Abstract
By enforcing specific pollinator interactions, Aquilegia petal nectar spurs maintain reproductive isolation between species. Spur development is the result of three-dimensional elaboration from a comparatively two-dimensional primordium. Initiated by localized, oriented cell divisions surrounding the incipient nectary, this process creates a pouch that is extended by anisotropic cell elongation. We hypothesized that the development of this evolutionary novelty could be promoted by non-mutually exclusive factors, including (i) prolonged, KNOX-dependent cell fate indeterminacy, (ii) localized organ sculpting and/or (iii) redeployment of hormone-signalling modules. Using cell division markers to guide transcriptome analysis of microdissected spur tissue, we present candidate mechanisms underlying spur outgrowth. We see dynamic expression of factors controlling cell proliferation and hormone signalling, but no evidence of contribution from indeterminacy factors. Transcriptome dynamics point to a novel recruitment event in which auxin-related factors that normally function at the organ margin were co-opted to this central structure. Functional perturbation of the transition between cell division and expansion reveals an unexpected asymmetric component of spur development. These findings indicate that the production of this three-dimensional form is an example of organ sculpting via localized cell division with novel contributions from hormone signalling, rather than a product of prolonged indeterminacy.
Keywords: organ shape, TCP4, virus-induced gene silencing, hormones, gene expression, Aquilegia
1. Background
Plant developmental studies commonly focus on organ identity. However, the most variable aspects of morphology relate to organ elaboration, a broad class of features downstream of organ identity that include organ fusion, colour, shape and other structural elaborations. We have learned a great deal about the genetic basis of complex lateral organ shape from studies of leaves, but the adaptive significance of leaf shape diversity is often difficult to quantify [1]. On the contrary, variation in petal elaboration has clear adaptive significance linked to pollinator interactions: for example, petals display UV pollinator guides, provide landing platforms, create nectar tubes and mimic female pollinators. All these features are thought to coevolve rapidly with pollinators (reviewed [2]).
Petal nectar spurs are a clear example of extreme petal shape modification and appear to contribute to high speciation rates in the New World clade of the genus Aquilegia [3,4]. In Aquilegia, punctuated evolution of longer spur lengths correlates with shifts to pollinators possessing longer tongues [5,6]. Early hypotheses suggested that these spurs are produced by the activity of ‘meristematic knobs’ flanking the petal attachment point [7,8]. On the contrary, a recent study tracking HISTONE4 expression demonstrated that cell divisions are not localized to the attachment point [9]. Instead, the spur develops in two distinct developmental phases: early cell divisions become concentrated in the area surrounding the nascent spur, producing a nectary cup (Phase I; figure 1a). These divisions cease when the spur is still a small fraction of its final length, meaning that highly oriented cell elongation drives the majority of spur growth (Phase II). Variation in this second phase, involving anisotropic cell elongation, is the basis for diversity in spur length across the genus [9].
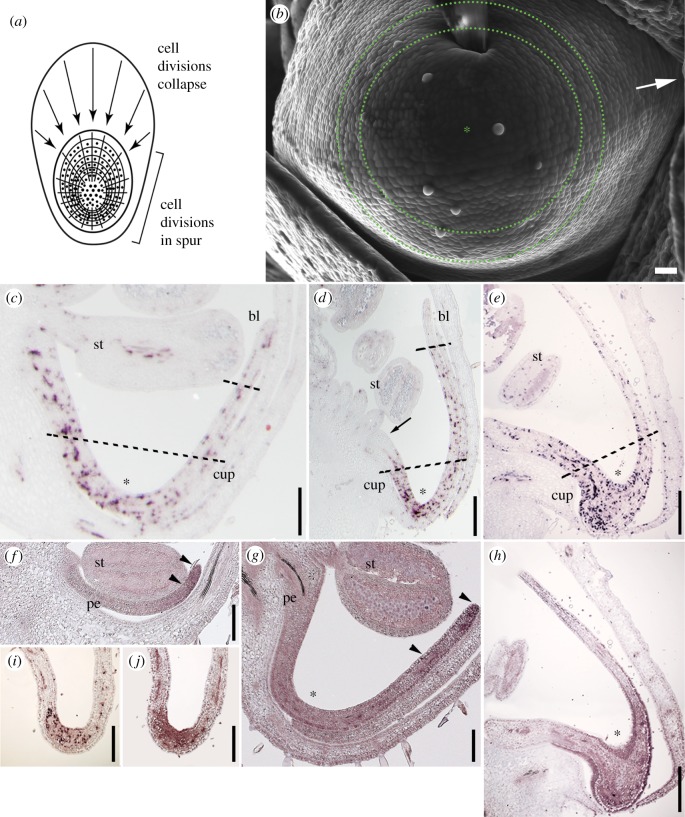
Figure 1.
Phase I of Aquilegia petal spur development. (a) Schematic two-dimensional projection of the spur showing modified cell arrest front with cell divisions localized to the nascent spur (based on [8]). Cell divisions (dots) cease in a wave that progresses circumferentially from the entire margin of the organ towards the presumptive nectary. (b) SEM of abaxial surface of 2 mm petal cup. Arrow points to petal attachment point. Zone of tangentially oriented cell walls is highlighted by dotted circles. (c–e) AqHIS4 marking cell division during petal development, which was used as a guide for tissue sampling as indicated by dashed lines. In each panel, asterisks mark the presumptive nectary; st, stamen; bl, blade sample; cup, cup sample. (c) Petal primordium from early stage 10 meristem (approx. 1 mm petal). Cell divisions are diffuse throughout the organ but just beginning to decline in the blade region. (d) Petal primordium from mid-stage 10 meristem (approx. 3 mm petal). Cell divisions have ceased in the proximal attachment region (arrow) and the distal blade region (bl) but are concentrated in the nascent spur (asterisk). (e) Petal primordium from late stage 10 meristem (approx. 5 mm petal). Cell divisions are becoming more concentrated in area surrounding the nectary. (f–h) AqTCP4 expression in early developing petals. In each panel, arrowheads indicate the extent of strongest expression. (f) Stamen (st) and petal (pe) primordia in stage 8 floral meristem. (g) Stamen (st) and petal (pe) primordia in early stage 10 floral meristem. (h) Petal primordium from late stage 10 meristem. (i–j) Spur tips from stage 11 meristem. (i) AqHIS4 expression shows that cell divisions have contracted to a domain only marking the nascent nectary. (j) Corresponding AqTCP4 expression overlaps with this domain. Scale bars, 30 µm in b, 200 µm in c, f and g, 0.5 mm in d, i and j, and 1 mm in e and h. (Online version in colour.)
With this framework, we ask the following question: what developmental modules underlie the early phase of spur formation? If we focus just on Phase I, in which cell divisions localize to a specific region, the question centres on how two factors—(i) spatial control of the transition from cell division to expansion and (ii) patterned orientation of new cell walls—direct three-dimensional organ sculpting. One attractive hypothesis is that maintenance of indeterminacy may contribute to petal spur development. Candidate genes for this mechanism include the type I KNOX gene SHOOTMERISTEMLESS (STM). Many homologues of STM play critical roles in the maintenance of the shoot apical meristem (SAM) by promoting cell division and suppressing differentiation [10]. Additionally, in taxa with compound leaves these genes play similar roles in prolonging indeterminate growth [11]. Two Antirrhinum mutants that develop spur-like structures were found to be caused by ectopic expression of homologues of Arabidopsis STM [12]. Recently, an investigation of the closely related, spurred genus Linaria found that the KNOX orthologues are expressed in petals, including the spur-producing ventral petal [13]. These findings suggest a model that places petal nectar spurs within the context of a broad spectrum of complex lateral organs that use KNOX genes to maintain indeterminacy in order to produce leaflets, lobes and even adventitious meristems (reviewed in [14,15]). While our previous study ruled out the existence of conspicuous ‘meristematic knobs’, it is still possible that KNOX genes are involved in nascent spur formation and promote a mechanism involving cell fate indeterminacy.
Alternatively, it is possible that genes controlling the late transition between cell proliferation and expansion could generate the extreme curvature of the petal spur without any contributions from the KNOX genes. In both simple and complex lateral organs, interacting genetic modules have been described that regulate laminar curvature (reviewed in [16]). The localization of cell division-promoting proteins, such as the GROWTH-REGULATING FACTORS (GRFs) and their interacting cofactors the GIFs, becomes delimited by division-repressing loci, including members of the TEOSINTE BRANCHED/CYCLOIDEA/PCF (TCP) transcription factor family. Broadly, these pathways govern the localization and timing of cell divisions as well as the subsequent pattern and orientation of cell elongation. The misregulation of either cell division or cell expansion results in curled, wrinkled or ruffled laminae (e.g. [17–19]). Theoretically, the observed localization of cell divisions in the nascent petal spur [9] could be sufficient to create the outpocketing of the spur cup from the lamina [20]. So, could spur development be simply owing to localized expression of genes that promote cell division in petals?
The broad goal of this study is to identify developmental modules associated with the localized cell divisions seen in the Aquilegia spur during its initial outpocketing. We use tissue dissection combined with RNA-seq to identify a number of candidate pathways. We do not detect any contribution from indeterminacy factors such as the type I KNOX transcription factors, but present both expression and functional evidence that localized tissue sculpting plays a critical role. Further, strong evidence for the relocalization of many auxin signalling-related factors suggests a novel role for this hormone in spur development. Together, these findings indicate that the Aquilegia petal spur is an example of extreme organ curvature rather than a product of prolonged meristematic indeterminacy.
2. Material and methods
(a). Plant material
Transcriptome sequencing, functional gene knockdown by virus-induced gene silencing (VIGS) and in situ hybridization were carried out on Aquilegia coerulea L. ‘Origami’, as previously described [21]. All plants were grown in 14 h photoperiods at 18°C during day and 15°C during night.
(b). RNA-sequencing and analysis
Triplicate pools of the distal 0.5 mm of petal spur cups and blades were removed under a dissecting microscope at three stages over petal spur development and immediately flash frozen in liquid nitrogen (1 mm, 3 mm and 6–7 mm spur length; figure 1c–e). For each bioreplicate, we dissected the entire relevant section from each of four to five spurs from a single flower, with separate bioreplicates drawn from different plants. Total RNA was isolated using the Qiagen RNeasy Micro kit (Qiagen, Valencia, CA, USA), including a repeated elution step. Intact RNA (as determined by Agilent Bioanalyzer) was used as input for Illumina TruSeq RNA (Illumina, San Diego, CA, USA) library generation. Residual primers and any primer dimer were eliminated using a second AMPure bead clean-up (Beckman Coulter, Danvers, MA, USA). All libraries were quality confirmed for correct size distributions by Bioanalyzer, quantified by QBIT (Life Technologies, Carlsbad, CA, USA) and quantitative PCR using the SYBR Fast Illumina Library Quantification Kit (Kapa Biosystems, Wilmington, MA, USA) and pooled in order to give equal coverage from each library over multiple HiSeq2000 lanes, using version 2 chemistry. All Illumina data were assessed for basic quality with FastQC v. 0.10.0 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were then trimmed all to a minimum of Q28 with FASTQ Quality Trimmer (FASTX-Toolkit v. 0.0.13; http://hannonlab.cshl.edu) or trimmed to 50 bp, whichever was shorter. Read mapping was performed with tophat v. 2.0.7, using the Aquilegia195 annotation as reference [22]. Transcripts were assembled for individuals using cufflinks v. 2.1.1 [23], using default parameters. Resultant files were converted, sorted and indexed with samtools v. 0.1.18 (http://samtools.sourceforge.net/). Uniquely mapped reads were counted in 24 823 gene models and converted to count-based data using HTSeq v. 0.5.3 in Python v. 2.7.3 (http://www-huber.embl.de/users/anders/HTSeq). Because counting variance is much higher for lowly expressed transcripts, we excluded all gene models with fewer than an average of 1 cpm in three samples for each blade/cup comparison. We assessed differential expression among the remaining genes using edgeR v. 3.3.8 [24,25] in Rstudio v. 0.98.501, using triplicate biological replicates for every tissue and time point.
(c). Identification and isolation of candidate genes
In order to further investigate mRNA expression patterns over time in selected genes, Aquilegia homologues of the Arabidopsis thaliana KNOX genes STM and KNAT1, as well as TCP4, were identified by using BLAST [26] of the A. thaliana nucleotide sequences against the publicly available Aquilegia coerulea L. ‘Origami’ genome sequence [22]. The Aquilegia homologue of HISTONE4 (HIS4) has been previously characterized [9]. For KNOX genes, nine annotated loci were identified. In two cases, the loci as annotated appeared to represent the separate 5′ and 3′ ends of a single gene. Examination of the genomic locus allowed prediction of contiguous reading frames; thus, a total of seven loci were analysed. For TCP genes, 13 annotated loci appear to represent TCP family members, but only three of these fall into class I of the TCP-C subfamily based on characteristics outlined in [27]. In order to determine the exact orthology of the putative Aquilegia TCP4 and KNOX loci, we performed phylogenetic analyses using a maximum-likelihood approach as implemented by RAxML in the CIPRES web portal [28–30]. In both cases, amino acid alignments were constructed using conserved domains of gene sequences from multiple core eudicot and monocot taxa. All accession numbers are displayed in electronic supplementary material, figure S2.
(d). In situ hybridization
Once orthology was established, fragments of AqTCP4, AqSTM and AqKN were designed that did not include highly conserved domains and were amplified from A. vulgaris cDNA using PCR (see electronic supplementary material, table S1 for primer sequences). Resultant PCR products were cloned using the TOPO-TA Cloning Kit (Life Technologies). Identity and orientation of the inserts was confirmed by sequencing. All in situ hybridization steps were conducted as described by Kramer [31]. Results were visualized in the Harvard Center for Biological Imaging on a Zeiss AxioImager Z2 microscope. Developmental floral stages were classified according to Ballerini & Kramer [32] as reported in electronic supplementary material, table S2. The position of the presumptive nectary can be determined by comparing serial sections and identifying the sections that contain the longest extent of the developing spur. Logically, this must represent a radial longitudinal section in which the distal tip is the nascent nectary.
(e). Virus-induced gene silencing
The VIGS inoculation of the TRV2-AqANS positive control plasmid was performed as previously described [33,34]. To generate the TRV2-AqTCP4-AqANS construct, a 300 bp fragment of AqTCP4 was amplified with primers that added EcoRI and Xba sites to the respective 5′ and 3′ ends of the PCR product (TCP4_EcoR1: CGGAATTCCGTATCAAGAAGGCAAAGGCTGC, TCP4_Xba 1: CGTCTAGACGAATGGGAAAGAAAGACTTAATGG). This PCR product was used to produce the TRV2-AqTCP4-AqANS construct as described by Kramer et al. [34]. Four sets of 100–110 Aquilegia coerulea ‘Origami’ plants at the four to six true leaf stage were vernalized at 4°C for four weeks; 1 d after the plants had been removed from vernalization, they were treated as described for seedlings in [33]. Seventy-eight control plants were treated with TRV1 and TRV2-AqANS. Flowers showing strong AqANS silencing were photo-documented and, on maturation, dissected. Individual organs were photographed using a Canon X type digital SLR camera (Canon, USA). For flowers showing silencing, petals were either frozen at −80°C for subsequent RNA expression analysis or fixed in freshly prepared, ice-cold formalin–acetic acid–alcohol for scanning electron microscopy (SEM) analysis. This process was also repeated for several unsilenced flowers from the TRV2-AqTCP4-AqANS cohort, as well as flowers that were treated with TRV2-AqANS as controls. SEM analysis and light microscopy was performed as described previously [34].
(f). Expression analysis of virus-induced gene silencing-treated organs
To confirm AqTCP4 silencing, total RNA was isolated and cDNA was prepared from floral organs as described in [35,36]. qRT-PCR analysis was carried out using PerfeCTa SYBR Green FastMix, Low ROX for qPCR (Quanta BioSciences, Gaithersburg, MD, USA) in a Stratagene (Santa Clara, CA, USA) Mx3005P QPCR System. A list of primers is included in electronic supplementary material, table S1. AqTCP4 transcript abundance was calculated relative to AqIPP2 (isopentyl pyrophosphate:dimethylallyl pyrophosphate isomerase) using the ddCt method [37]. Three biological replicates were performed with three technical replicates each.
(g). Scanning electron microscopy analysis
Images of spurs from VIGS-treated plants were obtained on a JEOL JSM-6010 LV SEM. Length and width of cells were manually measured using ImageJ software, and length/width was calculated for each cell. For wild-type, the regions selected for analysis were the blade, mid-spur and nectary cup. TCP4-VIGS spur analysis included the adjoining outgrowth and distal outgrowth regions. Twenty cells were measured in each region.
3. Results
(a). Developmental and molecular characterization of the Phase I petal spur
Previous work [9] defined a model of spur development in which a wave of cell division cessation progresses simultaneously from the entire margin of the organ and moves towards the nascent nectary, promoting a localized bulge that constitutes the first stage of spur development (figure 1a). The transition from cell division to expansion that marks Phase I begins when the petal is approximately 1 mm in length, as measured from the petal attachment point to the nascent spur tip, which corresponds to what we have defined as early stage 10 (staging given in electronic supplementary material, table S2; figure 1c). Close examination of this early stage 10 spur pocket with SEM reveals a distinct orientation of cell walls around the nascent nectary at the spur tip such that newly formed cell walls are tangentially arranged around the nectary (figure 1b). As the petal continues to differentiate throughout stage 10, a progressively narrowing region of cell division becomes restricted to the developing nectar spur (figure 1d–e). These cell divisions are last observed as a small crescent of activity in the nascent nectary during early stage 11, when the spur is approximately 7–8 mm in length (figure 1i).
In order to begin to assess which genes control this process, we conducted a developmental transcriptome study on microdissected regions of very young petals throughout Phase 1 using RNA-seq. We dissected terminal 4–16 mm2 tissue sections from regions of the cup and blade of both 1 mm and 3 mm spurs, and also spur tip tissue from 6–7 mm spurs, which are undergoing the transition to rapid anisotropic elongation (denoted by dashed lines in figure 1c–e). This sampling covers Phase I from its earliest through late stages and should encompass the entire period during which substantial cell divisions are localized to the spur cup. We aligned an average of 43 million reads from each of three biological replicates for every sample and time point to the 24 823 gene models in the JGI A. coerulea genome build [22]. After removing models to which less than half the bioreplicates gave confident mapping, 16 393 genes were included in the differential expression analysis at the 1 mm spur stage and 16 515 at the 3 mm spur stage. Of these, the number of genes that were significantly differentially expressed (DE) between any two tissues or time points was 4008. To determine which genes are consistently DE between the blade and cup regions, we sought the intersection of DE genes from each time point (1 mm stage DE, 653 genes; 3 mm stage DE, 1802 genes; electronic supplementary material, dataset 1). This list consisted of the majority of the DE genes from the early 1 mm contrast, 464 of which remained DE at the 3 mm stage. This reveals retention of programs activated early in spur development, but considerable elaboration and gene expression differentiation at the later stage (we observed approx. 3× the quantity of DE genes at 3 mm compared with 1 mm).
Several trends are evident among the group of genes that continue to be DE at both early stages. First, none of the Aquilegia type 1 KNOX homologues come up as DE. Second, the majority of type 1 KNOX loci fail default minimal expression filters for all tissues and time points. Third, we observe many factors governing cellular proliferation and expansion among the most highly expressed of the consistently DE genes. For instance, the cell division control factors AqTCP4 and AqANGUSTIFOLIA3 (AqAN3) stand out among the most highly expressed DE genes at both time points in the distal blade margin and spur cup, respectively. AqTCP4 shows very high expression in both tissues but is significantly higher in the blade at the 1 mm and 3 mm stages. By contrast, AqAN3 (also known as GRF-INTERACTING FACTOR1 or GIF1), while also highly expressed, is enriched in the spur cup (electronic supplementary material, figure S1). Many GRF homologues that might be expected to partner with AqAN3 are highly expressed but are not DE between the samples.
In addition to enrichment of AqAN3 in the early developing cup, our data implicate other candidate processes, with many significant DE genes associated with the hormone auxin (electronic supplementary material, figure S1). We see coordinated regulation of genes responsible for auxin synthesis; for example, downregulation of 3 YUCCA6 homologues in the cup at both time points while the Aquilegia CYP71A homologue, also implicated in auxin synthesis, is highly upregulated in all cup samples. Taking the intersection of DE genes at 1 mm and 3 mm, we note that the highest confidence DE gene in spur cups at both time points (lowest false discovery rate: 5.4 × 10−31 and 7.2 × 10−54, respectively) is an Aquilegia homologue of STYLISH1, a transcription factor that regulates auxin biosynthesis [38]. A second Aquilegia homologue of STY is also DE with enrichment in both cup time points. The two AqSTY genes show increasing expression over time in growing spur cups. Genes known to be downstream of both auxin and brassinosteroid (BR) signalling are also DE, including 12 Aquilegia homologues of SAUR (SMALL AUXIN UP RNA), which are upregulated in the latest spur time point (7 mm cups). In addition, several AUXIN RESPONSE FACTOR (ARF) family members show enrichment in the spur cup, particularly Aquilegia homologues of ARF3/ETTIN and ARF8. Both of these genes are very highly expressed in all tissues and time points sampled, but are significantly enriched in the 1 mm cup. In terms of the BR-signalling pathway, one of the most strongly DE genes in the 1 mm cup is the Aquilegia homologue of DWARF4, a cytochrome P450 that catalyses the flux-determining step in the BR synthesis pathway [39]. The Aquilegia homologue of BEH4, a member of the BZR1 family of BR response proteins [40], is also strongly enriched in the 3 mm cup.
(b). In situ hybridization confirms RNA-seq results for AqTCP4 and Aquilegia type I KNOX genes
Using orthologue-specific probes (electronic supplementary material, figure S2), we examined the expression of the Aquilegia orthologue of the cell division regulator TCP4, AqTCP4, across all stages of floral development. At later stages, when all floral organs are differentiating, AqTCP4 expression is most strongly detected in the petals. During stages 8–10, expression is relatively broad but is strongest in the petal margin (figure 1f–g). In older petals, the most intense expression moves down into the developing spur (figure 1h). Thus, in situ detection agrees with the RNA-seq data, showing a dynamic AqTCP4 expression profile during development, consistent with what has been observed for TCP4 in Arabidopsis thaliana [19]. If the AqHIS4 and AqTCP4 expression domains are compared, we see that AqTCP4 expression is highest in the region where cessation of cell division is under way, such that overlap between AqTCP4 and AqHIS4 can be observed (figure 1e,h). This is true even until the very last stages of the Phase I/II transition when cell divisions only persist in the presumptive nectary (figure 1i–j).
In order to confirm the lack of indeterminacy-mediating class I KNOX gene expression detected by the RNA-seq, we also examined Aquilegia homologues of the Arabidopsis class I KNOX gene subfamily, which includes the locus KNAT1/BP as well as STM (electronic supplementary material, figure S2). We observed strong association between AqHIS4 and AqSTM1 localization in the SAM and early developing compound leaves, as well as broad AqKN expression in young floral buds (electronic supplementary material, figure S3–S5). However, neither AqSTM1 nor AqKN were ever detected at any stage of petal development, including even broader temporal sampling through stage 11 (electronic supplementary material, figures S4–S5), although AqSTM could be detected in the developing carpels of the same sections (data not shown).
(c). AqTCP4 restrains proliferation in a restricted domain of the spur tube
Both our transcriptome and in situ expression data suggest that the dynamic expression of cell proliferation regulators is important for spur development. In order to disrupt this functional module, we used VIGS via the Tobacco rattle virus (TRV) system to knock down AqTCP4 mRNA expression. An approximately 300 bp fragment from the highly variable N-terminal region was used to specifically target AqTCP4 relative to other TCP family members. Aquilegia ANTHOCYANIDIN SYNTHASE (AqANS) was simultaneously targeted from the same TRV2 construct in order to visually identify plants exhibiting silencing.
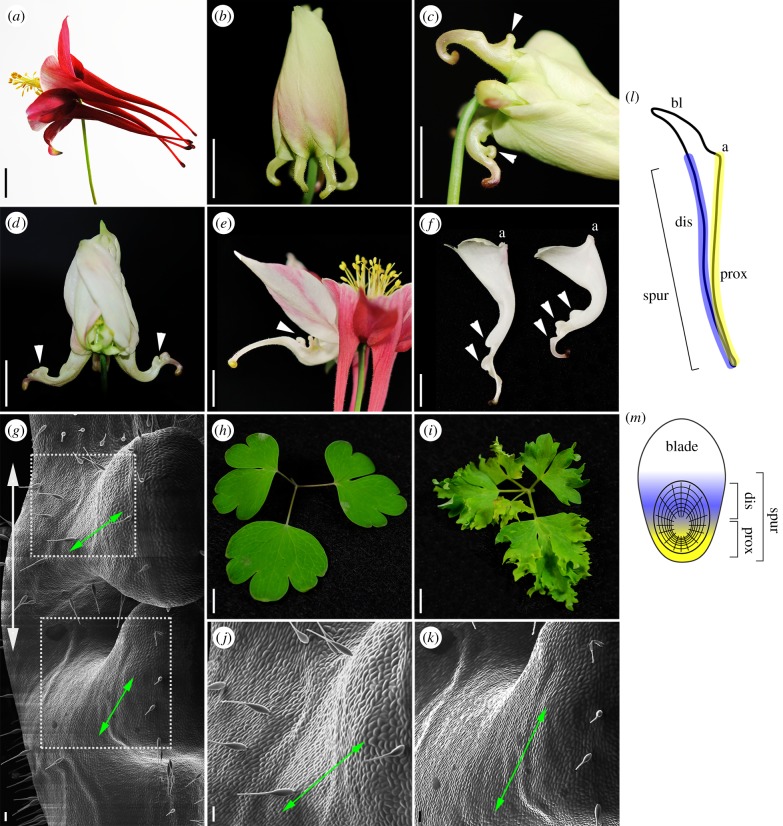
We generated a total of 44 independent plants that displayed silencing in a total of 79 flowers. The majority of these plants also exhibited some AqANS silencing in leaves. All observed perturbations of floral morphology affected the petals (figure 2b–g). Relative to control petals, AqTCP4 was downregulated in test petals exhibiting AqANS silencing an average of eightfold, as assessed by qRT-PCR (range 7–12-fold). As expected with VIGS, we recovered a range of phenotypes: in 43 flowers the petal phenotype was limited to increased spur curvature, while in 36 flowers we obtained strong silencing with a highly consistent phenotype (figure 2b–g). AqTCP4-silenced petals exhibited extreme morphological distortions on the side of the spur away from the floral pedicel (white arrowheads in figure 2c–f), which we term the distal compartment (figure 2l–m). This distortion includes localized ectopic outgrowths, which do not appear to be fully formed de novo spurs, as they never show any sign of nectary differentiation that is typical of Aquilegia spurs (figure 2d–f). Instead, we consider them to be buckles in the lamina owing to over-proliferation in the distal compartment relative to the proximal compartment, which never displayed such outgrowths. Over the course of development, the AqTCP4-silenced spurs first bent inward towards the stem (figure 2b), but then snapped outward owing to localized distortions in the lamina of the distal compartment (figure 2c–f). Furthermore, SEM examination of the epidermis in these outgrowths reveals many small, unexpanded cells, consistent with prolonged cell division (figure 2g; electronic supplementary material, figure S6). SEM analysis also revealed perturbation of both cell orientation and anisotropic expansion in AqTCP4-silenced petals (figure 2g,j–k; electronic supplementary material, figure S6). In unsilenced petal spurs, Phase I cell walls are tangentially oriented around the nascent nectary (figure 1a), a pattern that probably facilitates their later anisotropic expansion along the long axis of the spur. In the buckled regions of AqTCP4-silenced spurs, however, cell files are less organized and appear to be reoriented such that they run parallel to the new axis of outgrowth, which is itself perpendicular to the original spur axis (figure 2g,j–k). Aside from these phenotypes in the distal compartment of the spur tube, we additionally observed a much higher degree of dissection in the leaf margins of strongly silenced plants (figure 2i).
Figure 2.
AqTCP4 functions to restrain cell division in the distal compartment of the spur. (a) Untreated Aquilegia flower displaying essentially straight petal nectar spurs. (b–g) AqTCP4-silenced floral phenotypes. Arrowheads point to distal compartment outgrowths. (b–d) Strongly silenced flowers at increasing developmental stages (one spur is removed in d). Note how spurs bend inward in b but then outward in c and d. (e) Flower at anthesis, displaying one silenced spur (white) and four unsilenced (coloured). (f) Dissected petal spurs at anthesis displaying excess growth on distal (left) side. (g,j–k) SEM of outgrowths, showing reoriented cell files. Primary axis of spur is shown with vertical arrow, angled arrows indicate direction of reorientation of cell files in outgrowth regions. Dashed boxes indicate the higher magnification regions shown in j (top box) and k (bottom box). (h) Unsilenced leaf. (i) AqTCP4-silenced leaf. (l) Mature petal profile indicating blade (bl), spur and attachment point (a), as well as proximal (prox) and distal (dis) compartments. (m) Two-dimensional projection of spur in panel j showing proximal (prox) and distal (dis) compartments. Scale bars, 1 cm in a–f and h–i, 100 µm in g and j–k. (Online version in colour.)
4. Discussion
The production of the three-dimensional nectar spur from the comparatively two-dimensional Aquilegia petal primordium begins with cell divisions that progressively become more localized, collapsing down towards the nascent nectary (figure 1a). These oriented cell divisions create a patterned cup that achieves its final size and shape through highly oriented cell elongation [9]. After sampling stages throughout Phase I of spur development, we detected no differential expression of type I KNOX transcripts, of which there are five in Aquilegia (electronic supplementary material, figure S1). In fact, the expression of most KNOX homologues across all samples is below default filters for minimal reliable detectable expression using edgeR, and AqSTM average raw expression counts are thousands of times lower than those for other major transcription factors. Consistent with this, no expression of either AqSTM or AqKN was detected using in situ hybridization across an even broader range of developmental stages.
As discussed above, KNOX-dependent indeterminacy is not the only way to prolong cell divisions in a lateral organ. Many lateral organs appear to use roughly the same genetic module to control the spatial localization of cell divisions and their eventual cessation [41]. In our case, the single Aquilegia orthologue of the cell division suppressor TCP4 is expressed in the blade margin of 1–3 mm petals, as detected by both RNA-seq and in situ, but expression then shifts down into the petal spur at later stages (figure 1f–h). In the transition from the 1 mm spur cup to the 3 mm spur cup, we also observe a coordinated repression of 7 GRFs (in addition to the GIF AqAN3), suggesting orchestrated coregulation as cell divisions decline in this region. This degree of coregulation among homologues of a genetic module primarily known from Arabidopsis is notable, given the deep divergence, approximately 120 million years, between Aquilegia and Arabidopsis [42]. Taken together, these data support the hypothesis that Aquilegia petal spurs do not belong to the spectrum of complex lateral organs that use prolonged indeterminacy to generate varied form, but rather are better understood as extreme examples of organ curvature.
Studies in Arabidopsis have revealed that AtTCP4 plays a critical role in regulating petal development, most probably by repressing cell division [43], and modification of TCP4 homologue expression in petals causes ruffling and curvature in several core eudicot taxa [17,18]. We therefore targeted AqTCP4 for silencing in order to perturb the relative balance of cell proliferation and expansion, and to determine the effect on the developing petal spur. As confirmation that AqTCP4 functions in a similar manner to repress cell division in Aquilegia lateral organs, we recovered leaf phenotypes that are reminiscent of the A. thaliana jaw-1D phenotype, which overexpresses a microRNA that guides messenger RNA cleavage of several TCP genes controlling leaf development [19]. While we expected some distortion of spur shape, we unexpectedly discovered that AqTCP4 plays a specific role in suppressing cell divisions in the distal compartment of the spur; neither the petal blade nor the proximal compartment of the spur are ever effected. This independent development on the two sides of the spur seems surprising in A. coerulea, which has essentially straight spurs, but differential elongation has been implicated in other species that naturally have curved spurs (J. Puzey and E. M. Kramer 2012, unpublished data). Thus, our findings indicate that even in species with relatively simple, straight spurs, development of the proximal and distal compartments of the spur can be decoupled, a modularity that could be exploited to create curved spurs in other species. The existence of such modularity does raise another question, however, as to what mechanism is acting in the proximal compartment to regulate the proliferation-to-expansion transition in these cells.
Another intriguing component of the AqTCP4-silencing phenotype is the tendency towards reorientation of cell walls in association with the laminar buckles. Together with our examination of wild-type spurs (figure 1b), these data suggest that mechanical strain may play an important role in determining cell orientation in the developing spur. It is simple to imagine that during Phase I, proliferating cells in the spur cup experience a circumferential strain that feeds back onto both cytoskeletal organization and other developmental processes, such as auxin trafficking [44,45]. The observed tangential orientation of these cell walls may contribute to both the outpocketing of the spur cup and their proper positioning for the subsequent anisotropic elongation during Phase II. The formation of laminar distortions in the AqTCP4-silenced spurs would alter the orientation of this strain and result in comparable shifts in cell orientation. This system may, in fact, provide an excellent model for studying the interplay of mechanical strain, hormone signalling and the genetic pathways governing laminar development.
Our data reveal additional major expression classes associated with the early developing spur pocket. In particular, contingents of coregulated genes that mediate auxin synthesis or response are enriched in the cup at different stages of Phase I. Auxin plays important roles in diverse aspects of organ differentiation, including cell orientation, proliferation and expansion [14]. These loci include auxin biosynthesis genes such as CYP71A, as well as downstream factors like ARF8, ARF3/ETT and SAUR genes. The SAUR family has recently been shown to be involved in cellular expansion [46], while ARF8 influences both cell division and expansion, as well as mediating crosstalk between the BR and auxin pathways [47,48]. Consistent with this, in Arabidopsis BR regulates the proximal/distal cell proliferation gradient and the transition to cell expansion [49]. BR signalling is also among the represented processes with, for example, differential expression of AqBEH4, a close homologue of BZR1 that mediates BR-induced growth [40], along with DWF4, which promotes BR synthesis. On one level, these findings are not surprising, given the highly pleiotropic functions of the auxin pathway and the established role for BR in regulating the transition from cell division to expansion [50]. However, it is important to note that auxin synthesis and peak signalling are most commonly associated with the margins of lateral organs [51], suggesting that the centrally located petal spur represents a novel focal point for auxin responses. The extremely high and differential expression of Aquilegia STY homologues in the spur cup is significant in this regard since Arabidopsis studies have found these mRNAs to be strongly expressed at the very distal tips of developing organs [52]. Again, this points to a recruitment event in which an auxin-related factor that normally functions at the organ margin has been co-opted for development of a novel, centrally positioned structure. Furthermore, STY homologues are primarily known as activators of the YUCCA pathway of auxin synthesis [38], which our RNA-seq indicates is still deployed at the petal margin rather than in the spur. Therefore, it would seem that several aspects of this novel auxin-related focal point are distinct from what is known from core eudicot model systems and may be very recently evolved, given that the nectar spur itself evolved in the common ancestor of the genus Aquilegia, around 6 Ma [53].
Supplementary Material
Supplementary Material
Acknowledgements
We thank Kirsten Bomblies and Scott Hodges for helpful discussions, and Florian Jabbour together with one anonymous reviewer for constructive comments on the manuscript.
Data accessibility
All short read data are deposited in the Short Read Archive under BioProject ID PRJNA270946.
Author contributions
L.Y. conducted all experiments related to the RNA-seq and VIGS analysis of AqTCP4, participated in the design of the study and drafted the manuscript; S.C. conducted the in situ hybridization of AqTCP4, AqSTM, AqKN and AqHIS4; J.P. participated in the design of the study and conducted SEM analysis of early wild-type spur development; C.L. conducted SEM analysis of wild-type and AqTCP4-silenced petal cell morphology; E.M.K. conceived of the study, designed the study, coordinated the study and helped draft the manuscript. All authors gave final approval for publication.
Funding statement
This work was supported by a Ruth L. Kirschstein National Research Service Award from the National Institutes of Health to L.Y. (1F32GM096699) and NSF award IOS-1121005 to E.M.K.
Conflict of interests
The authors have no competing interests relative to this work.
References
- 1.Nicotra AB, Leigh A, Boyce CK, Jones CS, Niklas KJ, Royer DL, Tsukaya H. 2011. The evolution and functional significance of leaf shape in the angiosperms. Funct. Plant Biol. 38, 535–552. ( 10.1071/fp11057) [DOI] [PubMed] [Google Scholar]
- 2.Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD. 2004. Pollination syndromes and floral specialization. Ann. Rev. Ecol. Evol. Syst. 35, 375–403. ( 10.1146/annurev.ecolsys.34.011802.132347) [DOI] [Google Scholar]
- 3.Hodges SA. 1997. Floral nectar spurs and diversification. Int. J. Plant Sci. 158, S81–S88. ( 10.1086/297508) [DOI] [Google Scholar]
- 4.Bastida JM, Alcantara JM, Rey PJ, Vargas P, Herrera CM. 2010. Extended phylogeny of Aquilegia: the biogeographical and ecological patterns of two simultaneous but contrasting radiations. Plant Syst. Evol. 284, 171–185. ( 10.1007/s00606-009-0243-z) [DOI] [Google Scholar]
- 5.Whittall JB, Hodges SA. 2007. Pollinator shifts drive increasingly long nectar spurs in columbine flowers. Nature 447, 706–710. ( 10.1038/nature05857) [DOI] [PubMed] [Google Scholar]
- 6.Hodges SA, Fulton M, Yang JY, Whittall JB. 2004. Verne grant and evolutionary studies of Aquilegia. New Phytol. 161, 113–120. ( 10.1046/j.1469-8137.2003.00950.x) [DOI] [Google Scholar]
- 7.Tepfer SS. 1953. Floral anatomy and ontogeny in Aquilegia formosa var. truncata and Ranunculus repens. Univ. CA Publ. Bot. 25, 513–648. [Google Scholar]
- 8.Tucker SC, Hodges SA. 2005. Floral ontogeny of Aquilegia, Semiaquilegia, and Isopyrum (Ranunculaceae). Int. J. Plant Sci. 166, 557–574. ( 10.1086/429848) [DOI] [Google Scholar]
- 9.Puzey JR, Gerbode S, Hodges SA, Mahadevan L, Kramer EM. 2012. Evolution of spur-length diversity in Aquilegia petals is achieved solely through cell-shape anisotropy. Proc. R. Soc. B 279, 1640–1645. ( 10.1098/rspb.2011.1873) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barton MK. 2010. Twenty years on: the inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 341, 95–113. ( 10.1016/j.ydbio.2009.11.029). [DOI] [PubMed] [Google Scholar]
- 11.Koenig D, Sinha N. 2010. Evolution of leaf shape: a pattern emerges. Curr. Top. Dev. Biol. 91, 169–183. ( 10.1016/s0070-2153(10)91006-5) [DOI] [PubMed] [Google Scholar]
- 12.Golz JF, Keck EJ, Hudson A. 2002. Spontaneous mutations in KNOX genes give rise to a novel floral structure in Antirrhinum. Curr. Biol. 12, 515–522. ( 10.1016/S0960-9822(02)00721-2) [DOI] [PubMed] [Google Scholar]
- 13.Box MS, Dodsworth S, Rudall PJ, Bateman RM, Glover B. 2011. Characterization of Linaria KNOX genes suggests a role in petal-spur development. Plant J. 68, 703–714. ( 10.1111/j.1365-313X.2011.04721.x) [DOI] [PubMed] [Google Scholar]
- 14.Hay A, Tsiantis M. 2010. KNOX genes: versatile regulators of plant development and diversity. Development 137, 3153–3165. ( 10.1242/dev.030049) [DOI] [PubMed] [Google Scholar]
- 15.Moon J, Hake S. 2011. How a leaf gets its shape. Curr. Opin. Plant Biol. 14, 24–30. ( 10.1016/j.pbi.2010.08.012) [DOI] [PubMed] [Google Scholar]
- 16.Breuninger H, Lenhard M. 2010. Control of tissue and organ growth in plants. Curr. Top. Dev. Biol. 91, 185–220. ( 10.1016/s0070-2153(10)91007-7) [DOI] [PubMed] [Google Scholar]
- 17.Koyama T, Ohme-Takagi M, Sato F. 2011. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Sig. Behav. 6, 697–699. ( 10.4161/psb.6.5.14979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanaka Y, Yamamura T, Oshima Y, Mitsuda N, Koyama T, Ohme-Takagi M, Terakawa T. 2011. Creating ruffled flower petals in Cyclamen persicum by expression of the chimeric cyclamen TCP repressor. Plant Biotech. 28, 141–147. ( 10.5511/plantbiotechnology.10.1227a) [DOI] [Google Scholar]
- 19.Palatnik JF, Allen E, Wu XL, Schommer C, Schwab R, Carrington JC, Weigel D. 2003. Control of leaf morphogenesis by microRNAs. Nature 425, 257–263. ( 10.1038/nature01958) [DOI] [PubMed] [Google Scholar]
- 20.Kennaway R, Coen E, Green A, Bangham A. 2011. Generation of diverse biological forms through combinatorial interactions between tissue polarity and growth. PLoS Comp. Biol. 7, e1002071 ( 10.1371/journal.pcbi.1002071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sharma B, Kramer EM. 2013. Virus induced gene silencing in the rapid cycling Aquilegia coerulea ‘Origami’. In Virus-induced gene silencing: methods and protocols (ed. Becker A.), pp. 71–81. New York, NY: Springer Science+Business Media. [DOI] [PubMed] [Google Scholar]
- 22.Joint Genome Institute. 2013. Phytozome v9.1. See http://www.phytozome.net.
- 23.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotech. 28, 511–515. ( 10.1038/nbt.1621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCarthy DJ, Chen YS, Smyth GK. 2012. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. ( 10.1093/nar/gks042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. ( 10.1093/nar/25.17.3389) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Navaud O, Dabos P, Carnus E, Tremousaygue D, Herve C. 2007. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. ( 10.1007/s00239-006-0174-z) [DOI] [PubMed] [Google Scholar]
- 28.Stamatakis A, Ott M, Ludwig T. 2005. RAxML-OMP: an efficient program for phylogenetic inference on SMPs. In 8th International Conference on Parallel Computing Technologies (PaCT2005), pp. 288–302. Berlin, Germany: Springer. [Google Scholar]
- 29.Stamatakis A, Hoover P, Rougemont J. 2008. A fast bootstraping algorithm for the RAxML web-servers. Syst. Biol. 57, 758–771. ( 10.1080/10635150802429642) [DOI] [PubMed] [Google Scholar]
- 30.Miller MA, Holder MT, Vos R, Midford PE, Liebowitz T, Chan L, Hoover P, Warnow T. 2009. The CIPRES Portals. See http://www.phylo.org/sub_sections/portal.
- 31.Kramer EM. 2005. Methods for studying the evolution of plant reproductive structures: comparative gene expression techniques. In Molecular evolution: producing the biochemical data (eds Zimmer EA, Roalson EH.), pp. 617–635. San Diego, CA: Elsevier Academic Press. [DOI] [PubMed] [Google Scholar]
- 32.Ballerini ES, Kramer EM. 2011. The control of flowering time in the lower eudicot Aquilegia formosa. EvoDevo 2, 4 ( 10.1186/2041-9139-2-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gould B, Kramer EM. 2007. Virus-induced gene silencing as a tool for functional analyses in the emerging model plant Aquilegia (columbine, Ranunculaceae). Plant Meth. 3, 6 ( 10.1186/1746-4811-3-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kramer EM, Holappa L, Gould B, Jaramillo MA, Setnikov D, Santiago P. 2007. Elaboration of B gene function to include the identity of novel floral organs in the lower eudicot Aquilegia (Ranunculaceae). Plant Cell 19, 750–766. ( 10.1105/tpc.107.050385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer EM, Di Stilio VS, Schluter P. 2003. Complex patterns of gene duplication in the APETALA3 and PISTILLATA lineages of the Ranunculaceae. Int. J. Plant Sci. 164, 1–11. ( 10.1086/344694) [DOI] [Google Scholar]
- 36.Rasmussen DE, Kramer EM, Zimmer EA. 2009. One size fits all? Molecular evidence for a commonly inherited petal identity program in the Ranunculales. Am. J. Bot. 96, 1–14. ( 10.3732/ajb.0800038) [DOI] [PubMed] [Google Scholar]
- 37.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. ( 10.1006/meth.2001.1262) [DOI] [PubMed] [Google Scholar]
- 38.Eklund DM, et al. 2010. The Arabidopsis thaliana STYLISH1 protein acts as a transcriptional activator regulating auxin biosynthesis. Plant Cell 22, 349–363. ( 10.1105/tpc.108.064816) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Choe S. 2007. Signal-transduction pathways toward the regulation of brassinosteroid biosynthesis. J. Plant Biol. 50, 225–229. ( 10.1007/BF03030649) [DOI] [Google Scholar]
- 40.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120, 249–259. ( 10.1016/j.cell.2004.11.044) [DOI] [PubMed] [Google Scholar]
- 41.Powell AE, Lenhard M. 2012. Control of organ size in plants. Curr. Biol. 22, R360–R367. ( 10.1016/j.cub.2012.02.010) [DOI] [PubMed] [Google Scholar]
- 42.Moore MJ, Bell CD, Soltis PS, Soltis DE. 2007. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc. Natl Acad. Sci. USA 104, 19 363–19 368. ( 10.1073/pnas.0708072104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nag A, King S, Jack T. 2009. miR319a targeting of TCP4 is critical for petal growth and development in Arabidopsis. Proc. Natl Acad. Sci. USA 106, 22 534–22 539. ( 10.1073/pnas.0908718106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sampathkumar A, Krupinski P, Wightman R, Milani P, Berquand A, Boudaoud A, Hamant O, Jonsson H, Meyerowitz EM. 2014. Subcellular and supracellular mechanical stress prescribes cytoskeleton behavior in Arabidopsis cotyledon pavement cells. eLife 3, e01967 ( 10.7554/eLife.01967.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakayama N, Smith RS, Mandel T, Robinson S, Kimura S, Boudaoud A, Kuhlemeier C. 2012. Mechanical regulation of auxin-mediated growth. Curr. Biol. 22, 1468–1476. ( 10.1016/j.cub.2012.06.050) [DOI] [PubMed] [Google Scholar]
- 46.Spartz AK, et al. 2012. The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J. 70, 978–990. ( 10.1111/j.1365-313X.2012.04946.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jung J-H, Lee M, Park C-M. 2010. A transcriptional feedback loop modulating signaling crosstalks between auxin and brassinosteroid in Arabidopsis. Mol. Cells 29, 449–456. ( 10.1007/s10059-010-0055-6) [DOI] [PubMed] [Google Scholar]
- 48.Varaud E, Brioudes F, Szecsi J, Leroux J, Brown S, Perrot-Rechenmann C, Bendahmane M. 2011. AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH Transcription Factor BIGPETALp. Plant Cell 23, 973–983. ( 10.1105/tpc.110.081653) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzalez N, et al. 2010. Increased leaf size: different means to an end. Plant Phys. 153, 1261–1279. ( 10.1104/pp.110.156018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhiponova MK, et al. 2013. Brassinosteroid production and signaling differentially control cell division and expansion in the leaf. New Phyt. 197, 490–502. ( 10.1111/nph.12036) [DOI] [PubMed] [Google Scholar]
- 51.Mattsson J, Sung ZR, Berleth T. 1999. Responses of plant vascular systems to auxin transport inhibition. Development 126, 2979–2991. [DOI] [PubMed] [Google Scholar]
- 52.Eklund DM, Cierlik I, Staldal V, Claes AR, Vestman D, Chandler J, Sundberg E. 2011. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Phys. 157, 2069–2080. ( 10.1104/pp.111.182253) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fior S, Li MG, Oxelman B, Viola R, Hodges SA, Ometto L, Varotto C. 2013. Spatiotemporal reconstruction of the Aquilegia rapid radiation through next-generation sequencing of rapidly evolving cpDNA regions. New Phyt. 198, 579–592. ( 10.1111/nph.12163) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All short read data are deposited in the Short Read Archive under BioProject ID PRJNA270946.