Abstract
Predictive models of epidemic cholera need to resolve at suitable aggregation levels spatial data pertaining to local communities, epidemiological records, hydrologic drivers, waterways, patterns of human mobility and proxies of exposure rates. We address the above issue in a formal model comparison framework and provide a quantitative assessment of the explanatory and predictive abilities of various model settings with different spatial aggregation levels and coupling mechanisms. Reference is made to records of the recent Haiti cholera epidemics. Our intensive computations and objective model comparisons show that spatially explicit models accounting for spatial connections have better explanatory power than spatially disconnected ones for short-to-intermediate calibration windows, while parsimonious, spatially disconnected models perform better with long training sets. On average, spatially connected models show better predictive ability than disconnected ones. We suggest limits and validity of the various approaches and discuss the pathway towards the development of case-specific predictive tools in the context of emergency management.
Keywords: multilayer network model, spatially explicit model, ecohydrology, epidemic forecast, model calibration, model validation
1. Introduction
Cholera was reported in Haiti for the first time in recent history in October 2010, about nine months after the catastrophic earthquake that struck the country and damaged its poor infrastructures for healthcare, water and sanitation [1]. The source of the infection has been tracked back to the abrupt contamination of the Artibonite River from an external source, as unambiguously documented on both epidemiological and genetic grounds [2–6] (see also [7]). A first epidemic peak originated as cholera spread out from the Artibonite Valley. The disease was reported within weeks in all Haitian provinces, including the capital city Port-au-Prince. More than 150 000 cases and 3500 casualties were reported by the end of 2010. A second peak (spring 2011) was related to the revamping of disease transmission boosted by the rainy season [8]. One year after the beginning of the epidemic, the total toll of cholera in Haiti amounted to about 490 000 cases, with more than 6200 deaths. At that time, those figures already qualified the Haitian epidemic as the largest cholera outbreak in recent history. Cholera has not disappeared yet from Haiti about 4 years after its appearance in the country, as shown by the increasing counts of cases and casualties. As of December 2014, more than 720 000 cases and the death of 8700 people have been reported.
Several mathematical models of cholera transmission have been developed to describe the course of the Haitian epidemic [9–18]. Their application to the ongoing epidemic was made possible by the immediate release of epidemiological data, initially recorded by the disease-surveillance systems set up by the Haitian government in the aftermath of the earthquake and later by the National Cholera Surveillance System [1], as well as by the widespread availability of georeferenced environmental datasets. Those models were different in assumptions, spatial resolution and degrees of spatial coupling, but they all addressed the dynamics of susceptibles, infected individuals and bacterial concentrations in a discrete and geographically referenced set of local human communities. Mathematical modelling of the ongoing Haitian cholera epidemic has certainly provided important insights, especially concerning spatial transmission mechanisms [10–12,14], rainfall patterns [14,15,17], intervention strategies [9–12,16,17], local basic reproduction numbers [16], conditions for large-scale pathogen invasion [13] and the probability of epidemic extinction [18]. Most models provided predictions about the unfolding of the epidemic [9,10,14,17,18]. However, while predictive models have been successfully used in endemic settings [19–22], the predictive power of mathematical models of cholera transmission has never been evaluated formally and systematically in an epidemic setting (but see [14] for a reassessment of early predictions of the Haitian epidemic). Bridging this gap is obviously of paramount importance to understand whether (and, in case, to what extent) predictions drawn in the very course of an outbreak (i.e. in conditions of severe data limitation and uncertainty about the relevant epidemiological processes) can be trusted, and potentially used to aid real-time emergency management, allocate healthcare resources and evaluate the effects of alternative intervention strategies.
In this work, we aim at evaluating the predictive ability of mechanistic modelling for the Haitian cholera epidemic. To this end, we adopt the model proposed in [14] and later refined in [18]. This model has been shown to reproduce cholera transmission dynamics in Haiti in a fairly accurate and robust way. The model builds on a spatially explicit epidemiological framework [23,24] that has already been applied to both past [25,26] and ongoing cholera epidemics [10,17]. To study the role of spatial settings on the predictive potential of cholera modelling, we consider two different spatial scales, namely a fine-grained subdivision of the Haitian territory into hydrological units [14,18], and a coarse subdivision into administrative departments (e.g. [9,12,16]). The same set of epidemiological assumptions (detailed below) is retained for the two spatial scales, while different mechanisms of spatial propagation of the disease are considered. Specifically, a fine spatial resolution based on hydrological divides allows the model to account for both hydrological transport of pathogens and human mobility. Conversely, a coarser resolution based on administrative units cannot accommodate a proper description of hydrological connectivity. To evaluate the effects of the different assumptions on spatial processes that have been made in the literature at the latter spatial scale, we test a set-up of the model in which human mobility is the only driver of spatial disease spread between districts [12] and four set-ups in which no spatial transmission mechanisms are considered (i.e. each department's epidemiological dynamics is assumed to be independent of the other departments', e.g. [9]). Of these four set-ups, two (which differ from one another for initial conditions only) assume spatially heterogeneous epidemiological dynamics (different parameters across districts), while the other two (differing, again, for initial conditions only) assume homogeneous epidemiology (same parameters across districts).
The six model set-ups are calibrated against available epidemiological records by using progressively longer training sets. Validation (over different temporal windows) is performed to assess quantitatively the predictive ability of the calibrated models. Calibration and validation results are then analysed in a formal comparative framework to discuss strengths and limitations of the different approaches, as well as to identify the missing steps towards real-time use of the model. The intense computational effort made here is in fact meant to give practical (and possibly general) directions for management-prone modelling strategies in an actual epidemic context.
2. Material and methods
2.1. The model
The Haitian population is subdivided into n human communities spatially distributed within a domain that embeds both human mobility and hydrological networks (if applicable). Let Si(t), Ii(t) and Ri(t) be the local abundances of susceptible, symptomatic infected and recovered individuals in each node i of the network at time t, and let Bi(t) be the environmental concentration of Vibrio cholerae. Cholera transmission dynamics can be described by the following set of coupled differential equations [14,18]:
 |
The population of each node is assumed to be at demographic equilibrium, with μ and Hi being the human mortality rate and the population size of the local community. The force of infection Fi, which represents the rate at which susceptible individuals become infected because of contact with contaminated water, is expressed as
 |
where the parameter β represents the exposure rate (assumed to be constant over time; for a different hypothesis, [18]) and the fraction Bi/(K + Bi) is the probability of becoming infected because of exposure to a concentration Bi of V. cholerae (K being the half-saturation constant, [27]). Susceptible individuals can be exposed to the pathogen while travelling outside their home community. Parameter m represents the community-level fraction of individuals that travel outside their home site, while Qij represents the fraction of people travelling from their home community i to destination j. The force of infection in a given node thus depends not only on the local concentration of pathogens (for a fraction 1 − m of the susceptible hosts), but also on pathogen concentration in the connected communities (for the remaining fraction m, properly weighted by mobility patterns). Mobility fluxes are described through a gravity model [28,29], in which the attractiveness of node j for node i is directly proportional to the population size of j and inversely proportional to the distance between the two nodes (through an exponential kernel with scale factor D), i.e.
 |
Upon exposure to contaminated water, a fraction σ of infected individuals develops symptoms. Symptomatic infected individuals recover at rate γ, or die because of cholera or other causes at rates α or μ, respectively. Asymptomatic infected individuals shed V. cholerae bacteria at a much lower rate (about 1000 times lower, e.g. [30]) than symptomatic ones and recover more rapidly (in about 1 day instead of 5, according to [31]). Therefore, it is reasonable to assume that their contribution to environmental contamination is negligible with respect to that of symptomatic individuals [18]. Asymptomatic infections can still result in temporary immunity [20], thus contributing to the depletion of the pool of susceptibles and affecting the rate of occurrence of symptomatic infections. This assumption translates into a flux of asymptomatic infected individuals (1 − σ)FiSi entering directly the recovered compartment.
Recovered individuals lose their immunity and return to the susceptible compartment at rate ρ or die at rate μ. Infected individuals showing clinical symptoms are assumed to be non-mobile. Therefore, they contribute only to the local environmental concentration of V. cholerae at rate p/Wi, with p being the rate at which bacteria excreted by one infected individual reach and contaminate the local water reservoir of volume Wi (assumed to be proportional to population size, i.e. Wi = cHi as in [14]). Note that for the sake of parsimony a dimensionless bacterial concentration  can be introduced, along with a synthetic contamination rate θ = p/(cK). Bacteria die at a constant rate μB and undergo hydrologic dispersal at rate l. Cholera pathogens can move between nodes i and j with probability Pij. We assume Pij = 1 if j is the downstream nearest neighbour of node i and zero otherwise. To include the worsening of sanitation conditions due to rainfall-induced run-off, which causes additional loads of pathogens to enter the water reservoir because of the overflow of latrines and the washout of open-air defecation sites [8], the contamination rate is increased by local rainfall intensity Ji(t) via a proportionality coefficient ϕ [14].
can be introduced, along with a synthetic contamination rate θ = p/(cK). Bacteria die at a constant rate μB and undergo hydrologic dispersal at rate l. Cholera pathogens can move between nodes i and j with probability Pij. We assume Pij = 1 if j is the downstream nearest neighbour of node i and zero otherwise. To include the worsening of sanitation conditions due to rainfall-induced run-off, which causes additional loads of pathogens to enter the water reservoir because of the overflow of latrines and the washout of open-air defecation sites [8], the contamination rate is increased by local rainfall intensity Ji(t) via a proportionality coefficient ϕ [14].
2.2. Model set-ups
We consider six versions of this model: fine-grained spatially connected (M1), coarse-grained spatially connected (M2), coarse-grained spatially disconnected with heterogeneous parameters (M3 and M4) and coarse-grained spatially disconnected with homogeneous parameters (M5 and M6). All of them share the same basic epidemiological assumptions outlined above. Other possible mechanisms, such as bacterial hyperinfectivity [32] or latent infected stages [11], could obviously be accounted for, yet they have not been included in the present work to avoid potential confounding effects and limit the number of candidate models. This choice also follows from a previous comparative analysis of modelling assumptions in the context of the Haitian cholera epidemic [14]. The six model set-ups considered here differ from each other as regards the spatial scale of analysis, i.e. fine-grained (M1) versus coarse-grained (M2–M6), the inclusion of spatial coupling mechanisms, i.e. human mobility and hydrological transport (M1) versus human mobility only (M2, in which l = 0) versus no spatial coupling at all (M3–M6, in which l = 0 and m = 0), and the choice of initial conditions (spatially disconnected models M3–M6 only, see below).
The six set-ups of the model are also different in terms of parameter parsimony. As a matter of fact, spatially connected network models account for the spatial heterogeneity induced by spatial interactions, while spatially disconnected models do not. To circumvent this limitation, epidemiological parameters in the latter models are usually allowed to be different for different geographical units (see e.g. [9,15,16] for applications to the Haitian cholera epidemic). As a result, spatially disconnected models may be considerably less parsimonious than spatially connected ones, in which the epidemiological parameters are usually assumed to be spatially homogeneous (essentially to facilitate model calibration; see [14,18] and previous related applications) or linked to remotely acquired proxies [26]. Therefore, georeferenced spatially disconnected models could better account for local heterogeneities in disease transmission than parsimonious spatially connected ones. To investigate whether prodigality of structural parameters is actually compensated for by improved explanatory/predictive power, we thus consider two baseline disconnected models with spatially homogeneous epidemiological parameters (and, again, different assumptions regarding initial conditions). The main features of each model are summarized in table 1.
Table 1.
Summary of the main characteristics of the six model set-ups.
| model | spatial scale | spatial coupling | epidemiological parameters | initial conditions | calibration parameters |
|---|---|---|---|---|---|
| M1 | watershed | hydrological transport and human mobility | spatially homogeneous | estimated | 9 |
| M2 | department | human mobility | spatially homogeneous | estimated | 7 |
| M3 | department | none | spatially heterogeneous | estimated | 60 |
| M4 | department | none | spatially heterogeneous | calibrated | 70 |
| M5 | department | none | spatially homogeneous | estimated | 6 |
| M6 | department | none | spatially homogeneous | calibrated | 16 |
2.3. Application to the 2010 Haitian epidemic
As stated above, the model is run at two different spatial scales. At the finest resolution (model M1), the Haitian territory is subdivided into watersheds on the basis of hydrologic divides, inferred from drainage directions extracted from a digital terrain model (DTM, e.g. [33]). We use the DTM provided by the US Geological Survey (USGS, http://nationalmap.gov/viewer.html), with a grid resolution of 100 m and a precision of ±0.5 m in the elevation field (figure 1a). Following the procedure detailed in [14], the Haitian territory is subdivided into 365 hydrological units with an average extent of 76 km2 (figure 1b). The use of hydrologically defined units allows a straightforward identification of the hydrological connection from each watershed to its unique downstream neighbour (or to the ocean, for coastal watersheds). The hydrologic connectivity matrix [Pij] thus follows directly through nowadays standard extraction techniques [34]. Note that absorbing boundary conditions are assumed at the ocean outlets, i.e. pathogens that leave the river network from a coastal outlet cannot re-enter the system.
Figure 1.
Data for the Haitian cholera epidemic model. (a) DTM, (b) geomorphological subdivision in hydrological units and main river networks, (c) administrative departments and cumulative cholera cases reported in the first year of the epidemic, (d) high-resolution population distribution map, (e) main road network infrastructure and (f) remotely sensed rainfall intensity (average 1998–2012). See text for data sources and details on spatial manipulation of the georeferenced datasets.
At the coarsest resolution, the official Haitian subdivision in 10 administrative departments is retained. Note that this scale corresponds to that at which epidemiological records, consisting of daily counts of cholera cases reported in each of the 10 administrative departments, are currently made available by the Haitian Ministry of Health (http://mspp.gouv.ht/newsite/; figure 1c). At such a coarse resolution, however, the information on hydrological connectivity is lost (models M2–M6).
Human communities are defined as the population hosted within each computational unit (watershed or department, depending on the spatial scale considered), estimated by using a remotely sensed map of population distribution produced by the Oak Ridge National Laboratory (data http://www.ornl.gov/sci/landscan/index.shtml; figure 1d). The spatial resolution is 30 × 30 arc-seconds, resulting in cells of about 1 km2. Distances dij among communities in the fine-scale model are computed using the road network provided by the OpenStreetMap project (www.openstreetmap.org; figure 1e). Specifically, pairwise shortest distances along the road network are computed between the centroids of population distribution in each community. Great-circle distances between population centroids are used in the department-based connected model.
Systematic collection of rainfall through rain gauges has been relatively rare in post-earthquake Haiti, with on-the-ground rainfall measurements available only for Ouest (by USGS) and Sud (by Haiti Regeneration Initiative) departments (see fig. 1 in [15] for a map of the existing rain gauges). Therefore, daily rainfall Ji(t) for each community i is computed from satellite data collected by the NASA-JAXA's Tropical Rainfall Measuring Mission (TRMM_3B42 precipitation estimates, see http://trmm.gsfc.nasa.gov/ for details). Rainfall data have a spatial resolution of 0.25° of latitude and longitude (figure 1f). Precipitation fields are first downscaled to the resolution of the DTM with nearest neighbour interpolation and then averaged over the watershed/department area to obtain a representative value for the whole community. Because of the lack of surface measurements, it is not possible to perform a thorough comparison between remotely sensed estimates and on-the-ground data. Comparison of the surface data from Port-à-Piment (Sud, 2010–2012 data available online at http://blogs.cuit.columbia.edu/haitienvironment/environmental-monitoring/precipitation/monthly-rainfall/) to satellite estimates (at the scale of either the watershed or the administrative department that include the gauge station) suggests that remote sensing may consistently underestimate local rainfall measurements (electronic supplementary material, figure S1). This bias can be partly due to the complexity of Haitian orography, which can induce significant variations in local precipitation intensities within the spatial resolution of TRMM_3B42 estimates [15]. However, satellite-based rainfall estimates can qualitatively reproduce the rainfall patterns recorded on the ground (Pearsons's r = 0.63 (0.59) for satellite estimates at the watershed (department) scale).
As initial conditions for model simulations we assume that, as of the date of the reported beginning of the outbreak (around 20 October 2010; t = 0), the local numbers of infected people Ii(0) match the reported cases detailed in [3]. The locations of the first cases are tracked accurately at the fine spatial resolution (M1), while at the coarser scale (M2–M6) the reported cases can only be attributed to administrative departments. We note that just a few watersheds/departments were actually home to cholera cases as of 20 October 2010. This obviously represents an issue for the initialization of spatially disconnected models M3–M6. The lack of spatial coupling mechanisms, in fact, would keep the departments that were initially left untouched by the epidemic indefinitely in a cholera-free state. Suitable initial conditions are thus to be set also in these departments. A possible alternative to initialize the model would be to let t0 vary across departments following the observation of the first cases. However, this would require changing the calibration/validation windows across sites (see below). As such, we have decided not to follow this route. We test instead two different settings (for both spatially heterogeneous and homogeneous models), one in which one symptomatic carrier is placed in each of the cholera-free departments at t = 0 (M3 and M5), and one in which the initial number of infected individuals is calibrated in each department (including, for consistency, also those with cholera cases reported at the beginning of the epidemic, M4 and M6). As for the other state variables, we assume that the whole population is susceptible at the beginning of the epidemic, i.e. Si(0) = Hi − Ii(0) (and Ri(0) = 0), because of the lack of any pre-existing immunity, consistent with assessment of prior conditions in Haiti [3]. Local pathogen concentrations are assumed to be initially at equilibrium with the local number of infected cases, i.e. 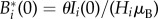 [14].
[14].
2.4. Model calibration
Some of the model parameters (namely μ, α and γ) can be reliably estimated from the literature or from epidemiological/demographic records, while numerical fitting is necessary to calibrate the remaining ones. To mimic quasi-real-time use of the model presented above in an ongoing epidemic context, we use 12 different calibration windows with increasing durations—ranging from one month (November 2010, when the first data became publicly available) to 1 year (from November 2010 to October 2011) with a one-month step.
Model fitting is performed via Markov chain Monte Carlo (MCMC) sampling [35,36]. Specifically, we use the algorithm DREAM (Differential Evolution Adaptive Metropolis [37], http://jasper.eng.uci.edu/software.html), an efficient implementation of MCMC that runs multiple different chains simultaneously to ensure global exploration of the parameter space, and adaptively tunes the scale and orientation of the jumping distribution using Differential Evolution [38] in addition to a Metropolis–Hastings update step [39,40]. We adopt the DREAMzs variant of the DREAM algorithm [41], which allows for an effective exploration of the target posterior distribution with just a few parallel chains. The algorithm is initialized with broad flat prior distributions for parameter values and is allowed to run up to convergence ( iterations).
iterations).
A Gaussian model is assumed for the likelihood of the epidemiological observations in MCMC sampling. Goodness of fit is measured as the residual sum of squares (RSS) between weekly reported cholera cases in each of the nd = 10 Haitian departments as recorded in the epidemiological bulletins and simulated by the model over the  weeks of the calibration window, i.e.
weeks of the calibration window, i.e.
 |
where Cr(i, j) and Cs(i, j) are the weekly reported cases in department i during week j according to records and model simulations, respectively. The estimation of weekly cases from the model output requires to compute
where tj marks the beginning of the jth week and Δt = 1 week. Note that the simulation results for the fine-grained version of the model (M1) are computed at the watershed level and need to be upscaled to the departmental level. The upscaling procedure is performed by accounting for the fraction of population of each watershed that belongs to a given administrative department.
We repeat the calibration procedure 12 times for each of the six model set-ups introduced above, each of which is characterized by a different number of tuning parameters. Specifically, M1 has nine calibration parameters (β, θ, σ, ρ, μB, ϕ, l, m, D), chosen as in [18] to ease across-study comparison. M2 has eight calibration parameters, the same as M1 except for l = 0. We have decided to fit the same set of epidemiological parameters for both spatially connected (M1 and M2) and spatially disconnected models (M3–M6). For disconnected models with spatially heterogeneous parameters (M3 and M4), calibration is performed department-by-department. Independent calibration runs (e.g. as done in [9]) in fact represent the simplest possibility when longitudinal data at different locations are available, but results in a large number of fitting parameters, namely 60 for M3 (β, θ, σ, ρ, μB, ϕ, calibrated independently for each of the 10 administrative departments) and 70 for M4 (same as M3, plus the initial number of infectives Ii(0) in each department). More parsimonious spatially disconnected models with heterogeneous parameters could actually be devised [9], also by constraining some epidemiological parameters across sites. Thus, the most parsimonious models are those with spatially homogeneous parameters (M5 and M6): the fitting parameters of M5 are six (the same as M1 with l = 0 and m = 0), while those of M6 are 16 (the same as M5, plus the initial number of infectives in each of the 10 departments).
To assess the explanatory power of the different versions of the model, for each calibration window we rank their performances according to Akaike's information criterion (AIC) [42] suitably corrected for small sample sizes [43]. AIC is a model-selection procedure that explicitly takes into account the trade-off between model accuracy and complexity, measured as the number of free parameters Θ (i.e. the structural parameters of the model, inclusive of initial conditions for M4 and M6, plus residual variance for each independent calibration run; [43]). After calibrating M1–M6 against the epidemiological data, for each best-fit model simulation, we compute
where 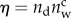 is the number of data points in the calibration window. The model with lowest AIC score is retained as the best candidate to explain the observed epidemic patterns in the relevant calibration window, with Akaike differences ΔAIC > 4 being required for a significant ranking [43]. Fitting is performed also for the shortest window (one month), but M3 and M4 are not included in AIC scoring because in that case the number of calibration parameters exceeds the available data points.
is the number of data points in the calibration window. The model with lowest AIC score is retained as the best candidate to explain the observed epidemic patterns in the relevant calibration window, with Akaike differences ΔAIC > 4 being required for a significant ranking [43]. Fitting is performed also for the shortest window (one month), but M3 and M4 are not included in AIC scoring because in that case the number of calibration parameters exceeds the available data points.
2.5. Model validation
Model validation is performed by extending model simulations outside the calibration windows. For each calibration window, we use 12 validation intervals with increasing durations (ranging from one month to 1 year with monthly steps), each of which starts right after the end of the relevant calibration window. Actual precipitation fields are fed to the model also during the validation periods. We simulate each model for the best-fit parameter combination found during calibration and for an ensemble of N = 100 000 sets from the posterior distribution of the model parameters obtained through MCMC sampling, including variance residual to account for total predictive uncertainty (observational errors are assumed to be additive and normally distributed as in [44]; see the electronic supplementary material, figure S2, for two examples of the evaluation of total predictive uncertainty). In this way, model predictions explicitly include parameter uncertainty and observational errors. Conversely, other possible sources of uncertainty, such as process noise [45], biases in the input rainfall patterns [46] and structural modelling errors [47] are not explicitly accounted for, and may thus be reflected in increased parameter/observational uncertainty.
Predictive power is assessed by using three different indicators, namely
 |
where  is the number of weeks in the validation window. The first indicator (V1) is the standard deviation of the prediction residuals and represents a natural modification of the goodness-of-fit score used in model calibration. The second and the third quantities are based on the cumulative number of cases predicted by the model in each department over the whole validation window, suitably averaged over the number of data points in the validation period to make it possible to compare the predictive power of the model over different windows. Although they may downplay temporal errors, these two quantities provide an easily readable measure of absolute (V2) or relative (V3) prediction errors, respectively. The three indicators are evaluated for the best-fit model simulation (
is the number of weeks in the validation window. The first indicator (V1) is the standard deviation of the prediction residuals and represents a natural modification of the goodness-of-fit score used in model calibration. The second and the third quantities are based on the cumulative number of cases predicted by the model in each department over the whole validation window, suitably averaged over the number of data points in the validation period to make it possible to compare the predictive power of the model over different windows. Although they may downplay temporal errors, these two quantities provide an easily readable measure of absolute (V2) or relative (V3) prediction errors, respectively. The three indicators are evaluated for the best-fit model simulation ( , x = 1, 2, 3), the ensemble of N simulations accounting for total predictive uncertainty (
, x = 1, 2, 3), the ensemble of N simulations accounting for total predictive uncertainty ( ; in this case, the three indicators are evaluated for each simulation, then average values are computed), and the median of the model predictions, that is the median of the weekly local cases predicted in the N model runs (
; in this case, the three indicators are evaluated for each simulation, then average values are computed), and the median of the model predictions, that is the median of the weekly local cases predicted in the N model runs ( ). This procedure is repeated for each of the six model set-ups, 12 calibration intervals and 12 validation windows.
). This procedure is repeated for each of the six model set-ups, 12 calibration intervals and 12 validation windows.
3. Results
All the six versions of the model are able to grasp qualitatively the evolution of the epidemic during the calibration phase (figure 2 and electronic supplementary material, figure S3), except for model M5, which consistently underestimates reported cholera cases. In terms of explanatory power, models M1 and M2 perform similarly in the shortest calibration window, but M1 significantly outperforms all the others for six calibration windows out of 12 (table 2), ranging from two to seven months. Model M6 is selected for the five longest calibration windows (8–12 months). Model M2 is the second-best for the four shortest calibration runs, after which it is outperformed by M6. Model M1 is also second-best for intermediate calibration windows (8–10 months), while M4 is second-best for the two longest calibration windows (11–12 months). The least supported models are M6 for the shortest calibration timespan, M3 and M4 for windows of two to six months, M5 for windows longer than six months.
Figure 2.
Fitting of models M1–M6 for different calibration windows. Shown are results for two- (a), four- (b), six- (c) and 12-month (d) calibration windows. Black dots indicate total weekly incidence in data, while coloured lines represent best-fit model simulations. Data and results are aggregated at the country level for an easier visual reference, but model fitting is performed at a higher resolution, i.e. that of the administrative departments shown in figure 1c. See the electronic supplementary material, figure S3, for the whole set of calibration intervals (1–12 months with monthly steps, starting 1 November 2010).
Table 2.
AIC differences for the six tested set-ups of the model for different calibration interval lengths.
| calibration length (months) | M1 | M2 | M3 | M4 | M5 | M6 |
|---|---|---|---|---|---|---|
| 1 | 0.0 | 1.3a | —b | —b | 29.3 | 55.4 |
| 2 | 0.0 | 59.7 | 711.1 | 1552.1 | 96.3 | 113.7 |
| 3 | 0.0 | 69.6 | 302.6 | 406.7 | 143.4 | 157.9 |
| 4 | 0.0 | 80.5 | 234.7 | 284.2 | 209.7 | 102.4 |
| 5 | 0.0 | 82.5 | 223.9 | 213.6 | 214.0 | 19.0 |
| 6 | 0.0 | 165.0 | 246.1 | 263.1 | 261.4 | 54.2 |
| 7 | 0.0 | 95.0 | 247.6 | 87.3 | 254.6 | 9.3 |
| 8 | 7.6 | 58.6 | 175.5 | 115.7 | 182.6 | 0.0 |
| 9 | 23.5 | 140.5 | 159.0 | 24.2 | 210.9 | 0.0 |
| 10 | 44.3 | 149.4 | 172.2 | 55.5 | 244.2 | 0.0 |
| 11 | 43.3 | 132.5 | 175.3 | 6.5 | 291.7 | 0.0 |
| 12 | 60.4 | 141.5 | 234.6 | 20.7 | 307.1 | 0.0 |
aThe candidate set-up cannot be safely discarded (ΔAIC < 4).
bThe model has not been included in model selection because the number of structural parameters exceeds the number of available data points.
As an example of the outcomes of the calibration procedure, figure 3 reports the best-fit parameter values of model M1 and the related uncertainties as obtained for different calibration windows. Local disease transmission, represented by the basic reproduction number (R0 = σβθ/μB/(μ + α + γ)); see [27], is found to be higher for long calibration windows (figure 3a). Conversely, higher fractions of moving people, shorter movement distances and higher hydrological dispersal rates are selected for short calibration intervals (figure 3b,c). Also, a longer immunity is selected for short calibration intervals, while higher values of the rainfall coefficient are selected for long calibration runs. As expected, the interquantile ranges of parameter uncertainty are quite large for short calibration windows, but progressively narrow for longer intervals, thus making it difficult to get reliable estimates of some of the model parameters (e.g. R0 and ρ) over relatively short timespans (figure 3a). Estimated initial conditions (M4 and M6) averaged over the different calibration runs are found to be well correlated with reported cases (Pearson's r = 0.50 (0.84) for M4 (M6)), although with large absolute differences (on average, ≈5400 (≈4500) cases per department for M4 (M6)).
Figure 3.

Parameter values of model M1 for different calibration windows (1–12 months). (a) Basic reproduction number (which depends on parameters as detailed in the text) and duration of acquired immunity (years); (b) community-average fraction of mobile susceptible individuals and scale factor of the exponential kernel of human mobility (km); (c) pathogen's hydrological dispersal rate (day−1) and rainfall coefficient (day mm−1). Lines and shaded areas represent best-fit model simulations and 5–95% credible intervals of parameter uncertainty. Non-calibrated parameters set as in [14]: γ = 0.2, α = 4.0 × 10−3, μ = 4.5 × 10−5 (all in (day−1)).
Sharp differences among the six versions of the model emerge in the validation trials (figure 4; see also the electronic supplementary material, figure S4, for the whole sequence of validation experiments). Spatially connected models (M1 and M2), even when tuned over very short calibration windows (say one to two months, figure 4a), are able to forecast at least the order of magnitude of the observed cholera cases for several weeks after the end of the calibration window. To a lesser extent, the same is true for parsimonious, spatially disconnected models M5 (one-month calibration window) and M6 (two-month calibration window). Conversely, spatially disconnected models with heterogeneous parameters (M3 and M4) largely overestimate cholera incidence. No model is actually able to predict consistently the peak of cholera cases recorded during spring 2011. Similar patterns can be found for relatively longer calibration windows (three to four months, figure 4b), although with better predictions of the 2011 spring peak (especially by M6), which however are robustly achieved (most notably by spatially connected models M1 and M2) only with calibration windows as long as six months (figure 4c), i.e. with a lead time of about one month. Validation performances greatly improve for longer calibration windows (7–10 months, electronic supplementary material, figure S4), especially for spatially connected models M1–M2 and parsimonious disconnected models M5–M6, which become able to predict actually observed peak and lull phases with a prediction lead time as long as 1 year. Note that model M1 tends to remarkably overestimate cholera incidence towards the end of the longest validation runs. Once tuned over even longer calibration windows (11–12 months, figure 4d) almost all models display good predictive ability at least for a few months after the end of the calibration window. The spring peak of 2012 is poorly captured by all models. The subsequent lull phase is better predicted by M2 among all.
Figure 4.
Model validation for different durations of the calibration window. Shown are results for two- (a), four- (b), six- (c) and 12-month (d) calibration windows. Validation intervals span from one month to 1 year, and begin right after the end of the relevant calibration window. Black dots indicate total weekly incidence in data, while coloured lines represent best-fit model simulations (thick line) or the medians (thin) of model predictions. See the electronic supplementary material, figure S4, for the whole set of calibration intervals.
An exhaustive investigation of the possible combinations of calibration/validation windows for performance indicator V1 (figure 4a–c) shows that the predictive ability of spatially connected models (M1 and M2) usually outperforms that of the spatially disconnected ones for short calibration windows or long validation intervals. Disconnected models with heterogeneous parameters (M3 and M4) are rarely found to be best, and only for long calibration and short validation periods. Disconnected models with homogeneous parameters (M5 and M6) may represent better predictive tools for intermediate calibration periods, or for long calibration/intermediate validation runs (best-fit parameters only). Similar patterns are found when considering V2, although with a larger share of validation trials in which model M6 performs best (figure 5d–f). A clear pattern emerges for V3: model M1 usually performs best for short calibration windows (with some exceptions), while model M6 represents the best predictive tool for long calibration windows (figure 5g–i). The best predictive performances are attained by either best-fit runs or median model projections for all the considered validation indicators (electronic supplementary material, figures S5–S7).
Figure 5.
Quantitative assessment of validation results. Panels (a–i) show the best-performing model for each combination of calibration/validation windows for different validation performance indicators (labels). Raw values of the validation indicators are given in electronic supplementary material, figures S5–S7.
The values of the validation performance indicators averaged over all the combinations of calibration and validation intervals (table 3) show that spatially connected models M1 and M2 have better overall predictive abilities than spatially disconnected ones. Averaging the validation performance indicators over the different validation windows (electronic supplementary material, figure S8) shows that spatially connected models actually are better predictive tools for either short or long calibration intervals, but not always for calibration windows of intermediate length. By averaging the validation performance indicators over the different calibration windows (electronic supplementary material, figure S9), it becomes evident that the predictive ability of the spatially connected models (M1 in particular) is higher than that of the spatially disconnected ones for almost all validation intervals.
Table 3.
Validation performance indicators averaged over all combinations of calibration and validation windows. See text for details on model validation and the computation of the different indicators.
| M1 | M2 | M3 | M4 | M5 | M6 | |
|---|---|---|---|---|---|---|
 |
659 | 674 | 5711 | 3788 | 780 | 1467 |
 |
963 | 1122 | 2192 | 1985 | 1195 | 1420 |
 |
674 | 712 | 1428 | 1084 | 836 | 801 |
 |
317 | 323 | 2326 | 1484 | 451 | 700 |
 |
552 | 705 | 1055 | 1073 | 760 | 759 |
 |
323 | 382 | 734 | 640.2 | 488 | 447 |
 |
6.7 | 6.3 | 54.2 | 12.8 | 7.3 | 7.2 |
 |
13.0 | 15.6 | 19.6 | 19.7 | 15.8 | 14.0 |
 |
6.5 | 7.3 | 12.1 | 9.5 | 7.8 | 6.7 |
4. Discussion
Almost all models are able to reproduce complex spatio-temporal epidemic patterns during calibration, provided that the algorithm used for parameter fitting is fed with a sufficient amount of epidemiological data (figure 2). Model M5 represents an exception in this respect, most probably because of its relatively poor structural properties (lack of spatial coupling mechanisms, homogeneous parameters, fixed initial conditions). Differences in the explanatory power of the different model set-ups do emerge, though. Interestingly, in fact, spatially connected models are consistently selected for short to intermediate calibration windows, while parsimonious, spatially disconnected models (with calibrated initial conditions, which could implicitly account for the disregarded spatial component of epidemic spread) perform better over long calibration intervals (table 2). We argue that spatially disconnected models cannot directly account for the spatial mechanisms of pathogen dissemination (i.e. hydrological transport and human mobility), which play a crucial role in the initial phase of the outbreak, when the infection starts propagating into disease-free regions [13,48]. Conversely, spatial coupling may become less important when looking at longer time horizons, once the epidemic has already spread all across the country. In this case, in fact, epidemiological dynamics is expected to be mostly controlled by local factors, which could indeed determine some spatial heterogeneity in the transmission processes [49]. The relatively good AIC scores of M4 for the longest calibration runs (11–12 months) seem to support the idea that spatially disconnected models with site-specific parameters could indeed account for this spatial heterogeneity, although the calibration windows used in this work may be too short to let this pattern emerge more clearly. The outcomes of model fitting in the fine-scale spatially connected model M1 (which provides the most detailed account of the spatial dissemination of the pathogen) further supports this explanation, with higher values of the dispersal rates/fractions selected for short calibration runs, and higher values of the basic reproduction number selected for long calibration intervals (figure 3). This study thus helps clarify which modelling tool might be best suited to describe outbreak unfolding depending on the stage of development of the epidemic.
As far as accurate spatio-temporal projections are concerned (e.g. as evaluated by validation indicator V1), spatially connected models seem to have greater predictive ability than spatially disconnected ones, especially for short calibration intervals (or long validation windows; figure 5). This finding may be particularly important in the early phase of an outbreak, when data are limited and the epidemic is undergoing rapid growth. We argue that the intrinsic lack of spatial coupling mechanisms and/or the prodigality of structural parameters determine the inability of spatially disconnected models to accurately project cholera dynamics in the first phase of the epidemic. On the one hand, in fact, the lack of spatial coupling mechanisms leads to biased estimations of initial conditions or local infection processes. On the other hand, the prodigality of structural parameters (which results in high uncertainty in parameter estimation) may act on top of the first shortcoming, thus leading to unsatisfying epidemiological projections even in the short run. Taken together (as in models M3 and M4), these two features may determine a sizeable excess of predicted cholera cases in the validation periods. Therefore, the use of spatially disconnected models with heterogeneous parameters for prediction purposes in the initial stage of an epidemic (i.e. when spatial coupling is of paramount importance and data availability is forcedly limited) seems hazardous at best. This remark is empirically confirmed by some early projections of cholera transmission dynamics in Haiti drawn from a spatially disconnected model with heterogeneous parameters published a few months after the beginning of the epidemic [9]. In that study, the authors predicted more than one million cases in the first year of the epidemic—a figure that fortunately has not been reached even 4 years after the beginning of the epidemic. By contrast, the first predictions drawn from a parsimonious, spatially connected model of cholera dynamics in Haiti [10] correctly predicted the unfolding of the epidemic with a prediction lead time of five months and were retrospectively judged quite robust in a later comparative reassessment [14]. Should coarser projections be of interest (e.g. as evaluated by validation indicator V2, which quantifies absolute errors in terms of cumulative cases; or V3, which measures relative errors and is less sensitive to deviations during high-prevalence periods), spatially disconnected models with homogeneous parameters would represent the tool of choice for intermediate-to-long calibration windows, while spatially connected models would still be best for short calibration runs (figure 5). This distinction could be relevant, for instance, to decision-makers interested in robust projections of total epidemic evolution over decisional timescales spanning from a few weeks to a few months.
As for the prediction of epidemic peaks, model validation outcomes are far from perfect also for the best-performing models. For instance, for calibration windows shorter than four months, none of the considered models is able to robustly predict the cholera peak observed during spring 2011, which has been linked to heavy seasonal precipitation [14]. As a matter of fact, these calibration windows correspond to the first, explosive phase of the epidemic, in which rapid transmission dynamics most probably beclouded the effect of rainfall (fairly scarce during this period, except for the passage of Hurricane Thomas at the beginning of November 2010) and possibly of other environmental drivers as well [8]. The outcomes of parameter calibration support this interpretation, with higher values of the rainfall coefficient being progressively selected for increasingly long calibration windows. The six model set-ups are not able to forecast the peak of spring 2012 either. However, in this case, other mechanisms could have played a role. A relatively large share of cases was in fact localized in the capital city Port-au-Prince during the peak. The mismatch could have thus been caused by a poor estimation of local rainfall intensity. Satellite-based precipitation estimates may be of limited utility when looking at local features [15,50], but represent a precious alternative to traditional ground measurements wherever the latter are rare, provided that they at least correlate with actual rainfall patterns (as our preliminary analyses seem indeed to confirm). This is true, in particular, for our country-scale model of cholera transmission, in which rainfall is considered as a forcing term for local contamination rates acting through a coefficient that has to be numerically calibrated. On the other hand, the errors possibly introduced in the model by inaccurate rainfall estimates are most likely translated into higher uncertainty in parameter values, and would perhaps deserve an explicit treatment (e.g. [46]; see also [14,17,18] on the use of stochastic rainfall generators for projections of the Haitian epidemic). All these considerations point to the importance of ground-truthing of remotely sensed rainfall patterns, and calls for a better integration of satellite estimates and co-located surface measurements. Further progress in this area will come through improved rainfall forecasting, which will also allow for real-time testing of our models. Another possible source of error could be represented by environmental and/or social factors not accounted for in the model. The analysis of epidemiological reports at the communal level (which are recorded but not yet available to the scientific community, see [1,8]) represents a possible way to reduce structural modelling errors. Such data could in fact shed light on highly localized transmission processes that may be difficult to describe when working at broad spatial scales [18]. However, increasing the spatial resolution of the model could make the application of a fully deterministic framework inappropriate (especially during lull phases in which disease transmission could be remarkably affected by demographic stochasticity [18]) and call for the inclusion of process noise in the modelling framework—which in turn would make numerical procedures much more involved (e.g. [45]).
As far as long-term predictions are concerned, a possible limitation of our approach is the underlying assumption that epidemiological processes do not change over time. Rainfall is in fact the only term in the model that depends explicitly upon time. However, local exposure/contamination rates could also have changed over the timescales considered in this study (as proposed in [8], where it was suggested that transmission patterns may have changed significantly in time and space). A first plausible mechanism to explain this change is the increasing awareness of the population about the mechanisms of cholera transmission, as a result of the campaigns for hygiene promotion set-up by Haitian authorities and non-governmental organizations [49,51]. Increased knowledge about cholera and its transmission dynamics may have prompted behavioural changes in the population at risk of infection, thus significantly influencing cholera dynamics [52]. As an example, [18] estimated that such increased awareness may have led to a 20% reduction of exposure risk just in a few months after the beginning of the epidemic. Another possible mechanism of change is the progressive improvement of living conditions, potentially linked to the return of internally displaced people to their original households after the earthquake that struck the country in January 2010 [53]. Structural interventions, aimed, for example, at improving access to safe water and basic sanitation, could also have helped reduce cholera transmission [54]. Because of the isolated and sporadic nature of such interventions, however, it is difficult to include their effects in a reasonably parsimonious mechanistic framework. Changes in human mobility patterns between the initial, explosive phase of the epidemic and the following phases, possibly triggered by the unfolding of the outbreak, could also have had non-trivial effects on disease dynamics [24]. All these missing features can be compensated for by biased parameter estimates, which in turn can contribute to the remarkable variations of parameters calibrated with different fitting windows.
The shortcomings of our approach do not preclude a quantitative assessment of the overall predictive ability of the different models. On average, in fact, the spatially connected models perform better than the others, although quantitative differences are relatively small for some indicators (table 3). Some practical observations are thus in order. First, because of their added information value, spatially connected models should represent the tool of choice for accurate predictions of the spatio-temporal patterns of cholera epidemics in their initial phases, when data are scant and the spatial spread of the pathogen is of paramount importance. Predictions drawn from current mathematical models of diseases like cholera must always be taken with care—but extra-care should be applied, in particular, whenever these predictions come from spatially disconnected models endowed with many structural parameters and calibrated over relatively short intervals. Second, although temporal, spatially aggregated models are usually much easier to fit to longitudinal data than spatio-temporal ones, spatially connected models for cholera epidemics are not necessarily doomed to be much more complex than spatially disconnected ones. This must be made clear not only to model makers, but also to consumers of models with diverse (and not always quantitative) backgrounds (e.g. general medical and public health readers as discussed in [55]). For modellers, some additional complexity clearly lies in the derivation of the connectivity structures for hydrological transport and human mobility; however, the increasing availability of georeferenced information on hydrological and transportation networks, human demography and mobility, sanitation infrastructure and treatment centre distribution, coupled with objective manipulation techniques, has allowed rapid progresses in the field of spatially explicit modelling of eco-epidemiological dynamics over the past few years. For model users, spatially connected models are often only seemingly more complex than spatially disconnected ones; while the mathematical notation might indeed look complicated at first sight, they offer a natural language to describe eco-epidemiological processes that are intrinsically rooted in spatial dynamics.
All these observations point consistently to the importance of being spatially explicit (and connected) when dealing with the early phases of a cholera epidemic. As a matter of fact, when only limited data are available, information about spatial coupling mechanisms proves essential for making reasonable predictions. One question could arise related to what spatial scale of analysis should be used. Our analysis has showed, in fact, that a clear distinction emerges between spatially disconnected versus connected models, while the predictive performances of the latter appear to be quite independent of the spatial scale of analysis. A possible explanation of this result is that river transport has played a limited role in disease propagation at the country scale after the initial contamination of the Artibonite River [8], and that neglecting hydrological connectivity can be compensated for by increased human mobility in model M2 (estimates of m are indeed systematically larger in M2 than in M1; see also [10], in which a fine-grained, spatially connected model without hydrological transport was successfully applied to the Haitian epidemic). An alternative explanation is the spatial resolution of the available epidemiological data, which are currently aggregated for administrative departments (i.e. the scale used in the coarse spatially connected model and in the spatially disconnected ones). As such, the current lack of more detailed epidemiological records may preclude a fair assessment of the actual explanatory/predictive power of the fine-scale spatially connected model. We thus suggest that the spatial scale of analysis should be suitably chosen so as to (i) match (at least) the resolution of the available epidemiological records; (ii) allow for the description of processes that are deemed important for the epidemiological dynamics (such as hydrological transport, whose inclusion usually requires a finer scale of analysis than that dictated by administrative boundaries); (iii) match the expectations of decision-makers with respect to the level of spatial detail required in the epidemiological projections (i.e. prediction of large-scale patterns versus local features) and in the assessment of intervention strategies. In this respect, coarse-grained spatially connected models (e.g. [12]) could represent a good trade-off between spatial accuracy, implementation effort and computational requirements, and could be used effectively to deploy robust epidemic predictions relatively soon after the beginning of an outbreak. These findings may have implications that extend beyond cholera dynamics, possibly being relevant to other diseases, not necessarily waterborne. As an example, multi-layer network models like the one presented in this work could be used, with the necessary modifications, to study vector-borne epidemics, although in this case a proper characterization of the spatio-temporal patterns of vector movement may not be straightforward (e.g. [56]). We also note that in endemic settings, provided that space–time data are available, other mathematical tools could be more appropriate than those used in this work, including time-series analysis [19,21,57,58], spatially implicit [20] and Markov chain models [22,59].
In conclusion, the wealth of data gathered during the Haiti cholera epidemic [1,3,8] paves the way for a new generation of epidemiological models, which will be required to accommodate real-time assimilation of epidemiological, hydrological and ecological information, as well as reliable projections of rainfall patterns, so as to improve epidemic forecasts and the evaluation of alternative interventions strategies (possibly within an adaptive management scheme; e.g. [60,61]). The inclusion of these features will turn tools that at their current stage are mostly descriptive into full-fledged decision-support systems for the prediction of the residual evolution of the epidemic and the design of optimal (ideally, in a multicriterial sense; e.g. [62]) intervention strategies. Such quantitative decision support tools will inform decision-makers towards sustainable choices, i.e. towards the design of public health policies and sanitary interventions linked to the territory where the measures are to be implemented. Real-time support to epidemic management will also allow timely decisions and a quantitative assessment of alternative intervention strategies, thus possibly contributing to the optimization of sanitary and humanitarian efforts.
Supplementary Material
Acknowledgements
The Authors wish to thank Prof. Mercedes Pascual (University of Michigan) and two anonymous reviewers for their insightful comments, and Dr Jasper A. Vrugt (University of California, Irvine) for making available the source code of the DREAMzs software. LM and RC are also grateful to the students of Alta Scuola Politecnica for promoting stimulating discussions during the annual courses on infectious disease dynamics.
Funding statement
L.M., E.B. and A.R. acknowledge the support provided by the European Research Council (ERC) advanced grant program through the project ‘River networks as ecological corridors for species, populations and waterborne disease’ (RINEC 227612). L.M., E.B., F.F. and A.R. acknowledge the support from the Swiss National Science Foundation (SNF/FNS) project ‘Dynamics and controls of large-scale cholera outbreaks' (DYCHO CR23I2_138104).
References
- 1.Barzilay EJ, et al. 2013. Cholera surveillance during the Haiti epidemic—the first 2 years. N. Engl. J. Med. 368, 599–609. ( 10.1056/NEJMoa1204927) [DOI] [PubMed] [Google Scholar]
- 2.Chin CS, et al. 2011. The origin of the Haitian cholera outbreak strain. N Engl. J. Med. 364, 33–42. ( 10.1056/NEJMoa1012928) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Piarroux R, Barrais R, Faucher B, Haus R, Piarroux M, Gaudart J, Magloire R, Raoult D. 2011. Understanding the cholera epidemic, Haiti. Emerg. Infect. Dis. 17, 1161–1168. ( 10.3201/eid1707.110059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hendriksen RS, et al. 2011. Population genetics of Vibrio cholerae from Nepal in 2010: evidence on the origin of the Haitian outbreak. mBio 2, e00157 ( 10.1128/mBio.00157-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frerichs RR, Keim PS, Barrais R, Piarroux R. 2012. Nepalese origin of cholera epidemic in Haiti. Clin. Microbiol. Infect. 18, E158–E163. ( 10.1111/j.1469-0691.2012.03841.x) [DOI] [PubMed] [Google Scholar]
- 6.Eppinger M, et al. 2014. Genomic epidemiology of the Haitian cholera outbreak: a single introduction followed by rapid, extensive, and continued spread characterized the onset of the epidemic. mBio 5, e01721-14 ( 10.1128/mBio.01721-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orata FD, Keim PS, Boucher Y. 2014. The 2010 cholera outbreak in Haiti: how science solved a controversy. PLoS Pathog. 10, e1003967 ( 10.1371/journal.ppat.1003967) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gaudart J, Rebaudet S, Barrais R, Boncy J, Faucher B, Piarroux M, Magloire R, Thimothe G, Piarroux R. 2013. Spatio-temporal dynamics of cholera during the first year of the epidemic in Haiti. PLoS Negl. Trop. Dis. 7, e2145 ( 10.1371/journal.pntd.0002145) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andrews JR, Basu S. 2011. Transmission dynamics and control of cholera in Haiti: an epidemic model. Lancet 377, 1248–1252. ( 10.1016/S0140-6736(11)60273-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bertuzzo E, Mari L, Righetto L, Gatto M, Casagrandi R, Rodriguez-Iturbe I, Rinaldo A. 2011. Prediction of the spatial evolution and effects of control measures for the unfolding Haiti cholera outbreak. Geophys. Res. Lett. 38, L06403 ( 10.1029/2011GL046823) [DOI] [Google Scholar]
- 11.Chao DL, Halloran ME, Longini IM. 2011. Vaccination strategies for epidemic cholera in Haiti with implications for the developing world. Proc. Natl Acad. Sci. USA 108, 7081–7085. ( 10.1073/pnas.1102149108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tuite AL, Tien J, Eisenberg M, Earns DJD, Ma J, Fisman DN. 2011. Cholera epidemic in Haiti, 2010: using a transmission model to explain spatial spread of disease and identify optimal control interventions. Ann. Internal Med. 154, 593–601. ( 10.7326/0003-4819-154-9-201105030-00334) [DOI] [PubMed] [Google Scholar]
- 13.Gatto M, Mari L, Bertuzzo E, Casagrandi R, Righetto L, Rodriguez-Iturbe I, Rinaldo A. 2012. Generalized reproduction numbers and the prediction of patterns in waterborne disease. Proc. Natl Acad. Sci. USA 48 19 703–19 708. ( 10.1073/pnas.1217567109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinaldo A, et al. 2012. Reassessment of the 2010–2011 Haiti cholera outbreak and rainfall-driven multiseason projections. Proc. Natl Acad. Sci. USA 109, 6602–6607. ( 10.1073/pnas.1203333109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenberg MC, Kujbida G, Tuite AR, Fisman DN, Tien JH. 2013. Examining rainfall and cholera dynamics in Haiti using statistical and dynamic modeling approaches. Epidemics 5, 197–207. ( 10.1016/j.epidem.2013.09.004) [DOI] [PubMed] [Google Scholar]
- 16.Mukandavire Z, Smith DL, Morris JG. 2013. Cholera in Haiti: reproductive numbers and vaccination coverage estimates. Nat. Sci. Rep. 3, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Righetto L, Bertuzzo E, Mari L, Schild E, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A. 2013. Rainfall mediations in the spreading of epidemic cholera. Adv. Water Resour. 60, 34–46. ( 10.1016/j.advwatres.2013.07.006) [DOI] [Google Scholar]
- 18.Bertuzzo E, Finger F, Mari L, Gatto M, Rinaldo A. In press On the probability of extinction of the Haiti cholera epidemic. Stochast. Environ. Res. Risk Assess.. ( 10.1007/s00477-014-0906-3) [DOI] [Google Scholar]
- 19.Pascual M, Rodó X, Ellner SP, Colwell RR, Bouma MJ. 2000. Cholera dynamics and El Niño Southern Oscillation. Science 289, 1766–1769. ( 10.1126/science.289.5485.1766) [DOI] [PubMed] [Google Scholar]
- 20.King AA, Ionides EL, Pascual M, Bouma MJ. 2008. Inapparent infections and cholera dynamics. Nature 454, 877–880. ( 10.1038/nature07084) [DOI] [PubMed] [Google Scholar]
- 21.Pascual M, Chaves LF, Cash B, Rodó X, Yunus M. 2008. Predicting endemic cholera: the role of climate variability and disease dynamics. Clim. Res. 36, 131–140. ( 10.3354/cr00730) [DOI] [Google Scholar]
- 22.Reiner RC, King AA, Emch M, Yunus M, Faruque ASG, Pascual M. 2012. Highly localized sensitivity to climate forcing drives endemic cholera in a megacity. Proc. Natl Acad. Sci. USA 109, 2033–2036. ( 10.1073/pnas.1108438109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertuzzo E, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A. 2010. On spatially explicit models of cholera epidemics. J. R. Soc. Interface 7, 321–333. ( 10.1098/rsif.2009.0204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mari L, Bertuzzo E, Righetto L, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A. 2012. On the role of human mobility in the spread of cholera epidemics: towards an epidemiological movement ecology. Ecohydrology 5, 531–540. ( 10.1002/eco.262) [DOI] [Google Scholar]
- 25.Bertuzzo E, Gatto M, Maritan A, Azaele S, Rodriguez-Iturbe I, Rinaldo A. 2008. On the space–time evolution of a cholera epidemic. Water Resour. Res. 44, W01424 ( 10.1029/2007WR006211) [DOI] [Google Scholar]
- 26.Mari L, Bertuzzo E, Righetto L, Casagrandi R, Gatto M, Rodriguez-Iturbe I, Rinaldo A. 2012. Modelling cholera epidemics: the role of waterways, human mobility and sanitation. J. R. Soc. Interface 9, 376–388. ( 10.1098/rsif.2011.0304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Codeço TC. 2001. Endemic and epidemic dynamics of cholera: the role of the aquatic reservoir. BMC Infect. Dis. 1, 1 ( 10.1186/1471-2334-1-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Erlander S, Stewart NF. 1990. The gravity model in transportation analysis—theory and extensions. Zeist, The Netherlands: VSP Books. [Google Scholar]
- 29.Truscott J, Ferguson NM. 2012. Evaluating the adequacy of gravity models as a description of human mobility for epidemic modelling. PLoS Comput. Biol. 8, e1002699 ( 10.1371/journal.pcbi.1002699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper JB, Morris JG, Levine MM. 1995. Cholera. Clin. Microbiol. Reviews 8, 48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nelson EJ, Harris JB, Morris JG, Calderwood SB, Camilli A. 2009. Cholera transmission: the host, pathogen and bacteriophage dynamic. Nat. Rev. Microbiol. 7, 693–702. ( 10.1038/nrmicro2204) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartley DM, Morris JG, Smith DL. 2006. Hyperinfectivity: a critical element in the ability of Vibrio cholerae to cause epidemics? PLoS Med. 3, 63–69. ( 10.1371/journal.pmed.0030007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarboton DG. 1997. A new method for the determination of flow directions and contributing areas in grid digital elevation models. Water Resour. Res. 33, 309–319. ( 10.1029/96WR03137) [DOI] [Google Scholar]
- 34.Rodriguez-Iturbe I, Rinaldo A. 1997. Fractal river basins. Chance and self-organization. New York, NY: Cambridge University Press. [Google Scholar]
- 35.Gilks WR, Richardson S, Spiegelharter DJ. 1995. Markov chain Monte Carlo in practice. New York, NY: Chapman and Hall. [Google Scholar]
- 36.Hamra G, MacLehose R, Richardson D. 2013. Markov chain Monte Carlo: an introduction for epidemiologists. Int. J. Epidemiol. 42, 627–634. ( 10.1093/ije/dyt043) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.ter Braak CJF, Vrugt JA. 2008. Differential evolution Markov chain with snooker updater and fewer chains. Stat. Comput. 18, 435–446. ( 10.1007/s11222-008-9104-9) [DOI] [Google Scholar]
- 38.Storn R, Price K. 1997. Differential evolution—a simple and efficient heuristic for global optimization over continuous spaces. J. Glob. Optim. 11, 341–359. ( 10.1023/A:1008202821328) [DOI] [Google Scholar]
- 39.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E. 1953. Equations of state calculations by fast computing machines. J. Chem. Phys. 21, 1087–1092. ( 10.1063/1.1699114) [DOI] [Google Scholar]
- 40.Hastings WK. 1970. Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57, 97–109. ( 10.1093/biomet/57.1.97) [DOI] [Google Scholar]
- 41.Vrugt JA, ter Braak CJF, Gupta HV, Robinson BA. 2009. Accelerating Markov chain Monte Carlo simulation by differential evolution with self-adaptive randomized subspace sampling. Int. J. Nonlinear Sci. Numer. Simul. 10, 271–288. ( 10.1515/IJNSNS.2009.10.3.273) [DOI] [Google Scholar]
- 42.Akaike H. 1974. A new look at the statistical model identification. IEEE Trans. Automatic Control 19, 716–723. ( 10.1109/TAC.1974.1100705) [DOI] [Google Scholar]
- 43.Burnham KP, Anderson DR. 2002. Model selection and multimodel inference: a practical information-theoretic approach. New York, NY: Springer. [Google Scholar]
- 44.Vrugt JA, ter Braak CJF, Diks CGH, Robinson BA, Hyman JM, Higdon D. 2009. Equifinality of formal (DREAM) and informal (GLUE) Bayesian approaches in hydrologic modeling? Stoch. Environ. Res. Risk Assess. 23, 1011–1026. ( 10.1007/s00477-008-0274-y) [DOI] [Google Scholar]
- 45.Ionides EL, Bret’ø C, King AA. 2006. Inference for nonlinear dynamical systems. Proc. Natl Acad. Sci. USA 103, 18 438–18 443. ( 10.1073/pnas.0603181103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vrugt JA, ter Braak CJF, Clark MP, Hyman JM, Robinson BA. 2008. Treatment of input uncertainty in hydrologic modeling: doing hydrology backwards with Markov chain Monte Carlo simulation. Water Resour. Res. 44, W00B09 ( 10.1029/2007WR006720) [DOI] [Google Scholar]
- 47.Kopec JA, et al. 2010. Validation of population-based disease simulation models: a review of concepts and methods. BMC Public Health 10, 710 ( 10.1186/1471-2458-10-710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gatto M, Mari L, Bertuzzo E, Casagrandi R, Righetto L, Rodriguez-Iturbe I, Rinaldo A. 2013. Spatially explicit conditions for waterborne pathogen invasion. Am. Nat. 182, 328–346. ( 10.1086/671258) [DOI] [PubMed] [Google Scholar]
- 49.Rebaudet S, et al. 2013. The dry season in Haiti: a window of opportunity to eliminate cholera. PLoS Curr. Outbreaks. ( 10.1371/currents.outbreaks.2193a0ec4401d9526203af12e5024ddc) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huffman GJ, Adler RF, Bolvin DT, Gu G, Nelkin EJ, Bowman KP, Hong Y, Stocker EF, Wolff DB. 2007. The TRMM multisatellite precipitation analysis (TMPA): quasi-global, multiyear, combined-sensor precipitation estimates at fine scales. J. Hydrometeorol. 8, 38–55. ( 10.1175/JHM560.1) [DOI] [Google Scholar]
- 51.de Rochars VEMB, et al. 2011. Knowledge, attitudes, and practices related to treatment and prevention of cholera, Haiti, 2010. Emerg. Infect. Dis. 17, 2158–2161. ( 10.3201/eid1711.110818) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Carpenter A. 2014. Behavior in the time of cholera: evidence from the 2008–2009 cholera outbreak in Zimbabwe. In Social computing, behavioral-cultural modeling and prediction, vol. 8393 (eds Kennedy WG, Agarwal N, Yang SJ.), pp. 237–244. Lecture Notes in Computer Science Berlin, Germany: Springer International Publishing. [Google Scholar]
- 53.Bengtsson L, Lu X, Thorson A, Garfield R, von Schreeb J. 2011. Improved response to disasters and outbreaks by tracking population movements with mobile phone network data: a post-earthquake geospatial study in Haiti. PLoS Med. 8, e1001083 ( 10.1371/journal.pmed.1001083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hubbard B, Lockhart G, Gelting RJ, Bertrand F. 2014. Development of Haiti's rural water, sanitation and hygiene workforce. J. Water Sanitation Hyg. Dev. 4, 159–163. ( 10.2166/washdev.2013.089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basu S, Andrews JR. 2013. Complexity in mathematical models of public health policies: a guide for consumers of models. PLoS Med. 10, e1001540 ( 10.1371/journal.pmed.1001540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wesolowski A, Eagle N, Tatem AJ, Smith DL, Noor AM, Snow RW, Buckee CO. 2012. Quantifying the impact of human mobility on malaria. Science 6104, 267–270. ( 10.1126/science.1223467) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Koelle K, Pascual M. 2004. Disentangling extrinsic from intrinsic factors in disease dynamics: a nonlinear time series approach with an application to cholera. Am. Nat. 163, 901–913. ( 10.1086/420798) [DOI] [PubMed] [Google Scholar]
- 58.Ruiz-Moreno D, Pascual M, Bouma M, Dobson A, Cash B. 2007. Cholera seasonality in Madras: dual role for rainfall in endemic and epidemic regions. EcoHealth 4, 52–62. ( 10.1007/s10393-006-0079-8) [DOI] [Google Scholar]
- 59.Finger F, Knox A, Bertuzzo E, Mari L, Bompangue D, Gatto M, Rodriguez-Iturbe I, Rinaldo A. 2014. Cholera in the Lake Kivu region (DRC): integrating remote sensing and spatially explicit epidemiological modeling. Water Resour. Res. 50, 5624–5637. ( 10.1002/2014WR015521) [DOI] [Google Scholar]
- 60.Holling CS. 1978. Adaptive environmental assessment and management. Chichester, UK: John Wiley & Sons. [Google Scholar]
- 61.Allan C, Stankey GH. 2009. Adaptive environmental management: a practitioner‘s guide. Collingwood, Australia: Springer/CSIRO Publishing. [Google Scholar]
- 62.Belton V, Stewart T. 2002. Multiple criteria decision analysis: an integrated approach. Dordrecht, The Netherlands: Kluwer Academic Publishers. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.






