Abstract
Enteric bacteria encounter a wide range of pHs throughout the human intestinal tract. We conducted experimental evolution of Escherichia coli K-12 to isolate clones with increased fitness during growth under acidic conditions (pH 4.5 to 4.8). Twenty-four independent populations of E. coli K-12 W3110 were evolved in LBK medium (10 g/liter tryptone, 5 g/liter yeast extract, 7.45 g/liter KCl) buffered with homopiperazine-N,N′-bis-2-(ethanosulfonic acid) and malate at pH 4.8. At generation 730, the pH was decreased to 4.6 with HCl. By 2,000 generations, all populations had achieved higher endpoint growth than the ancestor at pH 4.6 but not at pH 7.0. All evolving populations showed a progressive loss of activity of lysine decarboxylase (CadA), a major acid stress enzyme. This finding suggests a surprising association between acid adaptation and moderation of an acid stress response. At generation 2,000, eight clones were isolated from four populations, and their genomes were sequenced. Each clone showed between three and eight missense mutations, including one in a subunit of the RNA polymerase holoenzyme (rpoB, rpoC, or rpoD). Missense mutations were found in adiY, the activator of the acid-inducible arginine decarboxylase (adiA), and in gcvP (glycine decarboxylase), a possible acid stress component. For tests of fitness relative to that of the ancestor, lacZ::kan was transduced into each strain. All acid-evolved clones showed a high fitness advantage at pH 4.6. With the cytoplasmic pH depressed by benzoate (at external pH 6.5), acid-evolved clones showed decreased fitness; thus, there was no adaptation to cytoplasmic pH depression. At pH 9.0, acid-evolved clones showed no fitness advantage. Thus, our acid-evolved clones showed a fitness increase specific to low external pH.
INTRODUCTION
Enteric bacteria experience a wide range of pHs throughout the human intestinal tract: from pH 1.5 to 3.5 in the stomach, then a rise to pH 4 to 7 in the duodenum and pH 7 to 9 in the jejunum, and then a decline to pH 5 to 6 in the cecum (1, 2). Escherichia coli has evolved to grow over a relatively wide range of intestinal pHs from pH 4.5 to 9.0 (3–5) and to survive without growth for several hours at pH 2 (6–8; for reviews of acid stress and extreme-acid survival, see references 9 to 11). The ability to survive the low pH of the stomach—and enter rapid growth as the pH increases in the lower digestive tract—may contribute to the low infectious doses characteristic of enteric pathogens, such as E. coli O157:H7 (12).
The adaptations of E. coli to growth in mild acid (pH 5 to 6) have been studied previously (9–11). Growth in mild acid may be enhanced by components of metabolism and transport, such as the lysine and arginine decarboxylases (13, 14), and by cyclopropane fatty acid synthase (8, 15, 16). A transcriptomic study of growth at pH 5 versus pH 7 shows that acid upregulates proton pumps of the electron transport system and outer membrane proteins, the chemotaxis regulon for proton-driven motility, periplasmic acid chaperones hdeA and hdeB, the mar multidrug resistance regulon, oxidative stress genes, and numerous genes with unidentified products (17).
Nevertheless, we still know little about how E. coli grows below pH 5, at the lower limit of its pH range, just above the range of nongrowth survival (pH 1.5 to 4.0). The range of pHs occurring during the transition from nongrowth to growth conditions is experienced by bacteria as they pass through the stomach and enter the intestine. Understanding growth in and adaptation to this lower range of growth pH is particularly important, because the gut microbiome may include hundreds of competing species (18, 19) and the ability of bacteria to outgrow their neighbors at pH 4 to 5 could influence which strains colonize the gut. The same pH range also occurs in many fruit juices vulnerable to bacterial contamination (20).
To investigate genes that impact growth below pH 5, we used the approach of experimental evolution (21, 22). In an evolution experiment, multiple replicates of a single microbial lineage are repeatedly diluted and cultured under specific regulated conditions (23, 24). If the specified environment deviates from the optimal conditions for peak fitness, then there exists strong selective pressure for mutant alleles that enhance fitness in the new environment. E. coli has been the subject of a number of evolution studies involving adaptation to high temperatures (25), high ethanol concentrations (26), growth in complex media (27), and resistance to antibiotics (28).
Experimental evolution offers several advantages for the study of bacterial stress responses (29, 30). First, the approach allows the researcher to tightly regulate the microbe's environment for thousands of generations, which allows multiple selective sweeps of potentially unpredicted mutations to spread through the population. The evolving populations can be frozen regularly and stored indefinitely, allowing the researcher to set up direct comparisons between the ancestral strain and the adapted strains. Whole-genome resequencing can then permit identification of mutations in genes relevant to acid stress. Such an analysis might reveal surprising mechanisms of the stress response that differ from those identified by traditional gene-knockout investigation. Finally, experimental evolution allows us to determine whether acid adaptation comes at the price of a fitness trade-off in other environments.
Previous experimental evolution studies of acid stress are limited. E. coli experimental evolution in moderate acid (pH 5.3) yields an acid-dependent fitness increase, but genome sequence analysis has not been used to identify the underlying genetic changes (31). In another study, populations that evolved for 1,000 generations with 20 rounds of exposure to pH 2.5 showed 20 new mutations, including mutations in evgS alleles that relieve log-phase repression of extreme-acid resistance (32). Here we report on the isolation and characterization of eight E. coli strains from 4 of 24 populations cultured at pH 4.8 (for 730 generations) and then at pH 4.6 (2,000 generations in total). We found putative novel contributors to the acid stress response, including a strong selection for altered RNA polymerase.
MATERIALS AND METHODS
Bacterial strains and media.
Escherichia coli K-12 W3110 was the ancestral strain of all acid-adapted populations (33). Additional strains derived from E. coli K-12 W3110 were isolated during the course of the experiment (Table 1). For competition assays assessing the relative fitness of ancestral and adapted strains, a lacZ::kan insertion allele was introduced into each strain by P1 phage transduction from Keio collection strain GJ1886 (Coli Genetic Stock Center, Yale University) (34). The lacZ::kan insertion strains were maintained with 50 μg/ml kanamycin.
TABLE 1.
Strains isolated after 2,000 generationsa
| Strain | Population | Isolate no. |
|---|---|---|
| JLSE0079 | H9 | 1 |
| JLSE0080 | H9 | 2 |
| JLES0083 | B11 | 1 |
| JLSE0084 | B11 | 2 |
| JLSE0091 | F11 | 1 |
| JLSE0092 | F11 | 2 |
| JLSE0137 | F9 | 2 |
| JLSE0138 | F9 | 3 |
The sequence data are available in the NCBI SRA under accession number SRP041420.
All bacterial cultures were grown in media based on LBK medium (10 g/liter tryptone, 5 g/liter yeast extract, 7.45 g/liter KCl) (17). The culture media were buffered with either 100 mM homopiperazine-N,N′-bis-2-(ethanosulfonic acid) (HOMOPIPES; pKa = 4.55, 8.12), 100 mM 2-(N-morpholino)ethanesulfonic acid (MES; pKa = 5.96), or 100 mM 3-morpholinopropane-1-sulfonic acid (MOPS; pKa = 7.20), depending upon the desired pH range of 4.4 to 4.8, 5.5 to 6.5, or 7.0, respectively (17). The pKa values presented above are those found at 25°C. For media buffered to pH 4.5 to 4.8, the LBK medium was supplemented with 10 g/liter d/l-malic acid (pKa = 3.40, 5.11) (LBKmal). The pH of the medium was adjusted as necessary with either 5 M HCl or 5 M KOH. For all experiments, the pH of the medium was tested immediately before and after bacterial growth.
Experimental evolution.
A protocol based on successful evolution protocols described in the literature (22, 25, 27) was devised for experimental evolution of E. coli growth at external pH values below 5. Twenty-four populations were established from the same parental freezer stock of strain W3110. Evolving populations (200 μl each) were maintained in 96-well plates in a microplate reader. In each plate, 24 evolving populations occupied three columns of eight wells, with the columns being separated by an empty column to minimize the possibility of cross contamination. The initial inoculation of each well was with a 1:100 dilution of an overnight culture of the ancestral strain W3110. Populations were evolved in LBKmal with 100 mM HOMOPIPES at pH 4.8 (with daily dilutions of 1:4,000) for approximately 730 generations. The number of generations was estimated using the assumption that a dilution of 1:4,000 allows 12 generations of binary fission per day. After generation 730, the pH of the dilution medium was adjusted to 4.6 and the populations evolved for a further 1,270 generations (daily dilution, 1:100). A dilution of 1:100 allows 100-fold growth daily, producing 6.64 generations. The well plate was maintained at 37°C in a SpectraMax Plus384 MicroPlate reader (Molecular Devices). For the initial 22 h of the incubation, the well plate was shaken for 3 s every 15 min, after which the optical density at 450 nm (OD450) of each population was recorded. After 22 h, the plate was maintained at 37°C without shaking for 1 to 4 h until the next daily dilution.
For the first 730 generations, at the end of each daily cycle the endpoint cultures were supplemented with 100 μl glycerol (50%, vol/vol) and stored at −80°C. In the event that the cultures needed to be restarted from the freezer, aliquots of the most recent frozen cultures were diluted 1:50 into sterile growth medium on the first day and were then diluted as normal on the following days. After 730 generations (61 daily dilutions) of continuous growth in moderate acid, the pH of the LBKmal growth medium was lowered to 4.6. The daily dilution was decreased to 1:100, to decrease the population bottleneck (23). The glycerol solution for storing cultures was buffered with 100 mM MES, and the pH was adjusted to 6.5, to decrease the stress of the storage condition. All other aspects of the protocol remained unchanged.
After a cumulative 2,000 generations of evolution (61 dilutions at 1:4,000, 190 dilutions at 1:100), four populations were selected at random and streaked out on LBK agar. From each population, two clones were selected for genome sequencing (Table 1).
Endpoint growth measurement at pH 4.6.
As one measure of evolving fitness, the endpoint growth level (culture density at stationary phase) was measured for cultures derived from each of the 24 experimental populations. The reference populations were chosen after approximately 25, 730, and 2,000 generations. Reference populations were thawed, diluted 1:50 in LBKmal at pH 4.8 (100 mM HOMOPIPES), and placed into rows of wells (volume, 200 μl) in the same microplate. The cultures were then incubated in a well plate at 37°C in a SpectraMax Plus384 MicroPlate reader (Molecular Devices) for 22 h. Every 15 min for the duration of the growth period, the well plate was shaken for 3 s, after which the OD450 of each population was recorded. After 22 h, each population was diluted 1:100 into pH 4.6 medium for growth. A total of three serial dilutions at pH 4.6 (1:100) and 22-h incubations were performed. After each growth period, the endpoint values of the OD450 were recorded and averaged.
Lysine decarboxylase assays.
Isolated colonies were tested for lysine decarboxylase and other catabolic enzyme activities using an Entero-Pluri-Test indicator battery (Liofilchem). For large-scale screening, lysine decarboxylase was assayed by spotting an isolated colony into 200 μl Moeller decarboxylase broth, modified from the procedure of the company BD (BBL, 2011). The medium contained 5 g/liter l-lysine, 1 g/liter glucose, 3 g/liter yeast extract, and 15 mg/liter bromocresol purple. Samples were incubated at 37°C in the wells of a microplate; the plate was sealed for anaerobiosis. After 24 h, the plate was inspected visually. A purple color (alkalization) indicated positive lysine decarboxylase activity, whereas a yellow color indicated low or absent activity. Visual inspection was confirmed by analysis of the absorbance readings recorded for each well at 340 nm and 590 nm using the SpectraMax Plus384 microplate reader (Molecular Devices).
Whole-genome sequencing.
The genomes of the ancestral and evolved strains were sequenced. For each strain, the total DNA was extracted using a DNeasy DNA extraction kit (Qiagen). The DNA purity was confirmed by measuring the ratios of the absorbance at 260 nm/280 nm and 260 nm/230 nm. The genomes were sequenced with an Illumina MiSeq sequencer at the Michigan State University Research Technology Support Facility. Sequence reads were mapped to the E. coli K-12 W3110 reference genome (GenBank accession number NC_007779.1) (35). The quality scores (Q) for the sequences ranged from 25 to 33, with the average Q value being 30.8; this value corresponds to an error rate of 1/1,000. Sequences with 30-fold coverage or greater were accepted for mutation calls. Mutations were predicted with the breseq computational pipeline (http://barricklab.org/breseq) (22).
Competition assays.
Evolved clones were assayed for changes in fitness relative to that of the ancestral strain (36). Competitors were differentiated in coculture on the basis of lac marker states. Each evolved strain (lac-positive [lac+] phenotype) was cocultured with the lac-negative construct of the ancestral wild type (W3110 lacZ::kan) at pH 4.6. To ensure that the lac-negative phenotype did not confer a fitness advantage or disadvantage in the relevant environment, a competition assay between the lac-negative ancestor (W3110 lacZ::kan) and the original ancestor (W3110) was carried out simultaneously with each experiment. The two strains were cultured separately overnight at 37°C for 24 h with aeration in LBKmal at pH 4.8 (100 mM HOMOPIPES). The cultures were rediluted 1:100 into pH 4.8 media and incubated for an additional 16 to 18 h with aeration.
To commence competition, 125 μl of an overnight culture from a lac-negative strain and 125 μl of an overnight culture from a lac+ strain were combined and diluted (100-fold) in 25 ml of LBKmal at pH 4.6 (100 mM HOMOPIPES). Samples were immediately serially diluted in M63 minimal medium [0.4 g/liter KH2PO4, 0.4 g/liter K2HPO4, 2 g/liter (NH4)2SO4, 7.45 g/liter KCl] and plated on LBK agar supplemented with ChromoMax IPTG (isopropyl-β-d-thiogalactopyranoside)–X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) solution (4 μl/ml; Fisher). Cultures were incubated at 37°C with aeration for 24 h, and then the samples were again diluted and plated. The colony counts of white (lac-negative) and blue (lac+) colonies were used to calculate the total number of doublings for each competitor (31). The fitness of the evolved clone was compared to that of the ancestor by finding the difference between the two clones' number of doublings. The relative fitness (W) was calculated as [log2 100(Nt2) − log2(N02)]/[log2 100(Nt1) − log2(N01)], where N01 is the colony count for the ancestor in the mixed culture before dilution, Nt1 is the colony count for the ancestor after 100-fold dilution and 24 h of growth, N02 is the colony count for the evolved clone in the mixed culture before dilution, and Nt2 is the colony count for the evolved clone after 100-fold dilution and 24 h of growth (36). Our colony counts allowed calculation of W over values up to 3; values higher than 3 were cut off at 3, because the low colony counts of the outcompeted strain caused high levels of variation.
Competition assays were carried out with 4- to 6-fold replication. For each competition, we included an equal number of trials in which the ancestor contained lacZ::kan and an equal number of trials in which the acid-adapted strain contained lacZ::kan. Similar fitness competitions were performed using the growth media LBK medium with 100 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.5, and 15 mM benzoic acid and LBK medium with 150 mM TAPS (3-{[1,3-dihydroxy-2-(hydroxymethyl)-2-propanyl]amino}-1-propanesulfonic acid), pH 9.0.
Log-phase growth of isolates at pH 4.5.
Strains were cultured initially in LBKmal buffered to pH 4.8 (100 mM HOMOPIPES) at 37°C for 24 h with aeration. For initial overnight cultures, pH 4.8 was selected to allow physiological acclimation and obtain consistent culture densities for the adapted and ancestral clones. Overnight cultures were rediluted 1:100 in fresh overnight medium and grown for 16 to 18 h. Triplicate cultures of 15 ml LBKmal at pH 4.5 (100 mM HOMOPIPES) were inoculated via a 1:200 dilution of the overnight culture, and the initial OD600 of each culture was recorded. The cultures were incubated with aeration in a water bath at 37°C. The OD600 was measured after 1 h and then subsequently at approximately 20-min intervals until the cultures began to enter stationary phase. The growth rate of each culture was calculated with respect to the time period corresponding to log-phase growth (37).
Cotransductant strains and competition assays at pH 4.6.
A thiG761::kan insertion (80 to 90% linked to rpoC) was introduced into an evolved clone containing an rpoC mutation (clone B11-1) by P1 phage transduction using strain JW5549-1 (Keio collection). Cotransduction with the linked kan marker replaced the mutated region of the gene with the ancestral sequence. The replacement was confirmed by PCR amplification and sequencing of the gene. The forward primer sequence used was 5′-CCTCGGAGATGATTTCGTG-3′, and the reverse primer sequence used was 5′-GAGCTGTTCAAACCGTTCATC-3′ (844 bp between primers). The cotransductant strain was competed against the parental clone in LBKmal at pH 4.6 following the same protocol described above.
Nucleotide sequence accession number.
Sequence data have been deposited in the NCBI Sequence Read Archive (SRA) under accession number SRP041420.
RESULTS
Adaptation of 24 experimental E. coli populations at pH 4.6.
We established culture conditions for maintaining experimental E. coli lineages below pH 5. In LBK medium, E. coli catabolizes diverse metabolites to optimize growth at low pH (9). The added components, HOMOPIPES and d/l-malate (LBKmal), buffered the acidity below pH 5. Both d- and l-malate may be catabolized by reactions that consume acid (38, 39) and thus potentially supply the experimental lineages with a novel means of enhancing acid tolerance. Using this medium, LBKmal, we established that our laboratory strain of E. coli K-12 W3110 could grow consistently in a microplate with the medium adjusted to pH 4.8.
Twenty-four bacterial populations were evolved under a daily serial dilution regime in 96-well microtiter plates in LBK medium supplemented with HOMOPIPES and d/l-malate, buffered initially at pH 4.8, as described in Materials and Methods. After 730 generations of growth, trial experiments of three successive 1:100 dilutions revealed that cultures of all experimental populations could be maintained at pH 4.6. We therefore decreased the pH of the culture conditions to pH 4.6 to provide even greater selective pressure on the experimental populations. We also decreased the daily dilution factor to 1:100, in order to decrease the daily population bottleneck and thus sample a greater number of possible mutations.
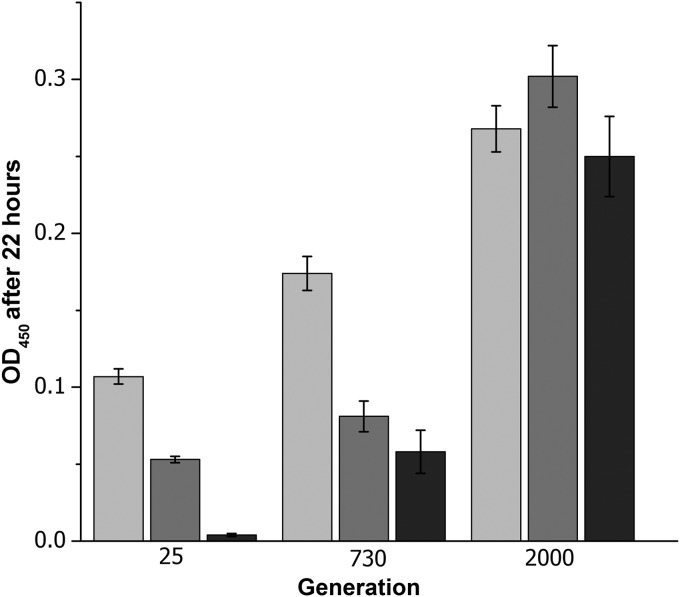
Over the course of the experiment, we observed significant increases in the endpoint culture density of all evolving populations, consistent with adaptation to low pH (Fig. 1). We measured the endpoint culture density of experimental populations sampled from populations frozen at several different generation numbers (generations 25, 730, and 2,000). The 25-generation plate (the second 4,000-fold dilution plate after the ancestor) was chosen instead of the ancestral strain, in order to test cultures frozen in plates under equivalent conditions. By generation 730, the acid-evolved populations showed an increased ability to grow at pH 4.6, although their endpoint densities declined by the third daily dilution. Comparison of the results by the t test to determine statistically significant differences (P < 0.001) verified the increase in endpoint growth between generations 25 and 730 and between generations 730 and 2,000 (evolved at pH 4.6). By generation 2,000, all populations reached endpoint densities higher than those at generations 25 and 730. Furthermore, the 2,000-generation populations grew consistently over three serial dilutions at pH 4.6; a t test showed no difference among the endpoint reads for the three successive dilutions (P = 0.54). Similar experiments conducted at pH 7.0 and at pH 9.0 showed no fitness advantage or disadvantage (data not shown). Thus, the growth advantage of the evolved populations was specific to pH 4.6, and the evolved populations incurred no fitness disadvantage at high pH.
FIG 1.
Growth of evolved populations at pH 4.6. The endpoint growth of the 24 populations from generations 25, 730, and 2,000 was compared. Frozen populations were thawed and diluted 1:50 in LBKmal, pH 4.8, for initial recovery and then incubated in a microplate reader for 22 h. Culture was conducted for three subsequent days by daily dilution 100-fold at pH 4.6. On each day, cultures were incubated at 37°C in a microplate reader with shaking every 15 min. The final OD450 was measured after 22 h. Error bars represent SEMs (n = 24).
Downregulation of lysine decarboxylase activity.
E. coli bacteria respond to acid stress through various shifts in metabolism, such as upregulation of the lysine and arginine decarboxylases (13, 40). We therefore screened isolates from our evolving populations for several metabolic enzyme activities, using the Entero-Pluri-Test. Several of our 2,000-generation clones tested negative for the activity of the degradative lysine decarboxylase (CadA). This was surprising, given that in the ancestor, the gene encoding CadA is one of the genes that is the most highly induced by acid (13, 41).
We investigated the loss of lysine decarboxylase activity by screening three generations of our 24 evolving populations for enzyme activity (Table 2). For each generation number (760, 1,400, 2,000), three isolates from each of the 24 populations were screened for lysine decarboxylase activity after 24 h growth in Moeller broth with lysine. In this assay, the ancestor W3110 consistently tested positive (purple), and isolates with a cadA deletion consistently tested negative (yellow). Among the sampled populations, we observed a progressive decline in the prevalence of decarboxylase-positive clones. At generation 760, only one isolate (one of three isolates from population B7) tested negative. At generation 1,400, 26 isolates from 15 populations tested negative for lysine decarboxylase function, and by generation 2,000, all but one isolate (an isolate from population A11) tested negative. Further screening of the populations revealed a minority of decarboxylase-positive isolates remaining in several populations, such as population F9 (data not shown).
TABLE 2.
Lysine decarboxylase activity of clones from acid-evolved populationsa
| No. of generations | No. of isolates in the following population testing negative for lysine decarboxylase activity: |
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A7 | B7 | C7 | D7 | E7 | F7 | G7 | H7 | A9 | B9 | C9 | D9 | E9 | F9 | G9 | H9 | A11 | B11 | C11 | D11 | E11 | F11 | G11 | H11 | |
| 760 | 1 | |||||||||||||||||||||||
| 1,400 | 1 | 2 | 3 | 2 | 1 | 1 | 2 | 2 | 3 | 1 | 2 | 3 | 1 | 1 | 1 | |||||||||
| 2,000 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
For each generation, three isolates from each population were assayed. The number of isolates of three isolates tested that lost lysine decarboxylase activity (indicated by a yellow color in Moeller broth) after 24 h of culture in a microplate is indicated.
Upon further incubation at 37°C, all sampled clones subsequently tested positive after 2 or 3 days. This observation suggests that the enzyme was present but downregulated. Indeed, genome sequencing subsequently confirmed that the cadA gene and its known regulators showed no mutations in the downregulated strains (see “Genomic analysis of acid-evolved isolates” below). The basis of cadA downregulation remains to be elucidated, but the occurrence of cadA downregulation in all 24 populations suggests a beneficial fitness effect under our acid stress conditions.
Genomic analysis of acid-evolved isolates.
To identify the genetic basis of the improved fitness of the evolved clones at low pH, we sequenced the genomes of eight clones from generation 2,000 (SRA accession number SRP041420). Two clones were isolated from each of four experimental populations, populations H9, B11, F11, and F9. Among the sequenced clones, two (clones F9-2 and F9-3 from population F9) displayed lysine decarboxylase activity comparable to that of the ancestor; the other six evolved clones tested negative.
For comparison with our acid-evolved clones, we resequenced the ancestral strain W3110 from our original reference freezer stock (D8-W), as well as a clone from a second freezer stock in our collection (D13-W) (see Table S1 in the supplemental material). Our two W3110 clones showed nearly identical sequences. However, several differences appeared between the genome of our lab strain and the sequenced E. coli K-12 W3110 reference genome (ASM1024v1). For example, our lab strain showed deletions of the insD-insC and insH-alsK regions. Also, a number of single nucleotide polymorphisms (SNPs) appeared in the icd coding sequence. The source of the unusual icd variability is unknown.
Mutations were identified in the sequenced genomes by aligning each isolate's raw reads with the E. coli K-12 W3110 reference genome (35) using the breseq computational pipeline (http://barricklab.org/breseq) (24). Mutation calls were accepted for sites showing 30-fold or greater coverage. Each acid-adapted strain had between two and six nonsynonymous mutations. Among the evolved genomes, we identified 24 mutations in coding regions (Table 3), which included 20 missense mutations, 2 single-base insertions (hepA, mppA), 1 9-bp insertion (yejM), and 1 silent mutation (yjiK). The high ratio of nonsynonymous mutations to synonymous mutations across the evolved isolates suggests that these mutations were likely to be adaptive under the experimental conditions (42). An additional 12 mutations in intergenic regions were also identified (Table 4). Pairs of clones isolated from the same population typically displayed both shared and unshared mutations; the shared mutations presumably arose earlier in the ancestor common to that population. Only one mutation (yejM insertion) that was clearly absent from the ancestor occurred in two acid-evolved populations. Multiple mutations were found in icd, but numerous SNPs in icd were found in the ancestral stock, suggesting a mutation frequency unrelated to the evolution experiment. The diversity of our evolved clones suggests that our remaining populations, of the original 24, contained further mutations of interest.
TABLE 3.
Mutations identified in coding regions in the 2,000th generation clonesa
| Gene | Function | Mutation | Identification of a mutation in clone: |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| H9-1 | H9-2 | B11-1 | B11-2 | F11-1 | F11-2 | F9-2 | F9-3 | |||
| adiY | Activator of adiA | Missense (R119P) | X | X | ||||||
| flu | AG43 transport (biofilm) | Missense (A162E) | X | X | ||||||
| ftsX | Cell division (IM transporter) | Missense (A101V) | X | X | ||||||
| fucI | l-Fucose isomerase | Missense (R363S) | X | X | ||||||
| gcvP | Glycine decarboxylase | Missense (T854A) | X | |||||||
| hepA | RNAP recycling factor | Insertion (1139, +G) | X | X | ||||||
| icd | Isocitrate dehydrogenase | Missense (D398E) | X | X | X | X | X | X | ||
| menH | Menaquinone biosynthesis | Missense (D79Y) | X | X | ||||||
| metE | Homocysteine transmethylase | Missense (R337S) | X | |||||||
| mhpB | Phenylpropionate degradation | Missense (D226Y) | X | |||||||
| mppA | Murein tripeptide transporter | Insertion (970, +A) | X | X | ||||||
| msbA | Lipid A transport IM to OM | Missense (G150C) | X | X | ||||||
| rlmE | 23S rRNA methyltransferase | Missense (P157L) | X | |||||||
| rpoB | RNAP beta subunit | Missense (A679V) | X | X | ||||||
| rpoC | RNAP beta prime subunit | Missense (I774S) | X | X | ||||||
| rpoC | RNAP beta prime subunit | Missense (V507L) | X | X | ||||||
| rpoD | RNAP sigma 70 | Missense (M273I) | X | X | ||||||
| spr | Murein endopeptidase | Missense (Q73K) | X | |||||||
| wzzE | ECA polysaccharide chain length | Missense (P314Q) | X | X | ||||||
| yeiH | IM protein | Missense (A245S) | X | |||||||
| yejM | IM hydrolase | Insertion (position 531) of 9 bp (TTTATCGCC) | X | X | X | X | ||||
| yfdI | IM protein | Missense (V7L) | X | |||||||
| yfjK | Ionizing radiation resistance | Missense (T400I) | X | |||||||
| yjiK | IM protein | Silent (TCG → TCA) | X | |||||||
The genomes of the experimental clones and the ancestral strain were sequenced and aligned with the sequence of the E. coli K-12 W3110 reference genome (35) using the breseq computational pipeline (http://barricklab.org/breseq). The mutations listed represent differences between the genome of the ancestor and the adapted clones, excluding intergenic mutations. An X signifies that the mutation is present in that clone. IM, inner membrane; OM, outer membrane; ECA, enterobacterial common antigen.
TABLE 4.
Intergenic mutations identified in the 2,000th generation acid-adapted clonesa
| Proximal gene(s) | Mutation (positions, nucleotide change) | Identification of a mutation in clone: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| H9-1 | H9-2 | B11-1 | B11-2 | F11-1 | F11-2 | F9-2 | F9-3 | ||
| matP →/← ompA | Intergenic (+40/+36, C to A) | X | |||||||
| racR ←/→ ydaS | Intergenic (−61/−63, C to A) | X | |||||||
| Y75_p4289 ←/→ eaeH | Intergenic (−545/−1035, C to A) | X | |||||||
| proL → | Noncoding (57/77 nt, C to A) | X | X | ||||||
| nuoA ←/← lrhA | Intergenic (−87/+544, C to A) | X | X | ||||||
| yfjL ←/← yfjM | Intergenic (−258/+102) +8 bp (GCACTATG) | X | |||||||
| rhaA →/→ rhaD | Intergenic (+308/−143, C to A) | X | X | ||||||
| rrsC ← | Noncoding (1417/1542 nt, +A) | X | X | ||||||
| pabA →/→ argD | Intergenic (+34/−52, A to G) | X | X | ||||||
| yjbI →/→ ubiC | Intergenic (+84/−139, G to T) | X | X | ||||||
| dcuA ←/← aspA | Intergenic (−202/+104, G to T) | X | X | ||||||
| acs ←/→ nrfA | Intergenic (−228/−165, G to A) | X | X | ||||||
The mutations listed indicate intergenic differences between the adapted clones and the ancestral wild type. A positive distance from a proximal gene represents the downstream location of the mutation relative to the gene's translation stop site, whereas a negative distance represents the mutation's upstream location relative to the gene's translation start site. nt, nucleotide position number.
Interestingly, only one of the sequenced mutations occurred in a gene with a well-known role in acid stress tolerance, adiY, the activator of the acid-inducible arginine decarboxylase (AdiA). The adiY mutation appeared in the two isolates of population B11 (B11-1 and B11-2). The other mutations that we found are likely to yield previously unrecognized mechanisms of acid stress response.
Every evolved genome had one mutation in a subunit of the RNA polymerase (RNAP) holoenzyme (43). The specific RNAP component that was affected differed among the populations. Population H9 isolates shared a mutation in rpoB (beta subunit; A679V), whereas B11 and F9 isolates each showed mutations in rpoC (beta prime subunit; V507L and I774S, respectively.) The rpoC beta prime subunit I774S mutation maps near the beta/beta prime interface and confers resistance to the CBR703 series of RNAP inhibitors (44). The F9 isolates shared a mutation in rpoD (housekeeping sigma 70; M273I). In each population, the RNAP mutation was shared by both clones, a result that would be consistent with early fixation in the evolution experiment. Besides the RNAP subunit mutations, the B11 strains also had a frameshift mutation in the HepA (RapA) helicase, which is involved in recycling RNAP and in UV resistance (45, 46).
Relative fitness of 2,000th generation isolates at pH 4.6.
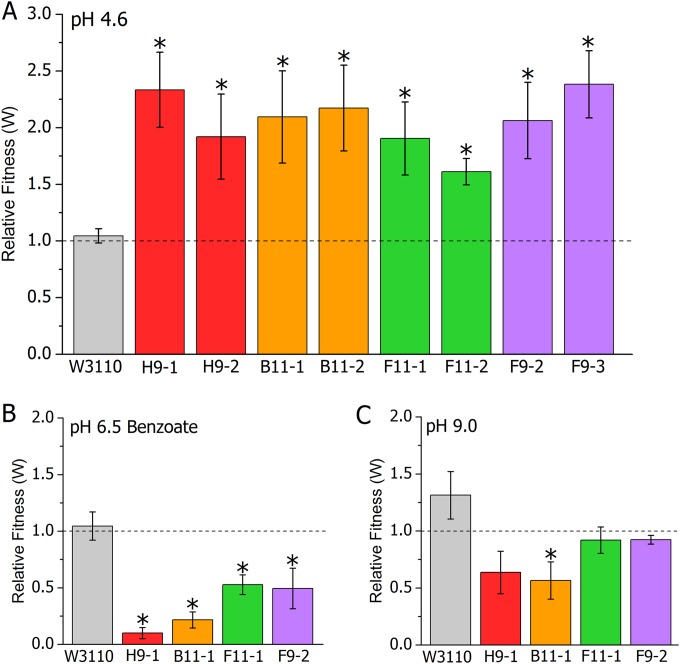
Fitness competitions were performed on our eight sequenced clones, in order to measure their relative fitness gains under acidic conditions. We cocultured the evolved clones against the ancestor over a 24-h growth period in LBKmal buffered to pH 4.6. Competitor colonies were counted before and after growth by plating on X-Gal agar, on which the different lac marker states could be discerned. The relative fitness of each evolved isolate was measured by determining the number of generations that each strain accumulated during the 24-h period and then dividing the isolate's number of doublings by that of the ancestor. Thus, a value of W of greater than 1 indicates that the lac+ isolate had a fitness advantage, whereas a value of W of less than 1 indicates that the lac-negative ancestor had a fitness advantage. For each isolate, equal numbers of competitions in which the lac phenotypes were reversed (lac+ isolate versus lac-negative ancestor) were performed. In order to confirm the neutrality of the lac-negative marker, the lac-negative ancestor was also competed with the lac+ ancestor. The mean relative fitness of W3110 versus W3110 lacZ::kan was not significantly different (one-sample t test) from 1.0 and was a mean of 1.045 ± 0.07 with 99% confidence.
All eight evolved clones outcompeted the ancestor and displayed relative fitness values of 1.5 or higher (Fig. 2A). The magnitude of the fitness advantage varied among the clones, with a somewhat lower fitness advantage being noted in the population F11 isolates, but all isolates outcompeted the ancestor. These fitness values are notably larger than those typically reported for bacterial evolution experiments, including those conducted at pH 5.3 (31).
FIG 2.
Fitness changes in acid-evolved clones. For each pair of strains, the relative fitness (W) was calculated as described in Materials and Methods. To control for any fitness effect of the transduced lac-negative allele, the original lac+ ancestral wild type was cocultured with the lac-negative wild type (gray bars). (A) pH 4.6. Clones were serially diluted (1:200) in LBKmal buffered to pH 4.8 and incubated overnight twice prior to each experiment. Each isolate was then cocultured with a lac-negative variant of the ancestral wild type (or else the lac-negative isolate was cocultured with a lac+ ancestor) for 24 h in LBKmal buffered to pH 4.6. Error bars represent SEMs (n = 6 for evolved strains, n = 18 for ancestral strain W3110). (B) Benzoate. Strains were serially diluted 1:200 and incubated overnight twice in LBK medium buffered to pH 6.5. For the competition, each isolate and ancestor were cocultured for 24 h in LBK medium with 15 mM benzoic acid buffered with 100 mM PIPES at pH 6.5 (n = 4 for evolved strains, n = 8 for W3110) (C) pH 9.0. Strains were serially diluted and incubated overnight twice in LBK medium buffered to pH 8.5. For the competition, each isolate and ancestor were cocultured for 24 h in LBK medium buffered with 150 mM TAPS at pH 9.0 (n = 4 for evolved strains, n = 8 for W3110). *, P ≤ 0.05 (t test) in comparison with the results for W3110.
Fitness with cytoplasmic pH depressed by benzoate.
The observed improved fitness of the evolved clones at pH 4.6 might be due to increased tolerance for depressed cytoplasmic pH, which occurs at values of external pH below 5 (9). To test this hypothesis, we measured the fitness of four evolved clones (one from each of the pairs described above) relative to that of the ancestor in a medium buffered to 6.5 but containing a permeant acid (benzoic acid, benzoate) which depresses the cytoplasmic pH (Fig. 2B). All four evolved clones showed a loss of fitness under these conditions, indicating that their improved fitness at pH 4.6 is not due to increased tolerance of the lowered cytoplasmic pH.
Fitness at pH 9.0.
Fitness improvement in one environment often comes at the cost of reduced fitness in another (23). We sought to determine if the improved fitness of the evolved clones at low pH carried the trade-off of reduced fitness at high pH. To test this possibility, we measured the fitness of four of the evolved clones relative to that of the ancestor in a medium buffered to pH 9 (Fig. 2C). The evolved clones showed insignificant to slightly negative fitness changes, in addition to displaying variation among replicates greater than that observed at low pH. This result shows that the improved fitness of our acid-evolved clones is specific to growth at low pH, with little to no correlated fitness change occurring at high pH.
Early-log-phase growth of 2,000th generation isolates at pH 4.6.
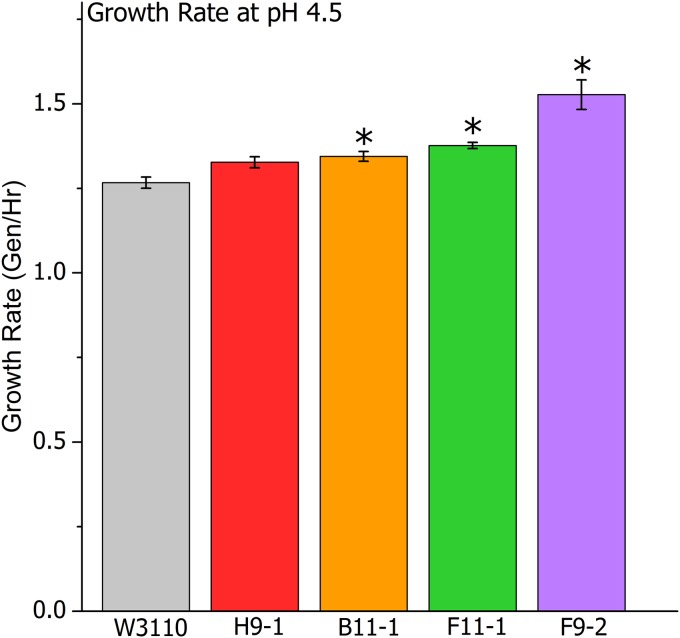
The relative fitness of bacterial strains evolved in our microplates could be due to various factors, including the culture density reached by stationary phase (Fig. 1) as well as the rate of growth in log phase. To separate these factors, we measured the growth rates of our acid-evolved isolates under conditions conducive to optimal log-phase growth (rotary shaking of baffled flasks). We cultured four isolates (representing the four populations) in LBKmal buffered to pH 4.5 and compared their early-log-phase growth rate to that of their ancestor (Fig. 3). The four isolates showed a significant increase in the log-phase growth rate compared to that of the ancestor. When cultured at pH 7, however, the log-phase growth rate of each strain was comparable to that of the ancestral wild type (data not shown).
FIG 3.
Early-log-phase growth rates of acid-evolved clones in LBKmal buffered to pH 4.5. Clones were twice serially diluted (100-fold) and cultured overnight in LBKmal buffered to pH 4.8. Cultures were then diluted 1:200 into LBK medium buffered to pH 4.5 and then grown until mid-log phase at 37°C under aerated conditions. Growth rates were calculated from OD600 measurements taken every 20 min throughout early-log-phase growth. Error bars indicate SEMs (n = 3). *, significant differences (P < 0.05) from the results for ancestor strain W3110, based on t test.
Relative fitness of evolved strain B11-1 with an rpoC mutation.
The evolved clone from population B11 had a missense mutation (V507L) in the rpoC gene (Table 3). In order to investigate the effect of the rpoC mutation on fitness, we transduced the linked W3110 thiG761::kan allele into strain B11-1 to produce a cotransductant strain in which the mutated rpoC is reverted to the ancestral sequence. The sequence reversion was confirmed by PCR. The cotransductant strain was competed against its parental mutant strain in LBKmal, pH 4.6. The mean fitness of B11-1 lacZ::kan (containing rpoC V507L) relative to that of B11-1 thiG761::kan (containing a reverted rpoC) was 1.358 ± 0.03 (99% confidence, n = 3, one-sample t test, P = 0.008). The decreased fitness of B11-1 thiG761::kan with reverted rpoC suggests that the rpoC mutation provides a competitive advantage for acid growth.
DISCUSSION
In this study, we examined the adaptation of E. coli to growth below pH 5, a condition relevant to understanding both enteric bacterial passage into the intestine (1, 18) and fruit juice contamination (20). Following 2,000 generations of experimental evolution at the lower limit of E. coli's tolerable pH range, we found that eight evolved clones isolated from 4 of our 24 populations displayed a substantially higher fitness than the ancestral clone under growth conditions of pH 4.6 (Fig. 2). Indeed, we observed a fitness improvement much greater than that which has been seen in previous long-term evolution experiments at pH 5.3 (31). Our experiment was performed in a complex medium, LBKmal, which broadened the range of possible metabolic responses and, hence, targets of selection to the acidic conditions (9, 27). Evolved populations showed both a significantly higher endpoint culture density (Fig. 1) and significantly higher early-log-phase growth rates (Fig. 3), suggesting that these conditions selected for improvements in both the early and late parts of the growth cycle.
Mutations in the RNA polymerase holoenzyme of acid-adapted strains.
All acid-adapted evolved clones that we examined have a missense mutation in a component of the RNAP holoenzyme (43, 47, 48). Our fitness competition assays suggest that the altered rpoC allele of evolved clone B11-1 confers a competitive advantage for acidic growth. In other reports, RNAP mutations arise in some but not all laboratory evolution experiments; the mechanism by which they alter fitness is poorly understood (23, 30).
For our acid-evolved strains, we proposed three possible mechanisms by which the observed RNAP mutations might have enhanced fitness at low pH. (i) The RNAP mutations could increase the stability of the complex at low pH, when the cytoplasm is slightly acidified by proton leakage from the external medium. In this case, we would expect the acid-evolved strains to show increased fitness in the presence of benzoic acid, which acidifies the cytoplasm (49). However, our acid-evolved clones showed decreased fitness in benzoate (Fig. 2B). Thus, under our conditions, acid-stressed bacteria did not evolve resistance to permeant acids.
(ii) The RNAP mutations could destabilize the complex and thus moderate the overexpression of acid stress genes. Such overexpression at the edge of the permissive pH range could be deleterious for growth, for example, by catabolizing amino acids needed for protein synthesis. Destabilizing mutations of RNAP have been described, although the details of their mechanism remain poorly understood (50). Downregulation via RNAP destabilization could be consistent with our observed downregulation of lysine decarboxylase in all evolving populations (Table 2). The gene cadA is one of the most highly expressed acid stress genes in E. coli (13, 17, 51).
(iii) The RNAP mutations could have caused the rapid adjustment of multiple regulons in small ways that cumulatively increase fitness under the selected condition. For example, under an experimental evolution involving adaptation to minimal glycerol medium, Herring et al. found that early RNAP substitutions conferred the greatest fitness advantage of all mutations isolated (52). Moreover, many other SNPs confer a fitness advantage only when combined with RNAP. The authors hypothesized that the RNAP mutations fixed very early in the experiment due to fitness benefits stemming from their manifold effects on gene regulation. They further hypothesized that subsequent mutations might have functioned to compensate for the inevitable negative side effects of altered RNAP function (47, 53).
Putative acid stress response genes.
In our analysis of acid-evolved genome sequences, we identified mutations in several genes known to be important for E. coli's acid stress response. The most notable example of these was in adiY (mutated in the B11 population of isolates). The adiY gene encodes an AraC-like regulatory protein that can induce expression of AdiA (arginine decarboxylase) and AdiC (arginine-agmatine antiporter), the essential components of the arginine-dependent acid resistance system (6, 40). The effect of the adiY mutation could be mediated by either an increased or a decreased rate of adiAC transcription.
Two other strains, F9-2 and F9-3, had a mutation in a regulatory upstream of the nuo operon, which codes for NADH:ubiquinone oxidoreductase (respiratory complex I), a component of the electron transport chain (54, 55). The respiratory complex I is a proton pump of the electron transport chain known to be upregulated at low external pH (17). If this mutation upstream of nuoA were to increase the rate of transcription of the nuo operon, it could potentially increase acid stress by increasing the bacterium's net proton efflux.
In one clone, H9-2, we identified a missense mutation in gcvP, which encodes glycine decarboxylase P, part of the glycine cleavage system (56). Transcription of gcvP is positively regulated by glycine (57) and is responsible for the reversible breakdown of glycine into carbon dioxide, ammonia, and a cofactor-bound methylene group (58). The glutamate, arginine, lysine, and ornithine decarboxylase reactions have been demonstrated to play a role in maintaining neutral cytoplasmic pH. It is possible that the catabolic glycine decarboxylase pathway similarly serves to buffer the cytoplasm via the release of ammonia during glycine decarboxylation. Indeed, Lu et al. recently demonstrated that the conversion of glutamine to glutamate, with the release of ammonia, can serve a protective role for E. coli during exposure at pH 2.5 (59).
The protein Ag43, encoded by flu, was mutated in clones isolated from population H9. Ag43 is an autotransporter protein involved in cellular aggregation and biofilm formation, and production of Ag43 in a planktonic culture is associated with bacterial autoaggregation (forming clumps) (60, 61). Expression of flu is subject to phase variation, in which individuals in the same clonal population vary qualitatively in their production of the protein (60, 61). The missense mutation (A162E) that we identified is located in the protein's passenger domain, α43, which protrudes from the outer membrane into the extracellular space (60). This suggests that the mutant strains might have an altered ability to adhere to other cells, perhaps by binding to each other more tightly within the culture. Aggregation could benefit acid-stressed cells by reducing the total area of the cell surface exposed to the acidic medium, thereby reducing the rate of proton influx from the environment across the membrane.
Another possible acid stress gene is msbA, which encodes an ATP-dependent inner membrane flippase responsible for lipid transport from the inner leaflet to the outer leaflet of the bacterium's inner membrane. Loss of function of MsbA causes phospholipids and lipopolysaccharides (LPSs) to collect at the cell's inner membrane, thereby compromising the cell's structural integrity (62). The two evolved clones from population F11 showed the presence of a G150C missense mutation in one of MsbA's transmembrane domains. It is possible that this mutation could alter membrane integrity, which could in turn impact acid sensitivity.
This study has established eight acid-adapted E. coli clones for future study, each of which shows significant, though varying, fitness improvements relative to their ancestor. Many exciting genetic and evolutionary questions about these strains remain to be addressed. In future studies, we intend to investigate the role of RNAP mutations in gene expression and relative fitness, as well as the putative contributions of the other potentially beneficial mutant alleles that we have identified.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Science Foundation grant MCB-1329815.
We are most grateful to Richard Lenski, Zachary Blount, and Irina Artsimovitch for valuable discussions of our data. We also thank Jeffrey Barrick, Rohan Maddamsetti, Peter Lund, and Houra Merrikh for stimulating discussions. We thank Harper Kerkhoff and Veronica De Pascuale for expert technical assistance.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03494-14.
REFERENCES
- 1.Evans D, Pye G, Bramley R, Clark A, Dyson T, Hardcastle J. 1988. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut 29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ibekwe VC, Fadda HM, McConnell EL, Khela MK, Evans DF, Basit AW. 2008. Interplay between intestinal pH, transit time and feed status on the in vivo performance of pH responsive ileo-colonic release systems. Pharm Res 25:1828–1835. doi: 10.1007/s11095-008-9580-9. [DOI] [PubMed] [Google Scholar]
- 3.Slonczewski JL, Rosen BP, Alger JR, Macnab RM. 1981. pH homeostasis in Escherichia coli: measurement by 31P nuclear magnetic resonance of methylphosphonate and phosphate. Proc Natl Acad Sci U S A 78:6271–6275. doi: 10.1073/pnas.78.10.6271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilks JC, Slonczewski JL. 2007. pH of the cytoplasm and periplasm of Escherichia coli: rapid measurement by green fluorescent protein fluorimetry. J Bacteriol 189:5601–5607. doi: 10.1128/JB.00615-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zilberstein D, Agmon V, Schuldiner S, Padan E. 1984. Escherichia coli intracellular pH, membrane potential, and cell growth. J Bacteriol 158:246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster JW. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol 2:898–907. doi: 10.1038/nrmicro1021. [DOI] [PubMed] [Google Scholar]
- 7.Lin JS, Lee IS, Frey J, Slonczewski JL, Foster JW. 1995. Comparative analysis of extreme acid survival in Salmonella typhimurium, Shigella flexneri, and Escherichia coli. J Bacteriol 177:4097–4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Riggins DP, Narvaez MJ, Martinez KA, Harden MM, Slonczewski JL. 2013. Escherichia coli K-12 survives anaerobic exposure at pH 2 without RpoS, Gad, or hydrogenases, but shows sensitivity to autoclaved broth products. PLoS One 8:e56796. doi: 10.1371/journal.pone.0056796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slonczewski JL, Fujisawa M, Dopson M, Krulwich TA. 2009. Cytoplasmic pH measurement and homeostasis in bacteria and archaea. Adv Microb Physiol 55:1–79. doi: 10.1016/S0065-2911(09)05501-5. [DOI] [PubMed] [Google Scholar]
- 10.Krulwich TA, Sachs G, Padan E. 2011. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanjee U, Houry WA. 2013. Mechanisms of acid resistance in Escherichia coli. Annu Rev Microbiol 67:65–81. doi: 10.1146/annurev-micro-092412-155708. [DOI] [PubMed] [Google Scholar]
- 12.Audia JP, Webb CC, Foster JW. 2001. Breaking through the acid barrier: an orchestrated response to proton stress by enteric bacteria. Int J Med Microbiol 291:97–106. doi: 10.1078/1438-4221-00106. [DOI] [PubMed] [Google Scholar]
- 13.Meng S-Y, Bennett G. 1992. Nucleotide sequence of the Escherichia coli cad operon: a system for neutralization of low extracellular pH. J Bacteriol 174:2659–2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neely MN, Olson ER. 1996. Kinetics of expression of the Escherichia coli cad operon as a function of pH and lysine. J Bacteriol 178:5522–5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chang YY, Cronan JE. 1999. Membrane cyclopropane fatty acid content is a major factor in acid resistance of Escherichia coli. Mol Microbiol 33:249–259. doi: 10.1046/j.1365-2958.1999.01456.x. [DOI] [PubMed] [Google Scholar]
- 16.Hayes ET, Wilks JC, Sanfilippo P, Yohannes E, Tate DP, Jones BD, Radmacher MD, BonDurant SS, Slonczewski JL. 2006. Oxygen limitation modulates pH regulation of catabolism and hydrogenases, multidrug transporters, and envelope composition in Escherichia coli K-12. BMC Microbiol 6:89. doi: 10.1186/1471-2180-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maurer LM, Yohannes E, Bondurant SS, Radmacher M, Slonczewski JL. 2005. pH regulates genes for flagellar motility, catabolism, and oxidative stress in Escherichia coli K-12. J Bacteriol 187:304–319. doi: 10.1128/JB.187.1.304-319.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T. 2010. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gevers D, Knight R, Petrosino JF, Huang K, McGuire AL, Birren BW, Nelson KE, White O, Methé BA, Huttenhower C. 2012. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol 10:e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W, Pan J, Xie H, Yang Y, Zhou D, Zhu Z. 2012. Pasteurization of fruit juices of different pH values by combined high hydrostatic pressure and carbon dioxide. J Food Prot 75:1873–1877. doi: 10.4315/0362-028X.JFP-12-127. [DOI] [PubMed] [Google Scholar]
- 21.Lee DH, Palsson BO. 2010. Adaptive evolution of Escherichia coli K-12 MG1655 during growth on a nonnative carbon source, l-1,2-propanediol. Appl Environ Microbiol 76:4158–4168. doi: 10.1128/AEM.00373-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blount ZD, Barrick JE, Davidson CJ, Lenski RE. 2012. Genomic analysis of a key innovation in an experimental Escherichia coli population. Nature 489:513–518. doi: 10.1038/nature11514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassen R. 2014. Experimental evolution and the nature of biodiversity. Roberts & Company, Greenwood Village, CO. [Google Scholar]
- 24.Dragosits M, Mattanovich D. 2013. Adaptive laboratory evolution—principles and applications for biotechnology. Microb Cell Fact 12:64. doi: 10.1186/1475-2859-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riehle MM, Bennett AF, Long AD. 2001. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci U S A 98:525–530. doi: 10.1073/pnas.98.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Goodarzi H, Bennett BD, Amini S, Reaves ML, Hottes AK, Rabinowitz JD, Tavazoie S. 2010. Regulatory and metabolic rewiring during laboratory evolution of ethanol tolerance in E. coli. Mol Syst Biol 6:378. doi: 10.1038/msb.2010.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Puentes-Tellez PE, Hansen MA, Sorensen SJ, van Elsas JD. 2013. Adaptation and heterogeneity of Escherichia coli MC1000 growing in complex environments. Appl Environ Microbiol 79:1008–1017. doi: 10.1128/AEM.02920-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laehnemann D, Peña-Miller R, Rosenstiel P, Beardmore R, Jansen G, Schulenburg H. 2014. Genomics of rapid adaptation to antibiotics: convergent evolution and scalable sequence amplification. Genome Biol Evol 6:1287–1301. doi: 10.1093/gbe/evu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet 4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 30.Conrad TM, Lewis NE, Palsson BO. 2011. Microbial laboratory evolution in the era of genome-scale science. Mol Syst Biol 7:11. doi: 10.1038/msb.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hughes BS, Cullum AJ, Bennett AF. 2007. Evolutionary adaptation to environmental pH in experimental lineages of Escherichia coli. Evolution 61:1725–1734. doi: 10.1111/j.1558-5646.2007.00139.x. [DOI] [PubMed] [Google Scholar]
- 32.Johnson MD, Bell J, Clarke K, Chandler R, Pathak P, Xia Y, Marshall RL, Weinstock GM, Loman NJ, Winn PJ, Lund PA. 2014. Characterization of mutations in the PAS domain of the EvgS sensor kinase selected by laboratory evolution for acid resistance in Escherichia coli. Mol Microbiol 93:911–927. doi: 10.1111/mmi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith MW, Neidhardt FC. 1983. Proteins induced by anaerobiosis in Escherichia coli. J Bacteriol 154:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol 2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayashi K, Morooka N, Yamamoto Y, Fujita K, Isono K, Choi S, Ohtsubo E, Baba T, Wanner BL, Mori H, Horiuchi T. 2006. Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110. Mol Syst Biol 2:2006.0007. doi: 10.1038/msb4100049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lenski RE, Rose MR, Simpson SC, Tadler SC. 1991. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat 138:1315–1341. [Google Scholar]
- 37.Deininger KNW, Horikawa A, Kitko RD, Tatsumi R, Rosner JL, Wachi M, Slonczewski JL. 2011. A requirement of TolC and MDR efflux pumps for acid adaptation and GadAB induction in Escherichia coli. PLoS One 6:e18960. doi: 10.1371/journal.pone.0018960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lukas H, Reimann J, Kim OB, Grimpo J, Unden G. 2010. Regulation of aerobic and anaerobic d-malate metabolism of Escherichia coli by the LysR-type regulator DmlR (YeaT). J Bacteriol 192:2503–2511. doi: 10.1128/JB.01665-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim OB, Reimann J, Lukas H, Schumacher U, Grimpo J, Duennwald P, Unden G. 2009. Regulation of tartrate metabolism by TtdR and relation to the DcuS-DcuR-regulated C4-dicarboxylate metabolism of Escherichia coli. Microbiology 155:3632–3640. doi: 10.1099/mic.0.031401-0. [DOI] [PubMed] [Google Scholar]
- 40.Stim-Herndon KP, Flores TM, Bennett GN. 1996. Molecular characterization of adiY, a regulatory gene which affects expression of the biodegradative acid-induced arginine decarboxylase gene (adiA) of Escherichia coli. Microbiology 142:1311–1320. doi: 10.1099/13500872-142-5-1311. [DOI] [PubMed] [Google Scholar]
- 41.Watson N, Dunyak D, Rosey E, Slonczewski J, Olson E. 1992. Identification of elements involved in transcriptional regulation of the Escherichia coli cad operon by external pH. J Bacteriol 174:530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang ZH, Bielawski JP. 2000. Statistical methods for detecting molecular adaptation. Trends Ecol Evol 15:496–503. doi: 10.1016/S0169-5347(00)01994-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Opalka N, Brown J, Lane WJ, Twist KA, Landick R, Asturias FJ, Darst SA. 2010. Complete structural model of Escherichia coli RNA polymerase from a hybrid approach. PLoS Biol 8:e1000483. doi: 10.1371/journal.pbio.1000483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artsimovitch I, Chu C, Lynch AS, Landick R. 2003. A new class of bacterial RNA polymerase inhibitor affects nucleotide addition. Science 302:650–654. doi: 10.1126/science.1087526. [DOI] [PubMed] [Google Scholar]
- 45.Muzzin O, Campbell EA, Xia LL, Severinova E, Darst SA, Severinov K. 1998. Disruption of Escherichia coli HepA, an RNA polymerase-associated protein, causes UV sensitivity. J Biol Chem 273:15157–15161. doi: 10.1074/jbc.273.24.15157. [DOI] [PubMed] [Google Scholar]
- 46.Jin DJ, Zhou YN, Shaw G, Ji XH. 2011. Structure and function of RapA: a bacterial Swi2/Snf2 protein required for RNA polymerase recycling in transcription. Biochim Biophys Acta 1809:470–475. doi: 10.1016/j.bbagrm.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. 2002. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature 417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 48.Malhotra A, Severinova E, Darst SA. 1996. Crystal structure of a σ70 subunit fragment from Escherichia coli RNA polymerase. Cell 87:127–136. doi: 10.1016/S0092-8674(00)81329-X. [DOI] [PubMed] [Google Scholar]
- 49.Kannan G, Wilks JC, Fitzgerald DM, Jones BD, BonDurant SS, Slonczewski JL. 2008. Rapid acid treatment of Escherichia coli: transcriptomic response and recovery. BMC Microbiol 8:37. doi: 10.1186/1471-2180-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baharoglu Z, Lestini R, Duigou S, Michel B. 2010. RNA polymerase mutations that facilitate replication progression in the rep uvrD recF mutant lacking two accessory replicative helicases. Mol Microbiol 77:324–336. doi: 10.1111/j.1365-2958.2010.07208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Auger EA, Bennett GN. 1989. Regulation of lysine decarboxylase activity in Escherichia coli K-12. Arch Microbiol 151:466–468. doi: 10.1007/BF00416608. [DOI] [PubMed] [Google Scholar]
- 52.Herring CD, Raghunathan A, Honisch C, Patel T, Applebee MK, Joyce AR, Albert TJ, Blattner FR, van den Boom D, Cantor CR, Palsson BO. 2006. Comparative genome sequencing of Escherichia coli allows observation of bacterial evolution on a laboratory timescale. Nat Genet 38:1406–1412. doi: 10.1038/ng1906. [DOI] [PubMed] [Google Scholar]
- 53.Hindre T, Knibbe C, Beslon G, Schneider D. 2012. New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol 10:352–365. doi: 10.1038/nrmicro2750. [DOI] [PubMed] [Google Scholar]
- 54.Weidner U, Geier S, Ptock A, Friedrich T, Leif H, Weiss H. 1993. The gene locus of the proton-translocating NADH ubiquinone oxidoreductase in Escherichia coli. Organization of the 14 genes and relationship between the derived proteins and subunits of mitochondrial complex I. J Mol Biol 233:109–122. doi: 10.1006/jmbi.1993.1488. [DOI] [PubMed] [Google Scholar]
- 55.Bongaerts J, Zoske S, Weidner U, Unden G. 1995. Transcriptional regulation of the photon translocating NADH dehydrogenase genes (nuoA–N) of Escherichia coli by electron acceptors, electron donors and gene regulators. Mol Microbiol 16:521–534. doi: 10.1111/j.1365-2958.1995.tb02416.x. [DOI] [PubMed] [Google Scholar]
- 56.Okamura-Ikeda K, Ohmura Y, Fujiwara K, Motokawa Y. 1993. Cloning and nucleotide sequence of the gcv operon encoding the Escherichia coli glycine cleavage system. Eur J Biochem 216:539–548. doi: 10.1111/j.1432-1033.1993.tb18172.x. [DOI] [PubMed] [Google Scholar]
- 57.Stauffer LT, Fogarty SJ, Stauffer GV. 1994. Characterization of the Escherichia coli gcv operon. Gene 142:17–22. doi: 10.1016/0378-1119(94)90349-2. [DOI] [PubMed] [Google Scholar]
- 58.Kikuchi G, Motokawa Y, Yoshida T, Hiraga K. 2008. Glycine cleavage system: reaction mechanism, physiological significance, and hyperglycinemia. Proc Jpn Acad Ser B Phys Biol Sci 84:246–263. doi: 10.2183/pjab.84.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lu PL, Ma D, Chen YL, Guo YY, Chen GQ, Deng HT, Shi YG. 2013. l-Glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res 23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van der Woude MW, Henderson IR. 2008. Regulation and function of Ag43 (Flu). Annu Rev Microbiol 62:153–169. doi: 10.1146/annurev.micro.62.081307.162938. [DOI] [PubMed] [Google Scholar]
- 61.Henderson IR, Meehan M, Owen P. 1997. Antigen 43, a phase-variable bipartite outer membrane protein, determines colony morphology and autoaggregation in Escherichia coli K-12. FEMS Microbiol Lett 149:115–120. doi: 10.1111/j.1574-6968.1997.tb10317.x. [DOI] [PubMed] [Google Scholar]
- 62.Doerrler WT, Raetz CRH. 2002. ATPase activity of the MsbA lipid flippase of Escherichia coli. J Biol Chem 277:36697–36705. doi: 10.1074/jbc.M205857200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.