Abstract
Dehalococcoides mccartyi 195 (strain 195) and Syntrophomonas wolfei were grown in a sustainable syntrophic coculture using butyrate as an electron donor and carbon source and trichloroethene (TCE) as an electron acceptor. The maximum dechlorination rate (9.9 ± 0.1 μmol day−1) and cell yield [(1.1 ± 0.3) × 108 cells μmol−1 Cl−] of strain 195 maintained in coculture were, respectively, 2.6 and 1.6 times higher than those measured in the pure culture. The strain 195 cell concentration was about 16 times higher than that of S. wolfei in the coculture. Aqueous H2 concentrations ranged from 24 to 180 nM during dechlorination and increased to 350 ± 20 nM when TCE was depleted, resulting in cessation of butyrate fermentation by S. wolfei with a theoretical Gibbs free energy of −13.7 ± 0.2 kJ mol−1. Carbon monoxide in the coculture was around 0.06 μmol per bottle, which was lower than that observed for strain 195 in isolation. The minimum H2 threshold value for TCE dechlorination by strain 195 in the coculture was 0.6 ± 0.1 nM. Cell aggregates during syntrophic growth were observed by scanning electron microscopy. The interspecies distances to achieve H2 fluxes required to support the measured dechlorination rates were predicted using Fick's law and demonstrated the need for aggregation. Filamentous appendages and extracellular polymeric substance (EPS)-like structures were present in the intercellular spaces. The transcriptome of strain 195 during exponential growth in the coculture indicated increased ATP-binding cassette transporter activities compared to the pure culture, while the membrane-bound energy metabolism related genes were expressed at stable levels.
INTRODUCTION
Groundwater contamination by trichloroethene (TCE), a potential human carcinogen, poses a serious threat to human health and can lead to the generation of vinyl chloride (VC), which is a known human carcinogen (1). Strains of Dehalococcoides mccartyi are the only known bacteria that can completely degrade TCE to the benign end product ethene. Biostimulation of indigenous Dehalococcoides spp. and bioaugmentation using Dehalococcoides-containing cultures are recognized as the most reliable in situ bioremediation technologies resulting in the complete dechlorination of TCE to ethene (2). However, the mechanisms that regulate the activity of D. mccartyi within natural ecosystems and shape its functional robustness in disturbed environments are poorly understood due to multiscale microbial community complexity and heterogeneity of biogeochemical processes involved in the sequential degradation (3, 4). D. mccartyi exhibits specific restrictive metabolic requirements for a variety of exogenous compounds, such as hydrogen, acetate, corrinoids, biotin, and thiamine, which can be supplied by other microbial genera through a complex metabolic network (1, 5–8). Therefore, the growth of D. mccartyi is more robust within functionally diverse microbial communities that are deterministically assembled than in pure cultures (5, 8, 9). Previous studies have shown that D. mccartyi uses hydrogen as its sole electron donor for TCE dechlorination and outcompetes other terminal electron-accepting processes such as methanogenesis and acetogenesis at low hydrogen partial pressures (10, 11). Interspecies hydrogen transfer between D. mccartyi and supportive organisms is a key process of electron flow that drives reductive dechlorination in the environment. Although dechlorinating microbial communities have been extensively studied over the past decades (4, 10, 12, 13), community assembly processes in the highly specialized ecological niche of Dehalococcoides and the network of interactions between D. mccartyi, its syntrophic partners and other coexisting community members have yet to be deciphered. The optimization of D. mccartyi-based bioremediation systems to treat TCE-contaminated groundwater would be facilitated by a system-level understanding of interspecies electron, energy and metabolite transfers that shape the structural and functional robustness of TCE-dechlorinating microbial communities.
To date, only a few studies of D. mccartyi-containing constructed cocultures and tricultures have been published (5, 7, 14–16). A study with D. mccartyi 195 (strain 195) revealed that its growth in defined medium could be optimized by providing high concentrations of vitamin B12 and that, over the short term, the strain could be grown to higher densities in cocultures and tricultures with the fermenters Desulfovibrio desulfuricans and/or Acetobacterium woodii that convert lactate to generate the hydrogen and acetate required by D. mccartyi (5). Recent studies demonstrated interspecies corrinoid transfer between Geobacter lovleyi and D. mccartyi strains BAV1 and FL2 (16) and interspecies cobamide transfer from a corrinoid-producing methanogen and acetogen to D. mccartyi (7). Another study showed that strain 195 can grow in a long-term sustainable syntrophic association with Desulfovibrio vulgaris Hildenborough (DvH) as a coculture, as well as with hydrogenotrophic methanogen Methanobacterium congolense (MC) as a triculture (15). The maximum dechlorination rates and cell yield of strain 195 were enhanced significantly in the defined consortia. Another unexpected syntrophic association has recently been discovered between carbon monoxide (CO)-producing strain 195 and CO-metabolizing anaerobes which enhance the growth and dechlorination activity of strain 195 by preventing the accumulation of toxic CO as an obligate by-product from acetyl coenzyme A (acetyl-CoA) cleavage (17). Although these studies demonstrated the robust growth of D. mccartyi-containing cocultures on tetrachloroethylene (PCE)/TCE with higher dechlorination activity and cell yields than isolates, a clear and mechanistic understanding of metabolic cross-feeding and electron transfer between D. mccartyi and its syntrophic partner(s) is needed to establish predictive models for dechlorination activity.
Butyrate fermentation is an endergonic reaction under standard conditions (Table 1) that can only be carried out for energy generation by syntrophic microorganisms growing with H2 consumers (18–20). Syntrophomonas wolfei is a model butyrate fermenter that grows syntrophically with hydrogenotrophic methanogens (19–21). S. wolfei can also grow without a syntrophic partner on crotonate as the sole energy source through its disproportionation to acetate and butyrate (Table 1) (22). In previous studies, Syntrophomonas spp. have been detected in butyrate-fed dechlorinating enrichment cultures in significant relative abundance (13, 23). In the present study, a TCE- and butyrate-fed syntrophic coculture of strain 195 with S. wolfei was established and maintained to study the physiology and transcriptome of syntrophically growing D. mccartyi. The spatial architecture and physical proximity of the cells were analyzed in an S. wolfei-strain 195 coculture and in another syntrophic DvH-strain 195 coculture (15). The knowledge gained from this study provides us with a more fundamental understanding of the metabolic exchange and energy transfer among the key players of TCE-dechlorinating communities.
TABLE 1.
Stoichiometric energy generating reactions in the syntrophic coculture
| Process | Redox reaction | ΔG°′ (kJ/mol) | Equation no. |
|---|---|---|---|
| Crotonate oxidation and reduction | 2C4H5O2− + 2H2O → 2C2H3O2− + C4H7O2− + H+ | –350 | 1 |
| Butyrate fermentation reaction | C4H7O2− + 2H2O → 2C2H3O2− + H+ + 2H2(g)b | 46.8a | 2 |
| TCE reduction to cis-DCE | C2HCl3 + H2 → C2H2Cl2 + H+ + Cl− | –133 | 3 |
| cis-DCE reduction to VC | C2H2Cl2 + H2 → C2H3Cl + H+ + Cl− | –144 | 4 |
| VC reduction to ETH | C2H3Cl + H2 → C2H4 + H+ + Cl− | –154 | 5 |
ΔG°′ (pH 7.0) was corrected to 307.15 K.
2H2(g), 2H2 gas.
MATERIALS AND METHODS
Bacterial cultures and growth conditions.
Bacterial cocultures of strain 195 and S. wolfei (5% [vol/vol] inoculation of each bacterium) were initially established in 160-ml serum bottles containing 100 ml of defined medium (5) with TCE supplied at a liquid concentration of 0.6 mM (corresponding to 78 μmol of TCE per bottle), 10 mM crotonic acid, vitamin B12 at 100 μg liter−1, and an N2/CO2 (90:10 [vol/vol]) headspace at 34°C with no agitation. Cultures were subsequently transferred (5% [vol/vol] inoculation) for growth on 4 mM butyric acid with 0.6 mM TCE as the electron acceptor. The established syntrophic coculture was continuously and stably maintained on butyrate in 100 ml of defined medium over 45 subculturing events before the experiments were performed. The electron donor-limited condition (on a hydrogen production/consumption basis) was achieved by feeding the coculture 0.25 mM butyrate (25 μmol per bottle that could theoretically generate 50 μmol of H2 based on stoichiometry) and 0.6 mM TCE (78 μmol per bottle that requires 234 μmol of H2 to reductively dechlorinate TCE to ethene). The strain 195 isolate was grown in defined medium with H2/CO2 (90:10 [vol/vol]) headspace, 0.6 mM TCE as an electron acceptor, and 2 mM acetate as a carbon source. Pure S. wolfei was grown on crotonate in 160-ml serum bottles as described previously (22). A DvH and strain 195 coculture was grown in the same medium with the substitutions of 5 mM lactate. Pure culture and coculture purities were routinely checked by phase-contrast microscopy and denaturing gradient gel electrophoresis.
Chemical analysis.
Chloroethenes and ethene were measured by flame ionization detector (FID) gas chromatography using 100-μl headspace samples, and hydrogen and carbon monoxide were measured by reducing gas detector (RGD) gas chromatography using a 300-μl headspace sample as described previously (23, 24). Organic acids, including butyrate and acetate, were analyzed with a high-performance liquid chromatograph as described previously (23). The mass of each compound was calculated based on the gas-liquid equilibrium using Henry's law constants at 34°C: mass (μmol/bottle) = (Cl × Vl) + (Cg × Vg) and kHcc = Cl/Cg, where Cl is the liquid phase concentration (in μM), Vl is the volume of liquid phase (in liters), Cg is the gas phase concentration (in μM), Vg is the volume of the gas phase (in liters), and kHcc is the Henry's law constant (unitless).
SEM.
The cocultures (S. wolfei and strain 195 grown on butyrate; DvH and strain 195 grown on lactate) provided with 0.6 mM TCE were collected when cell growth had ceased and treated according to the following standard protocols for scanning electron microscopy (SEM) observation (http://em-lab.berkeley.edu/EML/protocols/psem.php). Briefly, cultures were collected by slowly filtering 1 ml of fresh liquid sample through a 0.2-μm-pore-size GTTP filter (Isopore membrane, polycarbonate, hydrophilic, 25 mm in diameter [Millipore]), followed by chemical fixation, dehydration, critical point drying, and conductive coating (25). SEM images were obtained according to standard operation protocols (26). The maximum interspecies distances between strain 195 and S. wolfei that would enable the observed dechlorination rates for butyrate fermentation were estimated using Fick's diffusion law according to the procedure described by Ishii et al. (27):
| (1) |
where is the flux across the total surface areas (As,tot) of S. wolfei (pmol m−2 cell day−1) and As = 2.1 × 10−12 m2 cell−1, the S. wolfei surface area calculated based on the assumption of 0.25- by 2.5-μm cells (based on SEM observation). In addition, As,tot = As × the S. wolfei cell number; = 6.31 × 10−5 cm2 s−1, the molecular diffusion coefficient in water for hydrogen at 35°C (28); CH2-sw is the maximum H2 concentration enabling exergonic fermentation (expressed in μM; the highest H2 level at which S. wolfei can ferment butyrate); is the theoretical minimum H2 concentration useable by strain 195 for energy generation (expressed in μM; the lowest H2 level at which strain 195 can dechlorinate TCE); and dsw-195 is the allowed interspecies distance for accomplishing syntrophic oxidation at an observed substrate utilization rate (in μm).
DNA extraction and cell number quantification.
Liquid samples (1.5 ml) were collected for cell density measurements, and cells were harvested by centrifugation (21,000 × g, 10 min at 4°C). Genomic DNA was extracted from cell pellets using Qiagen DNeasy blood and tissue kit according to the manufacturer's instructions for Gram-positive bacteria. Quantitative PCR (qPCR) using SYBR green-based detection reagents was applied to quantify gene copy numbers of each bacterium with S. wolfei 16S rRNA gene primers (forward primer, 5′-GTATCGACCCCTTCTGTGCC-3′; reverse primer, 5′-CCCCAGGCGGGATACTTATT-3′) (19), and D. mccartyi tceA gene primers (forward primer, 5′-ATCCAGATTATGACCCTGGTGAA-3′; reverse primer, 5′-GCGGCATATATTAGGGCATCTT-3′), as previously described (29). The electron equivalents (in μmol) diverted to biomass were calculated based on empirical biomass formula C5H7O2N (113 g mol−1 cell, 20 e− eq mol−1 cells).
RNA preparation.
Cultures were sampled for RNA on day 6 during exponential growth when ca. 75% of 78 μmol of TCE was dechlorinated (∼20 μmol of TCE remained). In order to collect sufficient material for transcriptomic microarray analysis, 18 bottles of pure strain 195 and 18 bottles of the coculture were inoculated and grown from triplicate bottles of the isolate and coculture, respectively. For each biological triplicate, cells from six bottles were collected by vacuum filtration on day 6 during active dechlorination for the coculture and day 12 for the strain 195 pure culture (200 ml of culture per filter, 0.2-μm-pore-size autoclaved GVWP filter [Durapore membrane; Millipore, Billerica, MA]). Each filter was placed in a 2-ml orange-cap microcentrifuge tube, frozen using liquid nitrogen, and stored at −80°C until further processing.
RNA was extracted using the phenol-chloroform method described previously (30) with the following minor modifications. The ratio of phenol (pH 4.0)-chloroform-isoamyl alcohol used for extraction was 25:24:1 (vol/vol), and the RNA pellet was resuspended in 100 μl of nuclease-free water. RNA samples were purified according to the manufacturer's instructions with an AllPrep DNA/RNA minikit (Qiagen). Additional DNA contamination was removed with Turbo DNA free kit (Ambion, Austin, TX) according to the manufacturer's instructions. The quality of RNA samples was checked by electrophoresis (a 1.0-μl RNA sample), and the concentrations of RNA samples were quantified using a nanophotometer (Implen, Westlake Village, CA). The A260/A280 ratio for all samples was approximately between 1.80 and 2.0. Purified RNA was stored at −80°C prior to further use.
Transcriptomic microarray analysis.
A complete description of the Affymetrix GeneChip microarray used in the present study has been reported elsewhere (4). Briefly, the chip contains 4,744 probe sets that represent more than 98% of the open reading frames (ORFs) from four published Dehalococcoides genomes (strain 195, VS, BAV1, and CBDB1). cDNA was synthesized from 9 μg of RNA, and then each cDNA sample was fragmented, labeled, and hybridized to each array. All procedures were performed with minimal modifications to the protocols in section 3 of the GeneChip expression analysis technical manual (Affymetrix, Santa Clara, CA). The microarray data analysis methods were described previously (15, 31).
RESULTS
Syntrophic coculture degradation characteristics.
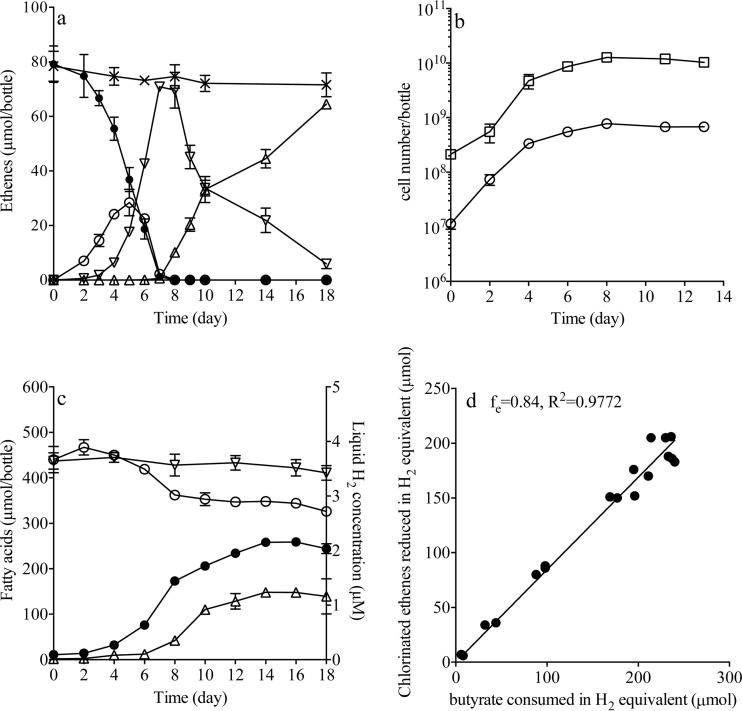
TCE did not inhibit growth of the S. wolfei isolate at concentrations up to 0.6 mM (see Fig. S1 in the supplemental material); therefore, cocultures were maintained with 0.6 mM TCE and 4 mM butyrate. After this coculture was subcultured 45 times, the maximum dechlorination rate of strain 195 was ∼2.6-fold (9.9 ± 0.1 μmol day−1) (Fig. 1a) greater than that of the strain 195 isolate (3.8 ± 0.1 μmol day−1) (15). The calculation of strain 195 cell yield was based on the metabolic reductive TCE dechlorination to VC, and the cell yield of strain 195 was 1.6 times greater in the coculture [(1.1 ± 0.3) × 108 cells μmol−1 Cl−] (Fig. 1b) compared to the pure culture [(6.8 ± 0.9) × 107 cells μmol−1 Cl−] (15), similar to results from a coculture containing a sulfate-reducing bacterium [(9.0 ± 0.5) × 107 cells μmol−1 Cl−] (15) (Table 2). The initial concentration of butyrate (from 1 to 20 mM) did not affect the dechlorination rate (data not shown), showing that high butyrate concentrations are not inhibitory to dechlorination. Cell numbers of strain 195 [(1.3 ± 0.2) × 108 cells ml−1] were consistently ∼16 times higher than those of S. wolfei [(7.7 ± 0.1) × 106 cells ml−1] in the cocultures growing on butyrate. In contrast, when the coculture was maintained on crotonate, the cell number ratio was about 1.3:1 (see the supplemental material, part 1, and Fig. S2 in the supplemental material). In the syntrophic coculture with DvH growing on lactate, the cell number ratio of strain 195 to DvH was reported to be ∼5:1 (15).
FIG 1.
Coculture S. wolfei with D. mccartyi strain 195 growing with 78 μmol of TCE and 4 mM butyrate amendment. (a) TCE dechlorination profile of coculture during the feeding cycle (●, TCE; ○, cis-DCE; ▽, VC; △, ETH; ×, control). (b) Cell numbers of coculture (□, strain 195; ○, S. wolfei). (c) H2 level and organic acid formation of coculture (●, acetate; ○, butyrate; △, hydrogen; ▽, control butyrate). (d) Graphical determination of fe values for strain 195 in the coculture in which the amounts of reducing equivalent H2 generated during butyrate fermentation were plotted against the amounts of electron acceptor reduced. The fe is indicated by the slope of the regression line. Values are averages of biological triplicates; error bars indicate the standard deviations.
TABLE 2.
Dechlorination rates and cell yields of D. mccartyi strains in various coculture studies
| Coculturea | Rate or yieldb |
Source or reference | ||
|---|---|---|---|---|
| Dechlorination rate (μmol day−1) | Specific dechlorination rate of strain 195 (μmol cell−1 day−1) | D. mccartyi cell yield (cells μmol−1 of Cl− released) | ||
| Strain 195 and Desulfovibrio desulfuricans* | 0.4 | 2.3 × 10−10 | 2.4 × 108 | 5 |
| Strain 195 and Sedimentibacter sp.† | 6.8 | NA | 4.2 × 108 | 14 |
| Strain 195 and Desulfovibrio vulgaris Hildenborough | 11 ± 0.01 | NA | (9.0 ± 0.5) × 107 | 15 |
| Strain BAV1 and Geobacter lovleyi‡ | 6.1 | 7.9 × 10−10 | 6.7 × 107 | 16 |
| Strain FL2 and Geobacter lovleyi | 4.2 | 1.5 × 10−9 | 3.3 × 107 | |
| Strain BAV1 and Sporomusa sp. strain KB1§ | 3.3 | 2.7 × 10−10 | (2.1 ± 0.2) × 108 | 7 |
| Strain GT and Sporomusa sp. strain KB1 | 2.1 | 1.4 × 10−10 | (2.1 ± 0.3) × 108 | |
| Strain FL2 and Sporomusa sp. strain KB1 | 2.5 | 7.2 × 10−10 | (9.0 ± 1.4) × 107 | |
| Strain 195 and S. wolfei | 9.9 ± 0.1 | 7.6 × 10−10 | (1.1 ± 0.3) × 108 | This study |
Dehalococcoides mccartyi strain 195, D. mccartyi strain BAV1, D. mccartyi strain GT, and D. mccartyi strain FL2 (strain FL2) were tested. *, calculated from data in Fig. 4B and Table 2 of reference 5; †, calculated from data in Fig. 1C and Fig. 3 of reference 14; ‡, calculated from data in Fig. 2 and Table 1 of reference 16; §, calculated from data in Fig. 6 and Table 1 of reference 7.
Values are given as means ± the standard deviations where applicable. NA, not applicable.
When the ratio of butyrate to TCE in the coculture was maintained under electron acceptor limitation with 79 ± 6.7 μmol of TCE and 440 ± 29 μmol of butyrate (measured by high-pressure liquid chromatography) as the electron donor, the H2 levels remained steady at about 24 to 180 nM during active dechlorination (from day 0 to day 7) and increased to 350 ± 20 nM right after TCE and cis-DCE were consumed on day 8 (Fig. 1c; see also the supplemental material, part 2, and Table S2 in the supplemental material). This H2 concentration was similar to the level (480 nM) that resulted in cessation of S. wolfei growth on butyrate in isolation (32). At the end of the experiment (day 18), 88.4% of the amended TCE could be accounted for in dechlorination products VC (7.4% ± 0.3% molar equivalents) and ethene (81.0 ± 0.1% molar equivalents). The missing 11.6% of initial TCE was likely due to the numerous sampling events during the experiment, as confirmed by losses in the abiotic controls (8.7% loss of initial TCE added). In addition, 115 ± 3.0 μmol of butyrate was consumed, while 234 ± 9 μmol of acetate and 3.9 ± 1.1 μmol of H2 remained in the bottles (in the abiotic control, the amount of butyrate decreased 26 μmol with no production of acetate or H2). This indicates that ca. 88.6% of electrons (0.886, corresponding to an fe° value [i.e., the proportion of electrons transferred to the electron acceptor for energy metabolism] of close to 0.84 [Fig. 1d]) in the form of H2 generated from butyrate fermentation (ca. 230 μmol of H2) supported dechlorination. A calculation based on biomass cell numbers (assumed 0.5 g of dry weight per g of cell, i.e., water accounts for 50% of the cell weight). The cell formula was C5H7O2N, with 20 electron equivalent per mol biomass, indicating that 7.4% (17 μmol of H2) of the electrons from butyrate fermentation (in the form of H2) were diverted to biomass production, giving a total electron recovery of ca. 98%. This result demonstrates that butyrate was efficiently used as the electron donor in the coculture with high electron transfer efficiency between the two bacteria.
The calculated Gibbs free energy of reaction (ΔGr; i.e., butyrate fermentation and associated hydrogen generation by S. wolfei) was −21.7 ± 0.3 kJ mol−1 on day 6 and decreased to −13.7 ± 0.2 kJ mol−1 on day 8 when TCE and cis-DCE were depleted. This number was smaller than the hypothetical minimum energy (−20 kJ mol−1) required by a bacterium to exploit the free-energy change in a reaction and support growth (33) (for detailed calculations, see the supplemental material, part 2). After TCE and cis-DCE depletion, we observed a slight decrease in butyrate (36.0 ± 6.6 μmol) during cometabolic VC dechlorination (Fig. 1c) with H2 concentrations increasing to a stable level of 1.2 ± 0.3 μM after day 10, while the cell numbers of strain 195 and S. wolfei were observed to decrease (Fig. 1b and c). The calculated ΔGr of butyrate fermentation during the cometabolic process was less negative and reached −5.7 ± 1.4 kJ mol−1 by the end of the experiment (see the supplemental material, part 2, and Fig. S3 in the supplemental material).
In order to determine the hydrogen threshold of strain 195 in the coculture, the strain was grown under electron donor-limited conditions (TCE fed in excess). The H2 concentrations dropped to thresholds of 0.6 ± 0.1 nM, at which point TCE was not further degraded by strain 195 and butyrate depletion ceased (see the supplemental material, part 3, and Fig. S4 in the supplemental material). In order to validate our method for quantifying the minimum H2-threshold concentration, we measured the hydrogen threshold of the sulfate reducing microorganism Desulfovibrio vulgaris Hildenborough (DvH) and obtained 15.2 ± 1.4 nM, a value that is in agreement with the literature results (14.8 nM) for this bacterium (34).
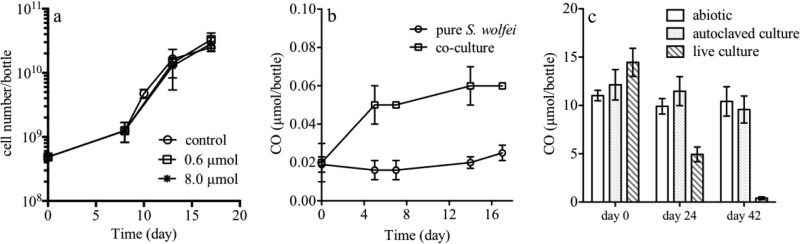
We tested the inhibitory effect of CO on S. wolfei by exposing cells to 0.6 to 8 μmol of CO per bottle and observed no significant effect on cell growth (Fig. 2a). CO accumulation for the S. wolfei isolate growing on crotonate was 0.02 μmol per bottle throughout the experiment (Fig. 2b), which was at the same level as that of the abiotic control (data not shown). In the coculture growing on butyrate, CO was measured at ∼0.06 μmol per bottle. This value was lower than that detected for the strain 195 isolate (0.5 ± 0.1 μmol per bottle after one dose of TCE [77 μmol] was depleted) (17). Furthermore, we found that pure S. wolfei could consume CO from 14.5 ± 1.5 μmol to 0.4 ± 0.1 μmol in 42 days (Fig. 2c) when grown on crotonate.
FIG 2.
(a) Inhibitory effect of different CO concentrations on S. wolfei cell growth. (b) CO accumulation for the S. wolfei isolate and in coculture with strain 195 on butyrate. (c) CO consumption by the S. wolfei isolate. Values are averages of biological triplicates; error bars indicate the standard deviations.
Formation of cell aggregates.
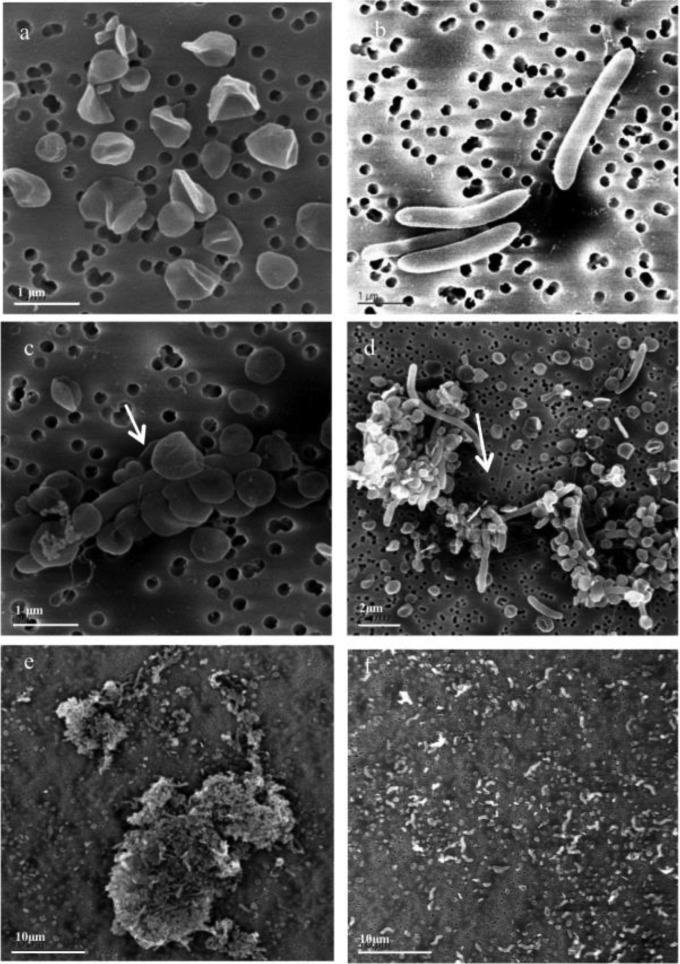
Strain 195 and S. wolfei isolates, as well as cocultures of S. wolfei and strain 195, maintained on butyrate were grown to late exponential to early stationary phase before samples were analyzed by SEM. Figure 3a and b show the specific cell morphology of the isolates with strain 195 growing in individual coccus shapes with diameters of approximately 0.5 to 1 μm, while most S. wolfei cells grew individually or in pairs as 0.25- to 0.5-μm by 2.5- to 5-μm rods, as reported in previous studies (1, 21). When butyrate was amended to the coculture as an electron donor with TCE as the electron acceptor, the cells form cell aggregates that ranged in size from 2 to 10 μm (Fig. 3c to e). In relatively small aggregates, cells were found connected with flagellum-like filaments (Fig. 3c and d). In large aggregates, extracellular polymeric substance (EPS)-like structures were observed (Fig. 3d and e). Cell aggregates were also observed during exponential growth phase (see the supplemental material, part 4, and Fig. S5 in the supplemental material). However, when strain 195 was grown as a coculture with DvH on lactate, the two strains grew together but did not form obvious cell aggregates (Fig. 3f).
FIG 3.
(a) Monoculture of strain 195 growing on H2 gas plus acetate and TCE. (b) Monoculture of S. wolfei growing on crotonate. (c to e) S. wolfei and strain 195 coculture growing on butyrate plus TCE at various magnifications. (f) DvH and strain 195 coculture growing on lactate plus TCE. Arrows indicate flagellum-like filaments of S. wolfei.
In order to determine whether the cocultures may be capable of sharing electrons via direct interspecies electron transfer (DIET), the crotonate and butyrate cocultures were sent to D. R. Lovley's laboratory for conductivity testing (35). The cocultures exhibited 3 orders of magnitude lower conductivity than Geobacter, indicating that H2 rather than DIET is the transfer mechanism. In addition, since S. wolfei could potentially use both hydrogen and formate as electron carriers for interspecies electron transfer (36), we tested the expression levels of several formate dehydrogenase genes (fdhA [Swol_0786, Swol_0800, and Swol_1825]) in cocultures grown on crotonate or butyrate and S. wolfei grown in isolation. The level of expression of fdhA subunits relative to that of the gyrB gene was generally in the range of 0.6 to 1.2 U for all three conditions (37), confirming that hydrogen rather than formate was the exclusive electron shuttle between these two bacteria.
The theoretical maximum interspecies distance for an observed rate of molecular transfer between two microorganisms can be calculated using Fick's diffusion law (20). Here, this distance was calculated for coculture growth on butyrate (Table 3, with detailed calculations in the supplemental material, part 4) by defining JH2 as the hydrogen flux between S. wolfei and strain 195. JH2 was calculated from the total surface areas of S. wolfei and substrate oxidation rate (hydrogen production rate = 2 × butyrate oxidation rate) measured in the coculture experiment using equation 2 in Table 1. (0.35 ± 0.02 μM) is the maximum H2 concentration immediately outside of an S. wolfei cell when fermentation by S. wolfei stops on butyrate and CH2-195 is the theoretical minimum H2 concentration above which strain 195 can gain energy, estimated in the present study to be 0.6 ± 0.1 nM. dsyn-195 is then the calculated maximum interspecies distance for accomplishing syntrophic oxidation at the observed substrate oxidation rate.
TABLE 3.
Calculation of cell-cell distance
| Day | Total no. of cellsa/ml | Mean cell-cell distanceb (μm) | Maximum avg interspecies distance for H2 transferc (μm) |
|---|---|---|---|
| 2 | 6.2 × 106 | 55.3 | 14 |
| 4 | 5.0 × 107 | 27.1d | 6.3d |
| 6 | 9.2 × 107 | 22.2 | 2.7 |
| 8 | 13.7 × 107 | 19.6 | 1.9 |
That is, for strain 195 plus S. wolfei.
The cell-cell distance was based on one sample and was calculated for suspended cells using the cell number quantified from quantitative PCR analysis.
That is, for the transfer of S. wolfei to strain 195 at the observed substrate oxidation rate. Values were calculated using Fick's diffusion law.
Detailed calculations for these values can be found in the supplemental material, part 4.
The theoretical mean cell-cell distance of randomly dispersed cells is calculated based on the total cell numbers (quantified using qPCR) suspended in the unstirred liquid culture (Table 3). In the present study, cell settling was not observed for the strain 195 isolate or cocultures growing on butyrate (strain 195 and S. wolfei) or lactate (strain 195 and DvH). Considering that the cell number ratio of strain 195 to S. wolfei was about 20:1 on day 4 (Fig. 1c), strain 195 cells account for the majority of all cells in the bottle, and the average cell-cell distance between suspended strain 195 and S. wolfei cells (i.e., ≫27.1 μm, Table 3) would therefore be larger than the calculated theoretical maximum cell-cell distance for achieving butyrate fermentation (Table 3). In order to accomplish syntrophic butyrate oxidation at the rate observed, the average interspecies distance must be much less than the distance between randomly dispersed cells (Table 3), necessitating the formation of aggregates. In addition, according to the thermodynamically consistent rate law, the calculated free energy available for S. wolfei growth on day 4 was −27.4 ± 1.1 kJ mol−1 which is small (see the supplemental material, part 2, and Fig. S3 in the supplemental material), indicating that close physical contact between the two species is particularly important for efficient syntrophic butyrate fermentation. In contrast, when strain 195 was grown with DvH as a coculture, the maximum calculated interspecies distance was large enough (755 μm) to enable interspecies hydrogen transfer between the two bacteria (see the supplemental material, part 4, and Table S3 in the supplemental material), and aggregates were not formed.
Strain 195 transcriptome during syntrophic growth.
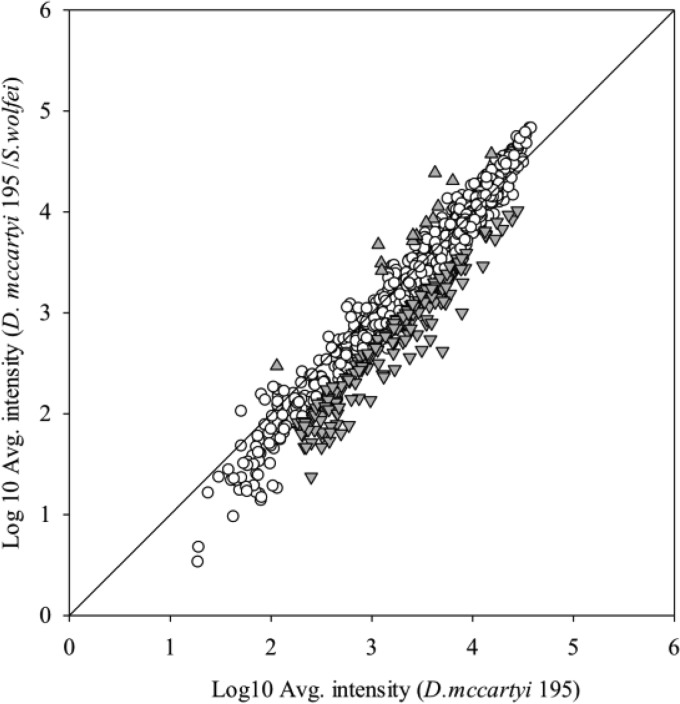
Transcriptomic microarray analysis comparing the coculture growing on butyrate and strain 195 growing in isolation identified 214 genes that were differentially transcribed (Fig. 4; see also Table S6 in the supplemental material). Among these differentially expressed genes, 18 were upregulated and 196 were downregulated in the syntrophic coculture compared to the strain 195 isolate. Among the upregulated genes, most significantly expressed genes (signal level, ca. 5,000 to 20,000) belonged to transport and metabolism functions, including several types of ATP binding cassette (ABC) transporters. Genes located within an operon of a Fec-type ABC transporter (from DET1173 to DET1176) which are involved in periplasmic iron-binding were upregulated 2.7 to 4.2 times, respectively. Genes (DET1491, DET1493) from a cluster encoding a peptide ABC transporter responsible for ATP binding were upregulated 2.1 and 2.3 times, respectively. Genes (DET0140, DET0141) encoding a phosphate ABC transporter and ATP-binding protein were upregulated 2.0 to 2.6 times, respectively. There were also a few genes with unknown functions upregulated at high signal levels compared to the isolate. DET1008, with highest homology to a gene encoding a cell division initiation protein (38), was upregulated 3.2 times, and acetyl-CoA synthase (DET1209) was upregulated 2.4 times at a high signal level (∼20,000).
FIG 4.
Microarray signal intensities of transcripts from strain 195 grown alone versus those grown in coculture with S. wolfei (gray-shaded points represent statistically significant differential transcription, average intensity >20 [P < 0.05], >2-fold difference, genes significantly upregulated [▲] or downregulated [▼] in coculture versus strain 195 monoculture). All measurements are averages from three biological replicates.
Genes associated with membrane-bound oxidoreductase complexes, which are related to energy metabolism, such as RDases, hydrogenases, molybdopterin oxidoreductases, and putative formate dehydrogenases, as well as NADH-oxidoreductases, showed no significant differential expression patterns between the coculture and pure culture throughout the experiment (see Table S6 in the supplemental material). Among the 196 downregulated genes in the coculture compared to the isolate, 75 were genes with signal levels lower than 1,000 in both treatments (suggesting low expression level), and most of them encode hypothetical proteins (see Table S6 in the supplemental material). Many of the downregulated genes were not related to energy metabolism, such as those encoding a phage domain protein (DET0354), mercuric reductase (DET0732), magnesium chelatase-like protein (DET0986), and virulence-associated protein E (DET1098).
DISCUSSION
In the strain 195 and S. wolfei syntrophic coculture studied here, strain 195 grew exponentially at the rate of 0.69 day−1, with a doubling time of 1.0 day, calculated from the cell numbers (tceA copies) from day 2 to day 6. This doubling time is shorter than previously reported for isolates of D. mccartyi (30, 39–41). The dechlorination rate was 9.9 ± 0.1 μmol day−1, which is similar to the value observed with strain 195 and DvH cocultures (11.0 ± 0.01 μmol day−1 [15]) and higher than the dechlorination rates observed with other D. mccartyi-containing cocultures (4.2 to 6.8 μmol day−1 [5, 7, 14–16]). The cell yield of strain 195 in this coculture [(1.1 ± 0.3) × 108 cells μmol−1 Cl− released] was similar to that observed in other D. mccartyi coculture studies, e.g., (9.0 ± 0.5) × 107 cells μmol−1 Cl−1 (15) and (9.0 ± 1.4) × 107 cells μmol−1 Cl− (7). Given that the minimum free energy change required for ATP synthesis is in the range of 15 to 25 kJ mol−1 for most syntrophic fermentations (42), only a small amount of energy saved as ATP is expected to be available during S. wolfei syntrophic butyrate fermentation (19). Indeed, S. wolfei in the coculture was growing very near the thermodynamic threshold (ΔG from −41.9 to −5.7 kJ mol−1), and consequently the cell numbers remained at low levels (1:16 ratio with strain 195 cells) when grown on butyrate compared to growth on crotonate. The ratio of calculated free energy available for strain 195 to reduce TCE to VC (−272.0 kJ mol−1) and S. wolfei (−43.6 kJ mol−1; see Table S2 in the supplemental material) growing on butyrate was about 6.2:1 at the beginning of the experiment. Considering that the cell size of S. wolfei is about 2.5 times larger than that of strain 195, the theoretical cell production ratio (based on free energy) of strain 195 to S. wolfei (15.6:1) is similar to our observation (16:1). While growing on crotonate, the ratio of available energy for strain 195 (−295.3kJ mol−1) and S. wolfei (−465.2 kJ mol−1) was 0.6, and the theoretical cell production ratio of 1.2:1 is similar to our observation of 1.3:1 (see the supplemental material, part 1). Roden and Jin (43) found a linear correlation between microbial growth yields (YXS, g cell mol−1 substrate) and estimated the catabolic ΔGr for the metabolism of short-chain fatty acids and H2-coupled metabolic pathways. Table 4 compares the growth yields calculated using the Roden and Jin (43) method (YXS,cal) with observed growth yields (YXS,obs) of syntrophs growing in a variety of cocultures. Our results confirmed that it is possible to estimate syntrophic cell yields by percentage error of less than 100% by applying an empirical equation for the listed syntrophic coculture studies (43), suggesting that the growth yield of syntrophic bacteria and the ratio maintained in the cocultures were mainly controlled by thermodynamics. Furthermore, we found H2 concentrations to be the key factor affecting the ΔGr value in the cocultures, which in turn affects predictions of microbial growth yields (see the calculations in Tables S2 and S5 in the supplemental material).
TABLE 4.
Estimation of syntrophic bacterial growth yields based on Gibbs free-energy (ΔGr) calculationsa
| Electron donor | Organisms | ΔGr (kJ/mol donor) | g of cells/mol |
Source or reference | |
|---|---|---|---|---|---|
| YXS,calb | YXS,obsc | ||||
| Lactate | DvH/Methanosarcina barkeri coculture | –61.4 | 3.4 | 6.7 (97)d | 62 |
| DvH/strain 195 coculture | –61.4 | 3.4 | 3.3 (3) | 15 | |
| DvH/strain 195/Methanobacterium congolense triculture | –61.4 | 3.4 | 5.3 (56) | 15 | |
| Butyrate | Syntrophomonas wolfei/Methanospirillum hungatei coculture | –9.1 | 2.3 | 1.36 (41) | 63 |
| Syntrophomonas wolfei/Desulfovibrio G11 coculture | –9.1 | 2.3 | 0.55 (76) | 63 | |
| Syntrophomonas wolfei/Methanospirillum hungatei/Methanosarcina barkeri triculture | –9.1 | 2.3 | 1.08 (53) | 63 | |
| Syntrophomonas wolfei/strain 195 coculture | –9.1 | 2.3 | 2.7 (17) | This study | |
ΔGr (Gibbs free-energy change) was calculated for H2 in the gaseous state at 1.3 Pa (∼10 nM in the aqueous phase). All other compounds were calculated at 10 mM.
Calculated cell yields (Ycal) were based on the equation Y = 2.08 + 0.0211 × (−ΔG′) (43).
Observed cell yields (Yobs) were based on directly measured cell masses (62) or masses converted from protein concentrations by assuming 2 g of cells/g of protein (63) or converted from cell numbers based on the assumption of 6 × 10−13 g per syntrophic cell (15, 43, 63). Percent error values are indicated in parentheses.
Absolute error E = 3.3 g of cells mol−1; relative error = 0.97; percent error = 97%.
When TCE was supplied in excess to the coculture (on an H2 production/consumption basis), the H2 level dropped to the threshold concentration of 0.6 ± 0.1 nM, and dechlorination ceased. Although the calculated ΔGr available for dechlorination by strain 195 was still negative (−145.5 kJ mol−1) at this concentration (see the supplemental material, part 5, and Table S5 in the supplemental material), it did not support growth. Thermodynamic calculations indicate that hydrogen concentrations would have to reach liquid concentrations of 10−27 nM to make the ΔGr for strain 195 positive, a value that is unrealistic in the environment (44). Therefore, the minimum H2 threshold for strain 195 cells is likely not based on thermodynamics but is rather based upon other factors such as enzyme binding affinity and specificity (45). This finding is consistent with previously published H2 thresholds for dechlorination by Dehalococcoides-containing communities (10, 41) and falls in the same range as the thresholds for sulfate reduction (46). Other dechlorinating isolates have been reported to have minimum H2 thresholds of around 0.04 to 0.3 nM (11, 46, 47). In previous studies of S. wolfei growing with H2-oxidizing hydrogenotrophic methanogens, sulfate reducers, and nitrate reducers, the estimated energy available for S. wolfei ranged from 0.9 to 15.0 kJ mol−1 (18) when butyrate fermentation ceased and the ratio of acetate to butyrate was ∼900. Although in the present study we observed that the estimated energy available for S. wolfei was −13.7 ± 0.2 kJ mol−1 with an acetate to butyrate ratio of 0.4:1, previous studies have shown that the calculated ΔGr value fell in the range (−10 to −15 kJ mol−1) observed in H2-dependent terminal electron-accepting processes under starvation conditions in sulfate-reducing bacteria and methanogenic archaea (48–50). Furthermore, the value obtained here was close to the average threshold value of butyrate metabolism (−13.8 ± 1.2 kJ mol−1) reported in coculture Syntrophus aciditrophicus and Desulfovibrio strain G11 at different acetate/butyrate ratios by Jackson and McInerney (51).
It is interesting that the amount of butyrate in the bottles continued to decrease slightly (36 μmol) after TCE was depleted and that ethene was being produced from VC (from day 8 to day 18). According to thermodynamics, microbial metabolism ceases when the ΔGr available from a reaction becomes positive. At the beginning of the experiment, the ΔGr available for S. wolfei butyrate fermentation was negative (−41.9 ± 1.5 kJ mol−1), and on day 6 this number increased to −21.7 ± 0.3 kJ mol−1 and then on day 8 it increased further to −13.7 ± 0.2 kJ mol−1 (see Fig. S3 in the supplemental material), which is less than the minimum energy (−20 kJ mol−1) required by a bacterium to synthesize ATP (33). During this time, the cell numbers decreased, indicating cell death and lysis. It has been reported that butyrate fermentation by S. wolfei stops when aqueous H2 concentrations reach 480 nM (32). In the present study, the H2 concentration was 350 ± 20 nM in the coculture on day 7 when all TCE was reduced to VC. However, the concentration of H2 steadily increased to 1,200 ± 300 nM by the end of the experiment on day 18, indicating that additional butyrate fermentation occurred, although cell growth had ceased, suggesting that regulation of the fermentation enzymes may not be stringent. During active dechlorination, H2 concentrations remained at levels below 200 nM (Fig. 1b), indicating that the H2 generation rate was at about the same level as the consumption rate (Fig. 1d) and that the growth rates of the two species were strictly coupled by hydrogen transfer.
In strain 195, acetyl-CoA is cleaved in an incomplete Wood-Ljungdahl pathway to provide the methyl group for methionine biosynthesis, whereby carbon monoxide (CO) is produced as a by-product that accumulates and eventually inhibits D. mccartyi growth and dechlorination (17, 52). However, in microbial communities, CO can serve as an energy source for many anaerobic microorganisms (53). A previous study showed a high CO concentration (15%) to have an inhibitory effect on S. wolfei (36). Here, we found that not only did low levels of CO not exhibit inhibitory effects on S. wolfei cell growth, but S. wolfei could consume CO (μmol per bottle) while growing on crotonate. CO levels in the coculture growing on butyrate were maintained at low levels during the feeding cycle rather than accumulating as it does with the 195 isolate. Therefore, it is possible that CO serves as a supplemental energy source for S. wolfei during syntrophic fermentation with strain 195. This finding is interesting since there were no anaerobic carbon monoxide dehydrogenase genes annotated in the S. wolfei genome (19). However, Swol_1136 and Swol_1818, which were originally annotated as iron-sulfur cluster binding domain-containing protein and 2Fe-2S binding protein in S. wolfei, were more recently annotated in the KEGG database (54) as carbon monoxide dehydrogenase small subunits for CO conversion to CO2, suggesting the genomic potential for CO metabolic ability in S. wolfei.
Formation of cell aggregates has been shown to be a distinctive feature of obligate syntrophic communities containing acetogenic bacteria and methanogenic archaea (20). Clustering of cells with decreasing intermicrobial distances leads to increased fluxes and increased specific growth rates (55). Previous studies reported that the syntrophic coculture of a propionate degrader and a hydrogenotrophic methanogen could form cell aggregates while growing on propionate for optimal hydrogen transfer (27, 56). A similar phenomenon was also observed in mixed cultures containing propionate degraders and methanogens enriched from an anaerobic biowaste digester where flocs formed showing that reducing the interspecies distances by aggregation was advantageous in complex ecosystems (57). In engineered dechlorinating systems, D. mccartyi species have been observed in biofilms within a membrane bioreactor (58) and in bioflocs maintained in a continuous flow bioreactor fed with butyrate (13). Based on the morphological observations of the present study, strain 195 cells grown in isolation are more likely to grow in planktonic form and, when grown with DvH as a syntrophic coculture on lactate, no significant aggregates were observed. However, when grown with S. wolfei on butyrate, the cocultures formed aggregates during syntrophic growth. Thermodynamic calculations show that syntrophic butyrate oxidation is endergonic unless the circumstantial H2 partial pressure is maintained very low (Table 1 and see Table S2 in the supplemental material). Furthermore, fdh dehydrogenase expression in S. wolfei under different growth conditions confirmed that these genes were not upregulated in the coculture grown on butyrate compared to growth on crotonate or in isolation (37). Ours is the first study to report D. mccartyi forming cell aggregates with a syntrophic partner. In addition, because the average intermicrobial distances are smaller for syntrophic aggregates, the H2 and metabolite flux between cells would be expected to increase, leading to higher material transfer efficiencies, which could partly explain the observed increased cell yields of strain 195. Another reason for the observed increased cell yields of strain 195 is likely due to the continuous removal of CO, which was shown to exert an inhibitory effect on D. mccartyi growth (17).
In a previous study of Methanococcus maripaludis growing in syntrophic association with Desulfovibrio vulgaris under hydrogen limitation in chemostats (59), M. maripaludis was growing close to the thermodynamic threshold for growth, and the acetyl-CoA synthase transcripts levels of M. maripaludis in the coculture decreased compared to the isolate. However, in our study, the acetyl-CoA synthase transcripts levels of strain 195 in the coculture increased compared to the isolate. A recent study (60) showed that a division of metabolic labor and mutualistic interactions were the reasons for increased rates of growth in a vast majority of cross-feeding strains. The benefit of cross-feeding was larger than the cost (60, 61).
Supplementary Material
ACKNOWLEDGMENTS
We thank Michael McInerney at University of Oklahoma for kindly providing the pure S. wolfei strain used for this study. We also thank Nikhil S. Malvankar and Derek Lovley at the University of Massachusetts for carrying out the conductivity tests for the cocultures.
This study was funded through research grants from the NIEHS (P42-ES04705-14) and the NSF (CBET-1336709).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03464-14.
REFERENCES
- 1.Maymó-Gatell X, Chien YT, Gossett JM, Zinder SH. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568–1571. doi: 10.1126/science.276.5318.1568. [DOI] [PubMed] [Google Scholar]
- 2.McCarty PL. 2010. Groundwater contamination by chlorinated solvents: history, remediation technologies, and strategies, p 1–24. In Stroo HF, Ward CH (ed), In situ remediation of chlorinated solvent plumes. Springer Science Publishing, New York, NY. [Google Scholar]
- 3.Lu X, Wilson JT, Kampbell DH. 2006. Relationship between Dehalococcoides DNA in ground water and rates of reductive dechlorination at field scale. Water Res 40:3131–3140. doi: 10.1016/j.watres.2006.05.030. [DOI] [PubMed] [Google Scholar]
- 4.Lee PKH, Warnecke F, Brodie EL, Macbeth TW, Conrad ME, Andersen GL, Alvarez-Cohen L. 2012. Phylogenetic microarray analysis of a microbial community performing reductive dechlorination at a TCE-contaminated site. Environ Sci Technol 46:1044–1054. doi: 10.1021/es203005k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He J, Holmes VF, Lee PK, Alvarez-Cohen L. 2007. Influence of vitamin B12 and cocultures on the growth of Dehalococcoides isolates in defined medium. Appl Environ Microbiol 73:2847–2853. doi: 10.1128/AEM.02574-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Men YJ, Seth EC, Yi S, Allen RH, Taga ME, Alvarez-Cohen L. 2014. Sustainable growth of Dehalococcoides mccartyi 195 by corrinoid salvaging and remodeling in defined lactate-fermenting consortia. Appl Environ Microbiol 80:2133–2141. doi: 10.1128/AEM.03477-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J, Im J, Yang Y, Löffler FE. 2013. Guided cobalamin biosynthesis supports Dehalococcoides mccartyi reductive dechlorination activity. Philos Trans R Soc Lond B Biol Sci 368:1–23. doi: 10.1098/rstb.2012.0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schipp CJ, Marco-Urrea E, Kublik A, Seifert J, Adrian L. 2013. Organic cofactors in the metabolism of Dehalococcoides mccartyi strains. Philos Trans R Soc Lond B Biol Sci 368:1–12. doi: 10.1098/rstb.2012.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziv-El M, Delgado AG, Yao Y, Kang D, Nelson KG, Halden RU, Krajmalnik-Brown R. 2011. Development and characterization of DehaloR^2, a novel anaerobic microbial consortium performing rapid dechlorination of TCE to ethene. Appl Microbiol Biotechnol 92:1063–1071. doi: 10.1007/s00253-011-3388-y. [DOI] [PubMed] [Google Scholar]
- 10.Yang YR, McCarty PL. 1998. Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture. Environ Sci Technol 32:3591–3597. [Google Scholar]
- 11.Löffler FE, Tiedje JM, Sanford RA. 1999. Fraction of electrons consumed in electron acceptor reduction and hydrogen thresholds as indicators of halorespiratory physiology. Appl Environ Microbiol 65:4049–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duhamel M, Wehr SD, Yu L, Rizvi H, Seepersad D, Dworatzek S, Cox EE, Edwards EA. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res 36:4193–4202. doi: 10.1016/S0043-1354(02)00151-3. [DOI] [PubMed] [Google Scholar]
- 13.Rowe AR, Lazar BJ, Morris RM, Richardson RE. 2008. Characterization of the community structure of a dechlorinating mixed culture and comparisons of gene expression in planktonic and biofloc-associated “Dehalococcoides” and Methanospirillum species. Appl Environ Microbiol 74:6709–6719. doi: 10.1128/AEM.00445-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng D, Chow WL, He J. 2010. A Dehalococcoides-containing coculture that dechlorinates tetrachloroethene to trans-1,2-dichloroethene. ISME J 4:88–97. doi: 10.1038/ismej.2009.90. [DOI] [PubMed] [Google Scholar]
- 15.Men YJ, Feil H, Verberkmoes NC, Shah MB, Johnson DR, Lee PK, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L. 2011. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J 6:410–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan J, Ritalahti KM, Wagner DD, Löffler FE. 2012. Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains. Appl Environ Microbiol 78:6630–6636. doi: 10.1128/AEM.01535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhuang WQ, Yi S, Bill M, Brisson VL, Feng X, Men Y, Conrad ME, Tang YJ, Alvarez-Cohen L. 2014. Incomplete Wood-Ljungdahl pathway facilitates one carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci U S A 111:6419–6424. doi: 10.1073/pnas.1321542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jin Q. 2007. Control of hydrogen partial pressures on the rates of syntrophic microbial metabolisms: a kinetic model for butyrate fermentation. Geobiology 5:35–48. doi: 10.1111/j.1472-4669.2006.00090.x. [DOI] [PubMed] [Google Scholar]
- 19.Sieber JR, Sims DR, Han C, Kim E, Lykidis A, Lapidus AL, McDonnald E, Rohlin L, Culley DE, Gunsalus R, McInerney MJ. 2010. The genome of Syntrophomonas wolfei: new insights into syntrophic metabolism and biohydrogen production. Environ Microbiol 12:2289–2301. doi: 10.1111/j.1462-2920.2010.02237.x. [DOI] [PubMed] [Google Scholar]
- 20.Stams AJM, Worm P, Sousa DZ, Alves MM, Plugge CM. 2012. Syntrophic degradation of fatty acids by methanogenic communities, p 127–142. In Hallenbeck PC. (ed), Microbial technologies in advanced biofuels production. Springer, New York, NY. [Google Scholar]
- 21.McInerney MJ, Bryant MP, Hespell RB, Costerton JW. 1981. Syntrophomonas wolfei gen. nov. sp. nov., an anaerobic, syntrophic, fatty acid-oxidizing bacterium. Appl Environ Microbiol 41:1029–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaty PS, McInerney MJ. 1987. Growth of Syntrophomonas wolfei in pure culture on crotonate. Arch Microbiol 147:389–393. doi: 10.1007/BF00406138. [DOI] [Google Scholar]
- 23.Freeborn RA, West KA, Bhupathiraju VK, Chauhan S, Rahm BG, Richardson RE, Alvarez-Cohen L. 2005. Phylogenetic analysis of TCE-dechlorinating consortia enriched on a variety of electron donors. Environ Sci Technol 39:8358–8368. doi: 10.1021/es048003p. [DOI] [PubMed] [Google Scholar]
- 24.Lee PKH, Johnson DR, Holmes VF, He J, Alvarez-Cohen L. 2006. Reductive dehalogenase gene expression as a biomarker for physiological activity of Dehalococcoides spp. Appl Environ Microbiol 72:6161–6168. doi: 10.1128/AEM.01070-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorby YA, Yanina S, McLean JS, Rosso KM, Moyles D, Dohnalkova A, Beveridge TJ, Chang IS, Kim BH, Kim KS, Culley DE, Reed SB, Romine MF, Saffarini DA, Hill EA, Shi L, Elias DA, Kennedy DW, Pinchuk G, Watanabe K, Ishii S, Logan B, Nealson KH, Fredrickson JK. 2006. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc Natl Acad Sci U S A 103:9536–9542. doi: 10.1073/pnas.0603786103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dykstra MJ, Reuss LE. 1992. Biological electron microscopy: theory, techniques, and troubleshooting, p 223–245. In Scanning electron microscopy, 2nd ed. ASM Press, Washington, DC. [Google Scholar]
- 27.Ishii S, Kosaka T, Hori K, Hotta Y, Watanabe K. 2005. Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus. Appl Environ Microbiol 71:7838–7845. doi: 10.1128/AEM.71.12.7838-7845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes WM. 2013. CRC handbook of physics and chemistry, 95th ed. CRC Press, Inc, Boca Raton, FL. [Google Scholar]
- 29.Johnson DR, Lee PKH, Holmes VF, Alvarez-Cohen L. 2005. An internal reference technique for accurately quantifying specific mRNAs by real-time PCR with application to the tceA reductive dehalogenase gene. Appl Environ Microbiol 71:3866–3871. doi: 10.1128/AEM.71.7.3866-3871.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DR, Brodie EL, Hubbard Alan E, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Temporal transcriptomic microarray analysis of Dehalococcoides ethenogenes strain 195 during the transition into stationary phase. Appl Environ Microbiol 74:2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.West KA, Lee PKH, Johnson DR, Zinder SH, Alvarez-Cohen L. 2013. Global gene expression of Dehalococcoides within a robust dynamic TCE-dechlorinating community under conditions of periodic substrate supply. Biotechnol Bioeng 110:1333–1341. doi: 10.1002/bit.24819. [DOI] [PubMed] [Google Scholar]
- 32.Wallrabenstein C, Schink B. 1994. Evidence of reversed electron transport in syntrophic butyrate and benzoate oxidation by Syntrophomonas wolfei and Syntrophus buswellii. Arch Microbiol 162:136–142. doi: 10.1007/BF00264387. [DOI] [Google Scholar]
- 33.Schink B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev 61:262–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cord-Ruwisch R, Seitz HJ, Conrad R. 1988. The capacity of hydrogenotrophic anaerobic bacteria to compete for traces of hydrogen depends on the redox potential of the terminal electron acceptor. Arch Microbiol 149:350–357. doi: 10.1007/BF00411655. [DOI] [Google Scholar]
- 35.Shrestha PM, Rotaru AE, Aklujkar M, Liu F, Shrestha M, Summers ZM, Malvankar N, Flores DC, Lovley DR. 2013. Syntrophic growth with direct interspecies electron transfer as the primary mechanism for energy exchange. Environ Microbiol Rep 5:904–910. doi: 10.1111/1758-2229.12093. [DOI] [PubMed] [Google Scholar]
- 36.Sieber JR, Le HM, McInerney MJ. 2014. The importance of hydrogen and formate transfer for syntrophic fatty, aromatic and alicyclic metabolism. Environ Microbiol 16:177–188. doi: 10.1111/1462-2920.12269. [DOI] [PubMed] [Google Scholar]
- 37.Crable BR. 2013. Enzyme systems involved in interspecies hydrogen and formate transfer between syntrophic fatty and aromatic acid degraders and Methanospirillum hungatei. Ph.D. dissertation. University of Oklahoma, Norman, OK. [Google Scholar]
- 38.Ueda K, Yamashita A, Ishikawa J, Shimada M, Watsuji TO, Morimura K, Ikeda H, Hattori M, Beppu T. 2004. Genome sequence of Symbiobacterium thermophilum, an uncultivable bacterium that depends on microbial commensalism. Nucleic Acids Res 32:4937–4944. doi: 10.1093/nar/gkh830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He J, Sung Y, Krajmalnik-Brown R, Ritalahti KM, Löffler FE. 2005. Isolation and characterization of Dehalococcoides sp. strain FL2, a trichloroethene (TCE)- and 1,2-dichlroethene-respiring anaerobe. Environ Microbiol 7:1442–1450. doi: 10.1111/j.1462-2920.2005.00830.x. [DOI] [PubMed] [Google Scholar]
- 40.Sung Y, Ritalahti KM, Apkarian RP, Löffler FE. 2006. Quantitative PCR confirms purity of strain GT, a novel trichloroethene-to-ethene-respiring Dehalococcoides isolate. Appl Environ Microbiol 72:1980–1987. doi: 10.1128/AEM.72.3.1980-1987.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton JAH, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63(Pt 2):625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 42.Stams AJM, Plugge CM. 2009. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol 7:568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 43.Roden EE, Jin Q. 2011. Thermodynamics of microbial growth coupled to metabolism of glucose, ethanol, short-chain organic acids, and hydrogen. Appl Environ Microbiol 77:1907–1909. doi: 10.1128/AEM.02425-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heimann AC, Jakobsen R. 2006. Experimental evidence for a lack of thermodynamic control on hydrogen concentrations during anaerobic degradation of chlorinated ethenes. Environ Sci Technol 40:3501–3507. doi: 10.1021/es052320u. [DOI] [PubMed] [Google Scholar]
- 45.Dolfing J. 2003. Thermodynamic considerations for dehalogenation, p 89–114. In Haggblom MM, Bossert ID (ed), Dehalogenation: microbial processes and environmental applications. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 46.Luijten ML, Roelofsen W, Langenhoff AA, Schraa G, Stams AJ. 2004. Hydrogen threshold concentrations in pure cultures of halorespiring bacteria and at a site polluted with chlorinated ethenes. Environ Microbiol 6:646–650. doi: 10.1111/j.1462-2920.2004.00608.x. [DOI] [PubMed] [Google Scholar]
- 47.Lu XX, Tao S, Bosma T, Gerritse J. 2001. Characteristic hydrogen concentrations for various redox processes in batch study. J Environ Sci Health A Tox Hazard Subst Environ Eng 36:1725–1734. doi: 10.1081/ESE-100106254. [DOI] [PubMed] [Google Scholar]
- 48.Hoehler TM, Alperin MJ, Alber DB, Martens CS. 2001. Apparent minimum free energy requirements for methanogenic Archaea and sulfate-reducing bacteria in an anoxic marine sediment. FEMS Microbiol Ecol 38:33–41. doi: 10.1111/j.1574-6941.2001.tb00879.x. [DOI] [Google Scholar]
- 49.Hoehler TM. 2004. Biological energy requirements as quantitative boundary conditions for life in the subsurface. Geobiology 2:205–215. doi: 10.1111/j.1472-4677.2004.00033.x. [DOI] [Google Scholar]
- 50.Heimann A, Jakobsen R, Blodau C. 2010. Energetic constraints on H2-dependent terminal electron accepting processes in anoxic environments: a review of observations and model approaches. Environ Sci Technol 44:24–33. doi: 10.1021/es9018207. [DOI] [PubMed] [Google Scholar]
- 51.Jackson BE, McInerney MJ. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454–456. doi: 10.1038/415454a. [DOI] [PubMed] [Google Scholar]
- 52.Tang YJ, Yi S, Zhuang W, Zinder SH, Keasling JD, Alvarez-Cohen L. 2009. Investigation of carbon metabolism in “Dehalococcoides ethenogenes” strain 195 by use of isotopomer and transcriptomic analyses. J Bacteriol 191:5224–5231. doi: 10.1128/JB.00085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Oelgeschläger E, Rother M. 2008. Carbon monoxide-dependent energy metabolism in anaerobic bacteria and archaea. Arch Microbiol 190:257–269. doi: 10.1007/s00203-008-0382-6. [DOI] [PubMed] [Google Scholar]
- 54.Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2014. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schink B, Thauer RK. 1988. Energetics of syntrophic methane formation and the influence of aggregation, p 5–17. In Lettinga G, Zehnder AJB, Grotenhuis JTC, Hulshoff LW (ed), Granular anaerobic sludge: microbiology and technology. Pudoc, Wageningen, The Netherlands. [Google Scholar]
- 56.Ishii S, Kosaka T, Hotta Y, Watanabe K. 2006. Simulating the contribution of coaggregation to interspecies hydrogen fluxes in syntrophic methanogenic consortia. Appl Environ Microbiol 72:5093–5096. doi: 10.1128/AEM.00333-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Felchner-Zwirello M, Winter J, Gallert C. 2013. Interspecies distances between propionic acid degraders and methanogens in syntrophic consortia for optimal hydrogen transfer. Appl Microbiol Biotechnol 97:9193–9205. doi: 10.1007/s00253-012-4616-9. [DOI] [PubMed] [Google Scholar]
- 58.Chung J, Krajmalnik-Brown R, Rittmann BE. 2008. Bioreduction of trichloroethene using a hydrogen-based membrane biofilm reactor. Environ Sci Technol 42:477–483. doi: 10.1021/es702422d. [DOI] [PubMed] [Google Scholar]
- 59.Walker CB, Redding-Johanson AM, Baidoo EE, Rajeev L, He Z, Hendrickson EL, Joachimiak MP, Stolyar S, Arkin AP, Leigh JA, Zhou J, Keasling JD, Mukhopadhyay A, Stahl DA. 2012. Functional responses of methanogenic archaea to syntrophic growth. ISME J 6:2045–2055. doi: 10.1038/ismej.2012.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pande S, Merker H, Bohl K, Reichelt M, Schuster S, de Figueiredo LF, Kaleta C, Kost C. 2014. Fitness and stability of obligate cross-feeding interactions that emerge upon gene loss in bacteria. ISME J 8:953–962. doi: 10.1038/ismej.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mee MT, Collins JJ, Church GM, Wang HH. 2014. Syntrophic exchange in synthetic microbial communities. Proc Natl Acad Sci U S A 111:2149–2156. doi: 10.1073/pnas.1405641111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Traore AS, Fardeau ML, Hatchikian CE, Legall J, Belaich JP. 1983. Energetics of growth of a defined mixed culture of Desulfovibrio vulgaris and Methanosarcina barkeri: interspecies hydrogen transfer in batch and continuous cultures. Appl Environ Microbiol 46:1152–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Beaty PS, McInerney MJ. 1989. Effects of organic-acid anions on the growth and metabolism of Syntrophomonas wolfei in pure culture and in defined consortia. Appl Environ Microbiol 55:977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.