Abstract
Although complex cooccurrence patterns have been described for microbes in natural communities, these patterns have scarcely been interpreted in the context of ecosystem functioning and stability. Here we constructed networks from species cooccurrences between pairs of microorganisms which were extracted from five individual aquatic time series, including a dystrophic and a eutrophic lake as well as an open ocean site. The resulting networks exhibited higher clustering coefficients, shorter path lengths, and higher average node degrees and levels of betweenness than those of random networks. Moreover, simulations demonstrated that taxa with a large number of cooccurrences and placement at convergence positions in the network, so-called “hubs” and “bottlenecks,” confer resistance against random removal of “taxa.” Accordingly, we refer to cooccurrences at convergence positions as system-relevant interdependencies, as they, like hubs and bottlenecks, determine network topology. These topology features of the cooccurrence networks point toward microbial community dynamics being resistant over time and thus could provide indicators for the state of ecosystem stability.
INTRODUCTION
Microorganisms underpin and drive aquatic biogeochemical cycles (1) and provide fuel for higher trophic levels (2, 3). Recent efforts, accelerated by access to novel sequencing technologies, have revealed the immense phylogenetic and metabolic diversity of microorganisms in aquatic environments and other biomes (4–9). While these studies have improved our understanding of the mechanisms determining community structure, the complexity of trophic interactions, competition, and other interdependencies among microorganisms has still not been studied extensively. To realize the full potential of the vast amount of microbial community data that are accumulating, novel methods are needed to explore and interpret cooccurrence patterns and thereby provide insights about such interaction patterns and their linkages to ecosystem features.
Thus far, most efforts to model interactions have focused on food webs where bacterial and microeukaryotic communities have not been resolved beyond very broad functional or taxonomic groups (10, 11). Knowledge of microbial interactions is also very fragmented when extended to a broader framework of ecological networks, which consider all kinds of taxon interactions, not only those that are strictly trophic. Several recent attempts to resolve interactions within microbial communities included the use of network analyses based on analysis of cooccurrence matrices (6, 12–16). However, with one exception (15), these studies neither included interactions between unicellular as well as multicellular eukaryotic microorganisms nor relied on absolute abundances to infer linkages. Nevertheless, these early studies provided important new information about the functioning of bacterial community interactions, where taxa with positive associations have been interpreted as functional guilds of organisms performing similar or complementary functions (6) or feature interactions shaped by interspecies cross-feeding (12). Analogously, negative associations have been suggested to reflect direct interactions, such as competition (6), or to be caused by niche partitioning (12) and/or resistance to losses by grazing (6).
We collected microbial taxon distribution patterns from five time series, including sample series collected from three water masses from a humic boreal lake (Alinen Mustajärvi) and surface samples from eutrophic Lake Erken and the Hawaiian Ocean Time (HOT) series (49). We used maximal information-based nonparametric exploration (MINE) (17) to capture relationships from temporal patterns of bacterioplankton taxa and, if available, also of phytoplankton and zooplankton, using both absolute and relative abundance matrices. This nonparametric approach also identified nonlinear relationships among pairs of taxa, providing some clues to the interactions among microbes. We complemented this widely used taxon-centric approach with graph theory, a concept currently making its way into the field of microbial ecology (see the work of Steele et al. [18] and Kara et al. [16]). In graph theory, networks are modeled as graphs, i.e., mathematical objects consisting of nodes and edges, which in our case represent taxa and significant cooccurrence relationships between pairs of taxa, respectively.
Our aim was to benchmark the analysis of network properties for ecological interpretations. In particular, we discuss the implications of real and simulated network properties for the resistance of aquatic microbial community patterns to taxon removals and of ecosystem functioning to environmental disturbances.
MATERIALS AND METHODS
Data sets.
Three of the data sets represent time series from the epi-, meta-, and hypolimnia of Lake Alinen Mustajärvi, southern Finland, collected during the open water seasons of 2006 to 2010. The epilimnion (here called “trophic”) data set consisted of 29 time series samples, while the meta- and hypolimnetic data sets comprised 35 samples each (Table 1). Characteristics of the lake and the sampling procedures have been described in detail by Peura et al. (7).
TABLE 1.
Network topology measures of absolute, Erdős-Rényi, and Barabási networks
| Parameter | Value or descriptiona |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Absolute network |
Erdős-Rényi network |
Barabási network |
|||||||||||||
| Trophic | Erken | HOT | Metalimnion | Hypolimnion | Trophic | Erken | HOT | Metalimnion | Hypolimnion | Trophic | Erken | HOT | Metalimnion | Hypolimnion | |
| No. of samples | 29 | 30 | 18 | 35 | 35 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Sample spacing (days) (mean ± SD) | 32 ± 29 | 11 ± 8.2 | 64 ± 23 | 29 ± 29 | 29 ± 27 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| P value | <0.05 | <0.05 | <0.05 | <0.05 | <0.05 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| No. of nodes | 298 | 146 | 308 | 203 | 258 | 298 | 146 | 308 | 203 | 258 | 298 | 145 | 307 | 203 | 258 |
| MIC | 0.40 | 0.44 | 0.56 | 0.44 | 0.44 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| No. of edges | 1,436 | 716 | 1,477 | 660 | 1,980 | 1,436 | 716 | 1,477 | 1,477 | 1,436 | 1,485 | 710 | 1,520 | 603 | 2,028 |
| No. of clusters | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Maximum degree | 50 | 49 | 54 | 36 | 87 | 19 | 19 | 19 | 14 | 27 | 147 | 85 | 150 | 82 | 161 |
| Density | 0.03 | 0.07 | 0.03 | 0.03 | 0.06 | 0.03 | 0.07 | 0.03 | 0.03 | 0.06 | 0.03 | 0.07 | 0.03 | 0.03 | 0.06 |
| Clustering coefficient | 0.26 | 0.36 | 0.35 | 0.24 | 0.30 | 0.03 | 0.07 | 0.03 | 0.03 | 0.06 | 0.08 | 0.14 | 0.08 | 0.07 | 0.14 |
| Avg path length | 3.04 | 2.79 | 3.61 | 3.21 | 2.52 | 2.76 | 2.42 | 2.77 | 3.03 | 2.32 | 2.22 | 2.10 | 2.22 | 2.50 | 1.97 |
| Diam | 7 | 7 | 9 | 7 | 6 | 5 | 4 | 5 | 6 | 4 | 4 | 3 | 4 | 4 | 3 |
| Degree distribution | PL/exp | PL/exp | PL/exp | PL/exp | PL/exp | Poisson | Poisson | Poisson | Poisson | Poisson | PL/Exp | PL/Exp | PL/Exp | PL/Exp | PL/Exp |
| Connectance | 0.02 | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 0.03 | 0.02 | 0.02 | 0.03 | 0.02 | 0.03 | 0.02 | 0.01 | 0.03 |
Values for the random networks are averages for 1,000 iterations. NA, not applicable.
In addition to 16S rRNA-based bacterioplankton community data, both phytoplankton and zooplankton data were available for the trophic data set. Zooplankton samples were preserved with formaldehyde, and the species composition and abundance of zooplankton were determined using an inverted microscope at a magnification of ×100 by counting the individual species. The phytoplankton organisms were counted by inverted microscopy, using a magnification of ×400 to ×600; at least 500 counting units (cells, colonies, or filaments) in total and at least 50 units of each of the most common taxa were counted. Phytoplankton organisms were identified to the species level if possible; otherwise, the genus or a higher taxonomic level was recorded. For the relative network, phytoplankton and zooplankton numbers were converted to proportions. Bacterioplankton diversity was assessed by 454 pyrosequencing as described by Peura et al. (7). Bacterial abundance was determined from 200-ml samples fixed with 1 ml of Lugol's solution. The samples were first decolorized with sodium thiosulfate and then stained with DAPI (4′,6-diamidino-2-phenylindole; Sigma) and filtered onto black polycarbonate filters (0.22-μm pore size; Osmonics). Ten random fields per filter were photographed with an epifluorescence microscope (Olympus BX60; Olympus Optical Co., Tokyo, Japan) at a magnification of ×1,000 and were analyzed with CellC software (19).
The two additional data sets included a time series from Lake Erken (59°51′N, 18°36′E) (described by Eiler et al. [6]) and the HOT series (22°30′N, 158°W [49]) ([http://vamps.mbl.edu]). Bacterial abundance data were obtained by flow cytometry for both the Lake Erken (50) and HOT (http://hahana.soest.hawaii.edu/hot/) series.
Network construction.
The 16S rRNA data from individual data sets were rarefied to equal sample sizes based on the sample with the fewest sequences, using the perl script daisychopper.pl (20). Absolute numbers for each bacterioplankton operational taxonomic unit (OTU) were approximated by estimating the products of relative OTU abundances and total prokaryotic counts under the assumptions that the archaeal fraction was negligible and that all bacterial taxa were targeted equally by PCR. For phytoplankton and zooplankton numbers in the trophic data, the absolute cell counts were available. Using absolute rather than proportional data avoids biased inferences associated with the use of relative numbers (21). For example, relative abundance changes are often driven by a few very abundant taxa, which means that even though the relative value changes, the actual number of an organism may remain the same. This can lead to false discoveries of significant covariations between taxa. To test the impact of relative versus absolute data, the network for trophic data was constructed using both approaches.
Relationships with P values of <0.05, as determined in the MINE package (17), were used to construct networks. Parameters for analysis were set to the defaults, and false discovery rates were below 0.04. In the trophic data set, all phytoplankton and zooplankton taxa and all bacterial OTUs with at least 20 reads and present in at least three samples in the total resampled data set were included in the analysis. For the bacterial data sets, the same data selection criteria as those used for OTUs in the trophic data set were used. The time lag between samples was not considered, as the sampling points were not evenly spaced. The chosen P value set the maximal information coefficient (MIC) (17) cutoff to 0.40 to 0.56, depending on the number of samples in the data set (Table 1). The MIC is a statistical measure, similar to R2 in general linear models, describing the goodness of fit between two variables (17). Another output of MINE used in our analyses, the nonlinearity statistic, describes the shapes of relationships, as values near 0 indicate linear relationships and large (up to 1) values indicate nonlinear relationships.
Network properties were additionally calculated for MIC cutoffs of 0.45 to 0.6 to study the stability of these properties in the trophic network. A MIC of 0.6 was chosen as the upper limit for the simulation because it decreased the number of edges in all data sets to below 30%. The resulting MIC matrices were translated into networks by using Cytoscape 2.6.3 (22). Cytoscape depicts data sets as nodes (plankton taxa and OTUs) connected by edges that denote the strength of the relationship. For visualization of the numbers of connection between different taxa in the trophic network, the number of connections for each taxon was normalized against the number of possible connections within each plankton compartment. Tnet (23) and igraph (24) as implemented in R (25) were used for exploring network topologies. R packages Vegan (26) and Bipartite (27) were used to scrutinize network properties according to classical community assembly rules (28). Community assembly was studied by a comparison of 1,000 random communities with the same number of species.
Network simulations.
The levels of robustness of the empirical networks and also of random networks constructed according to node and edge numbers of the empirical networks (Erdős-Rényi and Barabási networks) were tested for random removals and removals based on node degrees and clustering coefficients. The tested properties chosen for these simulations were average path length, clustering coefficient, connectance, and number of network clusters. The average path length represents the mean number of edges that need to be passed before linking any two (random) nodes, while the clustering coefficient describes the average fraction of pairs of nodes connected to the same nodes that are also connected to each other. Connectance is the proportion of possible links between species that are realized. In the simulations, 25% of nodes were removed, and network properties were calculated after every node removal. Random removals are reported as averages for 1,000 iterations (absolute network) or 100,000 iterations (random network; 1,000 random removals from each of 100 random networks). The stability of network properties with respect to species extinctions at contrasting MIC cutoff values (0.4 to 0.6, with 0.05-unit intervals) was tested with a simulation of random removals of up to 70 nodes, and these results are reported as averages for 1,000 iterations. Properties of Erdős-Rényi and Barabási random networks with numbers of nodes and edges equal to those in the absolute network are reported as means for 1,000 iterations.
Nucleotide sequence accession number.
The sequences obtained by 454 pyrosequencing have been deposited in the NCBI Sequence Read Archive under accession number SRP007933.
RESULTS
Comparing networks based on relative and absolute abundance matrices.
We constructed networks with two types of data: absolute abundances, generating five cooccurrence networks (for an example, see Fig. S1 in the supplemental material), and relative abundances, generating five “relative networks.” The details of the five data sets and the absolute and simulated networks are given in Table 1. Discrepancies between the absolute and relative networks for each data set are presented in Table 2, including differences in the numbers of nodes and edges. In most instances, we observed more edges (cooccurrences) in the absolute networks, with the exception of the hypolimnion data set, for which the relative network had over 200 edges more. Furthermore, edges turned from positive to negative or vice versa between the absolute and relative networks for all data sets. The proportion of nodes having edges with a changing interaction type (positive versus negative) ranged from 3.9% in the metalimnetic network to 21.5% in the trophic network.
TABLE 2.
Comparison of networks constructed based on absolute and relative abundances
| Data set | No. of edges |
No. of nodesb | No. of edgesb | ||
|---|---|---|---|---|---|
| Shared | Positive to negativea | Negative to positivea | |||
| Trophic | 629 | 25 | 27 | −5 | −34 |
| Erken | 391 | 3 | 6 | +1 | −82 |
| HOT | 549 | 5 | 5 | −46 | −688 |
| Metalimnion | 226 | 4 | 1 | −37 | −284 |
| Hypolimnion | 711 | 1 | 42 | −39 | +227 |
Number of edges changing the interaction type from an absolute to a relative network.
Number of more (+)/fewer (−) edges/nodes in relative than absolute network.
Dissecting the networks.
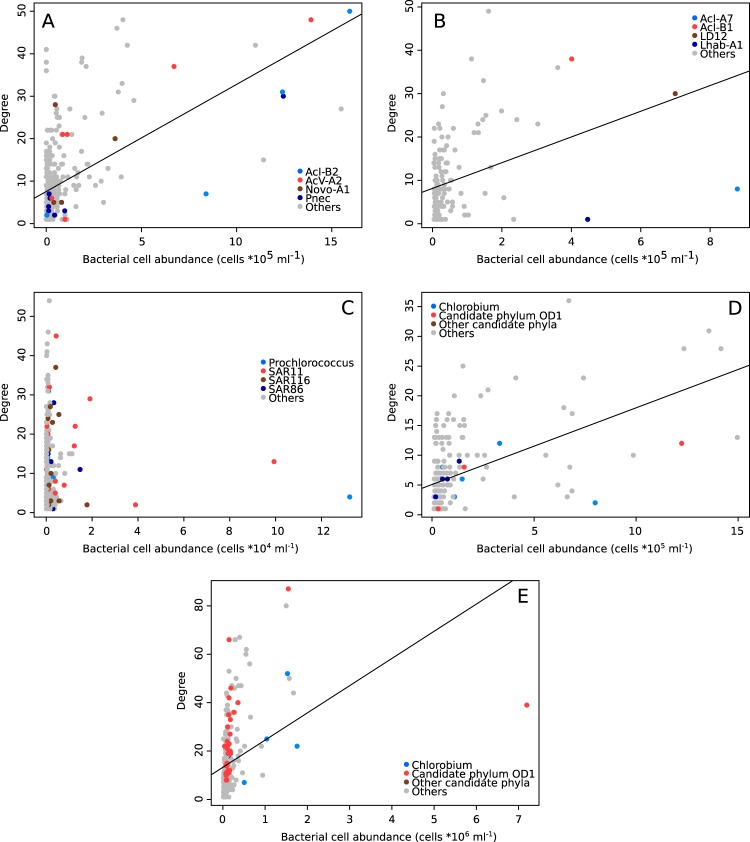
First, we scrutinized the taxonomic compositions of the five data sets. The epilimnetic lake bacterioplankton was dominated by typical freshwater taxa, such as acI-B2, Pnec, acV-A2, and Novo-A1, for the trophic data set (7). Lake Erken was dominated by another set of freshwater taxa, including acI-A7, LD12 (freshwater SAR11), acI-B1, and Limnohabitans (see reference 6 for details), while the major taxa in the marine bacterioplankton community were SAR11, SAR86, Prochlorococcus, and SAR116. Chlorobia and taxa annotated as candidate phyla dominated the communities in the suboxic meta- and hypolimnia of Lake Alinen Mustajärvi (7). These numerically dominant taxa often represented the highly connected nodes in the network, i.e., taxa covarying with many other taxa (Fig. 1). This relationship between each taxon's abundance and number of connections is also reflected in the significant linear regressions obtained with the four data sets (R2 > 0.13; P < 0.001), with the exception of the open ocean data set.
FIG 1.
Relationship between the number of edges and abundance for each node (taxon). Plots show the general linear models for the data sets collected from the epilimnion of Lake Alinen Mustajärvi (trophic data set) (A), Lake Erken (B), the Hawaiian Ocean Time (HOT) series (C), and the metalimnion (D) and hypolimnion (E) of Lake Alinen Mustajärvi. In panels A and B, bacterial tribes are defined as described by Newton et al. (51). In panel C, the most common marine alphaproteobacterial clades, SAR11 and SAR116, as well as the gammaproteobacterial clade SAR86, are highlighted.
In addition to the MIC, we observed that the nonlinearity statistic, representing another characteristic of the cooccurrence patterns among taxa, was highly variable. Its values ranged from −0.55 to 0.93, with the open ocean and Lake Erken networks having, on average, the most nonlinear cooccurrences of the five networks.
Network properties.
In all five networks, the average path lengths between nodes were shorter in empirical networks than in random networks, whereas the clustering coefficients were much higher in empirical than in random networks. Three of the networks were unfragmented (Table 1), whereas the other two (metalimnion and HOT) had second minor clusters, consisting of two and three OTUs, respectively. All of our networks also had a power law distribution of nodal degrees (number of connections a node shares with other nodes) (P < 0.001 for all). This results from the relative commonness of nodes with a degree that greatly exceeds the average. These highest-degree nodes are often referred to as network “hubs” and are thought to serve central purposes in their environments (6, 29), such that their loss is believed to cause fragmentation of the whole network.
Like the degree distribution of nodes, the levels of betweenness of nodes and edges followed a power law distribution (P < 0.001 for nodes and edges in all data sets). Node or edge betweenness is equal to the number of shortest paths from all nodes to all others that pass through that node/edge and is a measure of the centrality of the node/edge in the network, as most of the shortest paths in a network go through the nodes and edges with high degrees of betweenness. We labeled high-betweenness taxa “bottlenecks,” using terminology adopted from biochemical networks for proteins (30). We hypothesize that these bottlenecks, just like hubs, also play central roles in biological networks.
Network simulations.
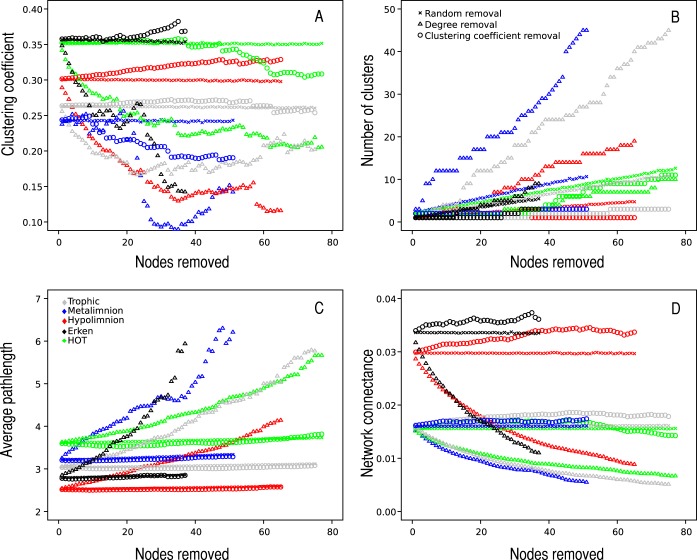
We simulated the impacts of removal of the most connected and random nodes on the properties of the cooccurrence networks (Fig. 2A to D). The removal of 25% of the most connected nodes resulted in fragmentation in all the networks and in a drop of the clustering coefficient. At the same time, random removals of 25% of nodes resulted, on average, in 66% fewer clusters than the removal of most connected nodes, without any change in clustering coefficient. An exception was found with the HOT data, where random removals resulted in more clusters than removals by degree. In all five networks, the average path length increased and network connectance decreased rapidly after the removal of the most connected nodes but not after random removals.
FIG 2.
Simulations of node (taxonomic unit) removals from the absolute, Erdős-Rényi, and Barabási networks. The panels show the impacts of removal of the highest-degree nodes and nodes with the highest levels of betweenness, as well as average values for removals of random nodes of 1,000 (absolute network) or 100,000 (random networks) random iterations, to the clustering coefficient (A), number of clusters (B), path length (C), and connectance (D). These simulations highlight the importance of the high-degree and high-betweenness nodes for the robustness of the microbial cooccurrence network in response to random taxon removal.
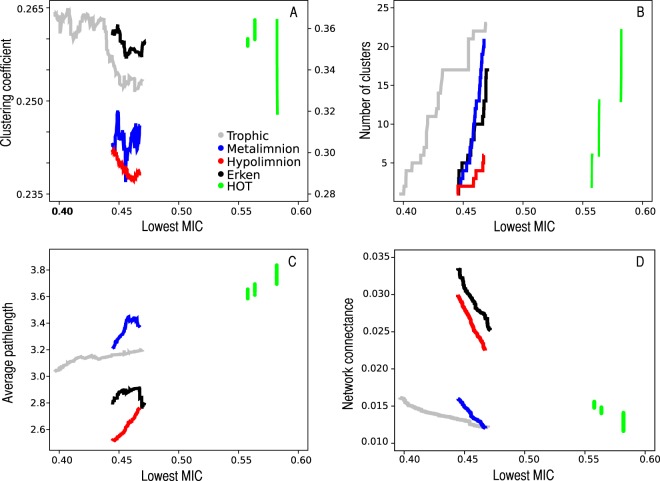
The impacts of removal of the edges with the lowest MICs were also simulated (Fig. 3) to explore the role of cooccurrence strength on network properties. There were clear responses to removal based on increasing MIC values in topology measures such as connectivity, number of clusters, and path length. The clustering coefficient dropped with the MIC cutoff increase, whereas the average path length increased in all data sets. Furthermore, the MIC simulations showed that the degree distributions of the networks followed the power law distribution, regardless of MIC values defining the edges (see Fig. S2 and S3 in the supplemental material for the trophic network; data for additional data sets not shown). We also report that the HOT data set behaved unlike the other data sets in the MIC simulations due to the small number of data points and resulting small numbers of discrete MIC values.
FIG 3.
Simulation of edge (statistical inference) removals based on MIC values in the absolute network. The edges were removed starting from the edge with the lowest MIC, and 25% of all edges were removed. The plots visualize the impacts of removals on the clustering coefficient (A), number of clusters (B), path length (C), and connectance (D). In panel A, the y axis on the left is for the trophic (gray) and metalimnetic (blue) data sets for Lake Alinen Mustajärvi, while the axis on the right is for Lake Erken (black), the HOT series (green), and the hypolimnion of Lake Alinen Mustajärvi (red).
DISCUSSION
Why absolute numbers should be used.
Bacterial community analyses have generally used relative proportions of taxonomic units to describe the phylogenetic community structure, as most approaches, such as next-generation sequencing (NGS), do not give absolute numbers. In contrast, the standard microscopic methods for describing phytoplankton and zooplankton communities typically provide absolute numbers for the abundances of taxonomic units, and even biomass estimates. Thus, the combined analysis of these different microbial compartments in a single analysis would either require a conversion of bacterial abundance data into absolute numbers or, conversely, a conversion of phytoplankton and zooplankton abundances into relative abundances.
Changing absolute abundance data to relative numbers resulted in pronounced differences in the cooccurrence patterns, even including shifts in the type of relationship, from positive to negative. For community data reported as relative abundances, an increase in one taxonomic unit must by necessity be accompanied by a similar decrease in others, leading to spurious correlations among nonindependent measurements (31). In addition, sparse sequence counts can cause artificial associations for low-abundance organisms with very few nonzero observations in relative abundance data (32). Friedman and Alm (21) also pointed to the key role of diversity in causing spurious correlations in relative abundance data sets, where data sets characterized by low richness and evenness are more affected than high-diversity data sets.
It is possible to avoid these biases by building networks based on absolute abundance data, and we therefore limit our discussion about community network properties to absolute changes in community structure. Nevertheless, the reader should be aware of statistical applications, such as linear Pearson correlations of relative compositional data (SparCC) (21), that were recently developed to compensate for spurious correlations. However, such statistics are limited to linear data exploration.
The ecology of hubs and bottlenecks.
Most of the hubs and bottlenecks were annotated as typical marine and freshwater taxa. Many of these bacterioplankton taxa are characterized by streamlined genome features and reduced metabolic capabilities resulting in auxotrophy for essential metabolites (i.e., see references 33 and 34). Such deficiencies have been implicated in fostering dependencies with other taxa that provide biosynthesis products (35) and may explain why streamlined bacteria often represent the most highly connected nodes in our analyzed cooccurrence networks.
In the trophic data set, several of the highly connected phytoplankton taxa are characterized as putative mixotrophs with an unresolved or unclear trophic status. An example of a highly connected phytoplankton was Chrysococcus (Chrysophyta), which is a heterotrophic phytoflagellate and often dominant bacterivore in boreal humic lakes (36). Through their photoautotrophic and organoheterotrophic capabilities, these phytoplankton taxa can use many different resources and thus compete for resources with many different organisms. The observed high connectivity for such taxa may thus provide some first indications that mixotrophs play an important role in the resistance of ecosystems.
The abundance of a taxon is often argued to be related to its importance, which is further emphasized by both betweenness and degree of taxa being related to their abundance. Still, as the relationship is weak, a few low-abundance taxa also hold central roles in the network. Whether the low-abundance bottlenecks and hubs represent so-called “keystone taxa” depends on the definition used (37). The term “keystone species” (38, 39) has been used to broadly define species that strongly interact and have large effects on communities and ecosystems. An operational definition has been proposed by Power and Mills (40), who suggested that “a keystone species is a species whose impacts on its community and ecosystems are much larger than would be expected from its abundance.” In any case, the aquatic cooccurrence networks presented here host some candidate nodes that likely fulfill even the stricter definition of a keystone species formulated by Power and Mills (40). One example is Kellicottia longispina, a rotifer that, despite its low abundance, was one of the most connected taxa (see Table S1 in the supplemental material). K. longispina has previously been reported to be an important regulator of phytoplankton and bacterial populations in freshwater (41), coinciding with our hypothesis that highly connected taxa have a key role in the community.
How to interpret overall network topology.
All networks exhibited properties typical of small-world networks, with high clustering coefficients, a short average path length, and a power law distribution of edge degrees and betweenness. In such networks with small-world properties, random loss of species is unlikely to jeopardize the overall properties of the cooccurrence network, at least up to a certain proportion of taxon and interdependency removal. This was also clear from the present simulations of node and edge removal. The minor impact of random removals on the network properties is explained by the higher likelihood of single nodes (taxonomic units) with a low degree or low level of betweenness being disconnected from the main network, rather than an event of network collapse into multiple clusters after a hub or bottleneck removal. Nevertheless, the low connectance (<0.07) and skewed degree distribution of the networks indicate that the temporal community patterns are particularly vulnerable to elimination of the most connected taxa (42). According to theory, the removal of fewer than 25% of the highly connected nodes in fragile networks should result in complete collapse of the network, whereas in more robust networks, even half of the nodes can be removed without changing network topology (42).
Combining simulations and theory, we suggest that the network topology of cooccurrence patterns indicates resistant temporal dynamics in natural aquatic communities. While it can be argued that bacterial communities feature a sufficiently high functional redundancy to fill any temporarily vacated niche over time, this may not be evident for phytoplankton and zooplankton. Previous results demonstrate that despite functional redundancy among zooplankton species, the effects of ecosystem-level variables, such as phytoplankton community structure, can be highly variable for different species combinations even within closely related organisms (43). Thus, taxa at different trophic levels may not be equal in terms of functional redundancy. For example, extinction of a bottleneck node such as K. longispina may have much more drastic consequences for the functioning of the microbial system than the extinction of a microbial bottleneck node. This would lead to a situation where seemingly equal nodes in the network would cause drastically different community impacts upon extinction.
Still, the process of network construction from observed interactions is very different from that of our network, which relies on temporal abundance data and the statistical identification of significant pairwise relationships over time. To date, most ecological networks that have been studied in the framework of graph (network) theory have been derived from observed interactions, i.e., predator-prey interactions (44), pollination (45), and physical associations, such as those identified in the human gut (46). In a predator-prey network, isolated nodes are usually outcompeted by the other (prey) taxa. In a cooccurrence network, where edges represent a statistical relationship and no biological dependencies, the consequences of being isolated might not be as severe for a taxon, and may even be beneficial for reasons other than losing an important mortality factor; not being linked to other taxa (showing no relationship to any other taxa over time) can be indicative that this particular taxon fills a specific niche space for which no direct competitor exists.
Conclusion.
Considering issues of core ecological interest, such as biodiversity and ecosystem function relationships, the existence of rich and abundant seed banks (47) and low-dispersal limitation (48) have been suggested to provide ecosystem stability for microbial communities. Based on the network properties we identified here, we add that the existence of hubs and bottlenecks creates intrinsic interdependency patterns in complex natural microbial systems. These interdependencies in temporal community patterns, we further argue, are indicative for high resistance to disturbances and may be used as indicators of ecosystem stability.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Academy of Finland (grant 114604 to R.I.J. and 265902 to S.P.) and the Swedish Research Council (grant 2012-4592 to A.E. and grant 2012-3892 to S.B.).
We thank Lammi Biological Station, University of Helsinki, for available facilities and for all personnel assisting with the experiments. The HOT team is acknowledged for making their data freely available to the scientific community. We also thank the Uppsala Multidisciplinary Center for Advanced Computational Science (UPPMAX) for providing resources for data storage and analysis and Minna Hiltunen for providing phytoplankton and zooplankton data.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03660-14.
REFERENCES
- 1.Cotner JB, Biddanda BA. 2002. Small players, large role: microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems 5:105–121. doi: 10.1007/s10021-001-0059-3. [DOI] [Google Scholar]
- 2.Jones RI. 1992. The influence of humic substances on lacustrine planktonic food chains. Hydrobiologia 229:73–91. doi: 10.1007/BF00006992. [DOI] [Google Scholar]
- 3.Pernthaler J. 2005. Predation on prokaryotes in the water column and its ecological implications. Nat Rev Microbiol 3:537–546. doi: 10.1038/nrmicro1180. [DOI] [PubMed] [Google Scholar]
- 4.Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, Beeson K, Tran B, Smith H, Baden-Tillson H, Stewart C, Thorpe J, Freeman J, Andrews-Pfannkoch C, Venter JE, Li K, Kravitz S, Heidelberg JF, Utterback T, Rogers Y-H, Falcón LI, Souza V, Bonilla-Rosso G, Eguiarte LE, Karl DM, Sathyendranath S, Platt T, Bermingham E, Gallardo V, Tamayo-Castillo G, Ferrari MR, Strausberg RL, Nealson K, Friedman R, Frazier M, Venter JC. 2007. The Sorcerer II global ocean sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol 5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dinsdale EA, Edwards RA, Hall D, Angly F, Breitbart M, Brulc JM, Furlan M, Desnues C, Haynes M, Li L, McDaniel L, Moran MA, Nelson KE, Nilsson C, Olson R, Paul J, Rodriguez Brito B, Ruan Y, Swan BK, Stevens R, Valentine DL, Vega Thurber R, Wegley L, White BA, Rohwer F. 2008. Functional metagenomic profiling of nine biomes. Nature 452:629–632. doi: 10.1038/nature06810. [DOI] [PubMed] [Google Scholar]
- 6.Eiler A, Heinrich F, Bertilsson S. 2012. Coherent dynamics and association networks among lake bacterioplankton taxa. ISME J 6:330–342. doi: 10.1038/ismej.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Peura S, Eiler A, Bertilsson S, Nykänen H, Tiirola M, Jones RI. 2012. Distinct and diverse bacterioplankton communities in boreal lakes dominated by candidate division OD1. ISME J 6:1640–1652. doi: 10.1038/ismej.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sogin ML, Morrison HG, Huber JA, Welch DM, Huse SM, Neal PR, Arrieta PM, Herndl GJ. 2006. Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc Natl Acad Sci U S A 103:12115–12120. doi: 10.1073/pnas.0605127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghiglione JF, Galand PE, Pommier T, Pedrós-Alió C, Maas EW, Bakker K, Bertilson S, Kirchman DL, Lovejoy C, Yager PL, Murray AE. 2012. Pole-to-pole biogeography of surface and deep marine bacterial communities. Proc Natl Acad Sci U S A 109:17633–17638. doi: 10.1073/pnas.1208160109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calbet A, Landry MR. 1999. Mesozooplankton influences on the microbial food web: direct and indirect trophic interactions in the oligotrophic open ocean. Limnol Oceanogr 44:1370–1380. doi: 10.4319/lo.1999.44.6.1370. [DOI] [Google Scholar]
- 11.Vargas CA, Martínez RA, González HE, Silva N. 2008. Contrasting trophic interactions of microbial and copepod communities in a fjord ecosystem, Chilean Patagonia. Aquat Microb Ecol 53:227–242. doi: 10.3354/ame01242. [DOI] [Google Scholar]
- 12.Fuhrman JA, Steele JA. 2008. Community structure of marine bacterioplankton: patterns, networks, and relationships to function. Aquat Microb Ecol 53:69–81. doi: 10.3354/ame01222. [DOI] [Google Scholar]
- 13.Barberán A, Bates ST, Casamayor EO, Fierer N. 2012. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J 6:343–351. doi: 10.1038/ismej.2011.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 15.Gilbert JA, Steele JA, Caporaso JG, Steinbrück L, Reeder J, Temperton B, Huse S, McHardy AC, Knight R, Joint I, Somerfield P, Fuhrman JA, Field D. 2012. Defining seasonal marine microbial community dynamics. ISME J 6:298–308. doi: 10.1038/ismej.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kara EL, Hanson PC, Hu YU, Winslow L, McMahon KD. 2013. A decade of seasonal dynamics and co-occurrences within freshwater bacterioplankton communities from eutrophic Lake Mendota, WI, USA. ISME J 7:680–684. doi: 10.1038/ismej.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reshef DN, Reshef YA, Finucane HK, Grossman SR, McVean G, Turnbaugh PJ, Lander ES, Mitzenmacher M, Sabeti PC. 2011. Detecting novel associations in large data sets. Science 334:1518–1524. doi: 10.1126/science.1205438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steele JA, Countway PD, Xia L, Vigil PD, Beman JM, Kim DY, Chow CET, Sachdeva R, Jones AC, Schwalbach MS, Rose JM, Hewson I, Patel A, Sun F, Caron DA, Fuhrman JA. 2011. Marine bacterial, archaeal and protistan association networks reveal ecological linkages. ISME J 5:1414–1425. doi: 10.1038/ismej.2011.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Selinummi J, Seppälä J, Yli-Harja O, Puhakka JA. 2005. Software for quantification of labeled bacteria from digital microscope images by automated image analysis. Biotechniques 39:859–863. doi: 10.2144/000112018. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert JA, Field D, Swift P, Newbold LK, Oliver AE, Smyth TJ, Somerfield PJ, Huse SM, Joint I. 2009. The seasonal structure of microbial communities in the western English Channel. Environ Microbiol 11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 21.Friedman J, Alm EJ. 2012. Inferring correlation networks from genomic survey data. PLoS Comput Biol 8:e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Opsahl T. 2009. Structure and evolution of weighted networks, p 104–122. University of London (Queen Mary College), London, United Kingdom. [Google Scholar]
- 24.Csardi G, Nepusz T. 2006. The igraph software package for complex network research. Interjournal Complex Systems:1695 http://igraph.org. [Google Scholar]
- 25.R Development Core Team. 2011. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 26.Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H. 2013. Vegan: community ecology package. R package version 2.0-6. http://CRAN.R-project.org/package=vegan. [Google Scholar]
- 27.Dormann CF, Gruber B, Fruend J. 2008. Introducing the bipartite package: analysing ecological networks. R News 8/2:8–11. [Google Scholar]
- 28.Diamond JM. 1975. Assembly of species communities, p 342–444. In Cody M, Diamond JM (ed), Ecology and evolution of communities. Harvard University Press, Cambridge, MA. [Google Scholar]
- 29.Montoya JM, Solé RV. 2002. Small world patterns in food webs. J Theor Biol 214:405–412. doi: 10.1006/jtbi.2001.2460. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. 2007. The importance of bottlenecks in protein networks: correlation with gene essentiality and expression dynamics. PLoS Comput Biol 3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aitchison J. 1982. The statistical analysis of compositional data. J R Stat Soc Series B Stat Methodol 44:139–177. [Google Scholar]
- 32.Aitchison J. 1981. A new approach to null correlations of proportions. Math Geol 13:175–189. doi: 10.1007/BF01031393. [DOI] [Google Scholar]
- 33.Tripp HJ, Schwalbach MS, Meyer MM, Kitner JB, Breaker RR, Giovannoni SJ. 2009. Unique glycine-activated riboswitch linked to glycine-serine auxotrophy in SAR11. Environ Microbiol 11:230–238. doi: 10.1111/j.1462-2920.2008.01758.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia SL, McMahon KD, Martinez-Garcia M, Srivastava A, Sczyrba A, Stepanauskas R, Grossart HP, Woyke T, Warnecke F. 2013. Metabolic potential of a single cell belonging to one of the most abundant lineages in freshwater bacterioplankton. ISME J 7:137–147. doi: 10.1038/ismej.2012.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giovannoni SJ, Thrash JC, Temperton B. 2014. Implications of streamlining theory for microbial ecology. ISME J 8:1553–1565. doi: 10.1038/ismej.2014.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isaksson A, Bergström AK, Blomqvist P, Jansson M. 1999. Bacterial grazing by phagotrophic phytophagellates in a deep humic lake in northern Sweden. J Plankton Res 21:247–268. doi: 10.1093/plankt/21.2.247. [DOI] [Google Scholar]
- 37.Kotliar NB. 2000. Application of the new keystone-species concept to prairie dogs: how well does it work? Conserv Biol 14:1715–1721. doi: 10.1046/j.1523-1739.2000.98384.x. [DOI] [PubMed] [Google Scholar]
- 38.Paine RT. 1966. Food web complexity and species diversity. Am Nat 100:65–75. doi: 10.1086/282400. [DOI] [Google Scholar]
- 39.Paine RT. 1969. A note on trophic complexity and community stability. Am Nat 103:91–93. doi: 10.1086/282586. [DOI] [Google Scholar]
- 40.Power ME, Mills LS. 1995. The keystone cops meet in Hilo. Trends Ecol Evol 10:182–184. doi: 10.1016/S0169-5347(00)89047-3. [DOI] [PubMed] [Google Scholar]
- 41.Lair N, Ali HO. 1990. Grazing and assimilation rates of natural populations of planktonic rotifers Keratella cochlearis, Keratella quadrata and Kellicottia longispina in a eutrophic lake (Aydat, France). Hydrobiologia 194:119–131. doi: 10.1007/BF00028413. [DOI] [Google Scholar]
- 42.Estrada E. 2007. Food webs robustness to biodiversity loss: the roles of connectance, expansibility and degree distribution. J Theor Biol 244:296–307. doi: 10.1016/j.jtbi.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 43.Norberg J. 2000. Resource-niche complementarity and autotrophic compensation determines ecosystem-level responses to increased cladoceran species richness. Oecologia 122:264–272. doi: 10.1007/PL00008855. [DOI] [PubMed] [Google Scholar]
- 44.Allesina S, Pascual M. 2007. Network structure, predator-prey modules, and stability in large food webs. Theor Ecol 1:55–64. doi: 10.1007/s12080-007-0007-8. [DOI] [Google Scholar]
- 45.Olesen JM, Bascompte J, Elberling H, Jordano P. 2008. Temporal dynamics in a pollination network. Ecology 89:1573–1582. doi: 10.1890/07-0451.1. [DOI] [PubMed] [Google Scholar]
- 46.Greenblum S, Tunrbaugh PJ, Borenstein E. 2012. Metagenomic systems biology of the human gut microbiome reveals topological shifts associated with obesity and inflammatory bowel disease. Proc Natl Acad Sci U S A 109:594–599. doi: 10.1073/pnas.1116053109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, Huber DH, Langenheder S, Lennon JT, Martiny JBH, Matulich KL, Schmidt TM, Handelsman J. 2012. Fundamentals of microbial community resistance and resilience. Front Microbiol 3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Karl DM, Church MJ. 2014. Microbial oceanography and the Hawaii Ocean Time-series programme. Nat Rev Microbiol 12:699–713. doi: 10.1038/nrmicro3333.. [DOI] [PubMed] [Google Scholar]
- 50.Heinrich F, Eiler A, Bertilsson S. 2013. Seasonality and environmental control of freshwater SAR11 (LD12) in a temperate lake (Lake Erken, Sweden). Aquat Microb Ecol 70:33–44. doi: 10.3354/ame01637. [DOI] [Google Scholar]
- 51.Newton RJ, Jones SE, Eiler A, McMahon KD, Bertilsson S. 2011. A guide to the natural history of freshwater lake bacteria. Microbiol Mol Biol Rev 75:14–49. doi: 10.1128/MMBR.00028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.