessment of the active methanogenic archaeal community composition in the transgenic and nontransgenic parental rice rhizospheres. FEMS Microbiol Ecol 87:746–756. doi: 10.1111/1574-6941.12261. [DOI] [PubMed] [Google Scholar]
- 53.Blazewicz SJ, Barnard RL, Daly RA, Firestone MK. 2013. Evaluating rRNA as an indicator of microbial activity in environmental communities: limitations and uses. ISME J 7:2061–2068. doi: 10.1038/ismej.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gilbert B, Frenzel P. 1998. Rice roots and CH4 oxidation: the activity of bacteria, their distribution and the microenvironment. Soil Biol Biochem 30:1903–1916. doi: 10.1016/S0038-0717(98)00061-3. [DOI] [Google Scholar]
- 55.Grosskopf R, Janssen PH, Liesack W. 1998. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl Environ Microbiol 64:960–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Andreesen JR, Schaupp A, Neurauter C, Brown A, Ljungdahl LG. 1973. Fermentation of glucose, fructose, and xylose by Clostridium thermoaceticum: effect of metals on growth yield, enzymes, and the synthesis of acetate from CO2. J Bacteriol 114:743–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang KH, Yue H, Blankenship RE. 2010. Energy metabolism of Heliobacterium modesticaldum during phototrophic and chemotrophic growth. BMC Microbiol 10:150. doi: 10.1186/1471-2180-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nielsen JL, Juretschko S, Wagner M, Nielsen PH. 2002. Abundance and phylogenetic affiliation of iron reducers in activated sludge as assessed by fluorescence in situ hybridization and microautoradiography. Appl Environ Microbiol 68:4629–4636. doi: 10.1128/AEM.68.9.4629-4636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Albuquerque L, França L, Rainey FA, Schumann P, Nobre MF, da Costa MS. 2011. Gaiella occulta gen. nov., sp. nov., a novel representative of a deep branching phylogenetic lineage within the class Actinobacteria and proposal of Gaiellaceae fam. nov. and Gaiellales ord. nov. Syst Appl Microbiol 34:595–599. doi: 10.1016/j.syapm.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 60.Yanagi M, Yamasato K. 1993. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and DNA sequencer. FEMS Microbiol Lett 107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 61.Fudou R, Jojima Y, Iizuka T, Yamanaka S. 2002. Haliangium ochraceum gen. nov., sp. nov. and Haliangium tepidum sp. nov.: novel moderately halophilic myxobacteria isolated from coastal saline environments. J Gen Appl Microbiol 48:109–116. doi: 10.2323/jgam.48.109. [DOI] [PubMed] [Google Scholar]
- 62.Boden R, Cunliffe M, Scanlan J, Moussard H, Kits KD, Klotz MG, Jetten MS, Vuilleumier S, Han J, Peters L, Mikhailova N, Teshima H, Tapia R, Kyrpides N, Ivanova N, Pagani I, Cheng JF, Goodwin L, Han C, Hauser L, Land ML, Lapidus A, Lucas S, Pitluck S, Woyke T, Stein L, Murrell JC. 2011. Complete genome sequence of the aerobic marine methanotroph Methylomonas methanica MC09. J Bacteriol 193:7001–7002. doi: 10.1128/JB.06267-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Semrau JD, DiSpirito AA, Yoon S. 2010. Methanotrophs and copper. FEMS Microbiol Rev 34:496–531. doi: 10.1111/j.1574-6976.2010.00212.x. [DOI] [PubMed] [Google Scholar]
- 64.Reim A, Lüke C, Krause S, Pratscher J, Frenzel P. 2012. One millimetre makes the difference: high-resolution analysis of methane-oxidizing bacteria and their specific activity at the oxic-anoxic interface in a flooded paddy soil. ISME J 6:2128–2139. doi: 10.1038/ismej.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Horz HP, Yimga MT, Liesack W. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl Environ Microbiol 67:4177–4185. doi: 10.1128/AEM.67.9.4177-4185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shrestha M, Shrestha PM, Frenzel P, Conrad R. 2010. Effect of nitrogen fertilization on methane oxidation, abundance, community structure, and gene expression of methanotrophs in the rice rhizosphere. ISME J 4:1545–1556. doi: 10.1038/ismej.2010.89. [DOI] [PubMed] [Google Scholar]
- 67.van Bodegom P, Stams F, Mollema L, Boeke S, Leffelaar P. 2001. Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl Environ Microbiol 67:3586–3597. doi: 10.1128/AEM.67.8.3586-3597.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Y, Conrad R. 2011. Stable isotope probing and plants, p 151–163. In Murrell JC, Whiteley A (ed), Stable isotope probing and related technologies. ASM Press, Washington, DC. [Google Scholar]
- 69.Xie CH, Yokota A. 2005. Azospirillum oryzae sp. nov., a nitrogen-fixing bacterium isolated from the roots of the rice plant Oryza sativa. Int J Syst Evol Microbiol 55:1435–1438. doi: 10.1099/ijs.0.63503-0. [DOI] [PubMed] [Google Scholar]
- 70.Choudhury ATMA, Kennedy IR. 2004. Prospects and potentials for systems of biological nitrogen fixation in sustainable rice production. Biol Fertil Soils 39:219–227. doi: 10.1007/s00374-003-0706-2. [DOI] [Google Scholar]
- 71.Ueki A, Akasaka H, Suzuki D, Ueki K. 2006. Paludibacter propionicigenes gen. nov., sp. nov., a novel strictly anaerobic, Gram-negative, propionate-producing bacterium isolated from plant residue in irrigated rice-field soil in Japan. Int J Syst Evol Microbiol 56:39–44. doi: 10.1099/ijs.0.63896-0. [DOI] [PubMed] [Google Scholar]
- 72.Inceoğlu O, Salles JF, van Overbeek L, van Elsas JD. 2010. Effects of plant genotype and growth stage on the betaproteobacterial communities associated with different potato cultivars in two fields. Appl Environ Microbiol 76:3675–3684. doi: 10.1128/AEM.00040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmalenberger A, Hodge S, Bryant A, Hawkesford MJ, Singh BK, Kertesz MA. 2008. The role of Variovorax and other Comamonadaceae in sulfur transformations by microbial wheat rhizosphere communities exposed to different sulfur fertilization regimes. Environ Microbiol 10:1486–1500. doi: 10.1111/j.1462-2920.2007.01564.x. [DOI] [PubMed] [Google Scholar]
- 74.Bergmann GT, Bates ST, Eilers KG, Lauber CL, Caporaso JG, Walters WA, Knight R, Fierer N. 2011. The under-recognized dominance of Verrucomicrobia in soil bacterial communities. Soil Biol Biochem 43:1450–1455. doi: 10.1016/j.soilbio.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sangwan P, Chen X, Hugenholtz P, Janssen PH. 2004. Chthoniobacter flavus gen. nov., sp. nov., the first pure-culture representative of subdivision two, Spartobacteria classis nov., of the phylum Verrucomicrobia. Appl Environ Microbiol 70:5875–5881. doi: 10.1128/AEM.70.10.5875-5881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sangwan P, Kovac S, Davis KE, Sait M, Janssen PH. 2005. Detection and cultivation of soil Verrucomicrobia. Appl Environ Microbiol 71:8402–8410. doi: 10.1128/AEM.71.12.8402-8410.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kant R, van Passel MW, Palva A, Lucas S, Lapidus A, Glavina del Rio T, Dalin E, Tice H, Bruce D, Goodwin L, Pitluck S, Larimer FW, Land ML, Hauser L, Sangwan P, de Vos WM, Janssen PH, Smidt H. 2011. Genome sequence of Chthoniobacter flavus Ellin428, an aerobic heterotrophic soil bacterium. J Bacteriol 193:2902–2903. doi: 10.1128/JB.00295-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chin KJ, Liesack W, Janssen PH. 2001. Opitutus terrae gen. nov., sp. nov., to accommodate novel strains of the division ‘Verrucomicrobia’ isolated from rice paddy soil. Int J Syst Evol Microbiol 51:1965–1968. doi: 10.1099/00207713-51-6-1965. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.