Abstract
Integrin-mediated cell adhesion is important for development, immune responses, hemostasis and wound healing. Integrins also function as signal transducing receptors that can control intracellular pathways that regulate cell survival, proliferation, and cell fate. Conversely, cells can modulate the affinity of integrins for their ligands a process operationally defined as integrin activation. Analysis of activation of integrins has now provided a detailed molecular understanding of this unique form of “inside-out” signal transduction and revealed new paradigms of how transmembrane domains (TMD) can transmit long range allosteric changes in transmembrane proteins. Here, we will review how talin and mediates integrin activation and how the integrin TMD can transmit these inside out signals. [BMB Reports 2014; 47(12): 655-659]
Keywords: Cell adhesion, Integrin, Nanodisc, Signal transduction, Talin, Transmembrane domain
INTRODUCTION
Integrins, heterodimeric type I transmembrane proteins consisting of α and β subunits, are a major class of receptors involved in adhesive events that control development and lead to pathologies such as cancer and thrombosis. Eighteeen integrin α subunits and 8 β subunits heterodimerize to form 24 different integrins (1). Each subunit contains a single transmembrane domain (TMD) and a short cytoplasmic tail. Besides mediating cell adhesion, integrins transmit signals across the plasma membrane that regulate cell migration, cell survival and growth (2). Conversely, signals from inside cells can increase the binding of integrin extracellular domains to ligands, a process operationally defined as integrin activation. Integrin activation encompasses both changes in affinity of individual integrins due to conformational changes and avidity increases due to integrin clustering (3-5). Precise regulation of integrin activation is particularly important in controlling platelet aggregation through integrin αIIbβ3 (6). Rapid activation of this integrin at the site of a wound is required for hemostasis (7); conversely inappropriate activation of αIIbβ3 can cause a platelet thrombus to occlude a blood vessel resulting in myocardial infarction or stroke. Here we will discuss recent progress in understanding how integrins are activated.
INTEGRIN TMD-THE CONDUIT FOR ALLOSTERIC REARRANGEMENTS
Changes in the conformation of integrin extracellular domains are responsible for the changes in integrin monomer affinity. These conformational changes have been the subject of several excellent reviews (3, 8-12) and will not be discussed here. Similarly, clustering of integrins can enchance the binding of multivalent ligands and kindlins have recently emerged as major players in clustering (13). The capacity of intracellular signals to change the conformation of the extracellular domain requires a remarkable transmemebrane allosteric change, a change that must traverse the integrin TMD. Truncation of the integrins at the C-termini of extracellular domains results in constitutively active integrins (14), indicating that TMDs and cytoplasmic tails limit the activation state of integrins. Furthermore, many activating mutations, map to the α or β TMD (15-18). Heterodimeric interactions between α and β TMDs and cytoplasmic tails have been observed by co-immunoprecipitation (19), cysteine crosslinking (20, 21) and by NMR (22) in phospholipid bicelles, but not in detergent micelles (23). Importantly, mutations in TMDs that activate integrins invariably inhibit α and β TMD interactions (19). Thus, physiological integrin activation is likely to require that intracellular signals disrupt integrin αβ TMD interactions.
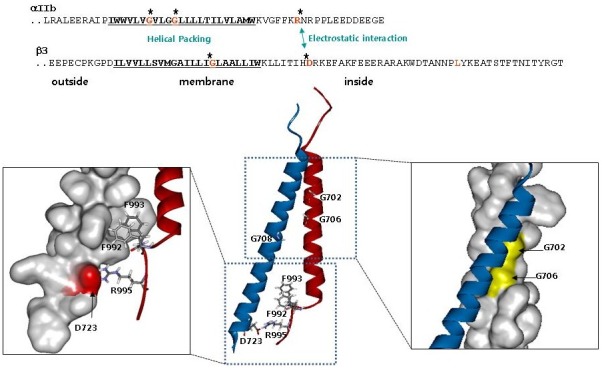
The structure of the αIIbβ3 TMD complex in a phospholipid bicelle (22) revealed the basis of association of the α and β through two interaction interfaces. The αIIb TMD helix is short, straight and broken at Gly991, the first residue of the highly-conserved Gly-Phe-Phe-Lys-Arg (GFFKR) motif in the membrane proximal region of the α subunits. The two Phe residues of the αIIb GFFKR motif do not form a continuous helix but instead make a sharp turn toward β3 (Fig. 1). In this way, the hydrophobic side chains of those residues reside in the hydrophobic core of the lipid bilayer and stack against hydrophobic residues in the β3 TMD, particularly Trp715 and Ile719. The turning of the membrane-proximal region of αIIb also permits the long-predicted (24) electrostatic interaction between αIIb Arg995 and β3 Asp723 by placing those residues in proximity (Fig. 1). The structure at the inner membrane interface is unique to and likely conserved in integrins and is termed the inner membrane clasp (IMC) (22). The second interface involves helical packing centered on β3 Gly708 and αIIb G972XXXG976 motif at the outer membrane region and is termed the outer membrane clasp (OMC) (Fig. 1). Integrin β3 TMD makes a long and continuous helix with a 25° tilting angle to enable the multipoint interactions with αIIb and accommodate the extra hydrophobic residues in the β3 TMD.
Fig. 1. Structure of integrin αIIbβ3 TMD (ribbon view; αIIb in red and β3 in blue. From PDB 2K9J) showing the two interaction interfaces. Right, outer membrane clasp (OMC) illustrating the helical packing involving αIIb Gly 702 and 706. Left, inner membrane clasp (IMC) showing the electrostatic interaction between αIIb Arg995 and β3 Asp723. Also depicted are the hydrophobic interactions of αIIb Phe992,933 with the β3 TMD. Adapted from reference (22).
As noted above, an important feature of the structure of the αIIbβ3 TMD dimer is that the helical β TMD must be precisely tilted to maintain simultaneous formation of the OMC and IMC. Precise tilt is maintained via β3 Lys716 whose alpha carbon resides in the hydrophobic region of the lipid bilayer but its positively charged ε-NH3+ is predicted to snorkel into the negatively charged phosphate head group region (25). Mutation of Lys716 any residue other than Arg (which also contains a snorkeling basic side chain) reduces α-β TMD interactions and dramatically increases integrin activation (25). The effects of Lys716 mutation can be ameliorated by breaking the continuous β TMD helix into two halves by introduction of a Pro mutation (A711P). The Pro mutation, introduces a flexible hinge that partially decouples the tilting angles of inner and outer helices favoring simultaneous formation of OMC and IMC (25).
TALIN “TILTS” THE INTEGRIN β TMD TO INDUCE ACTIVATION
Talin regulates integrin affinity and provides a mechanical link between integrins and the actin cytoskeleton. Talin comprises a 50-kDa N-terminal FERM domain (talin head domain or THD) that contains a high-affinity binding site for integrin β tails and a 220-kDa rod domain that contains multiple binding sites for actin and vinculin (26). The THD is further divided into F0, F1, F2 and F3 subdomains (26, 27). The F3 subdomain, contains the major integrin integrin β tail binding site (28, 29). The essential role of talin in regulating integrin affinity has been well documented in model cells (29-32), transgenic mice (33-36) and reconstituted systems with purified proteins (37). In in vitro systems, recombinant THD alone is sufficient to activate αIIbβ3 reconstituted in either liposomes or phospholipid nanodiscs, and activation is associated with a shift towards an αIIbβ3 extended conformation (37).
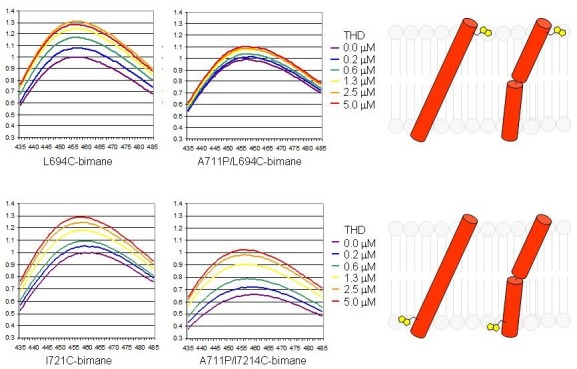
We now have considerable insight into how talin induces this allosteric rearrangement in integrins. Talin binds to two sites on integrin β tails: a strong binding site centered around the first NPxY motif that contributes most of the binding free energy and a weaker membrane proximal (MP) binding site (38). In addition, THD also binds to negatively charged phospholipids via positively-charged residues (38-40). The weak interaction with the MP region has two important effects: 1) it brings talin Lys324 close to Asp723 of the β3 tail, thus competing for the Arg995-Asp723 electrostatic interactions in IMC (40); 2) it stabilizes α-helical structure of the β MP region and to form a continuous helix with the β3 TMD (38, 40). As the simultaneous interaction with integrin β tails and phospholipids, can change the tilt angle of the β3 MP tail and thus of the contiguous β3 TMD (Fig. 2) (41). Such talin-induced motion was demonstrated by increased fluorescence of solvatochromic dyes attached to the N- or C-terminii of the β3 TMD in the presence of THD (41) and is further supported by molecular dynamic simulations (42). The change in tilting angle destabilizes α-β TMD interactions and shifts the equilibrium towards an activated integrin conformation. In further support of this model, introducing a flexible proline kink in the middle of the β3 TMD blocks THD-induced tilting of the outer membrane segment without blocking tilting of the inner membrane segment (Fig. 3). Integrins bearing this mutation are remarkably resistant to talin induced integrin activation (41).
Fig. 2. Talin changes the topology of the β3 TMD. A peptide containing the β3 TMD and cytoplasmic domain was labeled with environment sensitive bimanes at the outer edge of the TMD (N terminal labeling, Leu694) or at the TMD cytosol interface (C-terminal labeling, Ile721). The peptides were individually embedded in phospholipid nanodiscs and increasing concentrations of talin head domain (THD) were added and bimanes emission spectra were recorded. The increased fluorescence indicates that both sides of the β3 TMD were in a less polar environment suggesting that THD increased the tilting of the β3 TMD. Adapted from reference (41).
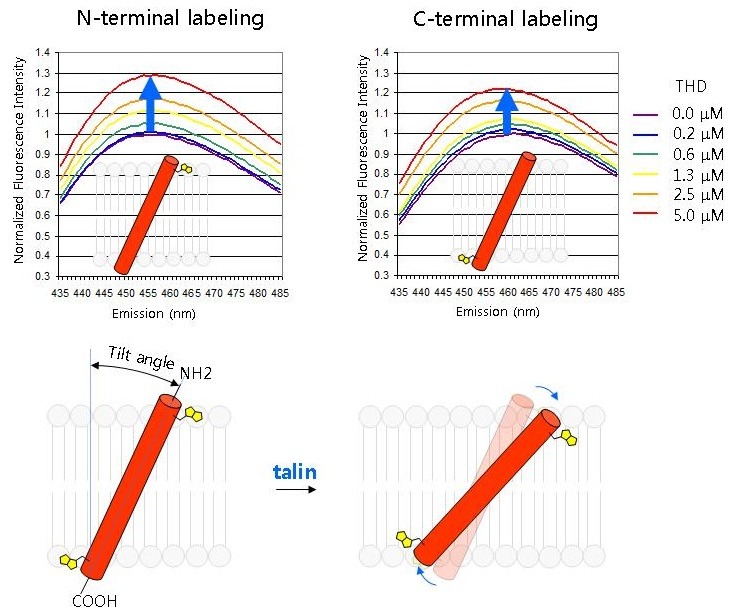
Fig. 3. A proline kink prevents transmission of altered tilt across the β3 TMD. In the left two panels The experimental design was identical to that in Fig. 2 and depicts the talin-induced increased embedding at both the inner (where THD binds) and outer edges of the TMD. Introduction of a flexible kink by β3 (A711P) mutation (right two panels) prevents the transmission of increased embedding of the inner TMD to the outer region. Adapted from reference (41).
SUMMARY AND CONCLUSIONS
Integrin activation was first observed in 1978 in integrin αIIbβ3 and it has proved to be a conserved property of β1, β2, and β3 integrins. As summarized here, our understanding of this unique form of transmembrane signal transduction has moved from a black box in which agonists, such as thrombin, caused a change in the affinity of integrin αIIbβ3. Today, cell biological and reverse genetic experiments have verified that talin binding to the integrin β cytoplasmic domain is a final common step in activation. Structural studies have revealed how two binding interfaces of talin with the integrin in combination with talin membrane binding sites can effect this form of transmembrane allostery. Studies have also revealed unique features of the heterodimeric integrin TMD that form a stable yet dynamic αβ TMD interaction that enables transmission of the activation signal across the phospholipid bilayer. The lessons learned in studying integrin transmembrane signaling, such as the importance of snorkeling basic residues in maintaining TMD topology, are likely to pertain to other examples of transmembrane signaling through transmembrane receptors.
References
- 1.Hynes R. O. Integrins: bidirectional, allosteric signaling machines. Cell. (2002);110:673–687. doi: 10.1016/S0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]
- 2.Giancotti F. G., Ruoslahti E. Integrin signaling. Science. (1999);285:1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 3.Kim C., Ye F., Ginsberg M. H. Regulation of integrin activation. Annu. Rev. Cell. Dev. Biol. (2011);27:321–345. doi: 10.1146/annurev-cellbio-100109-104104. [DOI] [PubMed] [Google Scholar]
- 4.Shattil S. J., Kim C., Ginsberg M. H. The final steps of integrin activation: the end game. Nat. Rev. Mol. Cell. Biol. (2010);11:288–300. doi: 10.1038/nrm2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shattil S. J., Newman P. J. Integrins: dynamic scaffolds for adhesion and signaling in platelets. Blood. (2004);104:1606–1615. doi: 10.1182/blood-2004-04-1257. [DOI] [PubMed] [Google Scholar]
- 6.Wagner C. L., Mascelli M. A., Neblock D. S., Weisman H. F., Coller B. S., Jordan R. E. Analysis of GPIIb/IIIa receptor number by quantification of 7E3 binding to human platelets. Blood. (1996);88:907–914. [PubMed] [Google Scholar]
- 7.Shattil S. J., Kashiwagi H., Pampori N. Integrin signaling: the platelet paradigm. Blood. (1998);91:2645–2657. [PubMed] [Google Scholar]
- 8.Luo B. H., Carman C. V., Springer T. A. Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. (2007);25:619–647. doi: 10.1146/annurev.immunol.25.022106.141618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arnaout M. A., Goodman S. L., Xiong J. P. Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell. Biol. (2007);19:495–507. doi: 10.1016/j.ceb.2007.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo B. H., Springer T. A. Integrin structures and conformational signaling. Curr. Opin. Cell. Biol. (2006);18:579–586. doi: 10.1016/j.ceb.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arnaout M. A., Mahalingam B., Xiong J. P. Integrin structure, allostery, and bidirectional signaling. Annu. Rev. Cell. Dev. Biol. (2005);21:381–410. doi: 10.1146/annurev.cellbio.21.090704.151217. [DOI] [PubMed] [Google Scholar]
- 12.Shimaoka M., Takagi J., Springer T. A. Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. (2002);31:485–516. doi: 10.1146/annurev.biophys.31.101101.140922. [DOI] [PubMed] [Google Scholar]
- 13.Ye F., Petrich B. G., Anekal P., Lefort C. T., Kasirer-Friede A., Shattil S. J., Ruppert R., Moser M., Fässler R., Ginsberg M. H. The Mechanism of Kindlin-mediated Activation of Integrin αIIbβ3. Curr. Biol. (2013);23:2288–2295. doi: 10.1016/j.cub.2013.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehta R. J., Diefenbach B., Brown A., Cullen E., Jonczyk A., Güssow D., Luckenbach G. A., Goodman S. L. Transmembrane-truncated alphavbeta3 integrin retains high affinity for ligand binding: evidence for an 'inside-out' suppressor? Biochem. J. (1998);330:861–869. doi: 10.1042/bj3300861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Partridge A. W., Liu S., Kim S., Bowie J. U., Ginsberg M. H. Transmembrane domain helix packing stabilizes integrin alphaIIbbeta3 in the low affinity state. J. Biol. Chem. (2005);280:7294–7300. doi: 10.1074/jbc.M412701200. [DOI] [PubMed] [Google Scholar]
- 16.Luo B. H., Carman C. V., Takagi J., Springer T. A. Disrupting integrin transmembrane domain heterodimerization increases ligand binding affinity, not valency or clustering. Proc. Natl. Acad. Sci. U. S. A. (2005);102:3679–3684. doi: 10.1073/pnas.0409440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W., Metcalf D. G., Gorelik R., Li R., Mitra N., Nanda V., Law P. B., Lear J. D., Degrado W. F., Bennett J. S. A push-pull mechanism for regulating integrin function. Proc. Natl. Acad. Sci. U. S. A. (2005);102:1424–1429. doi: 10.1073/pnas.0409334102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li R., Mitra N., Gratkowski H., Vilaire G., Litvinov R., Nagasami C., Weisel J. W., Lear J. D., DeGrado W. F., Bennett J. S. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science. (2003);300:795–798. doi: 10.1126/science.1079441. [DOI] [PubMed] [Google Scholar]
- 19.Kim C., Lau T. L., Ulmer T. S., Ginsberg M. H. Interactions of platelet integrin alphaIIb and beta3 transmembrane domains in mammalian cell membranes and their role in integrin activation. Blood. (2009);113:4747–4753. doi: 10.1182/blood-2008-10-186551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu J., Luo B. H., Barth P., Schonbrun J., Baker D., Springer T. A. The structure of a receptor with two associating transmembrane domains on the cell surface: integrin alphaIIbbeta3. Mol. Cell. (2009);34:234–249. doi: 10.1016/j.molcel.2009.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo B. H., Springer T. A., Takagi J. A specific interface between integrin transmembrane helices and affinity for ligand. PLoS Biol. (2004);2:e153. doi: 10.1371/journal.pbio.0020153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lau T. L., Kim C., Ginsberg M. H., Ulmer T. S. The structure of the integrin alphaIIbbeta3 transmembrane complex explains integrin transmembrane signalling. EMBO J. (2009);28:1351–1361. doi: 10.1038/emboj.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R., Babu C. R., Lear J. D., Wand A. J., Bennett J. S., DeGrado W. F. Oligomerization of the integrin alphaIIbbeta3: roles of the transmembrane and cytoplasmic domains. Proc. Natl. Acad. Sci. U. S. A. (2001);98:12462–12467. doi: 10.1073/pnas.221463098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes P. E., Diaz-Gonzalez F., Leong L., Wu C., McDonald J. A., Shattil S. J., Ginsberg M. H. Breaking the integrin hinge: a defined structural constraint regulates integrin signaling. J. Biol. Chem. (1996);271:6571–6574. doi: 10.1074/jbc.271.12.6571. [DOI] [PubMed] [Google Scholar]
- 25.Kim C., Schmidt T., Cho E. G., Ye F., Ulmer T. S., Ginsberg M. H. Basic amino-acid side chains regulate transmembrane integrin signalling. Nature. (2012);481:209–213. doi: 10.1038/nature10697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Critchley D. R. Biochemical and structural properties of the integrin-associated cytoskeletal protein talin. Annu. Rev. Biophys. (2009);38:235–254. doi: 10.1146/annurev.biophys.050708.133744. [DOI] [PubMed] [Google Scholar]
- 27.Elliott P. R., Goult B. T., Kopp P. M., Bate N., Grossmann J. G., Roberts G. C., Critchley D. R., Barsukov I. L. The Structure of the talin head reveals a novel extended conformation of the FERM domain. Structure. (2010);18:1289–1299. doi: 10.1016/j.str.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calderwood D. A., Fujioka Y., de Pereda J. M., García-Alvarez B., Nakamoto T., Margolis B., McGlade C. J., Liddington R. C., Ginsberg M. H. Integrin beta cytoplasmic domain interactions with phosphotyrosine-binding domains: a structural prototype for diversity in integrin signaling. Proc. Natl. Acad. Sci. U. S. A. (2003);100:2272–2277. doi: 10.1073/pnas.262791999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calderwood D. A., Yan B., de Pereda J. M., Alvarez B. G., Fujioka Y., Liddington R. C., Ginsberg M. H. The phosphotyrosine binding-like domain of talin activates integrins. J. Biol. Chem. (2002);277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 30.Lee H. S., Lim C. J., Puzon-McLaughlin W., Shattil S. J., Ginsberg M. H. RIAM activates integrins by linking talin to ras GTPase membrane-targeting sequences. J. Biol. Chem. (2009);284:5119–5127. doi: 10.1074/jbc.M807117200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han J., Lim C. J., Watanabe N., Soriani A., Ratnikov B., Calderwood D. A., Puzon-McLaughlin W., Lafuente E. M., Boussiotis V. A., Shattil S. J., Ginsberg M. H. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr. Biol. (2006);16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- 32.Calderwood D. A., Zent R., Grant R., Rees D. J., Hynes R. O., Ginsberg M. H. The Talin head domain binds to integrin beta subunit cytoplasmic tails and regulates integrin activation. J. Biol. Chem. (1999);274:28071–28074. doi: 10.1074/jbc.274.40.28071. [DOI] [PubMed] [Google Scholar]
- 33.Petrich B. G., Marchese P., Ruggeri Z. M., Spiess S., Weichert R. A., Ye F., Tiedt R., Skoda R. C., Monkley S. J., Critchley D. R., Ginsberg M. H. Talin is required for integrin-mediated platelet function in hemostasis and thrombosis. J. Exp. Med. (2007);204:3103–3111. doi: 10.1084/jem.20071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nieswandt B., Moser M., Pleines I., Varga-Szabo D., Monkley S., Critchley D., Fässler R. Loss of talin1 in platelets abrogates integrin activation, platelet aggregation, and thrombus formation in vitro and in vivo. J. Exp. Med. (2007);204:3113–3118. doi: 10.1084/jem.20071827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petrich B. G., Fogelstrand P., Partridge A. W., Yousefi N., Ablooglu A. J., Shattil S. J., Ginsberg M. H. The antithrombotic potential of selective blockade of talin-dependent integrin alpha IIb beta 3 (platelet GPIIb-IIIa) activation. J. Clin. Invest. (2007);117:2250–2259. doi: 10.1172/JCI31024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Haling J. R., Monkley S. J., Critchley D. R., Petrich B. G. Talin-dependent integrin activation is required for fibrin clot retraction by platelets. Blood. (2011);117:1719–1722. doi: 10.1182/blood-2010-09-305433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye F., Hu G., Taylor D., Ratnikov B., Bobkov A. A., McLean M. A., Sligar S. G., Taylor K. A., Ginsberg M. H. Recreation of the terminal events in physiological integrin activation. J. Cell. Biol. (2010);188:157–173. doi: 10.1083/jcb.200908045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wegener K. L., Partridge A. W., Han J., Pickford A. R., Liddington R. C., Ginsberg M. H., Campbell I. D. Structural basis of integrin activation by talin. Cell. (2007);128:171–182. doi: 10.1016/j.cell.2006.10.048. [DOI] [PubMed] [Google Scholar]
- 39.Goult B. T., Bouaouina M., Elliott P. R., Bate N., Patel B., Gingras A. R., Grossmann J. G., Roberts G. C., Calderwood D. A., Critchley D. R., Barsukov I. L. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. EMBO J. (2010);29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anthis N. J., Wegener K. L., Ye F., Kim C., Goult B. T., Lowe E. D., Vakonakis I., Bate N., Critchley D. R., Ginsberg M. H., Campbell I. D. The structure of an integrin/talin complex reveals the basis of inside-out signal transduction. EMBO J. (2009);28:3623–3632. doi: 10.1038/emboj.2009.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim C., Ye F., Hu X., Ginsberg M. H. Talin activates integrins by altering the topology of the beta transmembrane domain. J. Cell. Biol. (2012);197:605–611. doi: 10.1083/jcb.201112141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalli A. C., Wegener K. L., Goult B. T., Anthis N. J., Campbell I. D., Sansom M. S. The structure of the talin/integrin complex at a lipid bilayer: an NMR and MD simulation study. Structure. (2010);18:1280–1288. doi: 10.1016/j.str.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]


