Abstract
Aldehydes are a class of chemicals with many industrial uses. Several aldehydes are responsible for flavors and fragrances present in plants, but aldehydes are not known to accumulate in most natural microorganisms. In many cases, microbial production of aldehydes presents an attractive alternative to extraction from plants or chemical synthesis. During the past 2 decades, a variety of aldehyde biosynthetic enzymes have undergone detailed characterization. Although metabolic pathways that result in alcohol synthesis via aldehyde intermediates were long known, only recent investigations in model microbes such as Escherichia coli have succeeded in minimizing the rapid endogenous conversion of aldehydes into their corresponding alcohols. Such efforts have provided a foundation for microbial aldehyde synthesis and broader utilization of aldehydes as intermediates for other synthetically challenging biochemical classes. However, aldehyde toxicity imposes a practical limit on achievable aldehyde titers and remains an issue of academic and commercial interest. In this minireview, we summarize published efforts of microbial engineering for aldehyde synthesis, with an emphasis on de novo synthesis, engineered aldehyde accumulation in E. coli, and the challenge of aldehyde toxicity.
INTRODUCTION
The word “aldehyde” was coined in the early 19th century by Justin von Liebig, who formed a contraction using the Latin words “alcohol dehydrogenatus,” or “alcohol deprived of hydrogen” (1). Aldehydes have a variety of industrial uses, but they are perhaps most familiar for their effects on two of the mammalian senses: olfaction and gustation. Numerous aldehyde odorants are known to bind to G-protein-coupled receptors, triggering reaction cascades that ultimately result in mammalian perception (2–5). At dilute concentrations, fatty aldehydes such as hexanal, octanal, decanal, and dodecanal offer apple, citrus, orange peel, and violet scents, respectively (6). Aromatic aldehydes, such as benzaldehyde, anisaldehyde, vanillin, and cinnamaldehyde, are responsible for the natural fragrances of almond, sweet blossom, vanilla, and cinnamon, respectively (6, 7). Notable terpenoid aldehydes include citral, which provides lemon scent (6), and safranal, which is one of the primary molecules responsible for saffron aroma (8). Aldehydes play a role in other animal phyla as well. Certain aldehydes, such as trans-2-hexenal, phenylacetaldehyde, and nonanal, evoke responses in insects by serving as pheromones or attractants (9–11). The high reactivity of the carbonyl group of aldehydes enables many industrial uses beyond flavors and fragrances, such as precursors to pharmaceuticals (12–15). However, the high reactivity of aldehydes also contributes to their increased toxicity in microorganisms. Given the high-value applications and large markets for several aldehydes, commercial focus on microbial aldehyde synthesis has surged in recent years (16). This minireview summarizes published efforts of microbial engineering for aldehyde synthesis, with an emphasis on de novo aldehyde synthesis, engineered aldehyde accumulation in Escherichia coli, and the challenge of aldehyde toxicity.
ENGINEERING ALDEHYDE BIOSYNTHETIC REACTIONS AND PATHWAYS
Because most microbes do not naturally accumulate aldehydes, microbial production of these molecules from simple carbon sources requires at least two parallel approaches: pathway construction for product generation and strain engineering for product accumulation. A starting point for pathway construction is consideration of enzymatic reactions that can produce desired aldehydes from cellular metabolites. Carboxylic acids are found throughout cellular metabolism, and many can be converted to aldehydes with the aid of a single enzyme. Prior to the detailed characterization and cloning of enzymes capable of broadly catalyzing aldehyde formation, various natural organisms ranging from actinomycetes to white rot fungi were tested for the innate ability to convert carboxylic acids into their corresponding aldehydes or alcohols (17–21). A significant advance occurred roughly 1 decade ago, when a carboxylic acid reductase (CarNi) from Nocardia iowensis was cloned into Escherichia coli and shown to be active on several aromatic carboxylic acids in vitro (22). Later publications from Rosazza and colleagues demonstrated that CarNi requires one-time activation by a phosphopantetheinyl transferase and that CarNi has activity in vitro on a broader range of substrates that includes several citric acid cycle dicarboxylic acids (23, 24). Motivated by the activity of CarNi on diverse carboxylic acid substrates, we investigated its activity on straight-chain and branched-chain aliphatic acids ranging from C2 to C8 (25). A homolog of CarNi from Mycobacterium marinum was also demonstrated to have activity on straight-chain aliphatic acids ranging from C6 to C18 (26). A recent review describes a larger number of carboxylic acid reductases that could be harnessed for biosynthesis of a variety of aldehydes (27). The general stoichiometry for reactions catalyzed by carboxylic acid reductases is as follows (where “e−” represents a reducing equivalent):
| (1) |
Aliphatic aldehydes across a broad range of carbon lengths can also be formed by using fermentative aldehyde reductases or by using enzymes that act on activated forms of carboxylic acids (acyl-coenzyme A [CoA] or acyl-ACP). During anaerobic cultivation of E. coli, conversion of acetyl-CoA to acetaldehyde is catalyzed by a CoA-dependent acetaldehyde dehydrogenase (also known as acetaldehyde CoA dehydrogenase) (28). However, the same protein, encoded by adhE, has a second catalytic site that converts acetaldehyde into ethanol (29). In solvent-producing clostridial strains, acetaldehyde and butyraldehyde can be produced by CoA-acylating aldehyde dehydrogenases that are found as individual enzymes or as bifunctional enzymes (30–33). The conversion of acyl-CoA to aldehyde is as follows (for acyl-ACP substrates instead of acyl-CoA substrates, replace “S-CoA” and “CoASH” with “ACP”):
| (2) |
Synthesis of longer carbon-chain aliphatic aldehydes from acyl-ACP precursors can occur using enzymes from luminescent bacteria. In these bacteria, the multienzyme fatty acid reductase complex consisting of luxCDE is used to produce aldehydes that are immediate substrates for the light emission reaction (34). Note that the aldehyde biosynthetic reactions discussed so far use similar chemistries that primarily differ in the source of reducing equivalents and whether the carboxylic acid molecule or the reductase enzyme is activated first. In either case, activation requires the conversion of ATP to AMP and pyrophosphate and occurs because the energetics of converting a carboxylic acid to an aldehyde are ordinarily unfavorable.
Another set of nonoxidative aldehyde biosynthetic routes utilizes decarboxylation of 2-keto acid substrates. In these cases, no ATP is required because the irreversibility of CO2 formation provides the driving force for aldehyde formation. However, one carbon atom is lost per molecule of 2-keto acid substrate, which reduces the theoretical maximum yield. Two well-known enzymes in this category are pyruvate decarboxylase (PDC) and 2-ketoisovalerate decarboxylase (KivD). The native role of PDCs is to convert pyruvate to acetaldehyde, but their promiscuity and capability of catalyzing carboligation side reactions have led to their use in synthesis of chiral carboligation products (12). KivD is also promiscuous and has been utilized for synthesis of numerous nonnatural alcohols derived from amino acid intermediates (35). The 2-keto acid decarboxylation reaction is as follows:
| (3) |
Oxidative reactions can also be used for aldehyde synthesis, starting from either carboxylic acid substrates or primary alcohol substrates. Cn fatty acids can be converted to Cn−1 fatty aldehydes, as was shown using E. coli resting cells that expressed an α-dioxygenase from Oryza sativa (rice) (36). In this case, spontaneous decarboxylation of a Cn hydroperoxy fatty acid intermediate provides a driving force for aldehyde generation. The dioxygenase-catalyzed reaction is as follows:
| (4) |
In addition, aldehydes can be obtained by enzymatic oxidation of primary alcohols (37–40). From a de novo aldehyde synthesis perspective, these reactions are less relevant given that alcohols are typically produced via aldehyde intermediates. However, biocatalytic conversion of primary alcohols to aldehydes may provide an array of new opportunities for alcohols as starting materials and is revisited later in this review. Oxidation of alcohols to aldehydes generates a reducing equivalent as follows:
| (5) |
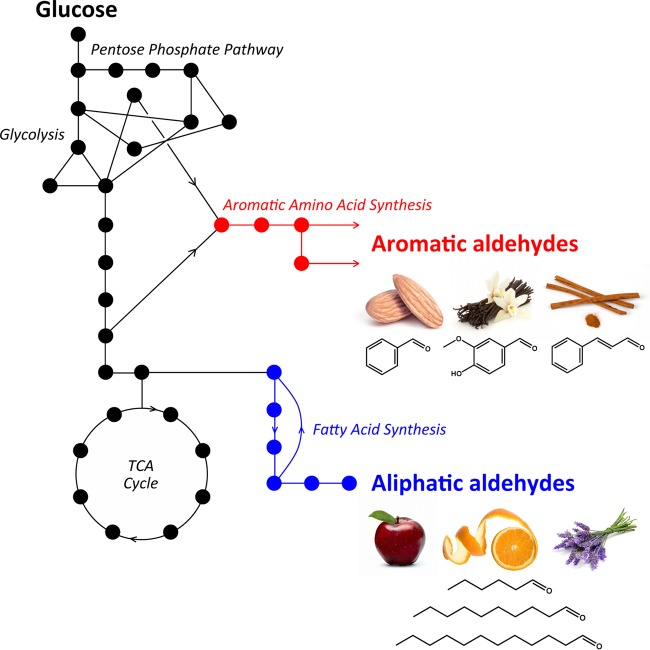
Natural and engineered pathways could be used to produce useful aldehydes from simple carbon sources via their corresponding carboxylic acids. Pathway selection leading to the relevant carboxylic acid precursor depends on the category of target aldehyde. Figure 1 illustrates known aromatic and aliphatic acid biosynthesis pathways that can be engineered to result in several familiar flavors and fragrances. In the case of vanillin, which has the largest annual market volume of any flavor compound, previous reports have described engineered heterologous pathways that use the natural aromatic amino acid precursor 3-dehydroshikimate as a branch-point metabolite for the heterologous reactions (41–43). Li and Frost constructed a system to produce vanillin from glucose that used an engineered strain of E. coli to produce vanillate from glucose, followed by extraction and reduction of vanillate to vanillin in vitro using purified carboxylic acid reductase from Neurospora crassa (41). De novo biosynthesis of vanillin and vanillin-β-d-glucoside was first demonstrated in both Saccharomyces cerevisiae and Schizosaccharomyces pombe and has since been optimized using flux balance analysis (42, 44, 45). In initial reports, titers of de novo vanillin-β-d-glucoside were roughly 50 mg/liter in batch flask cultures (42) and 500 mg/liter in 1.5-liter continuous cultures (44). The company Evolva has improved and commercialized this process (16).
FIG 1.
Overview of natural metabolic pathways that can be harnessed for the conversion of glucose to valuable aromatic and aliphatic aldehydes through carboxylic acid intermediates based on E. coli metabolism. Aldehydes can also be obtained from the 2-keto acid pathway (35, 55), terpenoid pathways (54), and other pathways. TCA, tricarboxylic acid.
Among flavor compounds, benzaldehyde has the second largest annual market volume after vanillin (46). Aromatic amino acid biosynthesis could also be used to engineer a microbial pathway to benzaldehyde, potentially from phenylalanine as the starting endogenous metabolite. Formation of benzaldehyde was reported after phenylalanine addition to a cell extract of Lactobacillus plantarum (47). In plants, benzaldehyde is derived from phenylalanine, potentially from β-oxidative and non-β-oxidative pathways (48). Recent work has uncovered key steps in the β-oxidative pathway that can lead to synthesis of benzoate, which could serve as the precursor to benzaldehyde in an engineered microbial pathway (49).
Aliphatic aldehydes can be obtained using pathways that result in free fatty acids (FFAs). Although microbial FFAs have been produced for decades, recent work has demonstrated the potential for obtaining advanced fuels or valuable chemicals as derivatives of FFAs (50–53). Based on the broad substrate range and known activities of carboxylic acid reductases, their addition to these pathways can result in production of C4 to C18 aliphatic aldehydes (25, 26). Microbial synthesis of other valuable aldehyde classes, such as terpenoid aldehydes, could potentially occur in E. coli using variations of previously engineered terpenoid pathways (54).
As mentioned earlier, commercial entities have actively pursued aldehyde biosynthesis routes using engineered microbes. Table 1 contains an overview of relevant published aldehyde biosynthesis patent applications during the past 30 years. These patents were grouped into three types of dominant routes of aldehyde biosynthesis. Although the third category is the most pertinent to the topic of this review, the other two categories of processes were included to provide context and perspective into chronological trends. For example, during the 1980s and 1990s, industry patents on biotransformation processes featured either isolated microbes or fruit homogenates. Commercial processes featuring fully de novo aldehyde synthesis using engineered microbes appear to have emerged only within the last decade. Of course, an overview of patent literature does not account for industrial advances that were retained as trade secrets.
TABLE 1.
Relevant published aldehyde biosynthesis patent applications
| Dominant aldehyde biosynthesis route | Applicant | Publication date | Publication no. | Patent name | Relevant claim(s)a | Grant (G) or application (A) |
|---|---|---|---|---|---|---|
| Biotransformation using homogenates or natural microorganisms | Takasago Perfumery | 6 Sep 1988 | US 4769243 A | Method for preparing green aroma compounds | Use of ground soybeans to convert unsaturated fatty acids to aliphatic aldehydes and alcohols | G |
| General Foods Corporation | 21 February 1989 | US 4806379 A | Process for producing a green leaf essence | Use of strawberry homogenate to convert linolenic acid to cis-3-hexanal and related aldehydes | G | |
| BASF | 17 October 1989 | US 4874701 A | Preparation of coniferylaldehyde by a microorganism | Use of Arthrobacter globiformis DSM 3597 to convert n-eugenol to coniferylaldehyde | G | |
| Haarmann & Reimer GmbH | 21 May 1991 | US 5017388 | Process for the preparation of vanillin | Use of certain species from the genus Serratia, Klebsiella, or Enterobacter to convert eugenol or isoeugenol to vanillin | G | |
| Kraft General Foods | 7 Jul 1992 | US 5128253 A | Bioconversion process for the production of vanillin | Use of ferulic acid-degrading microorganisms such as Aspergillus niger, Rhodotorula glutinis, or Corynebacterium glutamicum to convert ferulic acid to vanillin | G | |
| Firmenich | 7 Nov 1995 | US 5464761 A | Process for the enzymatic preparation of aliphatic alcohols and aldehydes from linoleic acid or a natural precursor | Use of lipoxygenase-containing soya flour and lyase-containing guava homogenate to convert linoleic acid to hexanal and related aldehydes | G | |
| BASF | 19 May 1998 | US 5753471 A | Biotechnological preparation of alcohols, aldehydes, and carboxylic acids | Use of isolated microorganisms capable of converting alkyl, alkenyl, aryl, and related compounds to their oxidized forms, including aldehydes | G | |
| In vitro conversion of acid substrates using purified carboxylic acid reductases | University of Iowa | 18 Aug 1998 | US 5795759 A | Carboxylic acid reductase, and methods of using same | A purified carboxylic acid reductase (Car) enzyme from Nocardia iowensis, and use of it to convert vanillic acid to vanillin | G |
| Michigan State University | 16 April 2002 | US 6372461 B1 | Synthesis of vanillin from a carbon source | Use of an engineered microbe expressing recombinant DHSD and COMT as part of a metabolic pathway from glucose to vanillic acid, followed by reduction of vanillic acid to vanillin using a purified Car | G | |
| University of Iowa | 16 September 2008 | US 7425433 B2 | Carboxylic acid reductase polypeptide, nucleotide sequence encoding same and methods of use | Use of Car to convert aromatic, aliphatic, and acyclic carboxylic acids to corresponding aldehydes | G | |
| Archer-Daniels-Midland | 17 February 2009 | US 7491854 B2 | Enzymatic method of making aldehydes from fatty acids | Use of Car to convert fatty acids ranging from C6-C32 to corresponding aldehydes | G | |
| De novo synthesis using engineered microbes harboring recombinant aldehyde biosynthetic genes (e.g., car, aar, and kivD) | DuPont | 29 August 2006 | US 7098000 B2 | Method for production of C30-aldehyde carotenoids | Use of an engineered microorganism to convert fermentable carbon sources to diaponeurosporene monoaldehyde, diapocarotene monoaldehyde, or diapocarotene dialdehyde | G |
| LS9 | 17 January 2012 | US 8097439 B2 | Methods and compositions for producing fatty aldehydes | Use of engineered microbes containing recombinant Car homologues to convert carbohydrates to aliphatic aldehydes | G | |
| LS9 | 18 September 2012 | US 8268599 B2 | Method for producing a fatty alcohol or fatty aldehyde | Use of acyl-ACP reductases to convert acyl-ACPs to aliphatic aldehydes | G | |
| International Flavors & Fragrances, and Evolva | 14 February 2013 | WO 2013022881 A1 | Compositions and methods for the biosynthesis of vanillin or vanillin-beta-d-glucoside | Use of a microbe expressing recombinant AROM and/or COMT to convert glucose to vanillin or vanillin-beta-d-glucoside | A | |
| University of California | 27 December 2013 | WO 2013192237 A1 | Escherichia coli engineered for isobutyraldehyde production | Use of an E. coli strain with reduced isobutyraldehyde reductase activity to accumulate isobutyraldehyde | A | |
| Easel Biotechnologies | 9 January 2014 | US 20140011231 A1 | Microbial synthesis of aldehydes and corresponding alcohols | Use of an engineered microbe to convert glucose to short fatty aldehydes, followed by removal of aldehydes from the fermentation medium and conversion to alcohols ex vivo | A | |
| Genomatica | 24 April 2014 | WO 2014062564 A1 | Microorganisms and methods for production of specific length fatty alcohols and related compounds | Use of a microbe expressing malonyl-CoA independent (or dependent) fatty acyl-CoA elongation pathways to produce fatty acids, aldehydes, and alcohols | A | |
| Evolva | 4 September 2014 | US 20140248668 A1 | Methods and materials for recombinant production of saffron compounds | Use of a microorganism expressing recombinant pathways to convert glucose to picrocrocin, safranal, crocin, crocetin, or crocetin esters | A |
AROM, arom multifunctional enzyme; COMT, catechol-O-methyltransferase; DHSD, 3-dehydroshikimate dehydratase.
MINIMIZING ENDOGENOUS CONVERSION OF ALDEHYDES TO ALCOHOLS
Despite known routes to a variety of aldehydes, microbial aldehyde production is hindered by the rapid endogenous conversion of nearly all aldehydes to their corresponding alcohols. For example, when expression of recombinant CarNi was first reported in E. coli, aromatic acids supplied to culture media were rapidly converted into aromatic alcohols (22). Even in E. coli, the genetically best-understood organism, numerous uncharacterized genes were thought to contribute to this activity. To our knowledge, explanations of how to significantly reduce endogenous conversion for any given aldehyde in E. coli became present in the public domain only very recently. It is worth highlighting here that, although oxidation of an aldehyde to a carboxylic acid is thermodynamically more favorable than reduction of a carboxylic acid to an aldehyde, endogenous aldehyde oxidation does not appear to be significant for most aldehydes of interest in model microbes. On the other hand, endogenous aldehyde reduction has been thoroughly documented in the literature and is the focus of this review.
In 2012, Rodriguez and Atsumi reported accumulation of isobutyraldehyde in E. coli by sequentially deleting eight genes (yqhD, adhP, eutG, yiaY, yjgB [now ahr], betA, fucO, and eutE) encoding putative isobutyraldehyde reductases (55). When individually overexpressed, five of these genes displayed activity toward isobutyraldehyde. The engineered deletion strain increased isobutyraldehyde production from 0.14 g/liter/optical density at 600 nm (OD600) to 1.5 g/liter/OD600 and decreased isobutanol production from 1.5 g/liter/OD600 to 0.4 g/liter/OD600. Although isobutanol formation still occurred, that study suggested that the number of gene deletions required to mitigate conversion of a particular aldehyde may be a manageable quantity.
We became interested in determining whether gene deletions could enable accumulation of aromatic aldehydes and believed that fewer gene deletions might be required for accumulation under aerobic conditions. After deletion of six genes that encode enzymes with confirmed activity on benzaldehyde in vitro (dkgA, dkgB, yeaE, yahK, ahr, and yqhD), the engineered E. coli strain accumulated benzaldehyde and vanillin with minimal alcohol formation and was thus dubbed “RARE” for displaying reduced aromatic aldehyde reduction (Addgene catalog no. 61440) (43). Each targeted gene was capable of causing reduction of benzaldehyde and vanillin in vivo when individually overexpressed in the RARE background. However, the use of deletion subset strains and quantitative reverse transcription-PCR (qRT-PCR) revealed that deletions of dkgB and yeaE did not contribute to aldehyde accumulation under the conditions tested due to low native expression of these genes (43).
Soon after aromatic aldehyde accumulation was reported, Rodriguez and Atsumi reported the construction of an E. coli strain that minimally converted exogenously supplied aliphatic aldehydes ranging from C2 to C12 to their corresponding alcohols (56). Their study examined 44 candidate aldehyde reductases in vivo by overexpressing candidates using the previously reported isobutraldehyde-accumulating strain (55). However, overexpression of genes encoding aldehyde reductases has been shown to lead to false positives when such genes are minimally expressed under relevant conditions (43). Rodriguez and Atsumi noted that fewer than the 13 genes deleted in their final strain (yqhD, adhP, eutG, yiaY, ahr, betA, fucO, eutE, yahK, dkgA, gldA, ybbO, and yghA) may be sufficient to create useful strains devoted to a specific set of aldehyde products (56). Given that the consequential gene deletions in the RARE strain form a subset of the genes deleted by Rodriguez and Atsumi, both strains are likely capable of accumulating most aromatic and aliphatic aldehydes of interest under aerobic conditions at the shake flask scale. Under other conditions, such as high-cell-density industrial fermentations that commonly feature anaerobic zones, it may be better to err on the side of inclusion of more deletions as long as cell health and stability are not significantly perturbed. Together, these studies should aid efforts to engineer aldehyde accumulation in other microbes.
ENHANCING BIOCONVERSION OF ALDEHYDES TO OTHER CHEMICAL CLASSES
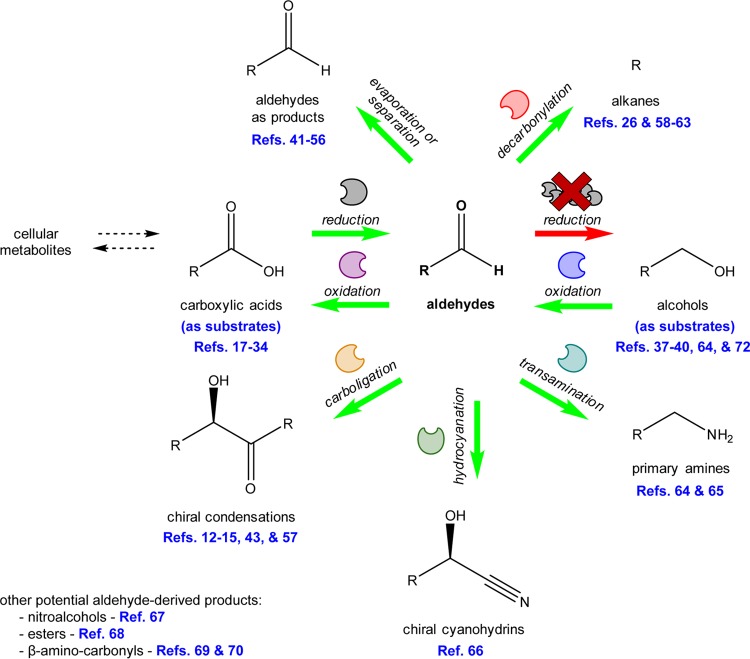
Microbial aldehyde accumulation enables biosynthesis of several previously problematic compounds that can be derived enzymatically from aldehyde intermediates (Fig. 2). In our report on aromatic aldehyde accumulation (43), we demonstrated this potential by using the RARE strain to produce l-phenylacetylcarbinol (L-PAC), a chiral precursor to the pharmaceutical ephedrine (12–15). Although whole-cell catalysts have been used for L-PAC synthesis for a long time, significant benzyl alcohol byproduct formation occurs from their use, resulting in low yields (12). Cultures of the RARE strain expressing a recombinant mutant PDC were able to produce L-PAC using exogenously supplied benzaldehyde and metabolized pyruvate with minimal benzyl alcohol formation. Under the conditions tested, the use of wild-type E. coli expressing the same PDC produced no detectable L-PAC (43). In addition to PDC, other enzymes capable of catalyzing chiral carboligations of aldehyde substrates have been discussed (57).
FIG 2.
Potential biocatalytic and metabolic engineering opportunities enabled by, or enhanced by, microbial aldehyde accumulation.
A similar challenge of limiting unwanted flux from aldehyde intermediates to alcohol byproducts has been encountered in the context of alkane production. The final step to alkane biosynthesis features the conversion of a Cn aldehyde to a Cn−1 alkane catalyzed by an aldehyde decarbonylase or aldehyde deformylating oxygenase (26, 58–62). Although the problem of alcohol byproduct formation has been described extensively, very few studies of alkane biosynthesis have used strains engineered with deletions of aldehyde reductases. Rodriguez and Atsumi discussed the relevance of their strain for alkane synthesis but did not demonstrate alkane production in their study (56). Production of propane was recently reported by Kallio and colleagues using engineered E. coli that displayed decreased endogenous conversion of butyraldehyde to butanol due to deletions of ahr and yqhD (63).
In addition to chiral carboligations and decarbonylations, aldehyde substrates can participate in numerous other enzyme-catalyzed reactions (Fig. 2), for example, transamination to form primary amines (64, 65), hydrocyanation to form chiral cyanohydrins (66), Henry reactions to form nitroalcohols (67), Baeyer-Villager oxidation to form esters (68), and Mannich reactions to form β-amino-carbonyl compounds (69, 70). Some of the aforementioned reactions have already been demonstrated to be functional in a cellular context using resting E. coli cells (66, 71). Microbial aldehyde accumulation enables potential synthesis of these compounds using metabolically active cells that can supply and regenerate expensive cofactors. Synthesis of some of these products may also be achieved using glucose or other simple sugars as the sole carbon source. In addition, biocatalytic oxidation of exogenously supplied alcohols (37–40, 64, 72) would be more effective in the absence of aldehyde reduction. In theory, any of the classes of aldehyde-derived compounds enabled in the absence of aldehyde reduction could also be obtained directly from the corresponding primary alcohols using a single engineered microbe.
ADDRESSING ALDEHYDE TOXICITY
Now that published reports have elucidated aldehyde accumulation in E. coli under laboratory-scale conditions, the next impediment to engineering microbial aldehyde synthesis is aldehyde toxicity. Observable toxicity is manifested by inhibition of microbial growth in the presence of aldehydes (43, 73), but morphological changes have also been reported (73). In most cases, the extent of toxicity seems to depend on the aldehyde but may also depend on the choice of microorganism. Cinnamaldehyde, for example, is known to be a potent antimicrobial (74). In the case of vanillin, Zaldivar et al. found that 1.5 g/liter of vanillin completely inhibited growth of the E. coli strains examined (73). The same study investigated the effect of exposing E. coli to several representative aromatic aldehyde products of hemicellulose hydrolysis and found that toxicity was directly related to the hydrophobicity of the aldehyde. The relationship with hydrophobicity suggested that a hydrophobic target, such as the cell membrane, may be involved. However, none of these aldehydes caused sufficient membrane damage to allow the leakage of intracellular magnesium (73). Another study investigated the toxicity of four aldehydes (furfural, 5-hydroxymethylfurfural, vanillin, and syringaldehyde) to Candida tropicalis and found that vanillin was the most toxic, followed by syringaldehyde, furfural, and 5-hydroxymethylfurfural (75). The influence of the structural elements of vanillin and related compounds on antifungal activity has also been examined, and differences in antifungal activity were found (76). However, when the effect of five aldehydes on the growth of the oleaginous yeast Trichosporon fermentans was investigated, no relationship was found between the hydrophobicity and toxicity of the aldehyde (77).
The E. coli strains investigated by Zaldivar et al. were not engineered to have minimal aldehyde reductase activity, and later studies from the same group suggested that growth inhibition may be caused by NADPH consumption resulting from aldehyde reduction (78, 79). Two genes (dkgA and yqhD) were found to be silenced in an evolved furfural-resistant strain. Expression of these genes, which encode enzymes with low Km values for NADPH, decreased furfural tolerance (78). In a separate investigation, transcriptome data were analyzed before and after exposure to furfural. Several lines of evidence suggested that cysteine and methionine biosynthesis was upregulated in order to combat a limitation in sulfur assimilation due to NADPH depletion (79). Although NADPH consumption may contribute to toxicity, our experience with aldehyde accumulation suggests that aromatic aldehydes remain toxic even when minimal endogenous reduction occurs (43).
A deeper understanding of precisely how aldehydes cause harm to cells may enable engineering strategies to surmount particular modes of toxicity. Certain aldehydes may be involved in mechanisms of toxicity that are far more detrimental than the mechanisms seen with others. For example, acetaldehyde has been shown to induce single-strand and double-strand breaks in DNA (80). Several aliphatic aldehydes are products of lipid peroxidation and have been implicated in the formation of adducts on a variety of biological macromolecules and as second messengers of reactive oxygen species (ROS) (81–83). However, the precise relationship between aldehydes and ROS is unclear. For example, it was recently shown that resistance of E. coli to exogenous methylglyoxal is conferred by decreased expression of sodC (84). This is a surprising result given that sodC encodes a superoxide dismutase, which breaks down ROS (85). There are numerous other potential mechanisms of aldehyde toxicity. Given the importance of lignocellulose utilization, potential mechanisms of toxicity for furfural in particular have been extensively reviewed and include mechanisms not described here (86, 87).
Until precise mechanisms of aldehyde toxicity are elucidated, there are some general engineering strategies that can be employed. Some bacteria have naturally evolved solutions to aldehyde toxicity beyond rapid reduction of aldehydes, such as protein microcompartments that feature aldehyde intermediates (88, 89). If control of selective metabolite transport through the protein shells were achieved, then the engineering of these compartments for biosynthesis of new aldehyde-derived products might aid in limiting the pool size of free aldehyde intermediates (90). Independently of the mode of toxicity, in situ separation using stripping (91), two-phase systems (92), or selective resins (93) may result in increased production of aldehydes as end products. Many aldehydes of interest are hydrophobic and volatile, which are properties that aid separation from water-based fermentation processes. In the event that precise mechanisms of aldehyde toxicity become known and prove to be insurmountable problems, then efforts should shift toward microbial engineering of aldehyde intermediates for synthesis of aldehyde-derived products.
CONCLUSION
In the past decade, research on microbial engineering for aldehyde synthesis has progressed from understanding how to synthesize aldehydes to understanding how to accumulate synthesized aldehydes. Given that advances in both of these areas apply to a broad range of societally relevant aldehydes, the work summarized here may serve as a foundation for future academic and commercial endeavors. The issue of aldehyde toxicity remains a major hurdle blocking improvement of commercial microbial processes for aldehyde production. Potential engineering solutions to this challenge are complicated by significant differences in the levels of toxicity among aldehydes and by the potential for each aldehyde to be deleterious due to multiple mechanisms acting at once. Regardless, given that aldehydes can now escape the fate of rapid reduction to their corresponding alcohols in living microbes, these molecules can serve as a gateway for synthesis of several previously challenging classes of biochemicals. For that reason at least, aldehydes should be of significant interest to practitioners of metabolic engineering and biocatalysis in the years ahead, even if the challenge of aldehyde toxicity has not been solved.
ACKNOWLEDGMENTS
This work was supported by the Synthetic Biology Engineering Research Center (SynBERC; grant no. EEC-0540879). A.M.K. is a recipient of a National Science Foundation Graduate Research Fellowship.
REFERENCES
- 1.Crosland MP. 2004. Historical studies in the language of chemistry. Dover Publications, Mineola, NY. [Google Scholar]
- 2.Zhao H, Ivic L, Otaki JM, Hashimoto M, Mikoshiba K, Firestein S. 1998. Functional expression of a mammalian odorant receptor. Science 279:237–242. doi: 10.1126/science.279.5348.237. [DOI] [PubMed] [Google Scholar]
- 3.Araneda RC, Kini AD, Firestein S. 2000. The molecular receptive range of an odorant receptor. Nat Neurosci 3:1248–1255. doi: 10.1038/81774. [DOI] [PubMed] [Google Scholar]
- 4.Araneda RC, Peterlin Z, Zhang X, Chesler A, Firestein S. 2004. A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium. J Physiol 555:743–756. doi: 10.1113/jphysiol.2003.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Soucy ER, Albeanu DF, Fantana AL, Murthy VN, Meister M. 2009. Precision and diversity in an odor map on the olfactory bulb. Nat Neurosci 12:210–220. doi: 10.1038/nn.2262. [DOI] [PubMed] [Google Scholar]
- 6.Fahlbusch K-G, Hammerschmidt F-J, Panten J, Pickenhagen W, Schatkowski D, Bauer K, Garbe D, Surburg H. 2000. Flavors and fragrances, Ullmann's encyclopedia of industrial chemistry. Wiley-VCH Verlag, Weinheim, Germany. [Google Scholar]
- 7.Hagedorn S, Kaphammer B. 1994. Microbial biocatalysis in the generation of flavor and fragrance chemicals. Annu Rev Microbiol 48:773–800. doi: 10.1146/annurev.mi.48.100194.004013. [DOI] [PubMed] [Google Scholar]
- 8.Tarantilis PA, Polissiou MG. 1997. Isolation and identification of the aroma components from saffron (Crocus sativus). J Agric Food Chem 45:459–462. [Google Scholar]
- 9.Raina AK, Kingan TG, Mattoo AK. 1992. Chemical signals from host plant and sexual behavior in a moth. Science 255:592–594. doi: 10.1126/science.255.5044.592. [DOI] [PubMed] [Google Scholar]
- 10.Dickens JC, Jang EB, Light DM, Alford AR. 1990. Enhancement of insect pheromone responses by green leaf volatiles. Naturwissenschaften 77:29–31. [Google Scholar]
- 11.Syed Z, Leal WS. 2009. Acute olfactory response of Culex mosquitoes to a human- and bird-derived attractant. Proc Natl Acad Sci U S A 106:18803–18808. doi: 10.1073/pnas.0906932106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tripathi CM, Agarwal SC, Basu SK. 1997. Production of L-phenylacetylcarbinol by fermentation. J Ferment Bioeng 84:487–492. [Google Scholar]
- 13.Shukla VB, Kulkarni PR. 2007. L-Phenylacetylcarbinol (L-PAC): biosynthesis and industrial applications. World J Microbiol Biotechnol: 16:499–506. [Google Scholar]
- 14.Yun H, Kim B-G. 2008. Enzymatic production of (R)-phenylacetylcarbinol by pyruvate decarboxylase from Zymomonas mobilis. Biotechnol Bioprocess Eng 13:372–376. doi: 10.1007/s12257-008-0030-7. [DOI] [Google Scholar]
- 15.Meyer D, Walter L, Kolter G, Pohl M, Müller M, Tittmann K. 2011. Conversion of pyruvate decarboxylase into an enantioselective carboligase with biosynthetic potential. J Am Chem Soc 133:3609–3616. doi: 10.1021/ja110236w. [DOI] [PubMed] [Google Scholar]
- 16.Hayden EC. 2014. Synthetic-biology firms shift focus. Nature 505:598. doi: 10.1038/505598a. [DOI] [PubMed] [Google Scholar]
- 17.Ježo I, Zemek J. 1986. Enzymatische Reduktion einiger aromatischer Carboxysäuren. Chem Pap 40:279–281. [Google Scholar]
- 18.Kato N, Konishi H, Uda K, Shimao M, Sakazawa C. 1988. Microbial reduction of benzoate to benzyl alcohol. Agric Biol Chem 52:1885–1886. doi: 10.1271/bbb1961.52.1885. [DOI] [Google Scholar]
- 19.Casey J, Dobb R. 1992. Microbial routes to aromatic aldehydes. Enzyme Microb Technol 14:739–747. [Google Scholar]
- 20.Arfmann H-A, Abraham W-R. 1993. Microbial reduction of aromatic carboxylic acids. Z Naturforsch C 48:52–57. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Rosazza JP. 1997. Purification, characterization, and properties of an aryl aldehyde oxidoreductase from Nocardia sp. strain NRRL 5646. J Bacteriol 179:3482–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He A, Li T, Daniels L, Fotheringham I, Rosazza JPN. 2004. Nocardia sp. carboxylic acid reductase: cloning, expression, and characterization of a new aldehyde oxidoreductase family. Appl Environ Microbiol 70:1874–1881. doi: 10.1128/AEM.70.3.1874-1881.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkitasubramanian P, Daniels L, Rosazza JPN. 2007. Reduction of carboxylic acids by Nocardia aldehyde oxidoreductase requires a phosphopantetheinylated enzyme. J Biol Chem 282:478–485. doi: 10.1074/jbc.M607980200. [DOI] [PubMed] [Google Scholar]
- 24.Venkitasubramanian P, Daniels L, Das S, Lamm AS, Rosazza JPN. 2008. Aldehyde oxidoreductase as a biocatalyst: reductions of vanillic acid. Enzyme Microb Technol 42:130–137. doi: 10.1016/j.enzmictec.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 25.Sheppard MJ, Kunjapur AM, Wenck SJ, Prather KLJ. 2014. Retro-biosynthetic screening of a modular pathway design achieves selective route for microbial synthesis of 4-methyl-pentanol. Nat Commun 5:5031. doi: 10.1038/ncomms6031. [DOI] [PubMed] [Google Scholar]
- 26.Akhtar MK, Turner NJ, Jones PR. 2013. Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities. Proc Natl Acad Sci U S A 110:87–92. doi: 10.1073/pnas.1216516110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Napora-Wijata K, Strohmeier GA, Winkler M. 2014. Biocatalytic reduction of carboxylic acids. Biotechnol J 9:822–843. doi: 10.1002/biot.201400012. [DOI] [PubMed] [Google Scholar]
- 28.Clark DP. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol Lett 63:223–234. [DOI] [PubMed] [Google Scholar]
- 29.Goodlove PE, Cunningham PR, Parker J, Clark DP. 1989. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene 85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 30.Palosaari NR, Rogers P. 1988. Purification and properties of the inducible coenzyme A-linked butyraldehyde dehydrogenase from Clostridium acetobutylicum. J Bacteriol 170:2971–2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nair RV, Bennett GN, Papoutsakis ET. 1994. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol 176:871–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Toth J, Ismaiel AA, Chen J-S. 1999. The ald gene, encoding a coenzyme A-acylating aldehyde dehydrogenase, distinguishes Clostridium beijerinckii and two other solvent-producing clostridia from Clostridium acetobutylicum. Appl Environ Microbiol 65:4973–4980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. 2002. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol 184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meighen EA. 1991. Molecular biology of bacterial bioluminescence. Microbiol Rev 55:123–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Atsumi S, Hanai T, Liao JC. 2008. Non-fermentative pathways for synthesis of branched-chain higher alcohols as biofuels. Nature 451:86–89. doi: 10.1038/nature06450. [DOI] [PubMed] [Google Scholar]
- 36.Kaehne F, Buchhaupt M, Schrader J. 2011. A recombinant α-dioxygenase from rice to produce fatty aldehydes using E. coli. Appl Microbiol Biotechnol 90:989–995. doi: 10.1007/s00253-011-3165-y. [DOI] [PubMed] [Google Scholar]
- 37.Gandolfi R, Ferrara N, Molinari F. 2001. An easy and efficient method for the production of carboxylic acids and aldehydes by microbial oxidation of primary alcohols. Tetrahedron Lett 42:513–514. doi: 10.1016/S0040-4039(00)02008-6. [DOI] [Google Scholar]
- 38.Romano D, Villa R, Molinari F. 2012. Preparative biotransformations: oxidation of alcohols. ChemCatChem 4:739–749. doi: 10.1002/cctc.201200042. [DOI] [Google Scholar]
- 39.Corberán VC, González-Pérez ME, Martínez-González S, Gómez-Avilés A. 2014. Green oxidation of fatty alcohols: challenges and opportunities. Appl Catal A 474:211–223. doi: 10.1016/j.apcata.2013.09.040. [DOI] [Google Scholar]
- 40.Duff SJB, Murray WD. 1989. Oxidation of benzyl alcohol by whole cells of Pichia pastoris and by alcohol oxidase in aqueous and nonaqueous reaction media. Biotechnol Bioeng 34:153–159. doi: 10.1002/bit.260340203. [DOI] [PubMed] [Google Scholar]
- 41.Li K, Frost JW. 1998. Synthesis of vanillin from glucose. J Am Chem Soc 120:10545–10546. doi: 10.1021/ja9817747. [DOI] [Google Scholar]
- 42.Hansen EH, Møller BL, Kock GR, Bünner CM, Kristensen C, Jensen OR, Okkels FT, Olsen CE, Motawia MS, Hansen J. 2009. De novo biosynthesis of vanillin in fission yeast (Schizosaccharomyces pombe) and baker's yeast (Saccharomyces cerevisiae). Appl Environ Microbiol 75:2765–2774. doi: 10.1128/AEM.02681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kunjapur AM, Tarasova Y, Prather KLJ. 2014. Synthesis and accumulation of aromatic aldehydes in an engineered strain of Escherichia coli. J Am Chem Soc 136:11644–11654. doi: 10.1021/ja506664a. [DOI] [PubMed] [Google Scholar]
- 44.Brochado A, Matos C, Moller B, Hansen J, Mortensen U, Patil K. 2010. Improved vanillin production in baker's yeast through in silico design. Microb Cell Fact 9:84. doi: 10.1186/1475-2859-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brochado AR, Patil KR. 2013. Overexpression of O-methyltransferase leads to improved vanillin production in baker's yeast only when complemented with model-guided network engineering. Biotechnol Bioeng 110:656–659. doi: 10.1002/bit.24731. [DOI] [PubMed] [Google Scholar]
- 46.Krings U, Berger RG. 1998. Biotechnological production of flavours and fragrances. Appl Microbiol Biotechnol 49:1–8. [DOI] [PubMed] [Google Scholar]
- 47.Nierop Groot MN, de Bont JAM. 1998. Conversion of phenylalanine to benzaldehyde initiated by an aminotransferase in Lactobacillus plantarum. Appl Environ Microbiol 64:3009–3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dudareva N, Klempien A, Muhlemann JK, Kaplan I. 2013. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol 198:16–32. doi: 10.1111/nph.12145. [DOI] [PubMed] [Google Scholar]
- 49.Qualley AV, Widhalm JR, Adebesin F, Kish CM, Dudareva N. 2012. Completion of the core β-oxidative pathway of benzoic acid biosynthesis in plants. Proc Natl Acad Sci U S A 109:16383–16388. doi: 10.1073/pnas.1211001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, McClure A, del Cardayre SB, Keasling JD. 2010. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 51.Dellomonaco C, Rivera C, Campbell P, Gonzalez R. 2010. Engineered respiro-fermentative metabolism for the production of biofuels and biochemicals from fatty acid-rich feedstocks. Appl Environ Microbiol 76:5067–5078. doi: 10.1128/AEM.00046-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dellomonaco C, Clomburg JM, Miller EN, Gonzalez R. 2011. Engineered reversal of the β-oxidation cycle for the synthesis of fuels and chemicals. Nature 476:355–359. doi: 10.1038/nature10333. [DOI] [PubMed] [Google Scholar]
- 53.Handke P, Lynch SA, Gill RT. 2011. Application and engineering of fatty acid biosynthesis in Escherichia coli for advanced fuels and chemicals. Metab Eng 13:28–37. doi: 10.1016/j.ymben.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 54.Martin VJJ, Pitera DJ, Withers ST, Newman JD, Keasling JD. 2003. Engineering a mevalonate pathway in Escherichia coli for production of terpenoids. Nat Biotechnol 21:796–802. doi: 10.1038/nbt833. [DOI] [PubMed] [Google Scholar]
- 55.Rodriguez G, Atsumi S. 2012. Isobutyraldehyde production from Escherichia coli by removing aldehyde reductase activity. Microb Cell Fact 11:90. doi: 10.1186/1475-2859-11-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodriguez GM, Atsumi S. 2014. Toward aldehyde and alkane production by removing aldehyde reductase activity in Escherichia coli. Metab Eng 25:227–237. doi: 10.1016/j.ymben.2014.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pohl M, Lingen B, Müller M. 2002. Thiamin-diphosphate-dependent enzymes: new aspects of asymmetric C-C bond formation. Chemistry 8:5288–5295. doi:. [DOI] [PubMed] [Google Scholar]
- 58.Schirmer A, Rude MA, Li X, Popova E, del Cardayre SB. 2010. Microbial biosynthesis of alkanes. Science 329:559–562. doi: 10.1126/science.1187936. [DOI] [PubMed] [Google Scholar]
- 59.Andre C, Kim SW, Yu X-H, Shanklin J. 2013. Fusing catalase to an alkane-producing enzyme maintains enzymatic activity by converting the inhibitory byproduct H2O2 to the cosubstrate O2. Proc Natl Acad Sci U S A 110:3191–3196. doi: 10.1073/pnas.1218769110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Howard TP, Middelhaufe S, Moore K, Edner C, Kolak DM, Taylor GN, Parker DA, Lee R, Smirnoff N, Aves SJ, Love J. 2013. Synthesis of customized petroleum-replica fuel molecules by targeted modification of free fatty acid pools in Escherichia coli. Proc Natl Acad Sci U S A 110:7636–7641. doi: 10.1073/pnas.1215966110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harger M, Zheng L, Moon A, Ager C, An JH, Choe C, Lai Y-L, Mo B, Zong D, Smith MD, Egbert RG, Mills JH, Baker D, Pultz IS, Siegel JB. 2013. Expanding the product profile of a microbial alkane biosynthetic pathway. ACS Synth Biol 2:59–62. doi: 10.1021/sb300061x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khara B, Menon N, Levy C, Mansell D, Das D, Marsh ENG, Leys D, Scrutton NS. 2013. Production of propane and other short-chain alkanes by structure-based engineering of ligand specificity in aldehyde-deformylating oxygenase. ChemBioChem 14:1204–1208. doi: 10.1002/cbic.201300307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kallio P, Pásztor A, Thiel K, Akhtar MK, Jones PR. 2014. An engineered pathway for the biosynthesis of renewable propane. Nat Commun 5:4731. doi: 10.1038/ncomms5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuchs M, Tauber K, Sattler J, Lechner H, Pfeffer J, Kroutil W, Faber K. 2012. Amination of benzylic and cinnamic alcohols via a biocatalytic, aerobic, oxidation-transamination cascade. RSC Adv 2:6262–6265. doi: 10.1039/c2ra20800h. [DOI] [Google Scholar]
- 65.Park E, Kim M, Shin J. 2012. Molecular determinants for substrate selectivity of ω-transaminases. Appl Microbiol Biotechnol 93:2425–2435. doi: 10.1007/s00253-011-3584-9. [DOI] [PubMed] [Google Scholar]
- 66.Scholz KE, Okrob D, Kopka B, Grünberger A, Pohl M, Jaeger K-E, Krauss U. 2012. Synthesis of chiral cyanohydrins by recombinant Escherichia coli cells in a micro-aqueous reaction system. Appl Environ Microbiol 78:5025–5027. doi: 10.1128/AEM.00582-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Purkarthofer T, Gruber K, Gruber-Khadjawi M, Waich K, Skranc W, Mink D, Griengl H. 2006. A biocatalytic Henry reaction—the hydroxynitrile lyase from Hevea brasiliensis also catalyzes nitroaldol reactions. Angew Chem Int Ed Engl 45:3454–3456. doi: 10.1002/anie.200504230. [DOI] [PubMed] [Google Scholar]
- 68.Moonen MJH, Westphal AH, Rietjens IMCM, van Berkel WJH. 2005. Enzymatic baeyer–villiger oxidation of benzaldehydes. Adv Synth Catal 347:1027–1034. doi: 10.1002/adsc.200404307. [DOI] [Google Scholar]
- 69.He T, Li K, Wu M-Y, Feng X-W, Wang N, Wang H-Y, Li C, Yu X-Q. 2010. Utilization of biocatalytic promiscuity for direct Mannich reaction. J Mol Catal B Enzym 67:189–194. doi: 10.1016/j.molcatb.2010.08.004. [DOI] [Google Scholar]
- 70.Li K, He T, Li C, Feng X-W, Wang N, Yu X-Q. 2009. Lipase-catalysed direct Mannich reaction in water: utilization of biocatalytic promiscuity for C-C bond formation in a “one-pot” synthesis. Green Chem 11:777–779. doi: 10.1039/b817524a. [DOI] [Google Scholar]
- 71.Mihovilovic MD, Kapitan P, Rydz J, Rudroff F, Ogink FH, Fraaije MW. 2005. Biooxidation of ketones with a cyclobutanone structural motif by recombinant whole-cells expressing 4-hydroxyacetophenone monooxygenase. J Mol Catal B Enzym 32:135–140. doi: 10.1016/j.molcatb.2004.11.009. [DOI] [Google Scholar]
- 72.Zambelli P, Pinto A, Romano D, Crotti E, Conti P, Tamborini L, Villa R, Molinari F. 2012. One-pot chemoenzymatic synthesis of aldoximes from primary alcohols in water. Green Chem 14:2158–2161. doi: 10.1039/c2gc35764j. [DOI] [Google Scholar]
- 73.Zaldivar J, Martinez A, Ingram LO. 1999. Effect of selected aldehydes on the growth and fermentation of ethanologenic Escherichia coli. Biotechnol Bioeng 65:24–33. doi:. [DOI] [PubMed] [Google Scholar]
- 74.Visvalingam J, Hernandez-Doria JD, Holley RA. 2013. Examination of the genome-wide transcriptional response of Escherichia coli O157:H7 to cinnamaldehyde exposure. Appl Environ Microbiol 79:942–950. doi: 10.1128/AEM.02767-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang L, Tang P, Fan X, Yuan Q. 2013. Effect of selected aldehydes found in the corncob hemicellulose hydrolysate on the growth and xylitol fermentation of Candida tropicalis. Biotechnol Prog 29:1181–1189. doi: 10.1002/btpr.1774. [DOI] [PubMed] [Google Scholar]
- 76.Fitzgerald DJ, Stratford M, Gasson MJ, Narbad A. 2005. Structure-function analysis of the vanillin molecule and its antifungal properties. J Agric Food Chem 53:1769–1775. doi: 10.1021/jf048575t. [DOI] [PubMed] [Google Scholar]
- 77.Huang C, Wu H, Liu Q-p, Li Y-y, Zong M-h. 2011. Effects of aldehydes on the growth and lipid accumulation of oleaginous yeast Trichosporon fermentans. J Agric Food Chem 59:4606–4613. doi: 10.1021/jf104320b. [DOI] [PubMed] [Google Scholar]
- 78.Miller EN, Jarboe LR, Yomano LP, York SW, Shanmugam KT, Ingram LO. 2009. Silencing of NADPH-dependent oxidoreductase genes (yqhD and dkgA) in furfural-resistant ethanologenic Escherichia coli. Appl Environ Microbiol 75:4315–4323. doi: 10.1128/AEM.00567-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Miller EN, Jarboe LR, Turner PC, Pharkya P, Yomano LP, York SW, Nunn D, Shanmugam KT, Ingram LO. 2009. Furfural inhibits growth by limiting sulfur assimilation in ethanologenic Escherichia coli strain LY180. Appl Environ Microbiol 75:6132–6141. doi: 10.1128/AEM.01187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Singh NP, Khan A. 1995. Acetaldehyde: genotoxicity and cytotoxicity in human lymphocytes. Mutat Res 337:9–17. doi: 10.1016/0921-8777(95)00006-6. [DOI] [PubMed] [Google Scholar]
- 81.Esterbauer H, Schaur RJ, Zollner H. 1991. Chemistry and biochemistry of 4-hydroxynonenal, malonaldehyde and related aldehydes. Free Radic Biol Med 11:81–128. doi: 10.1016/0891-5849(91)90192-6. [DOI] [PubMed] [Google Scholar]
- 82.West JD, Marnett LJ. 2006. Endogenous reactive intermediates as modulators of cell signaling and cell death. Chem Res Toxicol 19:173–194. doi: 10.1021/tx050321u. [DOI] [PubMed] [Google Scholar]
- 83.Grimsrud PA, Xie H, Griffin TJ, Bernlohr DA. 2008. Oxidative stress and covalent modification of protein with bioactive aldehydes. J Biol Chem 283:21837–21841. doi: 10.1074/jbc.R700019200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Warner JR, Reeder PJ, Karimpour-Fard A, Woodruff LBA, Gill RT. 2010. Rapid profiling of a microbial genome using mixtures of barcoded oligonucleotides. Nat Biotechnol 28:856–862. doi: 10.1038/nbt.1653. [DOI] [PubMed] [Google Scholar]
- 85.Gort AS, Ferber DM, Imlay JA. 1999. The regulation and role of the periplasmic copper, zinc superoxide dismutase of Escherichia coli. Mol Microbiol 32:179–191. doi: 10.1046/j.1365-2958.1999.01343.x. [DOI] [PubMed] [Google Scholar]
- 86.Mills T, Sandoval N, Gill R. 2009. Cellulosic hydrolysate toxicity and tolerance mechanisms in Escherichia coli. Biotechnol Biofuels 2:26. doi: 10.1186/1754-6834-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Almeida JM, Bertilsson M, Gorwa-Grauslund M, Gorsich S, Lidén G. 2009. Metabolic effects of furaldehydes and impacts on biotechnological processes. Appl Microbiol Biotechnol 82:625–638. doi: 10.1007/s00253-009-1875-1. [DOI] [PubMed] [Google Scholar]
- 88.Sampson EM, Bobik TA. 2008. Microcompartments for B12-dependent 1,2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J Bacteriol 190:2966–2971. doi: 10.1128/JB.01925-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Starai VJ, Garrity J, Escalante-Semerena JC. 2005. Acetate excretion during growth of Salmonella enterica on ethanolamine requires phosphotransacetylase (EutD) activity, and acetate recapture requires acetyl-CoA synthetase (Acs) and phosphotransacetylase (Pta) activities. Microbiology 151:3793–3801. doi: 10.1099/mic.0.28156-0. [DOI] [PubMed] [Google Scholar]
- 90.Kim EY, Tullman-Ercek D. 2013. Engineering nanoscale protein compartments for synthetic organelles. Curr Opin Biotechnol 24:627–632. doi: 10.1016/j.copbio.2012.11.012. [DOI] [PubMed] [Google Scholar]
- 91.Zhu H, Gonzalez R, Bobik TA. 2011. Coproduction of acetaldehyde and hydrogen during glucose fermentation by Escherichia coli. Appl Environ Microbiol 77:6441–6450. doi: 10.1128/AEM.05358-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jain AN, Khan TR, Daugulis AJ. 2010. Bioproduction of benzaldehyde in a solid-liquid two phase partitioning bioreactor using Pichia pastoris. Biotechnol Lett 32:1649–1654. doi: 10.1007/s10529-010-0353-2. [DOI] [PubMed] [Google Scholar]
- 93.Hua D, Ma C, Song L, Lin S, Zhang Z, Deng Z, Xu P. 2007. Enhanced vanillin production from ferulic acid using adsorbent resin. Appl Microbiol Biotechnol 74:783–790. doi: 10.1007/s00253-006-0735-5. [DOI] [PubMed] [Google Scholar]