Abstract
Mitochondrial dysfunction has been shown to contribute to age-related and proliferative retinal diseases. Over the past decade, the primary human fetal RPE (hfRPE) culture model has emerged as an effective tool for studying RPE function and mechanisms of retinal diseases. This model system has been rigorously characterized and shown to closely resemble native RPE cells at the genomic and protein level, and that they are capable of accomplishing the characteristic functions of a healthy native RPE (e.g., rod phagocytosis, ion and fluid transport, and retinoid cycle). In this review, we demonstrated that the metabolic activity of the RPE is an indicator of its health and state of differentiation, and present the hfRPE culture model as a valuable in vitro system for evaluating RPE metabolism in the context of RPE differentiation and retinal disease.
Keywords: primary cultured human fetal RPE, metabolism, differentiation
1. Introduction
Interposed between the neural retina and the choroidal blood supply, the retinal pigment epithelium (RPE) plays a critical role in maintaining metabolic homeostasis in the outer retina which is essential for normal photoreceptor activity and health (reviewed in (Adijanto and Philp, 2012)). The RPE regulates ion, pH, and fluid homeostasis in the inter-photoreceptor space (Hughes et al., 1998), transports nutrients to and metabolic waste products from the retina (Bergersen et al., 1999), processes retinol into 11-cis-retinal (Lamb and Pugh, 2004), phagocytizes shed photoreceptor outer segments (POS) (Kevany and Palczewski, 2010), secretes neurotrophic factors to maintain photoreceptor integrity (Steele et al., 1993), and forms the outer blood retina barrier (Rizzolo et al., 2011). All of these specialized functions of the RPE depend on the expression and polarized distribution of receptors, transporters, channels and enzymes, many of which are markers of differentiated RPE.
In various systems, cells undergo a metabolic shift from glycolytic to oxidative metabolism as they advance through stages of differentiation (Agathocleous and Harris, 2013; Agathocleous et al., 2012; De Pauw et al., 2009; Porter et al., 2011). A glycolytic phenotype allows the cells to rapidly generate metabolic intermediates and energy needed to actively proliferate and migrate (Lunt and Vander Heiden, 2011). However, as cells mature and differentiate, they rely on oxidative metabolism to sustain specialized cellular functions (Agathocleous and Harris, 2013). Disruption of mitochondrial biogenesis in the RPE of a postnatal mouse resulted in a shift in cell metabolism, from oxidative respiration to aerobic glycolysis, that was accompanied by a loss of RPE specific genes (Zhao et al., 2011). With the association between RPE mitochondrial dysfunction and many genetic and age-related ocular diseases (Feher et al., 2006; Nordgaard et al., 2008; Rath et al., 2008; Tyni et al., 2004; Udar et al., 2009), one may postulate that restoration of normal RPE metabolism could be a useful therapeutic strategy for these blinding diseases.
In this review, we demonstrate how RPE metabolism and differentiation can be evaluated in a primary human fetal RPE culture model. We show how siRNA-mediated gene knockdown (KD) can be used to study key metabolic pathways involved in RPE differentiation and highlight the tools that can be used to evaluate changes in RPE metabolism.
2. Human RPE cell culture as a model for studying RPE differentiation and metabolism
To date, there exist a number of primary cell culture systems (fetal and adult) (Blenkinsop et al., 2013; Gamm et al., 2008; Hu and Bok, 2001; Maminishkis et al., 2006; Sonoda et al., 2009; Valtink and Engelmann, 2009) and cell lines (e.g., RPE-J, D407, and ARPE-19) (Davis et al., 1995; Dunn et al., 1996; Nabi et al., 1993) that have been used to evaluate basic RPE function as well as the molecular mechanisms underlying RPE-associated diseases. While the RPE culture system is convenient because it can be easily mass-produced and experimentally manipulated, validation of these culture models for known properties of RPE function and physiology is essential. Among the most extensively characterized RPE cell culture model is the primary human fetal RPE first established in Dr. Dean Bok’s laboratory (Hu and Bok, 2001; Pfeffer, 1991) and later modified in Dr. Sheldon Miller’s laboratory (Maminishkis et al., 2006; Maminishkis and Miller, 2010). These two model systems differ in the methods used for isolation of fetal RPE cells and the culture medium used to grow these cells. Regardless, with both approaches, hfRPE cells re-establish pigmented epithelial monolayers with apical microvilli that express key mRNA, miRNA (Adijanto et al., 2012; Liao et al., 2010; Strunnikova et al., 2010; Wang et al., 2010), and proteins essential for RPE function (Adijanto et al., 2012), exhibit proper protein polarity of ion channels and transporters (Adijanto et al., 2009; Maminishkis et al., 2006), phagocytize rod outer segments (Castorino et al., 2011), metabolize retinol (Flannery et al., 1990; Hu and Bok, 2013), secrete growth and neurotrophic factors in a polarized manner (Castorino et al., 2011), establish tight and adherens junctions (Economopoulou et al., 2009; Peng et al., 2010; Peng et al., 2011), and mediate vectorial transepithelial transport of fluid (Castorino et al., 2011; Li et al., 2009; Maminishkis et al., 2006). Genomic analysis by microarray revealed that the mRNA expression profile of cultured human fetal RPE cells closely resembles native fetal RPE cells and adult native RPE cells (Strunnikova et al., 2010).
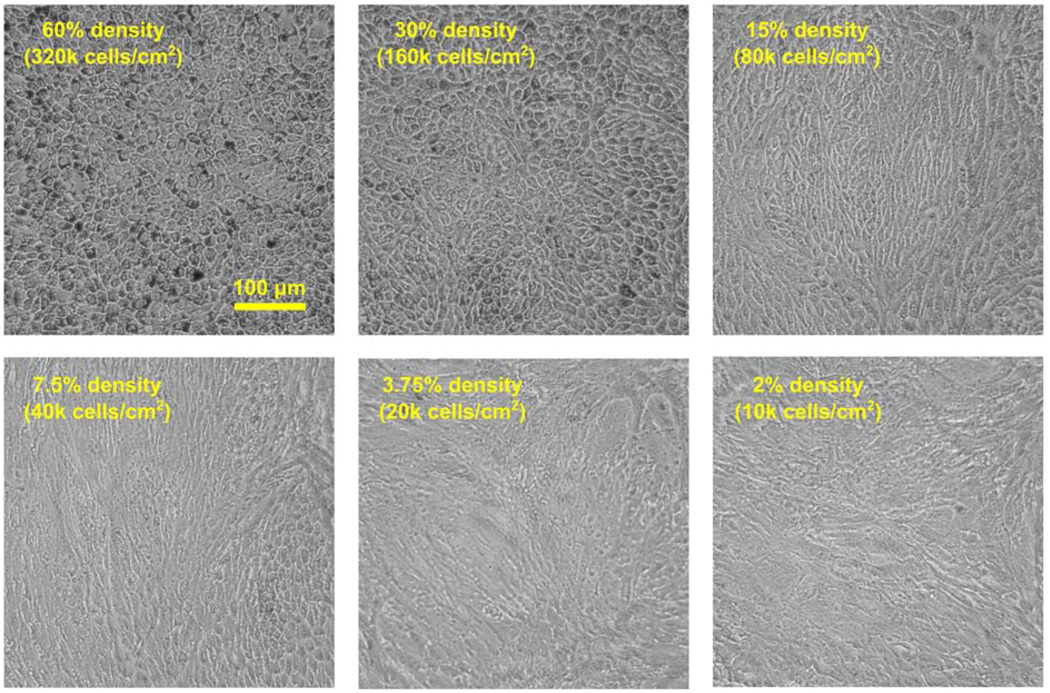
In Dr. Miller’s hfRPE culture model, native hfRPE cells from donors at 16–22 weeks of gestation were manually dissociated from the eyecup and trypsinized and seeded onto flasks (Passage 0; P0). hfRPE cells were cultured for 4 weeks during which time the cells re-established a highly pigmented monolayer before they were dissociated from the flasks and re-seeded onto 12mm transwell filters (Passage 1; P1 at 200k cells/well) where RPE cells again proliferate and migrate. Upon contact with adjacent cells, RPE cells initiate a “re-differentiation” program (or “re-morphogenesis”; see (Burke, 2008) for review) to establish tight junctions and begin expressing key RPE genes and proteins (e.g., bestrophin1 (BEST1), MCT3, RPE65) (Adijanto et al., 2012). This process typically takes 3–4 weeks, but can be accelerated or slowed depending on cell seeding density. Passage 2 (P2) primary hfRPE cells seeded at high density (30%; 160k cells/cm2 or more) can differentiate in 10–14 days, while cells seeded at lower densities (~15%; 80k cells/cm2) typically take 3–4 weeks to differentiate. While hfRPE cells from different donors exhibit some variability in the time it takes them to differentiate, it seldom deviates far from this range. It is important to note that primary RPE cells can only undergo a limited number of divisions before they lose their capacity to re-differentiate (Adijanto et al., 2012; Grisanti and Guidry, 1995) and hfRPE cells seeded at less than 5% density will not differentiate into a functional RPE monolayer, regardless of the time spent in culture. As a reference, Figure 1 presents the morphology of P2 hfRPE cells seeded at a range of densities (from 2% to 60%) cultured for 21 days in a 96-well plate. In this batch of cells, hfRPE cells seeded at 30% and 60% densities achieved proper cobblestone morphology. Cells seeded at 15% density require more time to differentiate, and hfRPE cells seeded at 7.5% and below had completely lost their ability to differentiate and are referred to as dedifferentiated hfRPE cells. Notably, P2 dedifferentiated hfRPE cells do not re-differentiate even when seeded at high densities (30–60%) in passage 3. In the experiments presented in this review, we compared the metabolism of P2 hfRPE seeded at or above 30% density and cultured over 2–3 weeks to achieve RPE differentiation to that of P2 hfRPE that dedifferentiated after seeding at or below 5% density and cultured over 2–3 weeks.
Figure 1. Morphology of hfRPE cells seeded at various densities.
Fully differentiated P1 hfRPE cells on transwells were trypsinized and re-seeded into 96-well plates at 60, 30, 15, 7.5, 3.75, and 2% densities. They were cultured in complete hfRPE culture medium (Advanced MEM (Gibco; cat# 12492) supplemented with Penicillin-Streptomycin, Glutamax®, THT, and 5% HIFBS). Media were replaced three times a week and images were taken at 10X magnification on 21 days post seeding.
3. The role of glycolysis and oxidative metabolism in RPE differentiation
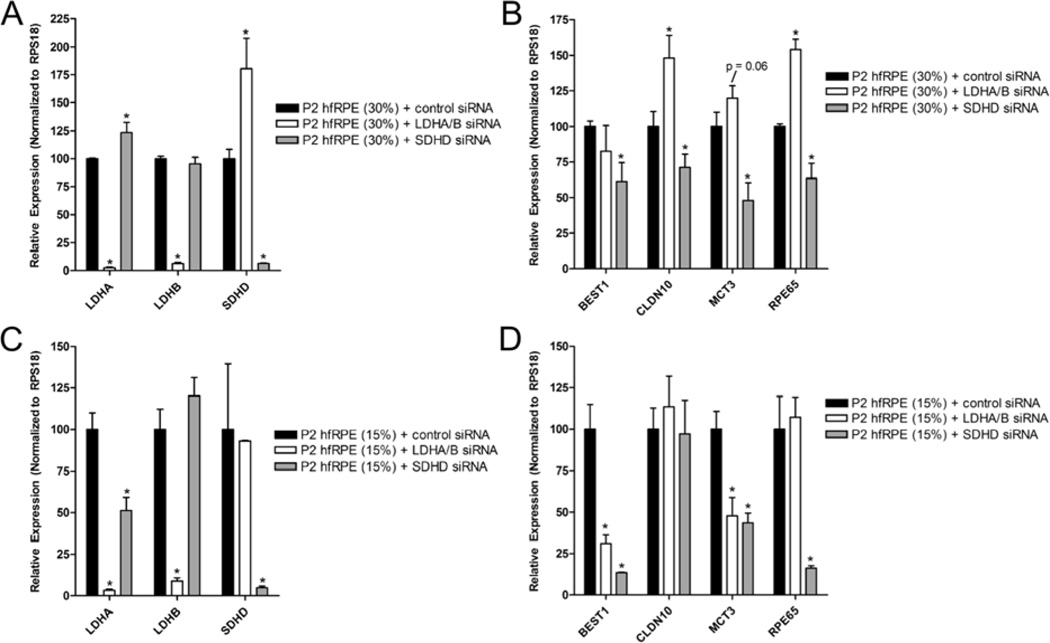
In the hfRPE culture model, we can take advantage of the difference in hfRPE seeding density to evaluate the metabolic profiles of early vs. late stage hfRPE differentiation. P1 hfRPE cells seeded at either high (30%) or low (15%) densities will both be fully confluent by the seventh day in culture, however hfRPE cells seeded at 30% density will be at a more advanced stages of differentiation than cells seeded at 15% density, as it more closely resembles the characteristic hexagonally-packed RPE morphology. Previous studies showed that cells at early stages of differentiation are highly glycolytic, whereas cells at late stage differentiation require oxidative metabolism to achieve maturity. To test these observations in the hfRPE model, we inhibited glycolysis in hfRPE cells by siRNA KD of lactate dehydrogenases, LDHA and LDHB, which catalyzes the conversion between pyruvate and lactate. This strategy was chosen because it does not block glucose metabolism in its entirety, instead it shunts pyruvate into the TCA cycle. To inhibit oxidative metabolism, we knocked down succinate dehydrogenase isoform D (SDHD) because it is an important enzyme for both the TCA cycle and electron transport chain (ETC). Silencing LDHA and LDHB or SDHD did not cause RPE cell death or alter epithelial morphology in 15% or 30% seeded hfRPE cells compared to control.
As expected, we found that KD of LDHA and LDHB in 30% density hfRPE cells (Figure 2A) resulted in increased RPE-specific gene expression (Claudin10 (CLDN10) and RPE65; Figure 2B), suggesting that inhibiting glycolytic activity promotes late-stage RPE differentiation. In 15% seeded RPE cells, which correspond to an earlier stage of RPE differentiation, KD of LDHA and LDHB (Figure 2C) resulted in a decrease in RPE-specific gene expression (BEST1 and MCT3; Figure 2D). The results of these experiments are consistent with the notion that glycolysis is important for early stage RPE differentiation, but its activity must be gradually decreased for late stage RPE differentiation to proceed. Inhibition of oxidative metabolism by siRNA KD of SDHD (Figures 2A & C) decreased RPE-specific gene expression in both 15% and 30% density hfRPE cells (Figures 2B & D), indicating that oxidative metabolism plays an important role in both early and late stage differentiation.
Figure 2. The transition from glycolytic to oxidative metabolism orchestrates the RPE differentiation process.
P1 hfRPE cells were seeded at (A) low (15%) or (B) high (30%) densities to mimic early and late stage differentiation, respectively. Cells were transfected with LDHA and LDHB (20 nM each) or SDHD (40 nM) siRNAs using Dharmafect 4 (0.2%), once upon seeding and another two days post seeding – this transfection protocol has been published in our previous work (Adijanto et al., 2012). Each condition was represented by three technical replicates. RNA was isolated seven days post-transfection, and the samples were evaluated for RPE-specific gene expression (BEST1, CLDN10, MCT3, and RPE65) using qRTPCR. RPS18 was used as endogenous control for delta-Ct-Ct analysis. P < 0.05 is considered statistically significant. Primer sequences for RPE-specific genes and RPS18 have been published (Adijanto et al., 2012).
4. Methods for evaluating RPE metabolism
4.1 Ringer Solution
Evaluation of RPE metabolism was achieved using a Ringer’s solution that did not contain other potential metabolic fuels (besides glucose) that are present in the MEM medium: vitamins, amino acids, phenol red (pH indicator), and supplements. While many Ringer solution recipes have been formulated for evaluating hfRPE physiology, the ideal Ringer solution is one that closely mimics the ionic composition, osmolality (300 mOsm/kg H2O), and pH (7.4) of the culture medium in which the cells have been cultured. For hfRPE cells cultured using Miller’s protocol, we use Ringer solution composed of NaCl (116.5 mM), NaHCO3 (26.2 mM), KCl (5 mM), MgCl2 (0.5 mM), CaCl2 (1.8 mM), HEPES-NMDG (12 mM HEPES dissolved in DI H2O in a separate beaker and titrated to pH 7.4 with a base, N-Methyl-Dglucamine (NMDG)), and glucose (5 mM). In practice, glucose is excluded from the Ringer and added separately prior to the experiment. Prior to sterile filtration, Ringer solution is equilibrated by passing the solution with 5% CO2 (balance air). When fully equilibrated, the final pH of the Ringer will be 7.4. Glucose-free Ringer can be stored at 4°C for up to 4 weeks. Variations of this Ringer’s solution are routinely used in electrophysiological and fluorescence imaging studies of the hfRPE culture model in the Miller lab (Adijanto et al., 2009). Furthermore, this Ringer’s solution can substitute for the basal MEM culture medium. HfRPE cells differentiate when cultured in this Ringer solution supplemented with insulin-transferrin-selenium (ITS), taurine-hydrocortisone-triiodo-thyronin (THT), glutamax®, nonessential amino acids, and heat-inactivated (HI) FBS (5%).
4.2 Evaluating hfRPE metabolism by measuring lactate release
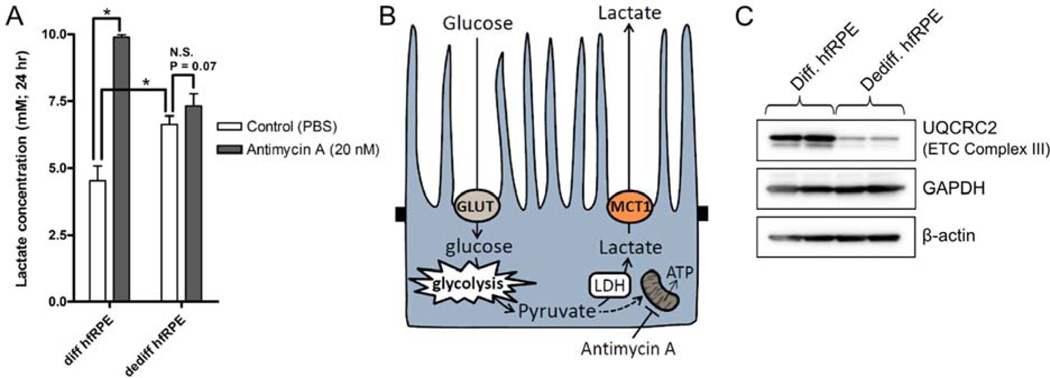
From previous studies, it is known that the state of RPE differentiation correlates with its metabolic activity – dedifferentiated hfRPE cells are glycolytic whereas differentiated hfRPE cells are oxidative. This difference in metabolism can be easily detected by measuring lactate concentration in RPE media using a colorimetric assay (Trinity Biotech PLC; Cat#: 735-10), and determining the change in lactate release by the RPE in response to a mitochondrial inhibitor, antimycin A. Cells that rely on oxidative metabolism will respond to antimycin A with a significant increase in lactate release, whereas cells that rely primarily on glycolysis will not. As expected, we found that addition of antimycin A (20 nM; an ETC inhibitor) to differentiated hfRPE cells (seeded at 30% and cultured over 21 days) significantly increased RPE lactate production and release (Figures 3 A & B), whereas addition of antimycin A to dedifferentiated hfRPE cells (seeded at 2% and cultured over 21 days) had a small but insignificant effect. This difference in metabolism is in part due to the lower mitochondrial density in dedifferentiated hfRPE cells, as indicated by the low expression of mitochondrial protein (complex III) (Figure 3C).
Figure 3. Differentiated hfRPE cells possess a high oxidative capacity.
P1 hfRPE cells were seeded on 96-well plate at 30% or 2% density and cultured over 21 days. (A) hfRPE cells incubated with antimycin A (20 nM; 24 hr) showed a dramatic increase in lactate release into the media whereas dedifferentiated hfRPE cells did not respond to antimycin A. (B) In differentiated hfRPE (but not in dedifferentiated hfRPE), inhibition of ETC with antimycin A drives conversion of pyruvate to lactate. MCT1 at the RPE apical membrane mediates lactate efflux. (C) Differentiated hfRPE cells expressed a higher level of UQCRC2 (complex III; Abcam Cat#: MS304), an indicator of mitochondrial density.
4.3 Evaluating hfRPE metabolism using Seahorse XF24 bioanalyzer
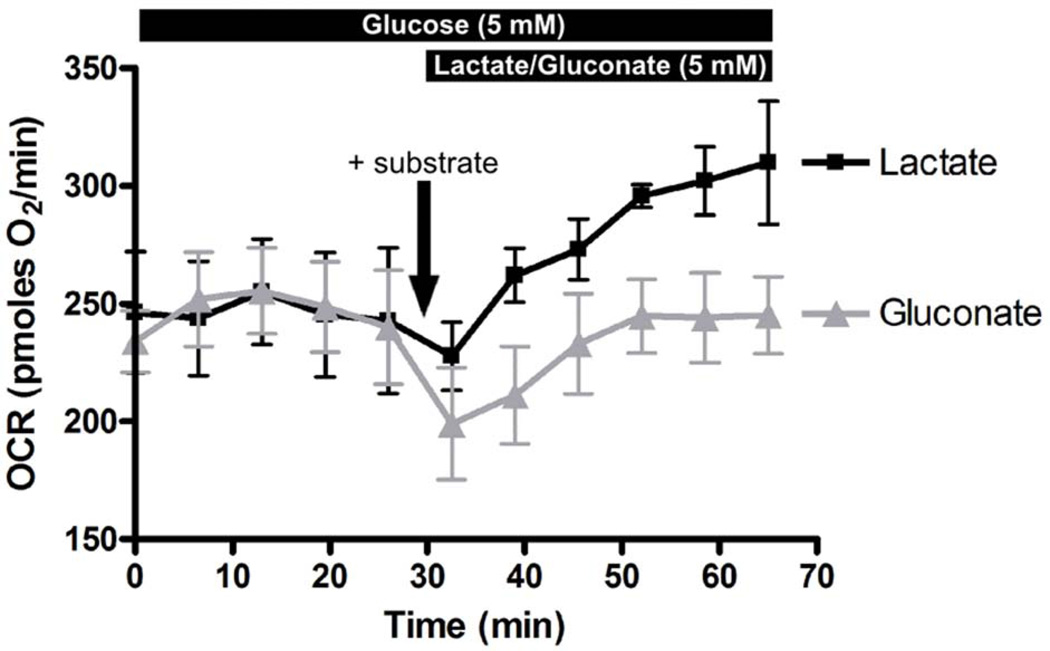
A more dynamic and direct measurement of RPE metabolism is by determining the rate at which RPE cells consume oxygen (which reflects oxidative metabolism) and release protons (which reflect glycolytic activity due to lactic acid release) using the Seahorse XF24 bioanalyzer (Seahorse Bioscience). With this technique, substrate utilization by RPE cells for oxidative metabolism is reflected by an increase in oxygen consumption rate (OCR) and decrease in extracellular acidification rate (ECAR). It is important to note that in this system, OCR and ECAR are measured in CO2/HCO3-free condition. This is achieved by replacing all NaHCO3 in the Ringer with an equimolar amount of Na-gluconate. The solution must not be equilibrated with CO2. HEPES in the Ringer helps maintain physiological pH at 7.4, although the strength of the buffer will render ECAR measurements ineffective. In our experience, hfRPE health deteriorate rapidly in the absence of HCO3 (signs of cell detachment can be observed after 1.5hr in CO2/HCO3-free Ringer), thus the Seahorse experiments with hfRPE cells should begin and end within an hour. In our studies, differentiated or dedifferentiated hfRPE cells were incubated in CO2/HCO3-free Ringer containing glucose to obtain a baseline OCR measurement (3–4 reads at 6.5 min intervals). Next, Ringer solution containing Na-lactate or Na-gluconate (50 mM) was injected into the medium for a 10-fold dilution to achieve 5 mM final lactate (or gluconate) concentration. Using this technique, we showed that differentiated hfRPE cells could use lactate as a substrate for oxidative metabolism, as shown by the dramatic increase in OCR by 31 ± 5% in Figure 4. In addition to lactate, RPE metabolism of other mitochondrial substrates (e.g., pyruvate, ketones, amino acids, fatty acids) can be evaluated with this technique.
Figure 4. Seahorse XF24 analysis of hfRPE metabolism.
P1 hfRPE cells were seeded on seahorse XF24 assay plate at 45% density and were cultured over 10 days to achieve RPE differentiation. On the day of the experiment, hfRPE cells were switched to CO2/HCO3-free HEPES-buffered Ringer (containing 5 mM glucose) and immediately transferred to the Seahorse analyzer and a baseline OCR measurement was obtained. Next lactate or gluconate (at a 10X concentrate) were injected into each well (n = 3 each) to achieve 5 mM final concentration and subsequent changes in OCR were recorded.
4.4 Evaluating RPE metabolism by measuring the rate of NADH and NADPH production
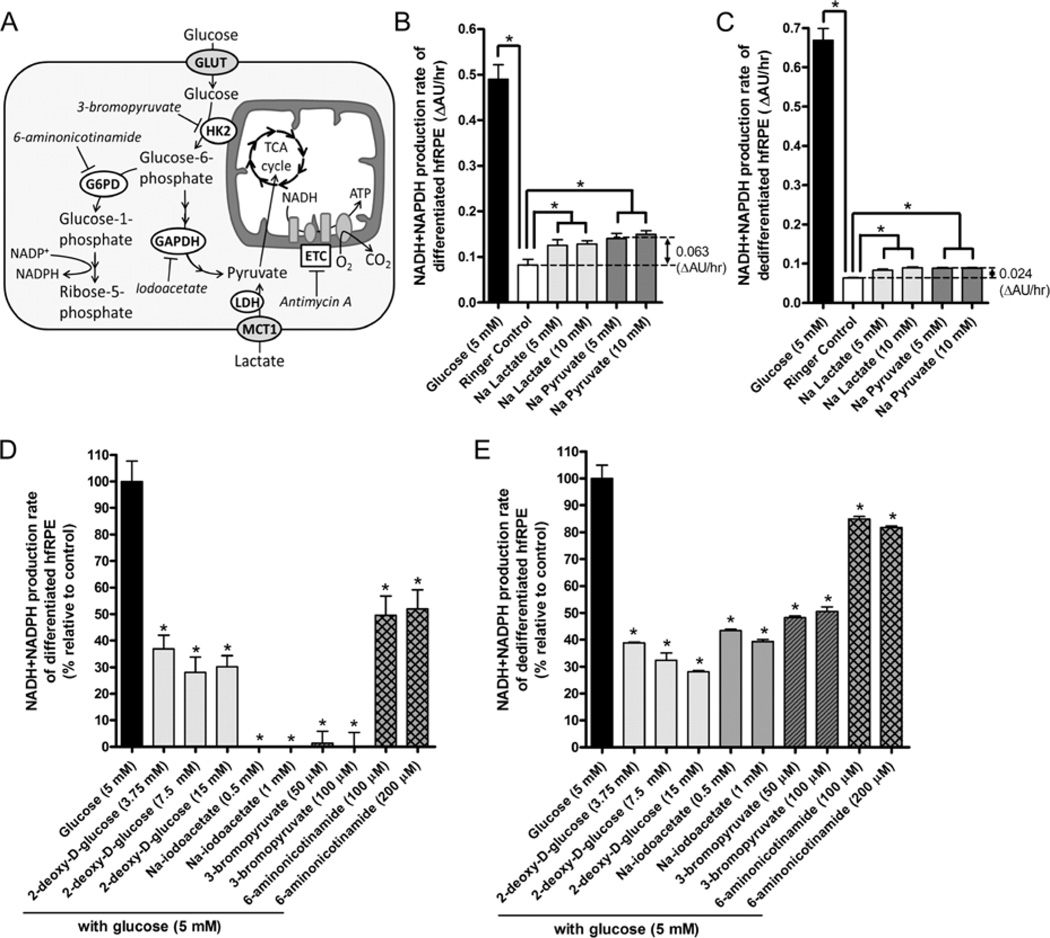
The Seahorse Bioanalyzer allows one to directly measure oxidative metabolism, but it does not provide any insights into how the RPE can utilize glucose in other metabolic pathways, such as the pentose phosphate pathway (PPP), which is responsible for synthesis of glutathione (an antioxidant), lipid, cholesterol, and ribose 5-phosphate (precursor for DNA and RNA). By measuring the rate of NADH and NADPH production (using a water-soluble form of the tetrazolium salt; purchased from Biolog Inc.) in the presence or absence of small molecule inhibitors of metabolic pathways (Figure 5A), we can indirectly determine how glucose (or other substrates; e.g., lactate) is utilized in differentiated vs. dedifferentiated hfRPE cells. Other tetrazolium salts may also be used in place of Biolog’s reagent, and resazurin (Alamar Blue Reagent) for example offers a highly sensitive alternative for measuring NADH production.
Figure 5. Water-Soluble MTT measurement of RPE metabolism (NADH/NADPH production rate).
(A) Glucose enters RPE cells via a facilitated glucose transporter (GLUT) and is metabolized to glucose-6-phosphate (G6P) by a mitochondria-associated hexokinase 2 (HK2). G6P are shunted into the PPP, a process regulated by G6PD (the first enzyme in PPP). The rest of G6P enters the glycolytic pathway, in which glyceraldehyde phosphate dehydrogenase (GAPDH) converts glyceraldehyde-3-phosphate into 1,3-biphosphoglycerate. Lactate that enters the cell via MCT1 bypasses the glycolytic pathway and is directly converted to pyruvate. Pyruvate enters the mitochondria where it is processed through the TCA cycle to generate NADH to fuel the ETC. 6-aminonicotinamide inhibits PPP by targeting G6PD. 2-deoxy D-glucose (not shown) inhibits glycolysis by acting as a competitive inhibitor for the production of G6P. 3-bromopyruvate and iodoacetate inhibits glycolysis by targeting HK2 and GAPDH, respectively. Antimycin A blocks complex III activity, thus shutting down the TCA cycle and ETC. (B) P2 Differentiated (30% density) and (C) dedifferentiated (2% density) hfRPE cells cultured over 21 days in 96-well plate were switched to glucose-free Ringer’s solution (100 µL/well) and incubated overnight at 37°C and 5% CO2. The next day, a mixture containing 10 µL of a 13X glucose, lactate, or pyruvate stock + 20 µL of 6X Biolog reagent was injected into each well (n = 5 each). Absorbance (AU) was measured at 590 nm over 2 hr at 30 min intervals and the rate of increase in absorbance (ΔAU/hr, which directly correlate with NADH and NADPH production rate) was calculated for each well. Glucose metabolism of (D) differentiated and (E) dedifferentiated hfRPE cells was evaluated in the presence of 2-deoxy-D-glucose, Na-iodoacetate, 3-bromopyruvate, and 6-aminonicotinamide (n = 5 for each condition). In these experiments, 5 µL of a 25X inhibitor stock was manually pipetted into each well, mix by tapping the plate. 30 min later, 20 µL of glucose + BiologReagent mixture (1.25 µL of a 0.5 M glucose stock + 18.75 µL of the 6X Biolog reagent) was injected into each well using a repeater pipette. Rate of increase in absorbance at 590nm was measured as previously described. Data is presented as % metabolism relative to glucose control (without any inhibitor; 100%) and Ringer control (no substrate; baseline reads set to 0%). Statistical analysis was performed using students t-test, p < 0.05 is considered statistical significant.
In these experiments, hfRPE cells were seeded (at 30% density; 50k cells/well) on 96-well plates and allowed to differentiate over 21 days. As a visual guide of RPE differentiation, one can look out for formation of “fluid-filled domes” on the RPE monolayer as indication of RPE fluid transport, which occurs only after hfRPE cells have established tight junctions and apical-basolateral polarity. One day prior to the experiment, hfRPE media was replaced with pre-warmed glucose-free CO2/HCO3-buffered Ringer’s solution containing ITS and 5% HI FBS and incubated overnight at 37°C and 5% CO2 – this step allows the RPE cells to equilibrate in a substrate-free environment. In a separate experiment with hfRPE cells on transwell filters, overnight incubation with the same glucose-free Ringer did not decrease transepithelial resistance compared to Ringer containing 5 mM glucose (data not shown). On the next day, hfRPE cells were pre-treated with various metabolic inhibitors for 30 min before injecting a mixture of glucose + 6X Biolog Reagent (Biolog Inc.; Cat: 74351). Other metabolic substrates such as lactate and pyruvate, can also be evaluated. The plate was incubated at 37°C and 5% CO2 and absorbance (AU) at 590 nm were recorded every 30 min over 2 hours (4 time points). Absorbance values were plotted against time to obtain metabolic rate (ΔAU/hr). From this experiment, we found that differentiated hfRPE cells produced NADH much more rapidly from glucose than mitochondrial substrates, lactate and pyruvate (Figure 5B), consistent with the faster kinetics of glycolysis compared to oxidative metabolism (Pfeiffer et al., 2001). Our results also suggest that dedifferentiated hfRPE cells do not possess a strong capacity for metabolizing lactate or pyruvate (Figure 5C), consistent with our findings with the Seahorse experiment.
While hfRPE cells can rely on oxidative metabolism as a source of energy, we found that differentiated hfRPE cells are exquisitely sensitive to iodoacetate (GAPDH inhibitor (Sabri and Ochs, 1971)), which effectively reduced the rate of NADH and NADPH production to zero (Figure 5D). Glucose metabolism in hfRPE cells was inhibited by 2-deoxy-D-glucose (an inhibitor of glycolysis; (Wick et al., 1957)) and its metabolism was completely blocked by 3-bromopyruvate, a specific Hexokinase 2 (HK2) inhibitor (Mathupala et al., 2006). On the other hand, dedifferentiated hfRPE cells were more resistant to iodoacetate and 3-bromopyruvate inhibition (Figure 5E), suggesting that dedifferentiated hfRPE cells possess metabolic flexibility that allows them to derive NADH through other pathways when glucose is limited. Another interesting observation is that inhibiting glucose-6-phosphate dehydrogenase (G6PD), the rate-limiting enzyme in the PPP, in differentiated hfRPE cells decreased the rate of NADH and NADPH production by ~50%. In contrast, dedifferentiated hfRPE cells responded to the same treatment with a smaller decrease in metabolic rate by ~20%, suggesting that a large fraction of glucose consumed by differentiated hfRPE cells enters the PPP. This is consistent with earlier findings in human RPE using 13C-labeled glucose in nuclear magnetic resonance (NMR) analysis (Miceli et al., 1990) and the role of RPE cells as an active producer of reduced glutathione (Davidson et al., 1994), a potent antioxidant that helps protect the RPE and retina from oxidative damage.
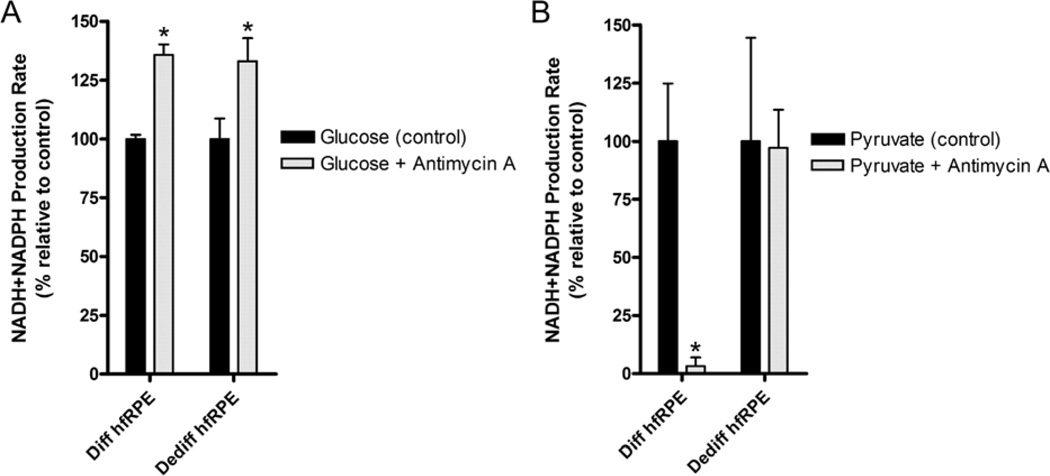
Although this technique can be used to evaluate how hfRPE cells utilize glucose or other metabolic substrates, it is important to note that cells possess a certain level of metabolic flexibility – inhibition of a specific metabolic pathway can induce a shift to other energy (and NADH) producing mechanisms that could potentially confound the results from these experiments. As an example, inhibition of oxidative metabolism in hfRPE cells using antimycin A increased the rate of NADH production (Figure 6A). This may seem counter-intuitive at first because inhibition of the TCA cycle should decrease NADH production, but instead hfRPE cells adapted by shifting its metabolism to rapidly metabolize glucose via glycolysis (as we know from its lactate release; Figure 3). On the other hand, antimycin A abolished NADH production in hfRPE cells given pyruvate, a direct mitochondrial substrate (Figure 6B). Dedifferentiated hfRPE cells did not effectively metabolize pyruvate (with barely detectable basal levels) and what little metabolism we measured, was unaffected by antimycin A.
Figure 6. Differentiated and dedifferentiated hfRPE cells metabolize glucose and pyruvate differently.
P2 differentiated (30%) or dedifferentiated (2%) hfRPE cells on 96-well plate cultured over 21 days were switched to glucose-free Ringer (containing 1X ITS and 5% HI FBS) and incubated overnight at 37°C and 5% CO2. On the day of the experiment, cells were pre-treated with 100 nM antimycin A (ETC complex III inhibitor) for 15 min before a 20 µL mixture of (A) glucose (5 mM) or (B) pyruvate (5 mM final conc.) + 6X Biolog reagent was added to each well (1.25 µL of 0.5 M substrate stock + 18.75 µL of 6X Biolog reagent). Absorbance at 590nm was obtained every 30 min over 2hr and the NADH and NADPH production rate was calculated.
Perhaps the most quantitative method for metabolic analysis of RPE cells can be achieved by using 13- carbon metabolic analysis. In this technique, RPE cells are incubated in Ringer solution containing 13C labeled substrates (e.g., glucose, lactate, lipids) over various time points (ranging from 5 min to 2 hr) before cells are harvested for 13C-labeled metabolite analysis using high performance liquid chromatography, gas-chromatography/mass spectrometry. From these experiments, it is possible to determine how glucose is incorporated into various metabolic pathways (Amaral et al., 2011): TCA cycle (e.g., citrate, succinate, fumarate) (Chertov et al., 2011), PPP (Crown et al., 2012), amino acid synthesis pathways (e.g., alanine, glutamate, aspartate) (Chertov et al., 2011). These techniques are wellestablished (see (Zamboni et al., 2009) for detailed protocol) and are routinely performed in many laboratories and metabolomics core facilities. Notably, this approach has been applied to study glucose metabolism in the neural retina (Chertov et al., 2011). 13C-labeled glucose analysis has been performed in cultured human RPE to evaluate 13C lactate release and metabolites in the PPP using NMR (Miceli et al., 1990).
5. Summary
We highlighted several tools (lactate measurement, Seahorse bioanalyzer, NADH and NADPH production rate) that can be used to evaluate RPE metabolism. From our experiments and those of others, it is apparent that RPE differentiation is associated with an increase in oxidative metabolism, and that interfering with oxidative metabolism inhibited RPE differentiation. However, beyond using the metabolic activity of the RPE as an indicator of its health and state of differentiation, the techniques highlighted here can be applied to evaluate how disease conditions (e.g., chronic oxidative stress and/or inflammation in age-related macular degeneration) could disrupt RPE metabolism, potentially causing a switch from oxidative to glycolytic metabolism. At the next level, one may perform a screening of promising compounds (e.g., antioxidants) in hfRPE cells to identify effective strategies that could prevent disease-induced metabolic switch, and better yet, reverse it. For these future studies, we present the hfRPE cell culture model as a well-established and extensively characterized in vitro system suitable for evaluating RPE metabolism in the context of retinal disease.
Highlights.
-
-
We show that primary human fetal RPE is a viable model for studying RPE metabolism.
-
-
We highlight various tools and techniques that can be used to evaluate RPE metabolism.
-
-
We show that RPE differentiation involves a shift from glycolytic to oxidative metabolism.
-
-
We demonstrate that blocking oxidative metabolism inhibits RPE differentiation.
Acknowledgements
This work was supported by NEI grant EY-012042 to NJP. The authors thank Drs. Sheldon Miller and Arvydas Maminishkis for providing us with hfRPE cells and Dr. Erin Seifert and Cynthia Moffat for providing expertise and use of the Seahorse FX24 bioanalyzer in their laboratory. The authors thank Drs. Erin Seifert and Arvydas Maminishkis for critically reviewing the manuscript.
Abbreviations
- hfRPE
Human Fetal Retinal Pigment Epithelium
- MCT
Monocarboxylate Transporter
- P0
Passage 0
- BEST1
Bestrophin1
- KD
Knock Down
- CLDN10
Claudin10
- LDH
Lactate Dehydrogenase
- SDHD
Succinate Dehydrogenase isoform D
- TCA
Tricarboxylic Acid
- ETC
Electron Transport Chain
- NMDG
N-Methyl-D-glucamine
- ITS
Insulin-Transferrin-Selenium
- THT
Taurine-Hydrocortisone-Triiodo-thyronin
- HI
Heat Inactivated
- OCR
Oxygen Consumption Rate
- ECAR
Extracellular Acidification Rate
- PPP
Pentose Phosphate Pathway
- AU
Absorbance Unit
- HK2
Hexokinase 2
- G6PD
Glucose-6-Phosphate Dehydrogenase
- NMR
Nuclear Magnetic Resonance.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adijanto J, Banzon T, Jalickee S, Wang NS, Miller SS. CO2-induced ion and fluid transport in human retinal pigment epithelium. J Gen Physiol. 2009;133:603–622. doi: 10.1085/jgp.200810169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adijanto J, Castorino JJ, Wang ZX, Maminishkis A, Grunwald GB, Philp NJ. Microphthalmia-associated transcription factor (MITF) promotes differentiation of human retinal pigment epithelium (RPE) by regulating microRNAs-204/211 expression. J Biol Chem. 2012;287:20491–20503. doi: 10.1074/jbc.M112.354761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adijanto J, Philp NJ. The SLC16A family of monocarboxylate transporters (MCTs)--physiology and function in cellular metabolism, pH homeostasis, and fluid transport. Current topics in membranes. 2012;70:275–311. doi: 10.1016/B978-0-12-394316-3.00009-0. [DOI] [PubMed] [Google Scholar]
- Agathocleous M, Harris WA. Metabolism in physiological cell proliferation and differentiation. Trends in cell biology. 2013;23:484–492. doi: 10.1016/j.tcb.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Agathocleous M, Love NK, Randlett O, Harris JJ, Liu J, Murray AJ, Harris WA. Metabolic differentiation in the embryonic retina. Nat Cell Biol. 2012;14:859–864. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral AI, Teixeira AP, Hakonsen BI, Sonnewald U, Alves PM. A comprehensive metabolic profile of cultured astrocytes using isotopic transient metabolic flux analysis and C-labeled glucose. Front Neuroenergetics. 2011;3:5. doi: 10.3389/fnene.2011.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergersen L, Johannsson E, Veruki ML, Nagelhus EA, Halestrap A, Sejersted OM, Ottersen OP. Cellular and subcellular expression of monocarboxylate transporters in the pigment epithelium and retina of the rat. Neuroscience. 1999;90:319–331. doi: 10.1016/s0306-4522(98)00427-8. [DOI] [PubMed] [Google Scholar]
- Blenkinsop TA, Salero E, Stern JH, Temple S. The culture and maintenance of functional retinal pigment epithelial monolayers from adult human eye. Methods in molecular biology. 2013;945:45–65. doi: 10.1007/978-1-62703-125-7_4. [DOI] [PubMed] [Google Scholar]
- Burke JM. Epithelial phenotype and the RPE: is the answer blowing in the Wnt? Prog Retin Eye Res. 2008;27:579–595. doi: 10.1016/j.preteyeres.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castorino JJ, Gallagher-Colombo SM, Levin AV, Fitzgerald PG, Polishook J, Kloeckener-Gruissem B, Ostertag E, Philp NJ. Juvenile cataract-associated mutation of solute carrier SLC16A12 impairs trafficking of the protein to the plasma membrane. Invest Ophthalmol Vis Sci. 2011;52:6774–6784. doi: 10.1167/iovs.10-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertov AO, Holzhausen L, Kuok IT, Couron D, Parker E, Linton JD, Sadilek M, Sweet IR, Hurley JB. Roles of glucose in photoreceptor survival. J Biol Chem. 2011;286:34700–34711. doi: 10.1074/jbc.M111.279752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crown SB, Ahn WS, Antoniewicz MR. Rational design of (1)(3)C-labeling experiments for metabolic flux analysis in mammalian cells. BMC systems biology. 2012;6:43. doi: 10.1186/1752-0509-6-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson PC, Sternberg P, Jr, Jones DP, Reed RL. Synthesis and transport of glutathione by cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1994;35:2843–2849. [PubMed] [Google Scholar]
- Davis AA, Bernstein PS, Bok D, Turner J, Nachtigal M, Hunt RC. A human retinal pigment epithelial cell line that retains epithelial characteristics after prolonged culture. Invest Ophthalmol Vis Sci. 1995;36:955–964. [PubMed] [Google Scholar]
- De Pauw A, Tejerina S, Raes M, Keijer J, Arnould T. Mitochondrial (dys)function in adipocyte (de)differentiation and systemic metabolic alterations. Am J Pathol. 2009;175:927–939. doi: 10.2353/ajpath.2009.081155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn KC, Aotaki-Keen AE, Putkey FR, Hjelmeland LM. ARPE-19, a human retinal pigment epithelial cell line with differentiated properties. Exp Eye Res. 1996;62:155–169. doi: 10.1006/exer.1996.0020. [DOI] [PubMed] [Google Scholar]
- Economopoulou M, Hammer J, Wang F, Fariss R, Maminishkis A, Miller SS. Expression, localization, and function of junctional adhesion molecule-C (JAM-C) in human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2009;50:1454–1463. doi: 10.1167/iovs.08-2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feher J, Kovacs I, Artico M, Cavallotti C, Papale A, Balacco Gabrieli C. Mitochondrial alterations of retinal pigment epithelium in age-related macular degeneration. Neurobiology of aging. 2006;27:983–993. doi: 10.1016/j.neurobiolaging.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Flannery JG, O'Day W, Pfeffer BA, Horwitz J, Bok D. Uptake, processing and release of retinoids by cultured human retinal pigment epithelium. Exp Eye Res. 1990;51:717–728. doi: 10.1016/0014-4835(90)90057-2. [DOI] [PubMed] [Google Scholar]
- Gamm DM, Melvan JN, Shearer RL, Pinilla I, Sabat G, Svendsen CN, Wright LS. A novel serum-free method for culturing human prenatal retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 2008;49:788–799. doi: 10.1167/iovs.07-0777. [DOI] [PubMed] [Google Scholar]
- Grisanti S, Guidry C. Transdifferentiation of retinal pigment epithelial cells from epithelial to mesenchymal phenotype. Invest Ophthalmol Vis Sci. 1995;36:391–405. [PubMed] [Google Scholar]
- Hu J, Bok D. A cell culture medium that supports the differentiation of human retinal pigment epithelium into functionally polarized monolayers. Mol Vis. 2001;7:14–19. [PubMed] [Google Scholar]
- Hu J, Bok D. The use of cultured human fetal retinal pigment epithelium in studies of the classical retinoid visual cycle and retinoid-based disease processes. Exp Eye Res. 2013 doi: 10.1016/j.exer.2013.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes BA, Gallemore RP, Miller SS. Transport mechanisms in the retinal pigment epithelium. In: Marmor MF, Wolfensberger TJ, editors. The retinal pigment epithelium. New York: Oxford University Press; 1998. pp. 103–134. [Google Scholar]
- Kevany BM, Palczewski K. Phagocytosis of retinal rod and cone photoreceptors. Physiology. 2010;25:8–15. doi: 10.1152/physiol.00038.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb TD, Pugh EN., Jr Dark adaptation and the retinoid cycle of vision. Progress in retinal and eye research. 2004;23:307–380. doi: 10.1016/j.preteyeres.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Li R, Maminishkis A, Banzon T, Wan Q, Jalickee S, Chen S, Miller SS. IFN{gamma} regulates retinal pigment epithelial fluid transport. Am J Physiol Cell Physiol. 2009;297:C1452–C1465. doi: 10.1152/ajpcell.00255.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao JL, Yu J, Huang K, Hu J, Diemer T, Ma Z, Dvash T, Yang XJ, Travis GH, Williams DS, Bok D, Fan G. Molecular signature of primary retinal pigment epithelium and stem-cell-derived RPE cells. Human molecular genetics. 2010;19:4229–4238. doi: 10.1093/hmg/ddq341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunt SY, Vander Heiden MG. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev Cell Dev Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- Maminishkis A, Chen S, Jalickee S, Banzon T, Shi G, Wang FE, Ehalt T, Hammer JA, Miller SS. Confluent monolayers of cultured human fetal retinal pigment epithelium exhibit morphology and physiology of native tissue. Invest Ophthalmol Vis Sci. 2006;47:3612–3624. doi: 10.1167/iovs.05-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maminishkis A, Miller SS. Experimental models for study of retinal pigment epithelial physiology and pathophysiology. Journal of visualized experiments : JoVE. 2010 doi: 10.3791/2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: cancer's double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miceli MV, Newsome DA, Schriver GW. Glucose uptake, hexose monophosphate shunt activity, and oxygen consumption in cultured human retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1990;31:277–283. [PubMed] [Google Scholar]
- Nabi IR, Mathews AP, Cohen-Gould L, Gundersen D, Rodriguez-Boulan E. Immortalization of polarized rat retinal pigment epithelium. J Cell Sci. 1993;104(Pt 1):37–49. doi: 10.1242/jcs.104.1.37. [DOI] [PubMed] [Google Scholar]
- Nordgaard CL, Karunadharma PP, Feng X, Olsen TW, Ferrington DA. Mitochondrial proteomics of the retinal pigment epithelium at progressive stages of age-related macular degeneration. Invest Ophthalmol Vis Sci. 2008;49:2848–2855. doi: 10.1167/iovs.07-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Adelman RA, Rizzolo LJ. Minimal effects of VEGF and anti-VEGF drugs on the permeability or selectivity of RPE tight junctions. Invest Ophthalmol Vis Sci. 2010;51:3216–3225. doi: 10.1167/iovs.09-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Rao VS, Adelman RA, Rizzolo LJ. Claudin-19 and the barrier properties of the human retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2011;52:1392–1403. doi: 10.1167/iovs.10-5984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer BA. Improved methodology for cell culture of human and monkey retinal pigment epithelium. In: Osborne NN, Chader GJ, editors. Prog Retin Res. Oxford: Pergamon Press; 1991. pp. 215–291. [Google Scholar]
- Pfeiffer T, Schuster S, Bonhoeffer S. Cooperation and competition in the evolution of ATP-producing pathways. Science. 2001;292:504–507. doi: 10.1126/science.1058079. [DOI] [PubMed] [Google Scholar]
- Porter GA, Jr, Hom J, Hoffman D, Quintanilla R, de Mesy Bentley K, Sheu SS. Bioenergetics, mitochondria, and cardiac myocyte differentiation. Progress in pediatric cardiology. 2011;31:75–81. doi: 10.1016/j.ppedcard.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rath PP, Jenkins S, Michaelides M, Smith A, Sweeney MG, Davis MB, Fitzke FW, Bird AC. Characterisation of the macular dystrophy in patients with the A3243G mitochondrial DNA point mutation with fundus autofluorescence. The British journal of ophthalmology. 2008;92:623–629. doi: 10.1136/bjo.2007.131177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzolo LJ, Peng S, Luo Y, Xiao W. Integration of tight junctions and claudins with the barrier functions of the retinal pigment epithelium. Prog Retin Eye Res. 2011;30:296–323. doi: 10.1016/j.preteyeres.2011.06.002. [DOI] [PubMed] [Google Scholar]
- Sabri MI, Ochs S. Inhibition of glyceraldehydes-3-phosphate dehydrogenase in mammalian nerve by iodoacetic acid. J Neurochem. 1971;18:1509–1514. doi: 10.1111/j.1471-4159.1971.tb00013.x. [DOI] [PubMed] [Google Scholar]
- Sonoda S, Spee C, Barron E, Ryan SJ, Kannan R, Hinton DR. A protocol for the culture and differentiation of highly polarized human retinal pigment epithelial cells. Nature protocols. 2009;4:662–673. doi: 10.1038/nprot.2009.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele FR, Chader GJ, Johnson LV, Tombran-Tink J. Pigment epithelium-derived factor: neurotrophic activity and identification as a member of the serine protease inhibitor gene family. Proc Natl Acad Sci U S A. 1993;90:1526–1530. doi: 10.1073/pnas.90.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikova NV, Maminishkis A, Barb JJ, Wang F, Zhi C, Sergeev Y, Chen W, Edwards AO, Stambolian D, Abecasis G, Swaroop A, Munson PJ, Miller SS. Transcriptome analysis and molecular signature of human retinal pigment epithelium. Human molecular genetics. 2010;19:2468–2486. doi: 10.1093/hmg/ddq129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyni T, Paetau A, Strauss AW, Middleton B, Kivela T. Mitochondrial fatty acid beta-oxidation in the human eye and brain: implications for the retinopathy of long-chain 3-hydroxyacyl-CoA dehydrogenase deficiency. Pediatric research. 2004;56:744–750. doi: 10.1203/01.PDR.0000141967.52759.83. [DOI] [PubMed] [Google Scholar]
- Udar N, Atilano SR, Memarzadeh M, Boyer DS, Chwa M, Lu S, Maguen B, Langberg J, Coskun P, Wallace DC, Nesburn AB, Khatibi N, Hertzog D, Le K, Hwang D, Kenney MC. Mitochondrial DNA haplogroups associated with age-related macular degeneration. Invest Ophthalmol Vis Sci. 2009;50:2966–2974. doi: 10.1167/iovs.08-2646. [DOI] [PubMed] [Google Scholar]
- Valtink M, Engelmann K. Culturing of retinal pigment epithelium cells. Developments in ophthalmology. 2009;43:109–119. doi: 10.1159/000223844. [DOI] [PubMed] [Google Scholar]
- Wang FE, Zhang C, Maminishkis A, Dong L, Zhi C, Li R, Zhao J, Majerciak V, Gaur AB, Chen S, Miller SS. MicroRNA-204/211 alters epithelial physiology. FASEB J. 2010;24:1552–1571. doi: 10.1096/fj.08-125856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wick AN, Drury DR, Nakada HI, Wolfe JB. Localization of the primary metabolic block produced by 2-deoxyglucose. J Biol Chem. 1957;224:963–969. [PubMed] [Google Scholar]
- Zamboni N, Fendt SM, Ruhl M, Sauer U. (13)C-based metabolic flux analysis. Nature protocols. 2009;4:878–892. doi: 10.1038/nprot.2009.58. [DOI] [PubMed] [Google Scholar]
- Zhao C, Yasumura D, Li X, Matthes M, Lloyd M, Nielsen G, Ahern K, Snyder M, Bok D, Dunaief JL, LaVail MM, Vollrath D. mTOR-mediated dedifferentiation of the retinal pigment epithelium initiates photoreceptor degeneration in mice. J Clin Invest. 2011;121:369–383. doi: 10.1172/JCI44303. [DOI] [PMC free article] [PubMed] [Google Scholar]