Abstract
Recently, spaCBA-encoded pili on the cell surface of Lactobacillus rhamnosus GG were identified to be key molecules for binding to human intestinal mucus and Caco-2 intestinal epithelial cells. Here, we investigated the role of the SpaCBA pilus of L. rhamnosus GG in the interaction with macrophages in vitro by comparing the wild type with surface mutants. Our results show that SpaCBA pili play a significant role in the capacity for adhesion to macrophages and also promote bacterial uptake by these phagocytic cells. Interestingly, our data suggest that SpaCBA pili also mediate anti-inflammatory effects by induction of interleukin-10 (IL-10) mRNA and reduction of interleukin-6 (IL-6) mRNA in a murine RAW 264.7 macrophage cell line. These pili appear to mediate these effects indirectly by promoting close contact with the macrophages, facilitating the exertion of anti-inflammatory effects by other surface molecules via yet unknown mechanisms. Blockage of complement receptor 3 (CR3), previously identified to be a receptor for streptococcal pili, significantly decreased the uptake of pilus-expressing strains in RAW 264.7 cells, while the expression of IL-10 and IL-6 mRNA by these macrophages was not affected by this blocking. On the other hand, blockage of Toll-like receptor 2 (TLR2) significantly reduced the expression of IL-6 mRNA irrespective of the presence of pili.
INTRODUCTION
Lactobacillus bacteria belong to the lactic acid bacteria (LAB) and are applied worldwide in the production of a variety of fermented food products. Besides their role in the food industry, some Lactobacillus species are naturally found throughout the human gastrointestinal tract (GIT) in different proportions, with the highest numbers being found in the proximal small intestine (1), yet their most important niche in the human body is probably within the vaginal cavity (2). Several lactobacilli have a documented probiotic function by providing health-promoting effects in humans upon administration. Among them, the well-studied probiotic Lactobacillus rhamnosus GG has been shown to prevent atopic diseases (3–5), decrease the risk of respiratory tract infections (6–8), prevent nosocomial gastrointestinal infections (7), prevent antibiotic-associated diarrhea (9), treat acute infectious diarrhea (10, 11), and be beneficial in the treatment of rotavirus-associated diarrhea (12), although there are always responders and nonresponders. For a more targeted approach, better knowledge of the underlying mechanisms is important. Because of its wide clinical use, L. rhamnosus GG is an important model probiotic strain for use in such targeted approaches.
During the last decade, incredible numbers of studies have focused on the possible immunomodulatory effects of probiotics. Probiotic bacteria are thought to confer such immunomodulatory effects by the microbe-associated molecular patterns (MAMPs) that are present on their cell surface or that are secreted in the environment by them (13). These MAMPs can be highly conserved, such as certain peptidoglycan motifs, or can be more strain specific, such as unique glyco- or lipoproteins. The interaction of a MAMP with its pattern recognition receptor (PRR) on host cells, such as Toll-like receptors (TLRs), results in the induction of a signaling cascade that modulates the expression of various response genes, such as cytokines and defensins (14). Increasing numbers of MAMPs that participate in the host interaction have been identified on the surface of probiotic lactobacilli (13, 15, 16). For instance, lipoteichoic acid (LTA) from L. rhamnosus GG was demonstrated to interact with the PRRs TLR2/6 (17), the S-layer protein SlpA in Lactobacillus acidophilus NCFM interacts with the dendritic cell (DC) receptor DC-SIGN (18), and specific peptidoglycan muropeptides in Lactobacillus salivarius Ls33 are recognized by NOD2 (19).
Recently, pili or fimbriae (i.e., proteinaceous heteropolymeric extracellular appendages) were discovered at the cell surface of L. rhamnosus GG (20). These pili are encoded by the spaCBA gene cluster (LGG_00441 to LGG_00444). The SpaC subunit, located at the tip and along the backbone structure, was shown to be responsible for the high mucus-binding capacity (21). A comparison of the retention times of exogenously applied piliated L. rhamnosus GG and the related nonpiliated strain L. rhamnosus Lc705 also suggests a key role for the pili in promoting residence in the human colon, since piliated L. rhamnosus GG could be detected for 7 days longer in fecal samples from healthy volunteers (21). By mutant analysis of the spaCBA genes, we could show that the loss of pili results in the loss of adhesion to the microvillus-containing Caco-2 intestinal epithelial cell (IEC) line and increased induction of the proinflammatory marker interleukin-8 (IL-8) in these nonphagocytic cells (22). Using recombinant lactococcal constructs, von Ossowski et al. (23) demonstrated that the SpaCBA pilus is a contributory factor in the activation of TLR2-dependent signaling in HEK cells by the recombinant lactococci as well as in the modulation of pro- and anti-inflammatory cytokine (tumor necrosis factor alpha, IL-6, IL-10, and IL-12) production in human monocyte-derived DCs (moDCs). However, a possible immunomodulatory role of the native pili present on the surface of live L. rhamnosus GG has not yet been explored. Interestingly, in the case of the pathogen Streptococcus pneumoniae, interaction of the pilus-associated RrgA adhesin with the complement receptor 3 (CR3), also named integrin CD11b/CD18 or Mac-1, was shown to enhance uptake of the bacteria by murine and human macrophages, on the basis of the results of experiments with a blocking antibody on the wild type and the isogenic mutant lacking RrgA (24). In addition, TLR2 was shown to mediate inflammatory responses to oligomerized RrgA (25).
In this study, we aimed to investigate the role of SpaCBA pili from the probiotic strain L. rhamnosus GG in the interaction with phagocytic innate immune cells. Intestine-resident DCs and macrophages are eager phagocytes capable of sampling bacteria in the intestinal lumen through the extension of transepithelial dendrites (26–28) and are important host cells for probiotic immunomodulatory applications. Here, we chose a simplified model with a murine macrophage-like (RAW 264.7) cell line. Cells of this cell line were coincubated with the L. rhamnosus GG wild-type strain and different pilus mutants of L. rhamnosus GG, so that the role of pili in their natural conformation and organization on live cells could be studied. Such in situ cellular experiments are important because host cellular responses against bacteria are always an integrated sum of the interactions of different MAMPs with different PRRs (13). In addition, we previously showed the importance of the zipper-like and nanospring properties of the pili in promoting close contacts of whole L. rhamnosus GG cells with substrates (29), which cannot be mimicked with isolated pili without their cellular anchor.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Wild-type L. rhamnosus GG and its mutant derivatives (Table 1) were routinely grown under nonshaking conditions in Man-Rogosa-Sharpe (MRS) medium (catalog number 288130; Difco, Becton, Dickinson, Erembodegem, Belgium) for 24 h at 37°C, and 1% of this preculture was inoculated into fresh MRS broth and grown overnight at 37°C. Bacteria were washed once with 1× phosphate-buffered saline (PBS; catalog number 14040-091; Invitrogen, Life Technologies, Ghent, Belgium), pH 7.4, and brought to a final concentration of 5 × 107 CFU/ml prior to incubation with macrophages. This concentration was selected on the basis of findings of preliminary tests in which different dose-response relationships (1:1, 1:10, 1:50, and 1:100 cells/bacteria) were evaluated.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant genotype and/or phenotype | Reference or source |
|---|---|---|
| Strains | ||
| Lactobacillus rhamnosus | ||
| GG (ATCC 53103) | Wild type; human isolate | 21 |
| CMPG5357 | ΔspaCBA::Tcr; mutant of L. rhamnosus GG without spaCBA-encoded pili | 22 |
| CMPG5351 | ΔwelE::Tcr; mutant of L. rhamnosus GG without long galactose-rich exopolysaccharides but with increased exposure of pili | 20 |
| CMPG1908 | CMPG5357 with chromosomal insertion of pMEC10 in the attB sequence of phage mv4; Eryr | This work |
| CMPG1907 | CMPG5351 with chromosomal insertion of pMEC10 in the attB sequence of phage mv4; Eryr | This work |
| FAJ1906 | L. rhamnosus GG with chromosomal insertion of pFAJ1934 in the attB sequence of phage A2; Eryr | 33 |
| Lactococcus lactis subsp. cremoris | ||
| MG1363 | Mesophilic lactic acid bacterium isolated from green plants | 30 |
| GRS1185 | L. lactis NZ9000 containing the nisin-inducible vector pKTH5080, which in turn contains the coding region of the spaCBA pilus operon; Cmr | 23 |
| Plasmids | ||
| pFAJ1934 | 2,634-bp PCR fragment (flanking EcoRI sites) carrying the nisRK genes of pMEC10 cloned into the EcoRI site of pEM40 downstream of the ery gene in the opposite orientation; Eryr Ampr | 33 |
| pMEC10 | Integration plasmid (attB located at the 3′ end of the tRNASer locus) containing the int-attP cassette (of phage mv4) of pMC1 cloned into pNZ9500 (a pUC19 derivative carrying a 2.7-kb chromosomal DNA fragment of L. lactis NZ9700 containing the 3′ end of nisP and nisRK); expression driven by ery read-through; Eryr | 34 |
| pMEC45 | L. lactis pSH71 replicon; pNZ8037 derivative with gfpuva cloned downstream of the nisA promoter from L. lactis NZ9800; Cmr | 36 |
gfpuv, synthetic GFP gene (with improved codon usage) that was subsequently subjected to recurrent cycles of DNA shuffling, thereby selecting for bright fluorescence that can be observed under UV light.
Wild-type strain Lactococcus lactis subsp. cremoris MG1363 (30) and the recombinant L. lactis subsp. cremoris clone GRS1185, itself having a nisin-inducible vector that in turn contains the coding region of the spaCBA pilus operon, were grown overnight in M17 broth (catalog number 218561; Becton, Dickinson, Erembodegem, Belgium) supplemented with 0.5% glucose at 30°C. A 1% aliquot taken from this preculture was used to further inoculate fresh M17 broth, and cells were kept overnight at 30°C. In the case of the recombinant lactococcal construct GRS1185, a similar procedure was followed, and the media were supplemented with 7.5 μg/ml chloramphenicol (catalog number C0378-25G; Sigma-Aldrich, St. Louis, MO, USA). After inoculation with 100 μl of preculture, the cultures were allowed to grow until the optical density at 600 nm (OD600) was 0.5. Subsequently, 3 ng/ml of nisin was added to the cultures to induce pilus expression. Nisin (catalog number N5764; Sigma-Aldrich, St. Louis, MO, USA) was prepared as previously described (31). Briefly, a stock solution of nisin (10 mg/ml) was made by suspending 100 mg of nisin per ml in 0.05% acetic acid, followed by 10-fold dilution with dimethyl sulfoxide.
Detection of SpaCBA pili on the L. rhamnosus GG wild type and mutants by immunofluorescence microscopy.
To phenotypically characterize the derivative mutants of L. rhamnosus GG, i.e., spaCBA mutant CMPG5357 and the exopolysaccharide (EPS)-deficient mutant CMPG5351, immunofluorescence assays were performed as described previously (32). Briefly, cells were grown to exponential phase and fixed for 1 h in 2.5% formaldehyde (catalog number VELC1374.1000; VWR International, Radnor, PA, USA) and 0.05% glutaraldehyde (catalog number G5882-50ML; Sigma-Aldrich, St. Louis, MO, USA). Cells were then washed and incubated in blocking buffer (0.5%; Boehringer Mannheim, Mannheim, Germany) for 10 min. The samples were further incubated in blocking buffer containing anti-SpaC rabbit polyclonal antiserum (21) or preimmune serum (1:100 and 1:400 dilutions, respectively) for 30 min and subsequently incubated in blocking buffer containing goat anti-rabbit IgG (1:200 dilution) conjugated to Alexa Fluor 488 (catalog number A-11070; Invitrogen, Life Technologies, Ghent, Belgium) for another 30 min. Samples were visualized with a Zeiss Axio Imager Z1 microscope equipped with an AxioCam MRm Rev 3 monochrome digital camera.
GFP labeling of L. rhamnosus GG pilus mutants.
The nisRK genes with their own promoter were integrated into the chromosome of the L. rhamnosus GG mutants, as previously described for the wild type (33). Site-specific integration was accomplished by electroporating the suicide plasmid pMEC10 (34) into the L. rhamnosus GG mutants. Integration of pMEC10 at the tRNASer locus was confirmed by PCR as previously described (35). The following primers were used: PRO-567 (5′-GTCGACACAGGATTTGAACC-3′; which corresponds to primer T2 [35]) and PRO-570 (5′-CAAGCCAACAGACGTGCAAGCA-3′; which binds to the 3′ end of the int gene [35]). Subsequently, pMEC45 (36), the expression vector containing gfp under the control of the nisA promoter, was electroporated into CMPG5357/pMEC10 (CMPG1908) and CMPG5351/pMEC10 (CMPG1907). Transformants were verified by PCR analysis. Finally, the expression of green fluorescent protein (GFP) driven by the nisA promoter (pMEC45) in the L. rhamnosus GG derivatives was optimally induced as described previously (33). Briefly, overnight cultures of L. rhamnosus GG FAJ1906/pMEC45, CMPG1908/pMEC45, and CMPG1907/pMEC45 were used to inoculate fresh prewarmed MRS medium diluted 1:50. After 30 min of incubation at 37°C without shaking, 500 ng/ml nisin was added to the cultures, which were then incubated for 2.5 to 3 h. Nisin was used to induce the transcription of the genes under the control of the nisA promoter.
Maintenance and culturing of cell lines.
The murine macrophage cell line RAW 264.7 (ATCC TIB-71) was purchased from the American Type Culture Collection (ATCC; Manassas, VA, USA). RAW 264.7 cells were maintained in Dulbecco's modified Eagle medium (DMEM; catalog number 41966-029; Life Technologies, Ghent, Belgium) containing 10% fetal bovine serum (FBS; catalog number SH30071.03; HyClone, USA) and 50 μg/ml gentamicin (catalog number G1397; Sigma-Aldrich, St. Louis, MO, USA) in a 5% CO2 humidified incubator at 37°C. Every 2 or 3 days, the cells were split at a subcultivation ratio of 1:3. For adhesion, phagocytosis, and immunomodulation experiments, RAW 264.7 cells were grown overnight in 12-well culture plates (catalog number 665180; Greiner Bio-One, Ltd., Solingen, Germany) at a concentration of 1 × 106 cells/ml.
Adhesion and phagocytosis assays with murine macrophages.
Experiments to assess the adhesion of various L. rhamnosus GG strains to RAW 264.7 cells were carried out as follows on the basis of the methods of Lebeer et al. (22). A 1-ml volume of L. rhamnosus GG and the corresponding mutants (5 × 107 CFU/ml) was added to tissue culture plate wells containing RAW 264.7 cells. The bacteria were allowed to adhere to the cells for 1 h at 37°C. The cells were then rinsed three times with 1× PBS prewarmed to 37°C and resuspended in 1 ml of 1× PBS, pH 7.4. Finally, the cells were detached using a cell scraper, and serial dilutions of bacterial suspensions were prepared and plated out on solid MRS medium for Lactobacillus strains and M17 medium for Lactococcus strains. The percentage of bacterial adhesion was calculated by comparing the total number of bacterial colonies counted after adhesion to the number of cells in the bacterial suspension originally added to the tissue culture plate wells. In addition, the total amount of macrophage-associated bacteria was determined. To this point the aforementioned protocol for adhesion had been followed, but macrophages were then lysed with 0.2% Triton X-100 (catalog number T8787; Sigma-Aldrich, St. Louis, MO, USA) for 15 min prior to preparation of the dilution series in order to liberate internalized bacteria. The percentage of total associated bacteria was then calculated by comparing the total number of bacterial colonies counted after the macrophages were lysed to the number of bacteria originally added. Furthermore, to determine the number of internalized bacteria, phagocytosis assays were performed as described previously (37), with slight modifications. Briefly, RAW 264.7 cells (1 × 106 cells) were incubated for 1 h at 37°C with L. rhamnosus GG or L. lactis subsp. cremoris pilus variants as described above. Bacterial internalization was stopped with the addition of ice-cold 1× PBS, pH 7.4, and macrophages were washed three times to remove nonadhered bacteria. The remaining extracellular bacteria were killed by addition of streptomycin (100 μg/ml) and penicillin (100 U/ml) for another hour. The cells were then washed three times with 1× PBS and lysed with 0.2% Triton X-100 for 15 min. Serial dilutions of bacterial suspensions were prepared and plated out on solid media. The percentage of internalized bacteria was determined by comparing the total number of bacterial colonies counted after the cells were lysed to the number of bacteria originally added. All experiments were repeated at least three times, and all strains were tested in triplicate.
Analysis of bacterium-macrophage interaction by epifluorescence microscopy and flow cytometry.
Epifluorescence microscopy was used to visualize the pattern of adhesion of the L. rhamnosus GG strains to RAW 264.7 cells. Macrophages were seeded onto coverslips in 6-well tissue culture plates (catalog number 657160; Greiner Bio-One, Ltd., Solingen, Germany) at a concentration of 1 × 106 cells per well and were grown overnight. An overnight culture of GFP-expressing (GFP+) L. rhamnosus GG (nisin-induced cultures of FAJ1906/pMEC45, CMPG1908/pMEC45, and CMPG1907/pMEC45) was used to inoculate fresh, prewarmed MRS medium diluted 1:50. After 30 min of incubation at 37°C without shaking, 500 ng/ml nisin was added to the culture, which was then incubated for 2.5 to 3 h. Fluorescently labeled L. rhamnosus GG strains were subsequently added to the macrophages at a ratio of 50 to 1 CFU/cell. After 2 h of incubation at 37°C in 5% CO2, the cells were washed three times with 1× PBS, pH 7.4, to remove nonadhered bacteria, and the interaction between the bacteria and the macrophages was examined by epifluorescence microscopy using a Zeiss Axio Imager Z1 microscope equipped with an AxioCam MRm Rev 3 monochrome digital camera (Carl Zeiss Microimaging GmbH, Germany). This assay was repeated at least five times.
For flow cytometric analysis, GFP-expressing L. rhamnosus GG strains were incubated with macrophages for 2 h at 37°C in 5% CO2. The cells were then fixed with 3% paraformaldehyde and resuspended in 1× PBS, and the fluorescence in the macrophages was examined by fluorescence-activated cell sorter (FACS) analysis with a BD Influx cell sorter (Becton, Dickinson, Erembodegem, Belgium). Data were analyzed using FlowJo software, version 7.6.4. The experiment was repeated at least three times, and all strains were tested in triplicate.
Induction of cytokine gene expression in RAW 264.7 cells.
Wild-type and mutant L. rhamnosus GG cells were grown overnight in MRS medium and then centrifuged at 2,000 × g for 15 min at 4°C. After washing with 1× PBS, pH 7.4, cells were resuspended in DMEM without FBS and adjusted to a final concentration of 5 × 107 CFU/ml. One milliliter of the bacterial suspension was added to the wells containing RAW 264.7 cells and incubated for 3 h at 37°C in 5% CO2 at 90% humidity. Then, the cells were rinsed three times with prewarmed 1× PBS, pH 7.4. Here, lipopolysaccharide (LPS; 1 μg/ml) from Escherichia coli O111:B4 (catalog number L4391; Sigma-Aldrich, St. Louis, MO, USA) was included as a positive control, and DMEM unsupplemented with FBS was used as a negative control. Additionally, nonpiliated L. lactis subsp. cremoris MG1363 and the recombinant SpaCBA-piliated lactococcal construct (GRS1185) were also included as controls. RNA was extracted from the RAW 264.7 cells by using an RNeasy minikit (Qiagen, MD, USA), following the manufacturer's protocol. The concentration was determined by spectrophotometric measurement of the OD260 (ND-1000; NanoDrop Technologies). In addition, OD260/OD280 and OD260/OD230 ratios were measured and found to range from 2.0 to 2.2, and the RNA quality and integrity were confirmed with an Experion RNA analysis kit (Bio-Rad Laboratories, Hercules, CA, USA). Values of the RNA integrity number (RIN) of between 9.8 and 10 were obtained, with a value of 10 indicating intact RNA (data not shown). Measurement of cytokine gene expression was determined by quantitative real-time PCR (qPCR) as described below. The experiment was repeated at least three times, and all strains were tested in triplicate. To determine whether direct cell-to-cell contact is required for cytokine induction, RAW 264.7 cells were incubated with the L. rhamnosus GG strains in a Transwell culture system (catalog number CLS3401-48EA; Sigma-Aldrich, St. Louis, MO, USA). RAW 264.7 cells were located in the lower chamber, and L. rhamnosus GG cells were located in the upper chamber. The cells were separated by a 0.4-μm-pore-size permeable filter membrane support, thereby avoiding any direct contact between bacterial cells and macrophages.
qPCR analysis.
Isolated total RNA (1 μg) was transcribed to cDNA with SuperScript II reverse transcriptase (catalog number 18064-071; Life Technologies, Ghent, Belgium) and oligo(dT) primers (catalog number N8080128; Life Technologies, Ghent, Belgium) according to the manufacturer's protocol. Afterward, nuclease-free water (catalog number AM9935; Life Technologies, Ghent, Belgium) was added to a volume of 100 μl. For each sample, 5 μl cDNA (containing 50 ng of transcribed RNA) was amplified in duplicate with the TaqMan universal PCR master mix (catalog number 4304437; Life Technologies, Ghent, Belgium) in a total reaction volume of 25 μl. We chose to evaluate eight reference genes commonly used as internal controls in expression studies, namely, TATAA box binding protein (TBP), succinate dehydrogenase complex subunit A (SDHA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β2-microglobulin (B2M), peptidyl-prolyl cis-trans-isomerase A (PPIA), ribosomal protein L13a (RPL13A), hypoxanthine phosphoribosyltransferase (HPRT), and β-actin. Interestingly, the TBP, SDHA, and GAPDH genes gave comparable results on the basis of RefFinder analyses (http://www.leonxie.com/referencegene.php) (data not shown). The GAPDH gene was then chosen as a reference gene, so that all results were normalized to those for the GAPDH gene to compensate for differences in the amount of starting material. qPCR was performed for IL-10, IL-6, and GAPDH in a StepOnePlus real-time PCR system (Applied Biosystems, Lennik, Belgium). cDNA plasmid standards, consisting of purified plasmid DNA specific for each cytokine, were used to quantify the target gene in the unknown samples, as described previously (38). For each cytokine, at least one primer or probe was designed to span an intron region of the matching cytokine gene. All primers and probes were designed on the basis of published sequences (39, 40) and chemically synthesized by Integrated DNA Technologies (IDT; Coralville, IA, USA) (Table 2). Each qPCR amplification was performed in duplicate wells in clear 96-well reaction plates (catalog number 4346906; Life Technologies, Ghent, Belgium) under the following conditions: 95°C for 10 min, followed by 40 cycles at 95°C for 15 s and 60°C for 1 min. qPCR data are presented as a ratio of the amount of cytokine mRNA to the amount of GAPDH mRNA. Nontemplate controls were included for each run.
TABLE 2.
Primers and probes sequences used in this study for mRNA measurementsa
| Target mRNA | Primer or probe | Sequence (5′–3′) |
|---|---|---|
| TBP | FW | TTGTACCGCAGCTTCAAAATATTGT |
| RV | GCAGCAAATCGCTTGGGAT | |
| TP | FAM-TTGACCTAAAGACCATTGCACTTCGTGCA-TAMRA | |
| B2M | FW | CCACTGAGACTGATACATACGCCT |
| RV | GATCACATGTCTCGATCCCAGTAG | |
| TP | FAM-TAAGCATGCCAGTATGGCCGAGCC-TAMRA | |
| SDHA | FW | ACATCAGAACTACGCCTAAACATGC |
| RV | AAACCATTCCCCTGTCGAATG | |
| TP | FAM-AACCATGCTGCAGTGTTCCGTGTGG-TAMRA | |
| RPL13A | FW | GCGCCTCAAGGTGTTGGAT |
| RV | CCCAGGTAAGCAAACTTTCTGG | |
| TP | FAM-TGGTCCCTGCTGCTCTCAAGGTTGTT-TAMRA | |
| HPRT | FW | TTATCAGACTGAAGAGCTACTGTAATGATC |
| RV | TTACCAGTGTCAATTATATCTTCAACAATC | |
| TP | FAM-TGAGAGATCATCTCCACCAATAACTTTTATGTCCC-TAMRA | |
| PPIA | FW | GCCGCGTCTCCTTCGAG |
| RV | GTAAAGTCACCACCCTGGCAC | |
| TP | FAM-TTGCAGACAAAGTTCCAAAGACAGCAGAAA-TAMRA | |
| β-Actin | FW | AGAGGGAAATCGTGCGTGAC |
| RV | CAATAGTGATGACCTGGCCGT | |
| TP | FAM-CACTGCCGCATCCTCTTCCTCCC-TAMRA | |
| GAPDH | FW | TCACCACCATGGAGAAGGC |
| RV | GCTAAGCAGTTGGTGGTGCA | |
| TP | FAM-ATGCCCCCATGTTTGTGATGGGTGT-TAMRA | |
| IL-6 | FW | GAGGATACCACTCCCAACAGACC |
| RV | AAGTGCATCATCGTTGTTCATACA | |
| TP | FAM-CAGAATTGCCATTGCACAACTCTTTTCTCA-TAMRA | |
| IL-10 | FW | GGTTGCCAAGCCTTATCGGA |
| RV | ACCTGCTCCACTGCCTTGCT | |
| TP | FAM-TGAGGCGCTGTCATCGATTTCTCCC-TAMRA |
TBP, TATAA box binding protein; B2M, β2-microglobulin; SDHA, succinate dehydrogenase complex subunit A; RPL13A, ribosomal protein L13a; HPRT, hypoxanthine phosphoribosyltransferase; PPIA, peptidyl-prolyl cis-trans-isomerase A; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; IL-6, interleukin-6; IL-10, interleukin-10; FW, forward primer; RV, reverse primer; TP, hydrolysis probe; FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine.
TLR2 and CD11b blocking assay.
The blocking monoclonal anti-human/mouse CD282 TLR2 purified antibody (clones T2.5 and 6C2) and monoclonal anti-mouse CD11b purified antibody (clone M1/70) were obtained from eBioscience (San Diego, CA, USA). RAW 264.7 cells (1 × 106 cells/ml) were incubated with TLR2 (5 to 10 μg/ml) or CD11b antibodies (5 μg/ml) for 30 min at 37°C in 5% CO2. Cells were washed three times with serum-free culture medium to wash off any free antibody. Subsequently, cells were cocultured with bacteria (5 × 107 CFU/ml). After 3 h of incubation, the RAW 264.7 cells were rinsed three times with prewarmed 1× PBS, pH 7.4. RNA was extracted, and measurement of cytokine gene expression was determined by qPCR as described above. Bacterial adhesion to and uptake by macrophages pretreated with CD11b antibodies were also analyzed as described above.
Statistical analysis.
Significant differences in the data between wild-type L. rhamnosus GG and its mutant derivatives were evaluated by the unpaired Student's t test. A P value of ≤0.05 was considered statistically significant. All data were also analyzed using SPSS Statistics (version 22) software. The normality of the distribution was assessed using the Shapiro-Wilk test in which significant values greater than 0.05 represented a normal distribution. Graphical tools were also used to test normality by analyzing histograms and quantile-quantile (Q-Q) plots of the sample data.
RESULTS
Detection of pili on the L. rhamnosus GG wild type and isogenic mutants by immunofluorescence microscopy.
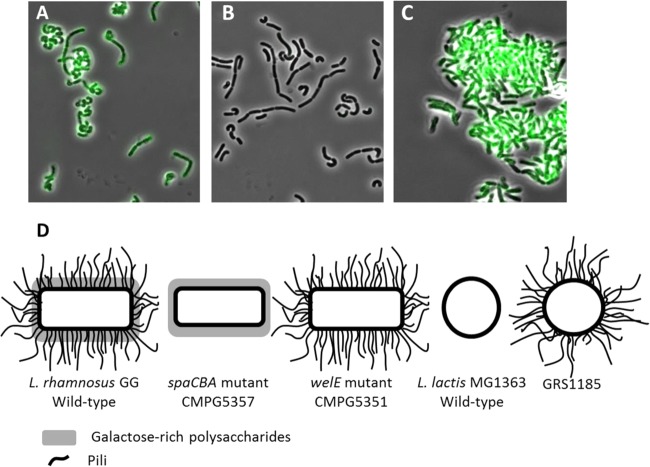
In addition to the L. rhamnosus GG wild type, the isogenic spaCBA mutant CMPG5357, which lacks the SpaCBA pili (22), and the welE mutant CMPG5351, which is deficient in the production of EPS but shows increased exposure of pili and glucose-rich polysaccharides (20), were the main pilus-expressing variants studied in this work (Fig. 1D). To phenotypically confirm the presence of pili in these derivative mutants of L. rhamnosus GG, indirect immunofluorescence microscopy with anti-SpaC serum was performed. Figure 1B confirms that the pilus mutant CMPG5357 does not contain SpaC monomeric adhesins, as no fluorescence was detected on the mutant cells stained with anti-SpaC serum. In contrast, we were able to visualize SpaC-containing pili on the L. rhamnosus GG wild type (Fig. 1A) and even observed an increased signal with the SpaC antiserum in the EPS-deficient mutant CMPG5351 (Fig. 1C). Staining with preimmune serum as a negative control showed no fluorescence in the wild type or mutants (data not shown).
FIG 1.
Presence of pili in L. rhamnosus GG and its derivative mutants. (A to C) Indirect immunofluorescence microscopy. Anti-SpaC rabbit antisera (1:100 dilution) were used on the L. rhamnosus GG wild type (A), pilus-deficient spaCBA mutant CMPG5357 (B), and EPS-deficient mutant CMPG5351 (C). SpaC-containing pili were then identified by anti-rabbit IgG antibodies (1:200 dilution) conjugated with Alexa Fluor 488. (D) Schematic representation of the bacterial strains used. The L. rhamnosus GG wild type, its derivative mutant strains, the L. lactis subsp. cremoris MG1363 wild type, and its recombinant SpaCBA-piliated lactococcal construct (GRS1185) were used in this study. The L. rhamnosus GG wild type has galactose-rich polysaccharides and pili on its surface. The spaCBA mutant CMPG5357 has galactose-rich polysaccharides but lacks the pili. The welE mutant CMPG5351 lacks the long galactose-rich polysaccharides but has increased exposure of pili and other surface molecules (20–23, 64–66).
SpaCBA pili promote L. rhamnosus GG adhesion to RAW 264.7 cells and modulate its internalization.
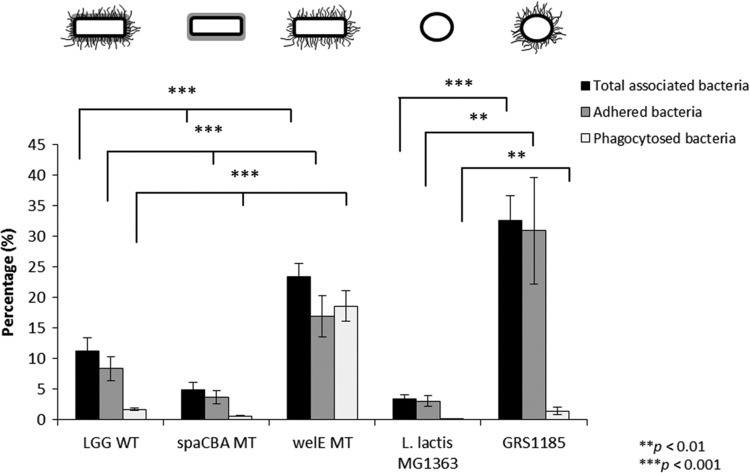
Since intestinal epithelial cells and monocytic cells have different cell surface properties, we subsequently aimed to investigate the role of pili in adherence to macrophages. Compared to the adhesion of the L. rhamnosus GG wild type, the pilus spaCBA-knockout mutant showed a ca. 2.3-fold reduction in adhesion to murine macrophage RAW 264.7 cells, while the EPS-deficient mutant CMPG5351 showed a ca. 2-fold increased adhesion capacity (Fig. 2). Likewise, an increase in the level of adhesion to RAW 264.7 cells of ca. 10-fold was observed for the nisin-induced GRS1185 lactococcal construct producing SpaCBA pili compared to the level observed for the nonpiliated L. lactis subsp. cremoris wild type. To specifically assess the role of SpaCBA pili on macrophage binding, the total amount of macrophage-associated bacteria was also determined, and when RAW 264.7 cells were stimulated with the SpaCBA pilus-deficient mutant CMPG5357, a reduction in the total amount of bacteria of ca. 2.3-fold compared to the total amount found when they were stimulated with L. rhamnosus GG was found. Conversely, the EPS-deficient mutant CMPG5351 showed an increase in the total amount of associated bacteria of ca. 2.1-fold (Fig. 2). Similarly, we could observe an increase in the total number of macrophage-associated bacteria of ca. 9.6-fold for the recombinant SpaCBA-piliated lactococcal cells compared to that for the nonpiliated L. lactis subsp. cremoris wild type.
FIG 2.
Adhesion to RAW 264.7 cells and phagocytosis of wild-type (WT) L. rhamnosus GG and its knockout mutant (MT) derivatives by RAW 264.7 cells. For adhesion assays, L. rhamnosus GG (5 × 107 CFU/ml) was coincubated for 1 h with 1 × 106 RAW 264.7 cells, and the percentage of adherent bacteria was determined. The total amount of macrophage-associated bacteria was also evaluated by lysis of the RAW 264.7 cells prior to preparation of the dilution series in order to liberate internalized bacteria. For phagocytosis assays, L. rhamnosus GG and its knockout mutant derivatives were added to RAW 264.7 cells at a ratio of 50:1 for 1 h. The remaining extracellular bacteria were killed by addition of streptomycin (100 μg/ml) and penicillin (100 U/ml) for an additional hour, and the percentage of internalized bacteria was then determined. Nonpiliated L. lactis subsp. cremoris MG1363 and the recombinant SpaCBA-piliated lactococcal construct (GRS1185) were included as controls. These experiments were done in triplicate, and error bars represent standard deviations. Asterisks represent statistically significant differences compared to the results for the L. rhamnosus GG wild type or L. lactis subsp. cremoris MG1363.
Furthermore, to examine whether SpaCBA pilus-mediated adhesion affects the uptake of L. rhamnosus GG by macrophages by phagocytosis and related uptake mechanisms, the L. rhamnosus GG wild type and its mutant derivatives were incubated with macrophages, and the number of internalized bacteria was counted. External bacteria were killed by antibiotic treatment consisting of a combination of streptomycin (100 μg/ml) and penicillin (100 U/ml). Previous experiments showed that this treatment resulted in a more than 99% decrease of bacterial survival after 1 h of incubation, so that external bacteria do not significantly bias the results (data not shown). We subsequently observed that the SpaCBA-deficient mutant CMPG5357 was internalized to a ca. 2.7-fold lower degree than the L. rhamnosus GG wild type in RAW 264.7 cells. Conversely, the EPS-deficient mutant CMPG5351 showed a ca. 10.8-fold increased uptake compared to the wild type (Fig. 2). Similarly, the recombinant SpaCBA-piliated lactococcal construct GRS1185 was internalized by RAW264.7 cells ca. 452-fold more than the nonpiliated L. lactis subsp. cremoris wild type.
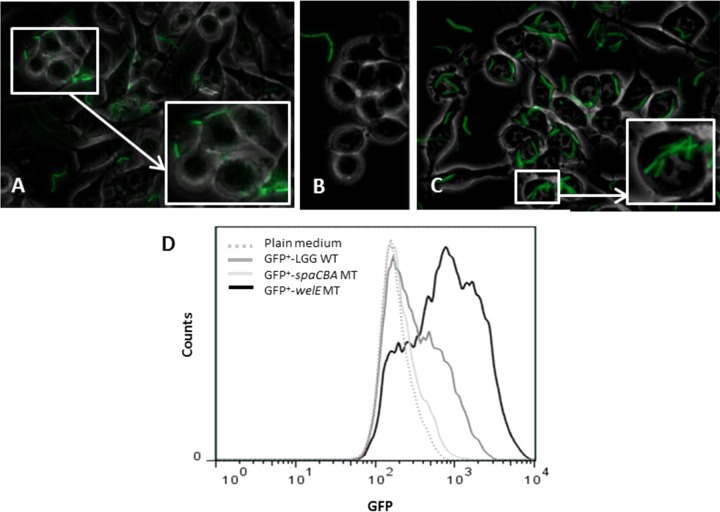
To further verify our findings, we also performed fluorescence microscopy analyses. First, the pilus mutant CMPG5357 and the EPS-deficient mutant CMPG5351 were transformed with a GFP-expressing plasmid as described in Materials and Methods. RAW 264.7 cells were then incubated with the GFP-expressing bacteria for 2 h, after which the interaction between bacteria and macrophages was analyzed. Notably, in five independent experiments we could barely detect the pilus-knockout mutant (CMPG1908/pMEC45) in the sample (Fig. 3B), while the EPS-deficient mutant (CMPG1907/pMEC45) showed a stronger capacity to interact with RAW 264.7 cells (Fig. 3C) than the L. rhamnosus GG wild type (Fig. 3A). Similarly, our flow cytometric data showed that the spaCBA pilus mutant (CMPG1908/pMEC45) had a lower capacity to interact with macrophages than the wild type, while the EPS-deficient mutant with an increased exposure of pili (CMPG1907/pMEC45) interacted with RAW 264.7 cells more than the L. rhamnosus GG wild type, showing a biphasic interaction curve indicating that the interaction is not synchronized (Fig. 3D). Collectively our data indicate that SpaCBA pili indeed play a crucial role in the adhesion of L. rhamnosus GG to murine macrophages and are also involved in its internalization process, while galactose-rich EPS molecules seem to prevent the adhesion capacity and further phagocytosis of L. rhamnosus GG by macrophages.
FIG 3.
Interaction of GFP-expressing L. rhamnosus GG WT and its mutant derivatives with macrophages visualized by fluorescence microscopy and flow cytometry. (A to C) For fluorescence microscopy assays, RAW 264.7 cells were incubated with L. rhamnosus GG FAJ1906/pMEC45 (A), CMPG1908/pMEC45 (GFP+ spaCBA mutant) (B), and CMPG1907/pMEC45 (GFP+ welE mutant) (C) for 2 h. Fluorescence micrographs are representative of those from 5 independent experiments. (D) FACS analysis. RAW 264.7 cells were incubated with the GFP-expressing L. rhamnosus GG wild type and mutants for 2 h at 37°C. The graph depicts the signal intensity of GFP (previously calibrated with fluorescently labeled beads [rainbow beads, catalog number RCP-30-5A, 8 peaks; Spherotech]) (x axis) versus the number of cells (y axis). The results represent the fluorescence intensity of 5 × 105 cells in one representative experiment.
Cytokine mRNA induction in RAW 264.7 cells is modulated by the presence of SpaCBA pili on L. rhamnosus GG.
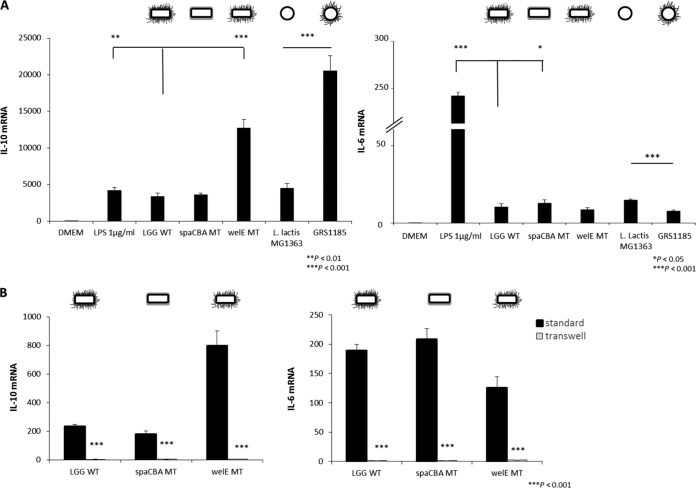
Subsequently, we investigated whether the SpaCBA pilus-mediated adhesion influences the induction of the pro- and anti-inflammatory cytokines IL-6 and IL-10, respectively, in RAW 264.7 cells. Macrophages were coincubated with bacteria for 3 h, on the basis of the findings of previous experiments in which different incubation times were tested, and 3 h was found to be the earliest time point with optimal expression of the mRNA of the cytokines under study (data not shown). Our qPCR analysis showed a ca. 4-fold increase in the expression of the anti-inflammatory cytokine IL-10 mRNA in macrophages stimulated with the EPS-deficient mutant CMPG5351 compared to that in macrophages stimulated with the L. rhamnosus GG wild type, whereas the pilus mutant CMPG5357 induced a similar level of IL-10 mRNA (Fig. 4A, left). We also analyzed the role of pili in a recombinantly engineered L. lactis subsp. cremoris strain that produces SpaCBA pili. Interestingly, GRS1185 cells producing SpaCBA pili induced the expression of IL-10 mRNA to levels ca. 4.5-fold greater than those for the nonpiliated L. lactis subsp. cremoris wild type. Taken these findings together, the presence of pili seems to moderately promote IL-10 mRNA induction by L. rhamnosus GG and L. lactis subsp. cremoris, but comparisons of the results for the mutants show that the pili per se are not required for IL-10 mRNA induction. It is also noteworthy that the level of induction of the expression of IL-10 by L. rhamnosus GG or the L. lactis subsp. cremoris wild type is comparable to that by the control, LPS (1 μg/ml) (Fig. 4A, left).
FIG 4.
Interleukin-10 mRNA and interleukin-6 mRNA response of RAW 264.7 macrophages. (A) Cells were stimulated with L. rhamnosus GG strains for 3 h. DMEM served as a negative control, and LPS at 1 μg/ml served as a positive control. Nonpiliated L. lactis subsp. cremoris MG1363 and the recombinant SpaCBA-piliated lactococcal construct (GRS1185) were included as additional controls. Bacteria were administered at a ratio of 1:50 cells to bacteria. Results were quantified by the use of a cDNA plasmid standard and expressed as the ratio of the cDNA copy numbers for IL-10 (left) or IL-6 (right) divided by the cDNA copy numbers for GAPDH multiplied by 104. Data are mean values ± standard deviations of triplicate values and represent those from one of three independent experiments. Asterisks represent statistically significant differences compared to the results for the L. rhamnosus GG wild type or L. lactis subsp. cremoris MG1363. (B) The role of direct cell-cell contact on L. rhamnosus GG-induced expression of IL-10 (left) and IL-6 (right) mRNA by RAW 264.7 cells was measured by qPCR after 3 h of coincubation with the different bacterial strains. In a Transwell system, direct cell-cell contact was prevented, and macrophages were grown in the basolateral compartment with apical addition of the respective bacterial strains. Data are mean values ± standard deviations of triplicate values and represent those from one of three independent experiments. Controls (no treatment) did not induce cytokine secretion.
We also tested the expression of IL-6 mRNA as a proinflammatory cytokine, considering its relation with inflammation. As shown in Fig. 4A (right), the expression of IL-6 mRNA was slightly increased by ca. 1.2-fold when macrophages were stimulated with the spaCBA pilus mutant CMPG5357, while the enhanced exposure of pili did not appear to impact IL-6 mRNA induction significantly when the results obtained with EPS-deficient mutant CMPG5351 and the L. rhamnosus GG wild type were compared (P < 0.05). However, stimulation with GRS1185 producing SpaCBA pili resulted in a decreased induction of IL-6 mRNA in RAW 264.7 cells by ca. 2-fold compared to that achieved with stimulation by the nonpiliated L. lactis subsp. cremoris wild type. Taken together, the presence of pili seems to moderately reduce IL-6 mRNA induction by L. rhamnosus GG and L. lactis subsp. cremoris. Importantly, LPS (1 μg/ml) induced IL-6 mRNA to much higher levels than all strains tested.
Subsequently, in order to investigate the role of cell contact in L. rhamnosus GG-mediated cytokine modulation in macrophages, Transwell experiments were performed to prevent direct contact between macrophages and bacteria. IL-10 mRNA expression was examined after 3 h using qPCR. Separation of macrophages from bacteria by a 0.4-μm-pore-size membrane indeed completely suppressed the induction of IL-10 mRNA (Fig. 4B, left). Similar results were found while investigating the expression of IL-6 mRNA (Fig. 4B, right).
Role of the host receptors CR3 and TLR2 in the interaction of L. rhamnosus GG with macrophages.
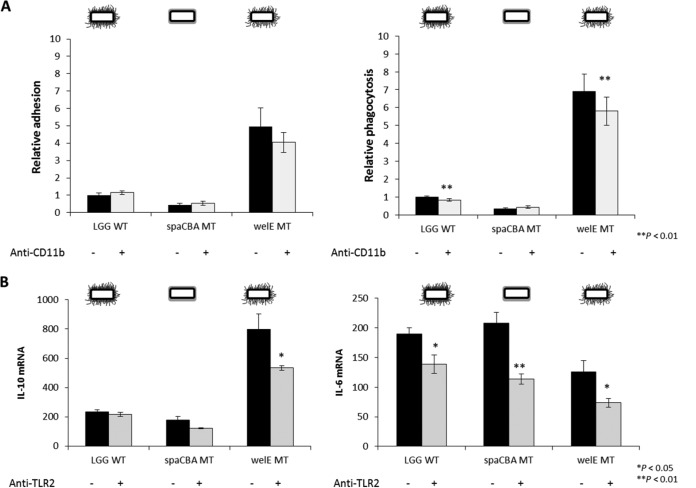
To identify putative receptors on macrophages that are involved in the recognition of L. rhamnosus GG, possibly via the SpaCBA pili, we used previously validated blocking antibodies (24, 41) against complement receptor 3 (CR3; consisting of the two subunits CD11b and CD18) and Toll-like receptor 2 (TLR2). First, we evaluated the effect of CR3 in SpaCBA pilus-mediated adhesion to RAW 264.7 cells by blocking its CD11b component. We observed no significant differences (P < 0.05) in the adhesion of the L. rhamnosus GG wild type, the pilus mutant CMPG5357 and the EPS-deficient mutant CMPG5351 to murine macrophages that had previously been treated with anti-CD11b antibodies (Fig. 5A, left). Even though the difference was not significant, a minor inhibition of the adhesion of the EPS-deficient mutant was found when RAW 264.7 cells were pretreated with the antibody. Furthermore, the level of macrophage ingestion of pilus-expressing strains was slightly but significantly inhibited by ca. 1.2-fold (P < 0.01) by anti-CD11b treatment (Fig. 5A, right) of both the L. rhamnosus GG wild type and the EPS-deficient mutant. However, when analyzing the role of CD11b blocking in the cytokine modulatory capacity of L. rhamnosus GG, we observed that the expression of IL-10 and IL-6 mRNA was not significantly affected by treatment with anti-CD11b antibodies (data not shown). In contrast, antibody blockage of TLR2 showed a significant decrease in the expression of IL-6 mRNA by macrophages after stimulation with all the L. rhamnosus GG strains, while for IL-10 mRNA, this decrease was significant only for the EPS-deficient mutant (P < 0.05) (Fig. 5B).
FIG 5.
Role of CD11b in adhesion and uptake of L. rhamnosus GG by RAW 264.7 cells and effect of TLR2 in the subsequent immunomodulatory response. (A) Role of CD11b in adhesion of the mutants to macrophages (left) and phagocytosis of the mutants by macrophages (right) relative to the results for the wild-type strain, which were set equal to 1. Bacterial strains (5 × 107 CFU/ml) were coincubated for 1 h with 1 × 106 RAW 264.7 cells that had previously been treated with anti-mouse CD11b monoclonal antibody (5 μg/ml) for 30 min. The data are presented as the mean relative adhesion or phagocytosis ± standard deviation. These experiments were done in triplicate. Asterisks represent statistically significant differences compared to the results for the controls (no antibody treatment). (B) The effect of TLR2 on the relative levels of IL-10 and IL-6 mRNA expression by RAW 264.7 macrophages was analyzed by pretreatment of macrophages with antibodies against TLR2 (5 to 10 μg/ml) for 30 min and then coincubation with L. rhamnosus GG strains for 3 h. Bacteria were administered at a ratio of 1:50 of cells to bacteria. The results are mean values ± standard deviations from three separate experiments. The values are normalized against those for GAPDH. Asterisks denote statistically significant differences from the results for the control (no antibody treatment).
DISCUSSION
Adherence is the first step by which components of the bacterial cell surface interact with receptors present on the host cells. Subsequently, surface-bound components and molecules secreted by the bacteria can trigger particular signaling pathways leading to the production of specific host proteins, resulting in specific immune responses. Thus, investigation of the molecular interactions between components on the bacterial cell surface and host cells is of great importance to unravel new mechanisms of regulation of the host immune responses (13). However, the role of bacterial adhesion in immunomodulation, especially in probiotic lactobacilli, remains unclear. Previous studies by our group and others have indicated that there are large differences in the adherence capacities of lactobacilli (reviewed in, e.g., reference 42). We recently showed that the long flexible pili of L. rhamnosus GG promote a strong interaction with substrates by a zipper-like mechanism involving SpaC adhesin subunits present at the tip but also along the pilus fibers (29). In addition, these SpaCBA pili seem to show a remarkable elasticity imparting nanospring properties that might help them to retract closer to substrates even under conditions of shear flow or peristalsis (29). Thus, given its high adherence capacity mediated by pili, L. rhamnosus GG is an interesting model strain with which to investigate the interplay between adhesion and immunomodulation. Here, we aimed to explore the importance of the presence of these pili on native L. rhamnosus GG cells in the interaction with phagocytic innate immune cells, i.e., macrophages. Interest in macrophages as important modulators of the interaction with gut microbiota and probiotics has been renewed, since it has been shown that they can also pass their dendrites through intestinal epithelial cells (26, 43). Moreover, macrophages can differentiate into classical (M1) or alternative (M2) subsets, with the first group promoting T helper 1 (Th1) immune responses and the second one being related to immunosuppression by having an anti-inflammatory phenotype (44, 45).
In this work, we demonstrated that SpaCBA pili of L. rhamnosus GG are crucial for its capacity to adhere to a murine macrophage (RAW 264.7) cell line and that this interaction stimulates bacterial uptake. Additionally, we observed that the pilus-mediated adhesion and enhanced cell-to-cell contact are coupled to an anti-inflammatory response, as monitored by the induction of IL-10 mRNA, since EPS-deficient mutant CMPG5351 with enhanced exposure of the pili and the L. lactis subsp. cremoris mutant with overexpression of the SpaCBA pili showed a markedly increased ability to induce this anti-inflammatory cytokine in RAW 264.7 cells compared to that of their respective wild-type strains. However, we could not observe any significant differences in the expression of IL-10 mRNA when the cells were stimulated with the nonpiliated spaCBA mutant CMPG5357 strain and the wild-type L. rhamnosus GG strain, suggesting that the pili per se are not required for the induction of IL-10 mRNA. IL-10 was selected as a biomarker for immunological activities due to its crucial role in the suppression and prevention of inflammatory responses (46, 47). This cytokine is produced by several immune cells, such as regulatory CD4+ T cells, which are key players in anti-inflammatory immune responses (48), and M2 macrophages, which express an IL-10high phenotype (49, 50). Interestingly, the level of IL-10 mRNA induced was similar to or even greater than that induced by LPS (1 μg/ml), a previously documented bacterial ligand shown by different groups to activate IL-10 gene expression in macrophages (51–53). Our findings after heterologous expression of the SpaCBA pili in L. lactis subsp. cremoris for IL-10 induction in murine macrophages are in agreement with those of previous work of von Ossowski and coworkers using moDCs (23). Given its zipper-like adhesion role, it is highly likely that the pili do not directly induce large amounts of IL-10 mRNA but, rather, mediate close interaction with the macrophages, resulting in an enhanced delivery of anti-inflammatory molecules. The exact IL-10-inducing molecules in L. rhamnosus GG thus remain to be determined, but our Transwell experiments suggest that they are located on the bacterial cell wall. Documented Lactobacillus molecules with anti-inflammatory properties include polysaccharide chains of rhamnose-rich EPSs produced by L. rhamnosus RW-9595M that induce immunosuppression by the production of anti-inflammatory IL-10 in macrophages (54), but they do not seem to be present in L. rhamnosus GG (20). Bernardo and coworkers identified a peptide (STp) secreted by L. plantarum BMCM12 that could induce intracellular IL-10 in human blood and intestinal DCs (67). Likewise, the S-layer protein A (SlpA) from L. acidophilus NCFM was found to mediate the interaction of bacteria with DCs by specifically binding their C-type lectin PRR DC-SIGN, resulting in the induction of IL-10 in the presence of LPS, but IL-10 was not as highly induced by LPS or SlpA alone (18). Furthermore, teichoic acids (TAs) from L. plantarum ATCC 14917 were found to be recognized by TLR2, activate the extracellular signal-regulated kinase pathway, and act synergistically with L. casei Shirota YIT9029 to induce IL-10 production in mouse peritoneal macrophages (55). However, other studies showed that expression of IL-10 was almost entirely unaffected in THP-1 cells after treatment with LTA isolated from L. plantarum KCTC10887BP (56) and that LTA from L. plantarum NCIMB8826 promotes IL-12 rather than IL-10 induction (57). Clearly, multiple Lactobacillus cell wall or secreted molecules could be directly involved in IL-10 mRNA induction.
Furthermore, our results indicate that the SpaCBA pili of L. rhamnosus GG do not directly influence the expression of proinflammatory cytokine IL-6 mRNA. We could observe an increased induction for the SpaCBA pilus mutant CMPG5357 compared to that for the wild type, whereas EPS-deficient mutant CMPG5351, showing an enhanced exposure of the pili, had the same levels of expression of IL-6 mRNA as the wild type. In support of the above-mentioned results, further experiments indicated that the SpaCBA-piliated lactococcus GRS1185 also reduced the level of IL-6 mRNA induction in RAW 264.7 cells compared to that achieved with nonpiliated L. lactis subsp. cremoris. However, these data are not in agreement with those from previous work using human moDCs (23), where the SpaCBA-piliated lactococcal constructs had increased levels of IL-6 production. It is worthy of mention that in the previous study (23), IL-6 production was measured according to the protein levels, while in the present work, gene expression at the mRNA level was evaluated. Moreover, different antigen-presenting cells (i.e., human monocyte-derived DCs and murine macrophages) were used in both studies, so that a variation in the pattern of cytokine production can be expected due to differential receptor expression (58). Taken together, our data suggest that the presence of pili probably indirectly prevents the induction of IL-6 mRNA by other surface molecules. Interestingly, a former study has demonstrated a role for pathogenic pneumococcal pili in the induction of proinflammatory cytokines, as higher levels of IL-6 were found in the serum of mice previously challenged with S. pneumoniae strain T4, which expresses pili, than in the serum of mice treated with its isogenic nonpiliated deletion mutant (59). Although the results from this in vivo study cannot be easily compared with those of our molecular interaction studies with cell lines, it is intriguing that pili from pathogenic bacteria induce proinflammatory immune responses in the host, while our data for the pili of the probiotic strain under study showed instead an indirect antipathogenic role. Nevertheless, future studies will have to investigate the role of pili on probiotic lactobacilli in modulating other anti- and proinflammatory biomarkers in different cell types and hosts with different disease states and genetic backgrounds.
To gain a better insight into the molecular interaction between piliated L. rhamnosus GG and macrophages, we also investigated the role of two PRRs, i.e., CR3 (CD11b/CD18) and TLR2. CR3 is a receptor present on the surface of most immune cells, including monocytes/macrophages, DCs, neutrophils, and natural killer (NK) cells, and plays an essential role in different immunological processes, including cell activation, chemotaxis, cytotoxicity, and phagocytosis (60) and tolerance induction (61). Previous studies demonstrated that pilus-associated adhesin RrgA from S. pneumoniae binds to CR3 and promotes bacterial uptake by murine and human macrophages (24). Using a similar approach with the same anti-CD11b antibodies, our data show that SpaCBA pili of L. rhamnosus GG promote bacterial phagocytosis by RAW 264.7 cells partially through CR3. However, the induction of IL-10 and IL-6 mRNA in these macrophages was not affected by the treatment with anti-CD11b antibodies, suggesting that L. rhamnosus GG-mediated cytokine modulation occurs in a CR3-independent manner. TLR2 has been found to be involved in the recognition of various MAMPs of Gram-positive bacteria. For instance, we recently demonstrated that LTA from L. rhamnosus GG stimulates TLR2/6-dependent activation of NF-κβ signaling in the HEK293T cell line and stimulates proinflammatory cytokines, at least under certain conditions (17). Interestingly, our present results show that induction of IL-6 mRNA by L. rhamnosus GG in macrophages is significantly influenced by the interaction with TLR2. This L. rhamnosus GG-TLR2 interaction is at least partially mediated by L. rhamnosus GG LTA, being a MAMP for TLR2/6, as determined on the basis of our previous work (17). Nevertheless, it is important to emphasize that LPS was still able to induce much higher levels of IL-6 mRNA than all L. rhamnosus GG strains tested, indicating that the induction of IL-6 by L. rhamnosus GG is subtler than that by the more immunostimulatory LPS of E. coli. Blockage of TLR2 also significantly affected the expression of IL-10 mRNA, but only when cells were treated with the EPS-deficient mutant, suggesting that molecules other than SpaCBA pili that are overexposed in this EPS-deficient mutant are signaling through TLR2 and that TLR2 could thus also modulate the anti-inflammatory responses. Interestingly, recent work in Staphylococcus aureus has also shown that its cell surface contains both proinflammatory and IL-10-inducing immunomodulatory molecules. These peptidoglycan-embedded IL-10-inducing molecules also bind to TLR2 on host antigen-presenting cells, activating the PI3K/Akt pathway, resulting in high levels of production of IL-10, which in turn downregulates the adaptive T cell response (62, 63). Thus, the exact receptor(s) by which the SpaCBA pili could mediate cytokine induction remains to be identified. For such studies, sufficient amounts of highly pure pili need to be obtained and preferentially studied in their native configuration. Such experiments are ongoing in our lab but are complicated.
Taken together, our data indicate that L. rhamnosus GG pili play a role in the host interaction with macrophages, mainly by promoting adhesion and internalization. As a result, other MAMPs, both pro- and anti-inflammatory molecules of L. rhamnosus GG, can interact more closely with their respective PRRs. Nevertheless, it remains to be substantiated in more complex models that the pilus-mediated high levels of adhesion of L. rhamnosus GG to host immune cells combined with a low inflammatory potential would be beneficial to humans.
ACKNOWLEDGMENTS
This work was supported in part through BOF program financing (spokesman, Jan Balzarini; PF/10/018) at KU Leuven and in part through the Fund for Scientific Research, Flanders (FWO; grants KaN 150711N and 1520114N), and IOF-SBO financing at the Universiteit Antwerpen. Dominique Bullens is a recipient of a senior researcher fellowship from the Fund for Scientific Research, Flanders (FWO). I.V.O. and A.P. acknowledge project financing from the Academy of Finland-funded Center of Excellence in Microbial Food Safety (CoE-MiFoSa) research program (grant no. 118602 and 141140) and an Academy of Finland general research grant (no. 118165).
Marijke Segers and Hanne Tytgat are acknowledged for many insightful discussions and suggestions. Lieve Coorevits is gratefully thanked for her invaluable help with the culture of RAW 264.7 cells. We also thank Stefanie Roberfroid, Louis Deforche, and David De Coster for their technical assistance with the fluorescence microscope, as well as Geert Schoofs for his assistance with the setup of the flow cytometry experiments.
REFERENCES
- 1.Bongaerts GPA, Severijnen RSVM. 2001. The beneficial, antimicrobial effect of probiotics. Med Hypotheses 56:174–177. doi: 10.1054/mehy.2000.1135. [DOI] [PubMed] [Google Scholar]
- 2.Petrova MI, van den Broek M, Balzarini J, Vanderleyden J, Lebeer S. 2013. Vaginal microbiota and its role in HIV transmission and infection. FEMS Microbiol Rev 37:762–792. doi: 10.1111/1574-6976.12029. [DOI] [PubMed] [Google Scholar]
- 3.Kalliomaki M, Salminen S, Arvilommi H, Kero P, Koskinen P, Isolauri E. 2001. Probiotics in primary prevention of atopic disease: a randomised placebo-controlled trial. Lancet 357:1076–1079. doi: 10.1016/S0140-6736(00)04259-8. [DOI] [PubMed] [Google Scholar]
- 4.Kalliomaki M, Salminen S, Poussa T, Arvilommi H, Isolauri E. 2003. Probiotics and prevention of atopic disease: 4-year follow-up of a randomised placebo-controlled trial. Lancet 361:1869–1871. doi: 10.1016/S0140-6736(03)13490-3. [DOI] [PubMed] [Google Scholar]
- 5.Kalliomaki M, Salminen S, Poussa T, Isolauri E. 2007. Probiotics during the first 7 years of life: a cumulative risk reduction of eczema in a randomized, placebo-controlled trial. J Allergy Clin Immunol 119:1019–1021. doi: 10.1016/j.jaci.2006.12.608. [DOI] [PubMed] [Google Scholar]
- 6.Hatakka K, Savilahti E, Ponka A, Meurman JH, Poussa T, Nase L, Saxelin M, Korpela R. 2001. Effect of long term consumption of probiotic milk on infections in children attending day care centres: double blind, randomised trial. Br Med J 322:1327–1329. doi: 10.1136/bmj.322.7298.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hojsak I, Abdovic S, Szajewska H, Milosevic M, Krznaric Z, Kolacek S. 2010. Lactobacillus GG in the prevention of nosocomial gastrointestinal and respiratory tract infections. Pediatrics 125:e1171–e1177. doi: 10.1542/peds.2009-2568. [DOI] [PubMed] [Google Scholar]
- 8.Hojsak I, Snovak N, Abdovic S, Szajewska H, Misak Z, Kolacek S. 2010. Lactobacillus GG in the prevention of gastrointestinal and respiratory tract infections in children who attend day care centers: a randomized, double-blind, placebo-controlled trial. Clin Nutr 29:312–316. doi: 10.1016/j.clnu.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 9.Vanderhoof JA, Whitney DB, Antonson DL, Hanner TL, Lupo JV, Young RJ. 1999. Lactobacillus GG in the prevention of antibiotic-associated diarrhea in children. J Pediatr 135:564–568. doi: 10.1016/S0022-3476(99)70053-3. [DOI] [PubMed] [Google Scholar]
- 10.Guandalini S, Pensabene L, Abu Zikri M, Dias JA, Casali LG, Hoekstra H, Kolacek S, Massar K, Micetic-Turk D, Papadopoulou A, de Sousa JS, Sandhu B, Szajewska H, Weizman Z. 2000. Lactobacillus GG administered in oral rehydration solution to children with acute diarrhea: a multicenter European trial. J Pediatr Gastroenterol Nutr 30:54–60. doi: 10.1097/00005176-200001000-00018. [DOI] [PubMed] [Google Scholar]
- 11.Szajewska H, Skorka A, Ruszczynski M, Gieruszczak-Bialek D. 2013. Meta-analysis: Lactobacillus GG for treating acute gastroenteritis in children—updated analysis of randomised controlled trials. Aliment Pharmacol Ther 38:467–476. doi: 10.1111/apt.12403. [DOI] [PubMed] [Google Scholar]
- 12.Guarino A, Canani RB, Spagnuolo MI, Albano F, DiBenedetto L. 1997. Oral bacterial therapy reduces the duration of symptoms and of viral excretion in children with mild diarrhea. J Pediatr Gastroenterol Nutr 25:516–519. doi: 10.1097/00005176-199711000-00005. [DOI] [PubMed] [Google Scholar]
- 13.Lebeer S, Vanderleyden J, De Keersmaecker SCJ. 2010. Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens. Nat Rev Microbiol 8:171–184. doi: 10.1038/nrmicro2297. [DOI] [PubMed] [Google Scholar]
- 14.Schlee M, Harder J, Koten B, Stange EF, Wehkamp J, Fellermann K. 2008. Probiotic lactobacilli and VSL#3 induce enterocyte beta-defensin 2. Clin Exp Immunol 151:528–535. doi: 10.1111/j.1365-2249.2007.03587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleerebezem M, Hols P, Bernard E, Rolain T, Zhou M, Siezen RJ, Bron PA. 2010. The extracellular biology of the lactobacilli. FEMS Microbiol Rev 34:199–230. doi: 10.1111/j.1574-6976.2009.00208.x. [DOI] [PubMed] [Google Scholar]
- 16.Sengupta R, Altermann E, Anderson RC, McNabb WC, Moughan PJ, Roy NC. 2013. The role of cell surface architecture of lactobacilli in host-microbe interactions in the gastrointestinal tract. Mediators Inflamm 2013:237921. doi: 10.1155/2013/237921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Claes IJJ, Segers ME, Verhoeven TLA, Dusselier M, Sels BF, De Keersmaecker SCJ, Vanderleyden J, Lebeer S. 2012. Lipoteichoic acid is an important microbe-associated molecular pattern of Lactobacillus rhamnosus GG. Microb Cell Fact 11:161. doi: 10.1186/1475-2859-11-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konstantinov SR, Smidt H, de Vos WM, Bruijns SC, Singh SK, Valence F, Molle D, Lortal S, Altermann E, Klaenhammer TR, van Kooyk Y. 2008. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc Natl Acad Sci U S A 105:19474–19479. doi: 10.1073/pnas.0810305105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Macho Fernandez EM, Valenti V, Rockel C, Hermann C, Pot B, Boneca IG, Grangette C. 2011. Anti-inflammatory capacity of selected lactobacilli in experimental colitis is driven by NOD2-mediated recognition of a specific peptidoglycan-derived muropeptide. Gut 60:1050–1059. doi: 10.1136/gut.2010.232918. [DOI] [PubMed] [Google Scholar]
- 20.Lebeer S, Verhoeven TLA, Francius G, Schoofs G, Lambrichts I, Dufrene Y, Vanderleyden J, De Keersmaecker SCJ. 2009. Identification of a gene cluster for the biosynthesis of a long, galactose-rich exopolysaccharide in Lactobacillus rhamnosus GG and functional analysis of the priming glycosyltransferase. Appl Environ Microbiol 75:3554–3563. doi: 10.1128/AEM.02919-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kankainen M, Paulin L, Tynkkynen S, von Ossowski I, Reunanen J, Partanen P, Satokari R, Vesterlund S, Hendrickx APA, Lebeer S, De Keersmaecker SCJ, Vanderleyden J, Hamalainen T, Laukkanen S, Salovuori N, Ritari J, Alatalo E, Korpela R, Mattila-Sandholm T, Lassig A, Hatakka K, Kinnunen KT, Karjalainen H, Saxelin M, Laakso K, Surakka A, Palva A, Salusjarvi T, Auvinen P, de Vos WM. 2009. Comparative genomic analysis of Lactobacillus rhamnosus GG reveals pili containing a human-mucus binding protein. Proc Natl Acad Sci U S A 106:17193–17198. doi: 10.1073/pnas.0908876106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lebeer S, Claes I, Tytgat HLP, Verhoeven TLA, Marien E, von Ossowski I, Reunanen J, Palva A, de Vos WM, De Keersmaecker SCJ, Vanderleyden J. 2012. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol 78:185–193. doi: 10.1128/AEM.06192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.von Ossowski I, Pietila TE, Rintahaka J, Nummenmaa E, Makinen VM, Reunanen J, Satokari R, de Vos WM, Palva I, Palva A. 2013. Using recombinant lactococci as an approach to dissect the immunomodulating capacity of surface piliation in probiotic Lactobacillus rhamnosus GG. PLoS One 8:e64416. doi: 10.1371/journal.pone.0064416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orrskog S, Rounioja S, Spadafina T, Gallotta M, Norman M, Hentrich K, Falker S, Ygberg-Eriksson S, Hasenberg M, Johansson B, Uotila LM, Gahmberg CG, Barocchi M, Gunzer M, Normark S, Henriques-Normark B. 2012. Pilus adhesin RrgA interacts with complement receptor 3, thereby affecting macrophage function and systemic pneumococcal disease. mBio 4(1):e00535-12. doi: 10.1128/mBio.00535-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Basset A, Zhang F, Benes C, Sayeed S, Herd M, Thompson C, Golenbock DT, Camilli A, Malley R. 2013. Toll-like receptor (TLR) 2 mediates inflammatory responses to oligomerized RrgA pneumococcal pilus type 1 protein. J Biol Chem 288:2665–2675. doi: 10.1074/jbc.M112.398875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Medina-Contreras O, Geem D, Laur O, Williams IR, Lira SA, Nusrat A, Parkos CA, Denning TL. 2011. CX3CR1 regulates intestinal macrophage homeostasis, bacterial translocation, and colitogenic Th17 responses in mice. J Clin Invest 121:4787–4795. doi: 10.1172/JCI59150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niess JH, Brand S, Gu X, Landsman L, Jung S, McCormick BA, Vyas JM, Boes M, Ploegh HL, Fox JG, Littman DR, Reinecker HC. 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science 307:254–258. doi: 10.1126/science.1102901. [DOI] [PubMed] [Google Scholar]
- 28.Rescigno M, Urbano M, Valzasina B, Francolini M, Rotta G, Bonasio R, Granucci F, Kraehenbuhl JP, Ricciardi-Castagnoli P. 2001. Dendritic cells express tight junction proteins and penetrate gut epithelial monolayers to sample bacteria. Nat Immunol 2:361–367. doi: 10.1038/86373. [DOI] [PubMed] [Google Scholar]
- 29.Tripathi P, Beaussart A, Alsteens D, Dupres V, Claes I, von Ossowski I, de Vos WM, Palva A, Lebeer S, Vanderleyden J, Dufrene YF. 2013. Adhesion and nanomechanics of pili from the probiotic Lactobacillus rhamnosus GG. ACS Nano 7:3685–3697. doi: 10.1021/nn400705u. [DOI] [PubMed] [Google Scholar]
- 30.Wegmann U, O'Connell-Motherway M, Zomer A, Buist G, Shearman C, Canchaya C, Ventura M, Goesmann A, Gasson MJ, Kuipers OP, van Sinderen D, Kok J. 2007. Complete genome sequence of the prototype lactic acid bacterium Lactococcus lactis subsp. cremoris MG1363. J Bacteriol 189:3256–3270. doi: 10.1128/JB.01768-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eichenbaum Z, Federle MJ, Marra D, de Vos WM, Kuipers OP, Kleerebezem M, Scott JR. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol 64:2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Claes IJ, Schoofs G, Regulski K, Courtin P, Chapot-Chartier MP, Rolain T, Hols P, von Ossowski I, Reunanen J, de Vos WM, Palva A, Vanderleyden J, De Keersmaecker SC, Lebeer S. 2012. Genetic and biochemical characterization of the cell wall hydrolase activity of the major secreted protein of Lactobacillus rhamnosus GG. PLoS One 7:e31588. doi: 10.1371/journal.pone.0031588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Keersmaecker SC, Braeken K, Verhoeven TL, Perea Velez M, Lebeer S, Vanderleyden J, Hols P. 2006. Flow cytometric testing of green fluorescent protein-tagged Lactobacillus rhamnosus GG for response to defensins. Appl Environ Microbiol 72:4923–4930. doi: 10.1128/AEM.02605-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pavan S, Hols P, Delcour J, Geoffroy MC, Grangette C, Kleerebezem M, Mercenier A. 2000. Adaptation of the nisin-controlled expression system in Lactobacillus plantarum: a tool to study in vivo biological effects. Appl Environ Microbiol 66:4427–4432. doi: 10.1128/AEM.66.10.4427-4432.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dupont L, Boizet-Bonhoure B, Coddeville M, Auvray F, Ritzenthaler P. 1995. Characterization of genetic elements required for site-specific integration of Lactobacillus delbrueckii subsp. bulgaricus bacteriophage mv4 and construction of an integration-proficient vector for Lactobacillus plantarum. J Bacteriol 177:586–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geoffroy MC, Guyard C, Quatannens B, Pavan S, Lange M, Mercenier A. 2000. Use of green fluorescent protein to tag lactic acid bacterium strains under development as live vaccine vectors. Appl Environ Microbiol 66:383–391. doi: 10.1128/AEM.66.1.383-391.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapot-Chartier MP, Vinogradov E, Sadovskaya I, Andre G, Mistou MY, Trieu-Cuot P, Furlan S, Bidnenko E, Courtin P, Pechoux C, Hols P, Dufrene YF, Kulakauskas S. 2010. Cell surface of Lactococcus lactis is covered by a protective polysaccharide pellicle. J Biol Chem 285:10464–10471. doi: 10.1074/jbc.M109.082958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giulietti A, Overbergh L, Valckx D, Decallonne B, Bouillon R, Mathieu C. 2001. An overview of real-time quantitative PCR: applications to quantify cytokine gene expression. Methods 25:386–401. doi: 10.1006/meth.2001.1261. [DOI] [PubMed] [Google Scholar]
- 39.Hafstad AD, Khalid AM, Hagve M, Lund T, Larsen TS, Severson DL, Clarke K, Berge RK, Aasum E. 2009. Cardiac peroxisome proliferator-activated receptor-alpha activation causes increased fatty acid oxidation, reducing efficiency and post-ischaemic functional loss. Cardiovasc Res 83:519–526. doi: 10.1093/cvr/cvp132. [DOI] [PubMed] [Google Scholar]
- 40.Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. 2003. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech 14:33–43. [PMC free article] [PubMed] [Google Scholar]
- 41.Kanaya S, Nemoto E, Ogawa T, Shimauchi H. 2009. Porphyromonas gingivalis fimbriae induce unique dendritic cell subsets via Toll-like receptor 2. J Periodontal Res 44:543–549. doi: 10.1111/j.1600-0765.2008.01149.x. [DOI] [PubMed] [Google Scholar]
- 42.Lebeer S, Vanderleyden J, De Keersmaecker SC. 2008. Genes and molecules of lactobacilli supporting probiotic action. Microbiol Mol Biol Rev 72:728–764. doi: 10.1128/MMBR.00017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peterson LW, Artis D. 2014. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol 14:141–153. doi: 10.1038/nri3608. [DOI] [PubMed] [Google Scholar]
- 44.Barros MH, Hauck F, Dreyer JH, Kempkes B, Niedobitek G. 2013. Macrophage polarisation: an immunohistochemical approach for identifying M1 and M2 macrophages. PLoS One 8:e80908. doi: 10.1371/journal.pone.0080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sica A, Mantovani A. 2012. Macrophage plasticity and polarization: in vivo veritas. J Clin Invest 122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraiva M, O'Garra A. 2010. The regulation of IL-10 production by immune cells. Nat Rev Immunol 10:170–181. doi: 10.1038/nri2711. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, Kim HJ, Yamamoto S, Kang XY, Ma XJ. 2010. Regulation of interleukin-10 gene expression in macrophages engulfing apoptotic cells. J Interferon Cytokine Res 30:113–121. doi: 10.1089/jir.2010.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Garra A, Vieira PL, Vieira P, Goldfeld AE. 2004. IL-10-producing and naturally occurring CD4+ Tregs: limiting collateral damage. J Clin Invest 114:1372–1378. doi: 10.1172/JCI200423215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martinez FO, Gordon S. 2014. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000prime Rep 6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tatano Y, Shimizu T, Tomioka H. 2014. Unique macrophages different from M1/M2 macrophages inhibit T cell mitogenesis while upregulating Th17 polarization. Sci Rep 4:4146. doi: 10.1038/srep04146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brightbill HD, Plevy SE, Modlin RL, Smale ST. 2000. A prominent role for Sp1 during lipopolysaccharide-mediated induction of the IL-10 promoter in macrophages. J Immunol 164:1940–1951. doi: 10.4049/jimmunol.164.4.1940. [DOI] [PubMed] [Google Scholar]
- 52.Iyer SS, Ghaffari AA, Cheng G. 2010. Lipopolysaccharide-mediated IL-10 transcriptional regulation requires sequential induction of type I IFNs and IL-27 in macrophages. J Immunol 185:6599–6607. doi: 10.4049/jimmunol.1002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu YW, Chen CC, Tseng HP, Chang WC. 2006. Lipopolysaccharide-induced transcriptional activation of interleukin-10 is mediated by MAPK- and NF-kappaB-induced CCAAT/enhancer-binding protein delta in mouse macrophages. Cell Signal 18:1492–1500. doi: 10.1016/j.cellsig.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 54.Bleau C, Monges A, Rashidan K, Laverdure JP, Lacroix M, Van Calsteren MR, Millette M, Savard R, Lamontagne L. 2010. Intermediate chains of exopolysaccharides from Lactobacillus rhamnosus RW-9595M increase IL-10 production by macrophages. J Appl Microbiol 108:666–675. doi: 10.1111/j.1365-2672.2009.04450.x. [DOI] [PubMed] [Google Scholar]
- 55.Kaji R, Kiyoshima-Shibata J, Nagaoka M, Nanno M, Shida K. 2010. Bacterial teichoic acids reverse predominant IL-12 production induced by certain Lactobacillus strains into predominant IL-10 production via TLR2-dependent ERK activation in macrophages. J Immunol 184:3505–3513. doi: 10.4049/jimmunol.0901569. [DOI] [PubMed] [Google Scholar]
- 56.Kim HG, Gim MG, Kim JY, Hwang HJ, Ham MS, Lee JM, Hartung T, Park JW, Han SH, Chung DK. 2007. Lipoteichoic acid from Lactobacillus plantarum elicits both the production of interleukin-23p19 and suppression of pathogen-mediated interleukin-10 in THP-1 cells. FEMS Immunol Med Microbiol 49:205–214. doi: 10.1111/j.1574-695X.2006.00175.x. [DOI] [PubMed] [Google Scholar]
- 57.Grangette C, Nutten S, Palumbo E, Morath S, Hermann C, Dewulf J, Pot B, Hartung T, Hols P, Mercenier A. 2005. Enhanced anti inflammatory capacity of a Lactobacillus plantarum mutant synthesizing modified teichoic acids. Proc Natl Acad Sci U S A 102:10321–10326. doi: 10.1073/pnas.0504084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu BS, Janssen HL, Boonstra A. 2011. IL-29 and IFNalpha differ in their ability to modulate IL-12 production by TLR-activated human macrophages and exhibit differential regulation of the IFNgamma receptor expression. Blood 117:2385–2395. doi: 10.1182/blood-2010-07-298976. [DOI] [PubMed] [Google Scholar]
- 59.Barocchi MA, Ries J, Zogaj X, Hemsley C, Albiger B, Kanth A, Dahlberg S, Fernebro J, Moschioni M, Masignani V, Hultenby K, Taddei AR, Beiter K, Wartha F, von Euler A, Covacci A, Holden DW, Normark S, Rappuoli R, Henriques-Normark B. 2006. A pneumococcal pilus influences virulence and host inflammatory responses. Proc Natl Acad Sci U S A 103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Springer TA. 1990. Adhesion receptors of the immune-system. Nature 346:425–434. doi: 10.1038/346425a0. [DOI] [PubMed] [Google Scholar]
- 61.Ehirchiou D, Xiong Y, Xu GW, Chen WJ, Shi YF, Zhang L. 2007. CD11b facilitates the development of peripheral tolerance by suppressing Th17 differentiation. J Exp Med 204:1519–1524. doi: 10.1084/jem.20062292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chau TA, McCully ML, Brintnell W, An G, Kasper KJ, Vines ED, Kubes P, Haeryfar SM, McCormick JK, Cairns E, Heinrichs DE, Madrenas J. 2009. Toll-like receptor 2 ligands on the staphylococcal cell wall downregulate superantigen-induced T cell activation and prevent toxic shock syndrome. Nat Med 15:641–648. doi: 10.1038/nm.1965. [DOI] [PubMed] [Google Scholar]
- 63.Peres AG, Madrenas J. 2013. The broad landscape of immune interactions with Staphylococcus aureus: from commensalism to lethal infections. Burns 39:380–388. doi: 10.1016/j.burns.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 64.Francius G, Lebeer S, Alsteens D, Wildling L, Gruber HJ, Hols P, De Keersmaecker S, Vanderleyden J, Dufrene YF. 2008. Detection, localization, and conformational analysis of single polysaccharide molecules on live bacteria. ACS Nano 2:1921–1929. doi: 10.1021/nn800341b. [DOI] [PubMed] [Google Scholar]
- 65.Reunanen J, von Ossowski I, Hendrickx AP, Palva A, de Vos WM. 2012. Characterization of the SpaCBA pilus fibers in the probiotic Lactobacillus rhamnosus GG. Appl Environ Microbiol 78:2337–2344. doi: 10.1128/AEM.07047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tripathi P, Dupres V, Beaussart A, Lebeer S, Claes IJ, Vanderleyden J, Dufrene YF. 2012. Deciphering the nanometer-scale organization and assembly of Lactobacillus rhamnosus GG pili using atomic force microscopy. Langmuir 28:2211–2216. doi: 10.1021/la203834d. [DOI] [PubMed] [Google Scholar]
- 67.Bernardo D, Sánchez B, Al-Hassi HO, Mann ER, Urdaci MC, Knight SC, Margolles A. 2012. Microbiota/host crosstalk biomarkers: regulatory response of human intestinal dendritic cells exposed to Lactobacillus extracellular encrypted peptide. PLoS One 7(5):e36262. doi: 10.1371/journal.pone.0036262. [DOI] [PMC free article] [PubMed] [Google Scholar]