Significance
Bacterial chemoreceptors are a key system for understanding how conformational signals propagate over large distances in transmembrane signaling. We have applied pulsed dipolar ESR spectroscopy of spin-labeled receptors to correlate conformation and dynamics with activity state. We find that the receptor cytoplasmic domain behaves as one large dynamically coupled system, in which activation signals destabilize membrane proximal regions but stabilize the most distal protein interaction tip. Inhibitory signals or adaptations of the receptor through chemical modification produce the opposite changes in conformational properties. This reciprocal coupling of conformational stability provides a versatile mechanism for sending signals throughout large modular proteins.
Keywords: chemotaxis, transmembrane signaling receptor, allostery, dynamics, adaptation
Abstract
Dynamics are hypothesized to play an important role in the transmission of signals across membranes by receptors. Bacterial chemoreceptors are long helical proteins that consist of a periplasmic ligand-binding domain; a transmembrane region; a cytoplasmic HAMP (histidine kinase, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases) domain; and a kinase-control module (KCM). The KCM is further composed of adaptation, hinge, and protein interaction regions (PIRs), the latter of which binds the histidine kinase CheA and adaptor CheW. Fusions of the Escherichia coli aspartate receptor KCM to HAMP domains of defined structure (H1-Tar vs. H1-2-Tar) give opposite responses in phosphotransfer and cellular assays, despite similar binding to CheA and CheW. Pulsed dipolar ESR spectroscopy (PDS) of these isolated on and off dimeric effectors reveals that, in the kinase-on state, the HAMP is more conformationally destabilized compared with the PIR, whereas in the kinase-off state, the HAMP is more compact, and the PIR samples a greater breadth of conformations. On and off HAMP states produce different conformational effects at the KCM junction, but these differences decrease through the adaptation region and into the hinge only to return with the inverted relationship in the PIR. Continuous wave–ESR of the spin-labeled proteins confirms that broader PDS distance distributions correlate with increased rates of dynamics. Conformational breadth in the adaptation region changes with charge alterations caused by modification enzymes. Activating modifications broaden the HAMP conformational ensemble but correspondingly, compact the PIR. Thus, chemoreceptors behave as coupled units, in which dynamics in regions proximal and distal to the membrane change coherently but with opposite sign.
The ability of localized dynamics to modulate the function of transmembrane receptors is an emerging theme in signal transduction (1–4). These ideas have been largely supported by computational studies (2), although direct measurements also correlate dynamics with activity (1–4). Nonetheless, we are only beginning to address the link between conformational heterogeneity and signal propagation in complex proteins. Bacterial chemotaxis, the process by which cells modulate their motility in response to the chemical environment, provides an important model system to explore receptor dynamics experimentally (5, 6). During chemotaxis, attractant-bound chemoreceptors cause counterclockwise (CCW) flagella rotation and smooth swimming, whereas repellant-bound receptors cause clockwise flagella rotation and cell tumbling. Chemoreceptors, also termed methyl-accepting chemotaxis proteins, form extended arrays in the cytoplasmic membrane to communicate ligand binding (6) to the histidine kinase CheA and the coupling protein CheW. Great progress has been made in understanding how receptors communicate ligand-binding events across the cytoplasmic membrane, but how these changes affect CheA is not well-understood (5–7).
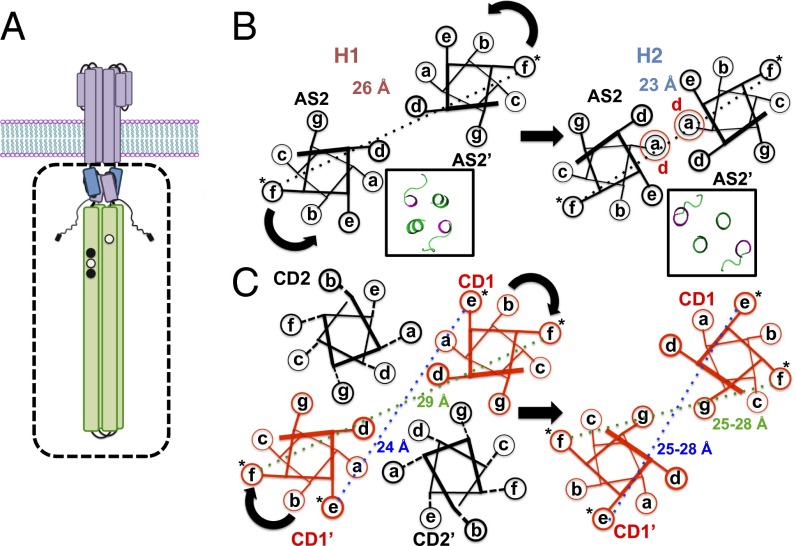
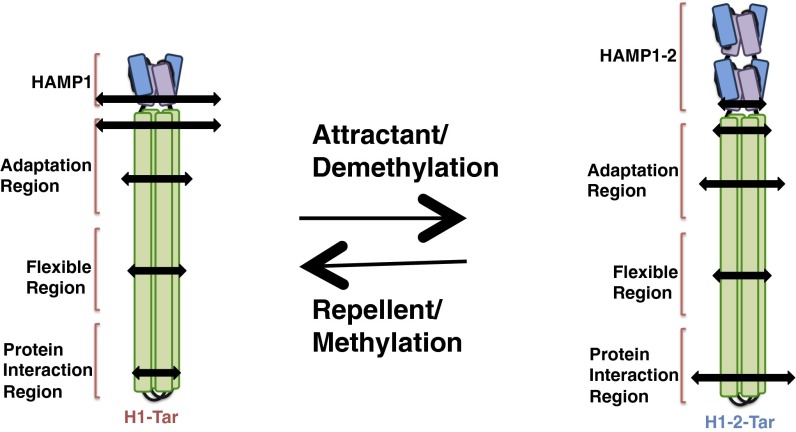
Homodimeric chemoreceptors have a modular architecture. Each subunit supplies a periplasmic ligand-binding domain, a helical transmembrane region, and one-half of two cytoplasmic four-helix bundles that extend from the membrane to engage CheA and CheW in the cytoplasm (8). The transmembrane region contains four antiparallel helices (TM1/TM2 and TM1′/TM2′) and connects to the membrane-proximal HAMP (histidine kinase, adenylyl cyclases, methyl-accepting chemotaxis proteins, and phosphatases) domain through TM2 and TM2′. The HAMP domain comprises a parallel four-helix bundle (AS1/AS2 and AS1′/AS2′) with two helices supplied from each subunit (7). HAMP joins to the kinase-control module (KCM), which forms a long, antiparallel four-helix bundle, with two helices supplied by each subunit (CD1/CD2 and CD1′/CD2′) (6). The KCM can be further divided into an adaptation region, a flexible bundle with a glycine hinge, and a protein interaction region (PIR) at the receptor tip. The adaptation region contains conserved Glu residues that undergo reversible methylation/demethylation by the methylase CheR and methylesterase CheB, respectively. For the well-studied aspartate (Tar) and serine (Tsr) receptors of Escherichia coli, chemoattractant binding inhibits CheA activity (kinase-off, CCW flagellar rotation), and this effect is countered by methylation, which reactivates the kinase and weakens affinity for attractant. In contrast, binding of repellants (or release of attractant) activates CheA (kinase-on, clockwise flagellar rotation) and is similarly countered by demethylation. Substitution of specific Glu residues to Gln functionally mimics methylation (9).
Conformational changes associated with ligand binding to chemoreceptors are well-described in the periplasm and transmembrane regions (6, 8, 10) but less well-described in the cytoplasm. Attractant binding to the periplasmic domain of Tar causes a vertical displacement of the α4-helix that directly attaches to TM2 (10). TM2 responds with a piston displacement that has been characterized by a variety of experimental and computational methods (6, 8, 11, 12). The TM2 piston motion relays to the HAMP domain through a small control cable on the cytoplasmic side of the membrane (13). HAMP is then proposed to undergo a change in both structure and dynamics (7, 14). Mutagenesis studies suggest that HAMP in kinase-off states is less dynamic than in kinase-on states (7, 15, 16). Molecular dynamics simulations on membrane-embedded Tar provide a different picture, wherein ligand binding produces only modest changes in dynamics, with motions of AS2 slightly increasing in the kinase-off state (12). Dynamical changes in HAMP may be mirrored by dynamical changes in the KCM: mutations that destabilize packing in the PIR lock in kinase-on states, whereas destabilization of the adaptation region produces kinase-off states (17). Rates of disulfide cross-linking indicate that kinase-off mutations are, indeed, more dynamic (14, 17). This so-called yin-yang model (14) represents the cytoplasmic regions of the receptor as two units—the adaptation and PIRs—that alternate their degree of conformational fluctuation depending on signaling state (14). Between these modules, a flexible bundle region facilitates conformational coupling (18).
Structural changes within the KCM have also been implicated in kinase regulation. The HAMP AS2 helices may undergo a scissor-opening motion when transitioning from kinase off to kinase on (14, 19). Attractant-sensitive disulfide bond formation between CheA and receptor suggests that the N-terminal CD1 helices rotate in the PIR on ligand binding (20). Rotations (21, 22) and longitudinal shifts of helices (23) within HAMP have been implicated as regulatory switches, and such transitions may interconvert states with different dynamical properties. Long-duration, all-atom molecular dynamics simulations indicate that separations of one KCM N-terminal helix (CD1) and the C-terminal helix of the other subunit (CD2′) are coupled to the flipping of a conserved Phe residue at the receptor tip and that these changes correlate with modification state (24). Nonetheless, these observations have yet to be combined in an encompassing model of receptor activation.
We have shown that, when two structurally distinct HAMP modules (H1 or H1-2) taken from Pseudomonas aerurginosa Aer2 are fused to the Tar KCM, the resulting chimeras produce exclusive kinase-on and kinase-off states in cells (25). Pulsed dipolar ESR spectroscopy (PDS) in concert with site-specific spin labeling showed that the two HAMP modules maintained their structures in the respective fusions and further enforced their AS2 arrangements across the junction into the KCM (25). The activating HAMP (H1), indeed, displayed much broader distributions of spin separations than the inactivating module (H1-2), and these dynamical features were mirrored in the attached KCM. Here, we further use PDS and continuous wave (CW) -ESR to probe the KCM dynamics of these kinase-on (H1-Tar) and kinase-off (H1-2-Tar) effector modules (Fig. 1). The widths of PDS distance distributions correlate well with spin-label dynamics. Together, they reveal that, in a given effector, the HAMP domain enforces a similar helical separation and dynamic state across the junction into the KCM, whereas the PIR assumes the opposite behavior. The conformational dynamics and activation state of all modules coherently switch when H1 is replaced by H1-2, and these changes can be mostly reversed by covalent modification in the adaptation region.
Fig. 1.
Schematic representations of spin separations generated by different helix packing and labeling locations. (A) Schematic of the bacterial chemoreceptor is shown with the effector module, studied in this report, depicted in the box. (B) Differences in spin separations on AS2 between the H1 and H1-2 conformations predicted from their crystal structures. Insets display the crystal structures of the C-terminal planes of the two HAMPs viewed from the N-terminal end (AS2 and linker in green). (C) Differences in spin separations within the four-helix bundle of the KCM for an f site and an e site and the hypothetical effect of a clockwise CD1 rotation on these distances. *Spin label.
Results
CheA Activity and Ternary Complex Stability.
Aer2 contains a poly-HAMP module of three concatenated HAMP domains (H1-2-3). The structure of the H1-2-3 poly-HAMP revealed that H2 has a different conformation compared with H1 and H3, which are similar in structure (26). In H2, the bundle is rhombically distorted, such that the AS2 helices interact more closely and approximate a two-helix coiled coil (Fig. 1B). As previously reported (25), chimeras that fuse H1 to the Tar KCM (here, H1-Tar) (named H1s in ref. 25) and H1-2 to the Tar KCM (here, H1-2-Tar) (named H1-2s in ref. 25) produce very different output signals when expressed in E. coli cells devoid of all other receptors (Fig. S1). In cells lacking the adaptation system (CheRB−), H1-Tar is 100% clockwise (kinase on), and H1-2-Tar is almost exclusively CCW (kinase off). In the presence of the methylation system (CheRB+), kinase activation increases with H1-2-Tar, although a converse inhibition is not seen with H1-Tar.
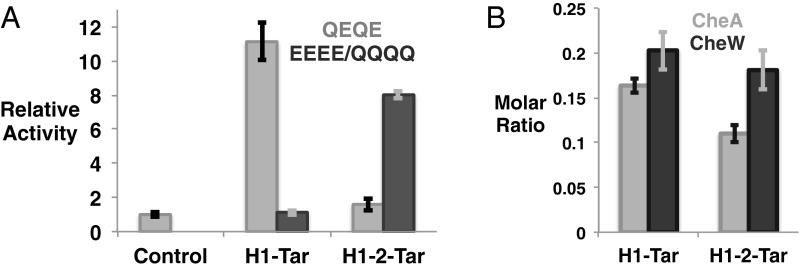
In vitro assays show the ability of H1-Tar and H1-2-Tar to differentially regulate CheA phosphotransfer activity without appreciably altering binding to the kinase or CheW (Fig. 2). H1-Tar activates phosphotransfer to CheY over 10-fold, whereas H1-2-Tar does not (Fig. 2A). These effects are largely reversed by modification in the adaptation region: when all Gln sites in H1-Tar are changed to Glu (EEEE), H1-Tar no longer activates CheA, and when the H1-2-Tar sites are changed to Gln (QQQQ), phosphotransfer increases substantially (Fig. 2A). Pull-down experiments with affinity-tagged effectors and purified E. coli CheA and CheW show that the opposing in vitro and cellular activities of H1-Tar and H1-2-Tar in the same modification state (QEQE) do not derive from the ability of only one protein to produce ternary complexes. Both effectors bind similar amounts of CheA and CheW, with H1-Tar binding slightly more of both proteins (Fig. 2B). The molar subunit ratios in the pull downs of receptor:CheW:CheA are 6:1.2:1 for H1-Tar and 6:1.1:0.7 for H1-2-Tar, which in both cases, are quite close to the stoichiometry predicted for the membrane arrays (6:1:1) (27, 28).
Fig. 2.
Properties of the H1-Tar and H1-2-Tar effectors. (A) In vitro activity of H1-Tar and H1-2-Tar. H1-Tar (gray) stimulates the phosphotransfer activity of CheA to CheY relative to CheA and CheW alone (control), whereas H1-2-Tar (gray) does not. This difference in activity is largely reversed on covalent modification in H1-Tar EEEE (black) or H1-2-Tar QQQQ (black). Error bars represent SEMs calculated from at least four independent experiments (4 ≤ n ≤ 6). (B) Interaction of H1-Tar and H1-2-Tar with CheA and CheW. Purified CheA and CheW were pulled down with S-tagged effectors. Molar subunit ratios of bound CheA and CheW to effector were calculated as described in SI Materials and Methods and averaged from five independent experiments. Error bars are shown as SEMs (n = 5; P = 0.01 and P = 0.06 for differential binding of CheA and CheW, respectively).
Conformational Differences in the KCM.
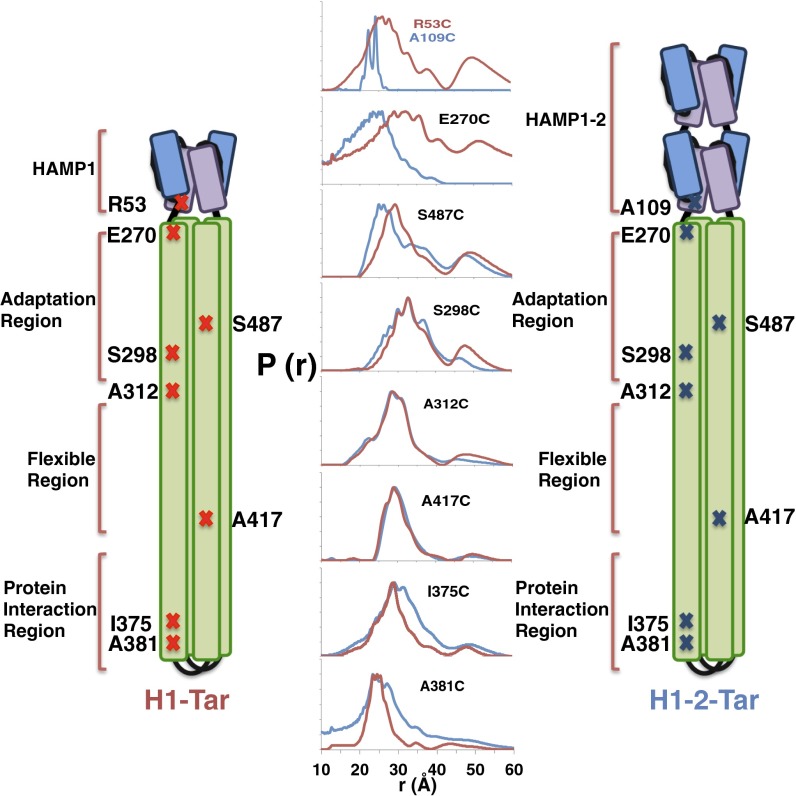
PDS and site-specific spin labeling were used to report on conformational dynamics at the HAMP junction and throughout the KCM (Fig. 3). Engineered Cys residues were spin-labeled with nitroxide moieties on HAMP AS2 and at seven sites in the KCM (Fig. 3). Distance distributions between each spin pair generated by effector dimerization were measured for both H1-Tar and H1-2-Tar by the PDS technique of double-electron electron resonance spectroscopy (Fig. 3, Fig. S2, and Table S1) (29). Broader spatial distributions represent larger amplitude motions of the spin labels. Residues in the H1 and H1-2 Aer2 HAMP domains have their Aer2 numbering, whereas residues in KCM have their E. coli Tar values. R53 of H1-Tar and A109 of H1-2-Tar are equivalent positions at the C-terminal end of AS2 in the HAMP modules. E270 in the N-terminal helix CD1 resides across the junction into the KCM at the HAMP proximal end of adaptation region. A298 resides in CD1 and A487 resides in CD2 of the adaptation region. A312 (in CD1) and A417 (in CD2) border the flexible bundle region. I375 and A381 (in CD1/CD1′) are in the N-terminal helix of the PIR. These positions were chosen to be mostly f sites in the heptad repeats of the effector coiled coils to minimize any structural perturbation by the label (although A381 holds an e site) (Fig. 1C). Indeed, spin-labeling the effectors in the HAMP and adaptation region does not affect their activities (Fig. S3).
Fig. 3.
Dynamical properties of the HAMP-KCM inferred by PDS. Schematic diagrams of H1-Tar and H1-2-Tar are marked with the residues where spin labels have been introduced. Because of dimeric symmetry, each spin-site position produces two spin sites from which distance distributions across the helix bundle are measured by PDS. Distance distributions [P(r)] measured by double-electron electron resonance spectroscopy are shown for H1-Tar (red) and H1-2-Tar (blue; both in the QEQE adaptation state).
The PDS distance distributions report on conformational differences in the helices through their mean values and reflect on dynamics through their breadth (Fig. 1). For example, because the H2 structure approximates a two-helix coiled coil, with the AS2 helices more closely associated than in H1, PDS finds that the 53 distribution has a greater mean (26 Å) than the 109 distribution (23 Å) (Figs. 1B and 3). Indeed, the short H2 distribution is sharply bimodal because of either distinct helix or spin-label conformations. The spin separations are mirrored across the junction into the KCM: the 270 position in H1-Tar shows a broad distribution centered at ∼30 Å, characteristic of an f position in a four-helix bundle, but 270 in H1-2-Tar, instead, shows a short, sharp separation characteristic of the two-helix packing in H2 AS2. In the adaptation region, the CD2 site (487) has a wider distribution in H1-2-Tar than in H1-Tar, but the CD1 site (298) is similar for both and overall, narrower. Through the flexible bundle region, the H1-Tar and H1-2-Tar distributions for 312 (CD1) and 417 (CD2) are nearly superimposable and typical of a dynamic four-helix bundle. Some distributions (particularly within H1-Tar) show a second peak of lower amplitude at approximately two times the primary peak distance. These longer separations are partly caused by a small contribution from disulfide cross-linked dimers (Fig. S4) but may also arise from a second family of conformational stated where the helices are far separated (see below). Interestingly, the second peak arises only in the HAMP domain (in some cases) and adaptation region, both of which are less well-ordered in cellular arrays viewed by cryo-EM (27).
As in HAMP, the sites in the PIR (375 and 381) show quite different behavior for H1-Tar and H1-2-Tar. Unlike HAMP, the trend in dynamics reverses, with H1-Tar having the more stable, narrow distributions and H1-2-Tar generating broader spin–spin separations indicative of increased dynamics. Thus, the measurements reveal yin-yang behavior between HAMP and the PIR, with a more mobile HAMP (kinase on) producing a tighter PIR and vice versa (Fig. 3). In H1-Tar, the ∼25-Å mean of the 381 distribution (e position) is notably shorter than the ∼30-Å mean of the 375 distribution (f position), which would be predicted by the structure of a four-helix bundle (Fig. 1C). Targeted disulfide cross-linking studies indicate that the CD1 helices may rotate in the kinase-off state (20) (i.e., for H1-2-Tar), which should increase the 381 separation and decrease the 375 separation in H1-2-Tar relative to those in H1-Tar (Fig. 1C). Interestingly, the 381 distribution does skew toward a longer distance in H1-2-Tar (Fig. 3), but a corresponding decrease in the 375 mean is not observed. Nonetheless, both the 375 and 381 distributions broaden in H1-2-Tar, thereby indicating that increased dynamics in the PIR are likely the dominant factor in transitioning to the kinase-off state. However, given that the sites in the Gly hinge and adaptation region also do not show changes consistent with CD1 rotation, it is unlikely that such a concerted motion propagates down the entire KCM.
Effector Modification Inverts Conformational Dynamics.
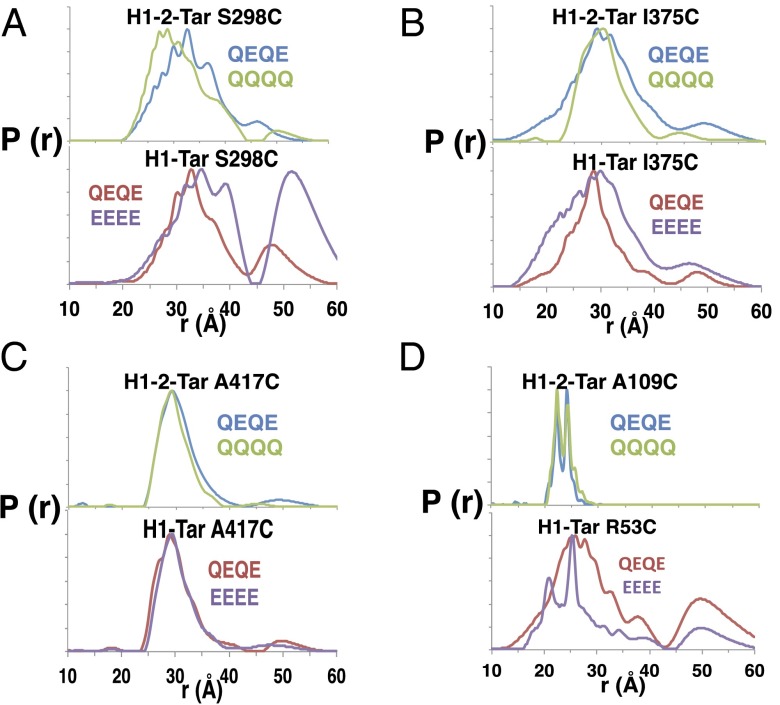
Modification impacts CheA activation (Fig. 2A), and thus, we tested if spin–spin separations at different regions of the effectors respond to modification state. QQQQ and QEQE states were monitored in H1-2-Tar (QEQE = kinase off), and the QEQE and EEEE states were monitored in H1-Tar (QEQE = kinase on) (Fig. 4 and Table S2). Increasing charge (and thus, local coulomb repulsion) in H1-Tar (QEQE→EEEE) shifts the distribution at S298 toward longer distance and greater width (Fig. 4A). Furthermore, a second major peak appears in the distribution centered at ∼50 Å. Examination of samples after PDS revealed only minor disulfide cross-linking (Fig. S4). Thus, the larger distances represent a conformation, where the CD1 helices no longer pack as a four-helix bundle. In contrast, decreasing charge in H1-2-Tar (QEQE→QQQQ) skews the distribution at S298 toward smaller separations (Fig. 4A) with slightly smaller breadth (Table S2). This measurement provides direct evidence that increased charge caused by demethylation or deamidation locally increases dynamics and destabilizes the adaptation region away from a compact four-helix bundle. In the PIR, deamidation (QEQE→EEEE) of H1-Tar causes I375 to become more dynamic and the HAMP site to order; indeed, the distributions now resemble those of H1-2-Tar QEQE (Fig. 4 B and D). Remarkably, the sharp bimodal distribution of H1-2 (which represents two closely related conformations of protein and/or spin label) is reproduced in H1-Tar EEEE. Thus, adaptation switches the H1 conformation toward that of H2. In contrast, charge neutralization (i.e., methylation QEQE→QQQQ) in H1-2-Tar causes the I375 site to become more rigid (Fig. 4B). However, in H1-2-Tar, the HAMP site does not correspondingly shift to a more mobile state (Fig. 4D). This lack of change is likely because modification cannot easily overcome the high stability of the dual HAMP H1-2 unit. Surprisingly, modification in the adaptation region does not influence conformation or dynamics at the A417 site in either of the two effectors (Fig. 4C). Overall, adaptation mostly reverses the dynamical properties both up and down the receptor, thereby underscoring the conformational coupling throughout the entire HAMP KCM.
Fig. 4.
Modification of adaptation sites reveals allosteric coupling throughout the HAMP-KCM. Methylation of the effector adaptation regions was mimicked by glutamine substitution. (A) Increased modification and decreasing charge in H1-2-Tar (QQQQ) shift the distribution at the adaptation region toward smaller distances, whereas decreased modification (EEEE) and increasing charge in H1-Tar shift the distribution to longer distances and amplify the peak centered around ∼50 Å. (B) Increased modification of H1-2-Tar decreases the distance distribution [P(r)] breadth and stabilizes the PIR, whereas in H1-Tar, decreased modification increases the P(r) breadth and destabilizes the PIR. (C) Modification at the adaptation region has no effect on PDS distributions in the hinge region for either effector. (D) Increased modification of H1-2-Tar has little effect on spin distributions in AS2 of the HAMP domain; however, decreased modification stabilizes the HAMP domain of H1-Tar toward the sharp bimodal distribution as seen in H1-2-Tar.
Dynamic Measurements by CW-ESR.
Pulsed dipolar ESR must be performed on flash-cooled samples to lengthen spin-dephasing times (29). Thus, the distance distributions measure the heterogeneity of conformational states in cold-trapped, static samples, from which dynamics before flash-cooling are inferred. To corroborate the relationship between dynamics and distribution width, we also measured CW-ESR on liquid samples at three temperatures: 4 °C, 20 °C, and 30 °C. The line shapes of the solution CW-ESR spectra reflect dynamics of the nitroxides on the nanosecond timescale (30, 31). CW-ESR spectra of spin labels at the HAMP (53 and 109 positions) and PIR (375) all clearly show two-component spectra (Table 1 and Fig. S5). One component corresponds to a relatively immobilized state and closely approximates a rigid limit spectrum (2Azz is equal to ∼69G). The rigid-like component was simulated (32), and its variable fraction was subtracted from the experimental spectrum to obtain a smooth line without outer features. The subtraction reveals the shape of the second major component with faster molecular motion parameters (Fig. S5 and Table S3). The fractions of these components were then calculated from double integrals. An isotropic diffusion parameter set to Rll = Rprp = 8 × 106 s−1 is sufficient to produce the correct splitting value and allows for robust subtraction of the rigid, slow component to reveal the mobile fraction. The slow diffusion could reflect spin labels associated with relatively rigid helices (33). In contrast, the state with higher mobility indicates relatively free motion of the tethered spin label and thus, little constraint by the attached protein (30, 31, 34).
Table 1.
CW-ESR spectra for spin labels in the HAMP (109, 53) and the PIR (375) were fit to two components [one representing a rigid nitroxide (slow component) and one representing freely rotating nitroxide (fast component)] as described in the text
| Two-component analysis | 4 °C (%) | 20 °C (%) | 30 °C (%) | |||
| Slow | Fast | Slow | Fast | Slow | Fast | |
| H1-2-Tar A109C | 52.5 | 47.5 | 28.5 | 71.5 | 20 | 80 |
| H1-Tar R53C | 47 | 53 | 26.5 | 73.5 | 16 | 84 |
| H1-2-Tar I375C | 52 | 48 | 31 | 69 | 24 | 76 |
| H1-Tar I375C | 61 | 39 | 41 | 59 | 28 | 72 |
The overall behavior of the spin labels most likely reflects an equilibrium between states where the spin label interacts with the surface of a relatively rigid substructure, such as receptor helices (33, 35), and states where the label tumbles unencumbered by the protein (31). The increased motion could be caused by the nitroxide tether releasing from the protein surface or may involve conformational transitions of the protein itself. In either case, the underlying cause of the increased mobile fraction reflects greater protein dynamics in these regions. Increase in the mobile component with temperature for all variants indicates that the rigid component is not an artifact related to, for example, partial denaturation and/or aggregation of the protein (Table 1 and Fig. S6). At lower temperature, the sites with broader PDS distributions (H1-2 375 and H1 53) show greater contributions from the more mobile conformation (Fig. 5 and Table 1). As temperature increases, the mobile state dominates at all sites, and the fractions become similar in the two effectors (Table 1). At the highest temperature, the HAMP is generally more dynamic than the PIR, and the two effectors only produce small differences at either position. Thus, the broader PDS distributions represent an ensemble of conformational states, some of which contain restrained nitroxides and others contain released nitroxides. At lower temperature, this ensemble coalesces into two distinct states with different spin-label dynamics on the nanosecond timescale. As the temperature increases, a range of intermediate states becomes occupied, most with relatively fast spin-label dynamics. Those variants with narrower PDS distributions (e.g., H1-375 and H1-2-109) have access to a greater proportion of states having constrained nitroxides.
Fig. 5.
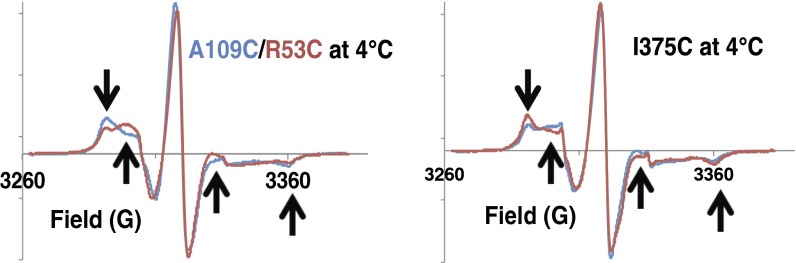
CW-ESR spectra reflect dynamic differences in HAMP and KCM. Spectra of spin labels at (Left) the HAMP domain and (Right) the PIR reveal differences in the dynamics in H1-2-Tar (blue) and H1-Tar (red). Consistent with the PDS distributions (Fig. 3), the PIR is more rigid but the HAMP domain is more dynamic in H1-Tar. Black arrows mark the spectral regions where the differences are prominent.
Discussion
Changes in local dynamics have been linked to functionally distinct states in G protein-coupled receptors (1, 2) and receptor tyrosine kinases (3). Our results confirm the modular structure of the chemoreceptor cytoplasmic domain. Although the measurements were made in the absence of membrane arrays, full-length dimeric receptors do transmit conformational signals between the ligand binding and the adaptation regions (36–38). Furthermore, H1-Tar and H1-2-Tar differentially regulate CheA phosphotransfer in vitro and respond appropriately to adaptation. Thus, the intrinsic properties of these effectors influence ternary complex activity, with summed dynamics that may differ from but must be a function of those of the individual receptors. Overall, the system displays dynamic coupling throughout, with kinase-on and kinase-off states set by either HAMP conformation or covalent modification in the adaptation region. HAMP enforces the opposite dynamical state on the PIR (Fig. 6). When the HAMP is conformationally destabilized (high dynamics), it is unable to influence the PIR, which sets into a more rigid state; however, when the HAMP module stabilizes, the receptor tip destabilizes, and its dynamics increase. In the adaptation region, CD2 becomes more dynamic in the kinase-off state, but somewhat surprisingly, CD1 shows similar distributions in both H1-Tar and H1-2-Tar. Lack of change is also found for sites in the flexible bundle region, where the spin labels give similar distributions in H1-Tar and H1-2-Tar. Chemical modifications of the adaptation sites produce large local effects. The EEEE state of H1-Tar markedly disorders the 298 position, but the QQQQ state of H1-2-Tar compacts the same region. CheR and CheB recognize different conformational states of the receptor (36). The large helical separation reported by the 298 site for the EEEE state is then likely CheR-specific, whereas the more compact structure of the QQQQ state provides a better substrate for CheB. Although modification generally compensates for HAMP-enforced dynamics in these systems, reversal is not complete, which may reflect multistate behavior in switching (39).
Fig. 6.
Dynamical properties of the HAMP KCM. The HAMP and PIR show opposing dynamical properties depending on receptor activation state (length of arrow correlates with conformational breadth). Activating HAMP or increased modification produces a dynamic HAMP and static CheA/CheW-coupling tip. In contrast, an inhibitory HAMP or decreased modification produces a conformationally stable HAMP but dynamic CheA/CheW-coupling tip. Inversion of dynamics occurs at the Gly hinge, which does not change properties in any of the states tested.
H1-Tar and H1-2-Tar cannot be distinguished by simple conformational transitions that involve rigid motions of the helices. Uniform helical rotations, vertical displacements, or lateral shifts would produce systematic changes to the spin-label separations inconsistent with those observed. The switch from a primarily four-helix bundle in H1 to a two-helix coiled coil in H1-2 propagates directly across the bundle into the KCM, but this feature does not continue into the adaptation region. Rearrangements that propagate through the Gly hinge do not affect the helix separations across the bundle at all. A relatively modest change in hydrophobic packing seems a likely candidate for such a transition, but other plausible motions could involve helical rotation coupled to changes in helix separation, a scissors motion, or a bend with the Gly hinge as the pivot point. In the PIR, increased dynamics are the dominant change measured by ESR, although changes in mean position could be masked by increased conformational breadth.
Changes in the dynamical state of separate KCM modules have been proposed in the context of a yin-yang model of receptor activation (14). These data were derived from the activity effects of Cys substitutions, disulfide cross-links, and removal of hydrophobic side chains that stabilize the bundle core (socket positions) (14). Assessments of dynamics in the adaptation region show several engineered Cys pairs to have increased rates of cross-linking when local negative charge increases (17). Mutagenesis data on the HAMP domain also suggest that destabilization of HAMP (or in fact, its entire removal) produces a kinase-on state, whereas changes that stabilize hydrophobic packing at the base of AS2/AS2′ produce a kinase-off state (15, 16, 40). It follows that HAMP dynamics likely influence KCM conformational properties. Despite agreement with these general ideas, our data reveal a more rigid PIR and dynamic HAMP in the kinase-on state and the opposite for kinase-off. Evidence that the PIR becomes more dynamic in the kinase-on state largely comes from the lock-on effect of socket mutations that destabilize hydrophobic packing in this region (14). Notably, similar residue substitutions in the adaptation region stimulate CheR methylase activity and prevent kinase activation, despite the receptors binding CheA/CheW (lock off) (14). Despite the segregation of lock-on socket mutants to the PIR and lock-off socket mutants to the adaptation region, not all socket-altering substitutions comply with the yin-yang model (only 54% of mutants in the adaptation region and 62% of mutants in the PIR). Furthermore, although socket mutants in the receptor tip prevent deactivation by attractant, they only activate the kinase to normal levels (and sometimes less so). Thus, these substitutions may primarily short circuit deactivation signals that rely on proper transmission of packing changes within the bundle core. Likewise, disruption of packing interactions in the adaptation region by socket mutations may remove a stabilizing influence over the kinase-on conformation in the PIR. It is important to note that PIR dynamics in the ternary complex and the isolated receptors could differ, even with the former depending on the latter. For example, in the kinase-off state, CheA and CheW may selectively stabilize a minor conformation of the PIR that is only accessible because of increased dynamics (destabilization) enforced by HAMP. How the signaling particle responds to changes in receptor dynamics is the next important question. Recent data indicate that CheA activity depends on dynamics that involve both subtle coupling of domains through linkers (41) and large-scale sequestration of the substrate (P1) domain (42).
CW-ESR spectra convolute the dynamic behavior of the spin-labeled side chain with that of the protein backbone (31, 32, 34). The inverse of the central line width (ΔH−1) and overall spectral breadth (31) were used to assess the mobility of TM1 in the ligand-free chemoreceptor Trg (43) and the C-terminal CheR-targeting peptide of Tar (44). Our CW-ESR data on the HAMP KCM module indicate an equilibrium of two conformational states. In one state, the label is rigid on the ESR timescale (τc > tens of nanoseconds), and in the other state, it is highly dynamic. These states may represent rigid helices that interact with the nitroxide and fluctuating chains that favor independent tumbling. For sites on both the HAMP and KCM, an increase in the rigid-state fraction correlates with narrower PDS distributions. At higher temperature, the ESR timescale no longer discriminates the motional states as well, but preferences for the rigid component persist.
Many bacterial receptors and signal transduction proteins have a modular construction (7, 45). A general strategy for propagating conformational signals through such architectures may involve adjacent domains that influence the stability of each other through connections, which are often helical. For linear domain arrangements, changes in hydrophobic packing and helix orientations can be effectively communicated through the connecting coiled coils. At the extreme, these structural changes could involve near unfolding of the respective modules, relieving the influence of one domain over the other (4, 7). Altering output domain stability could impact enzymatic function or the ability to engage partners. The inputs, such as ligand binding or change in cofactor state, alter stability of the binding domains to modulate their effects on downstream components. Signaling mechanisms based on competition for stability between adjacent domains can, thereby, allow different structural perturbations to achieve the same result. Conformational changes in a given context may well be specific, but they need not be unique. Thus, competition between the dynamics of coupled protein modules may help explain the ease at which domain fusions have been elaborated into the many signaling architectures observed in nature.
Materials and Methods
SI Materials and Methods describes the materials and methods followed in this article.
Briefly, proteins were recombinantly produced from E. coli and purified by affinity-label chromatography; pull-down assays were carried out with affinity-labeled purified proteins; phosphotransfer reactions were monitored by following 32P incorporation, pulsed ESR was performed at 17 GHz on a home-built spectrometer, and CW-ESR was performed at 9 GHz on a Bruker instrument with variable temperature control. For ESR data analysis, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work supported by National Institutes of Health Grants P41GM103521 (to J.H.F.) and GM066775 (to B.R.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414155112/-/DCSupplemental.
References
- 1.Nygaard R, et al. The dynamic process of β(2)-adrenergic receptor activation. Cell. 2013;152(3):532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Preininger AM, Meiler J, Hamm HE. Conformational flexibility and structural dynamics in GPCR-mediated G protein activation: A perspective. J Mol Biol. 2013;425(13):2288–2298. doi: 10.1016/j.jmb.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiesner S, et al. A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 2006;25(19):4686–4696. doi: 10.1038/sj.emboj.7601315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schultz JE, Natarajan J. Regulated unfolding: A basic principle of intraprotein signaling in modular proteins. Trends Biochem Sci. 2013;38(11):538–545. doi: 10.1016/j.tibs.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 5.Sourjik V, Wingreen NS. Responding to chemical gradients: Bacterial chemotaxis. Curr Opin Cell Biol. 2012;24(2):262–268. doi: 10.1016/j.ceb.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazelbauer GL, Falke JJ, Parkinson JS. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci. 2008;33(1):9–19. doi: 10.1016/j.tibs.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkinson JS. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu Rev Microbiol. 2010;64:101–122. doi: 10.1146/annurev.micro.112408.134215. [DOI] [PubMed] [Google Scholar]
- 8.Falke JJ, Hazelbauer GL. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem Sci. 2001;26(4):257–265. doi: 10.1016/s0968-0004(00)01770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunten P, Koshland DE., Jr Tuning the responsiveness of a sensory receptor via covalent modification. J Biol Chem. 1991;266(3):1491–1496. [PubMed] [Google Scholar]
- 10.Chervitz SA, Falke JJ. Molecular mechanism of transmembrane signaling by the aspartate receptor: A model. Proc Natl Acad Sci USA. 1996;93(6):2545–2550. doi: 10.1073/pnas.93.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall BA, Armitage JP, Sansom MSP. Transmembrane helix dynamics of bacterial chemoreceptors supports a piston model of signalling. PLOS Comput Biol. 2011;7(10):e1002204. doi: 10.1371/journal.pcbi.1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park H, Im W, Seok C. Transmembrane signaling of chemotaxis receptor tar: Insights from molecular dynamics simulation studies. Biophys J. 2011;100(12):2955–2963. doi: 10.1016/j.bpj.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kitanovic S, Ames P, Parkinson JS. Mutational analysis of the control cable that mediates transmembrane signaling in the Escherichia coli serine chemoreceptor. J Bacteriol. 2011;193(19):5062–5072. doi: 10.1128/JB.05683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Swain KE, Gonzalez MA, Falke JJ. Engineered socket study of signaling through a four-helix bundle: Evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry. 2009;48(39):9266–9277. doi: 10.1021/bi901020d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou Q, Ames P, Parkinson JS. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol Microbiol. 2009;73(5):801–814. doi: 10.1111/j.1365-2958.2009.06819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Q, Ames P, Parkinson JS. Biphasic control logic of HAMP domain signalling in the Escherichia coli serine chemoreceptor. Mol Microbiol. 2011;80(3):596–611. doi: 10.1111/j.1365-2958.2011.07577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Starrett DJ, Falke JJ. Adaptation mechanism of the aspartate receptor: Electrostatics of the adaptation subdomain play a key role in modulating kinase activity. Biochemistry. 2005;44(5):1550–1560. doi: 10.1021/bi048089z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coleman MD, Bass RB, Mehan RS, Falke JJ. Conserved glycine residues in the cytoplasmic domain of the aspartate receptor play essential roles in kinase coupling and on-off switching. Biochemistry. 2005;44(21):7687–7695. doi: 10.1021/bi0501479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swain KE, Falke JJ. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: A disulfide mapping study. Biochemistry. 2007;46(48):13684–13695. doi: 10.1021/bi701832b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piasta KN, Ullimann CJ, Slivka PF, Falke JJ. Defining a key receptor-CheA kinase contact and elucidating its function in the membrane-bound bacterial chemosensory array: A disulfide mapping and TAM-IDS Study. Biochemistry. 2013;52(22):3866–3880. doi: 10.1021/bi400385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferris HU, et al. The mechanisms of HAMP-mediated signaling in transmembrane receptors. Structure. 2011;19(3):378–385. doi: 10.1016/j.str.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 22.Hulko M, et al. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell. 2006;126(5):929–940. doi: 10.1016/j.cell.2006.06.058. [DOI] [PubMed] [Google Scholar]
- 23.Gushchin I, Gordeliy V, Grudinin S. Two distinct states of the HAMP domain from sensory rhodopsin transducer observed in unbiased molecular dynamics simulations. PLoS ONE. 2013;8(7):e66917. doi: 10.1371/journal.pone.0066917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ortega DR, et al. A phenylalanine rotameric switch for signal-state control in bacterial chemoreceptors. Nat Commun. 2013;4:2881. doi: 10.1038/ncomms3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Airola MV, et al. HAMP domain conformers that propagate opposite signals in bacterial chemoreceptors. PLoS Biol. 2013;11(2):e1001479. doi: 10.1371/journal.pbio.1001479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Airola MV, Watts KJ, Bilwes AM, Crane BR. Structure of concatenated HAMP domains provides a mechanism for signal transduction. Structure. 2010;18(4):436–448. doi: 10.1016/j.str.2010.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briegel A, et al. Bacterial chemoreceptor arrays are hexagonally packed trimers of receptor dimers networked by rings of kinase and coupling proteins. Proc Natl Acad Sci USA. 2012;109(10):3766–3771. doi: 10.1073/pnas.1115719109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu J, et al. Molecular architecture of chemoreceptor arrays revealed by cryoelectron tomography of Escherichia coli minicells. Proc Natl Acad Sci USA. 2012;109(23):E1481–E1488. doi: 10.1073/pnas.1200781109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Borbat P, Freed J. Pulse dipolar electron spin resonance: Distance measurements. Structural information from spin-labels and intrinsic paramagnetic centres in the biosciences. Struct Bonding. 2014;152:1–82. [Google Scholar]
- 30.Barnes JP, Liang Z, Mchaourab HS, Freed JH, Hubbell WL. A multifrequency electron spin resonance study of T4 lysozyme dynamics. Biophys J. 1999;76(6):3298–3306. doi: 10.1016/S0006-3495(99)77482-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liang Z, Lou Y, Freed JH, Columbus L, Hubbell WL. A multifrequency electron spin resonance study of T4 lysozyme dynamics using the slowly relaxing local structure model. J Phys Chem B. 2004;108(45):17649–17659. [Google Scholar]
- 32.Budil DE, Lee S, Saxena S, Freed JH. Nonlinear-least-squares analysis of slow-motion EPR spectra in one and two dimensions using a modified Levenberg–Marquardt algorithm. J Magn Reson A. 1996;120(2):155–189. [Google Scholar]
- 33.Guo Z, Cascio D, Hideg K, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Solvent-exposed sites in helix B of T4 lysozyme. Protein Sci. 2008;17(2):228–239. doi: 10.1110/ps.073174008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J Mol Biol. 2010;400(3):335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo Z, Cascio D, Hideg K, Kalai T, Hubbell WL. Structural determinants of nitroxide motion in spin-labeled proteins: Tertiary contact and solvent-inaccessible sites in helix G of T4 lysozyme. Protein Sci. 2007;16(6):1069–1086. doi: 10.1110/ps.062739107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boldog T, Grimme S, Li M, Sligar SG, Hazelbauer GL. Nanodiscs separate chemoreceptor oligomeric states and reveal their signaling properties. Proc Natl Acad Sci USA. 2006;103(31):11509–11514. doi: 10.1073/pnas.0604988103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bogonez E, Koshland DE., Jr Solubilization of a vectorial transmembrane receptor in functional form: Aspartate receptor of chemotaxis. Proc Natl Acad Sci USA. 1985;82(15):4891–4895. doi: 10.1073/pnas.82.15.4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Borkovich KA, Simon MI. Coupling of receptor function to phosphate-transfer reactions in bacterial chemotaxis. Methods Enzymol. 1991;200:205–214. doi: 10.1016/0076-6879(91)00140-r. [DOI] [PubMed] [Google Scholar]
- 39.Bornhorst JA, Falke JJ. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: Development of a heterogeneous two-state model. J Mol Biol. 2003;326(5):1597–1614. doi: 10.1016/s0022-2836(03)00026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ames P, Zhou Q, Parkinson JS. HAMP domain structural determinants for signalling and sensory adaptation in Tsr, the Escherichia coli serine chemoreceptor. Mol Microbiol. 2014;91(5):875–886. doi: 10.1111/mmi.12443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, et al. The linker between the dimerization and catalytic domains of the CheA histidine kinase propagates changes in structure and dynamics that are important for enzymatic activity. Biochemistry. 2014;53(5):855–861. doi: 10.1021/bi4012379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Briegel A, et al. The mobility of two kinase domains in the Escherichia coli chemoreceptor array varies with signalling state. Mol Microbiol. 2013;89(5):831–841. doi: 10.1111/mmi.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barnakov A, Altenbach C, Barnakova L, Hubbell WL, Hazelbauer GL. Site-directed spin labeling of a bacterial chemoreceptor reveals a dynamic, loosely packed transmembrane domain. Protein Sci. 2002;11(6):1472–1481. doi: 10.1110/ps.0202502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bartelli NL, Hazelbauer GL. Direct evidence that the carboxyl-terminal sequence of a bacterial chemoreceptor is an unstructured linker and enzyme tether. Protein Sci. 2011;20(11):1856–1866. doi: 10.1002/pro.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Uings IJ, Farrow SN. Cell receptors and cell signalling. Mol Pathol. 2000;53(6):295–299. doi: 10.1136/mp.53.6.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.