Significance
Many lemon-shaped double-stranded DNA viruses have been observed to infect archaeal cells in both extreme and moderate environments. We used cryo-electron tomography with subtomogram classification and averaging to reveal the three-dimensional structures of a lemon-shaped haloarchaeal virus, His1. Although the His1 exhibited size and shape heterogeneity, its tail structure was found to be constant. Extensive biochemical studies show that, while extremely stable, under certain conditions the capsid can transform into a tube without the genome. These observations demonstrate that the capsid proteins are able to perform a remarkable surface lattice transformation.
Keywords: cryo-ET, lemon-shaped virus, subtomogram averaging, virus tail organization, dissociation
Abstract
Lemon-shaped viruses are common in nature but so far have been observed to infect only archaea. Due to their unusual shape, the structures of these viruses are challenging to study and therefore poorly characterized. Here, we have studied haloarchaeal virus His1 using cryo-electron tomography as well as biochemical dissociation. The virions have different sizes, but prove to be extremely stable under various biochemical treatments. Subtomogram averaging of the computationally extracted virions resolved a tail-like structure with a central tail hub density and six tail spikes. Inside the tail there are two cavities and a plug density that separates the tail hub from the interior genome. His1 most likely uses the tail spikes to anchor to host cells and the tail hub to eject the genome, analogous to classic tailed bacteriophages. Upon biochemical treatment that releases the genome, the lemon-shaped virion transforms into an empty tube. Such a dramatic transformation demonstrates that the capsid proteins are capable of undergoing substantial quaternary structural changes, which may occur at different stages of the virus life cycle.
There seems to be only a limited number of different virus particle architectures (virion-based structural lineages) due to the limited protein-fold space (1–3). Examples of the uncommon architectures are spindle-, bottle-, and droplet-shaped virions, so far found only in archaeal viruses (4). Archaea, organisms forming the third domain of cellular life, are known to thrive in both moderate and extreme environments (5–7). Interestingly, archaeal viruses are morphologically diverse, resembling eukaryotic viruses in this respect (4). Of these morphotypes, the spindle-shaped (also known as lemon-shaped) viruses are the most common ones in archaea-dominated habitats (8–15). It appears that this architecture is unique. However, deeper structural and biochemical analyses are needed to confirm this claim.
Lemon-shaped virions are wider in the middle and narrow toward the ends. Three types of such viruses have been described based on the virion appearance: (i) those with one very short tail, (ii) those with one long tail, and (iii) those having two long tails (16–18). However, based on the comparison of the structural proteins, it was recently proposed that all known lemon-shaped viruses could be classified into two evolutionary lineages or viral families: Fuselloviridae, containing type i viruses, and Bicaudaviridae, containing type ii and iii viruses (19). Most of the isolated lemon-shaped viruses belong to the family Fuselloviridae, whose type species is the Sulfolobus spindle-shaped virus 1 (SSV1). Fuselloviruses infect hyperthermophilic crenarchaea and have one short tail with tail fibers (4, 20). The physical properties of the studied spindle-shaped viruses have recently been summarized (21). However, 3D structural understanding of lemon-shaped viruses remains limited.
His1 is the only high-salinity lemon-shaped virus isolate, and it infects an extremely halophilic euryarchaeon, Haloarcula hispanica, and morphologically resembles fuselloviruses (22). However, unlike fuselloviruses, which have a circular double-stranded DNA (dsDNA) genome, His1 has a linear dsDNA genome encoding a putative type-B DNA polymerase; consequently, His1 has been classified in the floating genus Salterprovirus (20, 23). The 14,462-bp genome of His1 is predicted to have 35 ORFs, and 4 of these have been shown to encode structural proteins of the virion (21, 23). His1 virion contains one major capsid protein (MCP), VP21. In addition, a few minor ones have been detected: VP11, VP26, and VP27. Interestingly, VP21 exists in two forms. One form is lipid modified, although there is no detectable lipid bilayer in the His1 virion (21). The His1 MCP is 47% similar to the MCP of SSV1, indicating that His1 and fuselloviruses may share a common ancestor, and it has been proposed that His1 could be classified into the family Fuselloviridae (19, 21). In addition to the DNA polymerase, the His1 genome is predicted to encode an ATPase and a glycosyltransferase (21, 23). His1, like the other lemon-shaped viruses, is nonlytic, and many of them also encode an integrase (16, 17, 21, 24–26). Despite its typically hypersaline environment, His1 tolerates a variety of salinities, from 50 mM up to 4 M NaCl (21).
Few attempts have been made to determine the structures of nonicosahedral viruses, mainly due to the fact that their pleomorphic nature introduces great challenges in particle structure classification and averaging. Recent advances in image-processing methods in cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) have been demonstrated to be a useful tool in the structural studies of nonicosahedral viruses, such as Tula hantavirus (27) and the immature capsid in HIV-1 virus (28), and some archaeal viruses, such as pleomorphic, two-tailed spindle-shaped, and linear viruses (17, 21, 29) and bottle-shaped and filamentous archaeal viruses (30–32). Recent structural studies of the spindle-shaped virus SSV1 revealed its 3D structure with a spindle body and a short tail at one end (33).
Here, we used cryo-ET and symmetry-free and model-free subtomogram averaging to reveal the previously unrecognized tail organization of the lemon-shaped virus His1, with a central tail hub and six surrounding tail spikes. Further analysis of a larger population of subtomograms showed variable dimensions of the lemon particles but a constant structure of the tail. Biochemical analysis of the virion under different chemical conditions revealed unexpected biochemical properties of the His1 virion, which may be relevant to the life cycle of this virus.
Results
Identifying the Tail Structure of the Lemon-Shaped Virus His1.
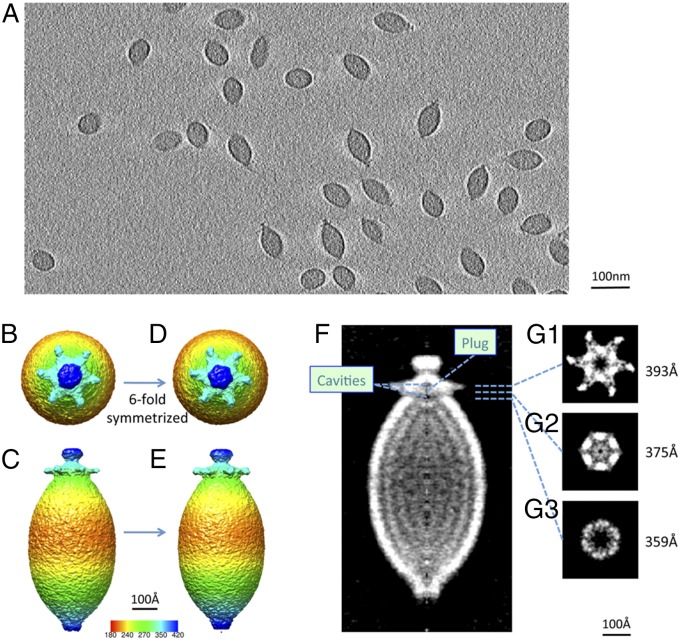
Cryo-ET, including alignment and classification of subtomograms in three dimensions, was used to resolve the structures of the lemon-shaped capsid and the tail. Tilt series images of virus particles embedded in vitreous ice were collected (Movie S1), and reconstructions of the tilt series yielded 3D volumes containing ∼100 virus particles in each tomogram, followed by extraction and analysis of individual virus subtomograms (Fig. 1A and Movie S2). Since not much prior knowledge of the 3D structures of the lemon viruses exists, a de novo approach with a completely model-free and symmetry-free algorithm (34) was used for the classification and averaging of individual subtomograms (details described in Materials and Methods and SI Appendix, Fig. S1). Based on computationally sorting a small set of 100 subtomograms from a single high-quality tomogram, a symmetry-free map of His1 virus particles was derived from the average of 30 subtomograms having a long axis of ∼920 Å and a short axis of ∼400 Å. The map also revealed an ∼120-Å tail-like structure with a central hub density that sits on one end of the capsid with six tail spikes attached to it, whereas at the other end, an extra density covers the tip (SI Appendix, Fig. S2, and Movie S3).
Fig. 1.
Refined tail structure at 20-Å resolution with sixfold average. (A) Single-slice view through a tomogram of His1 viruses embedded in vitreous ice. The radially colored symmetry-free map is shown in top view (B) and side view (C), and the sixfold symmetrized map is shown in D and E. The central slice of the refined sixfold average map is shown in gray scale in F; two cavities and a plug density are labeled accordingly. (G1–G3) Slice views normal to the long axis of the virion running through the tail spikes to the inner plug at the levels indicated by dashed lines in F. The cut-away views of the cavities are shown at the hollow centers of G1 and G3 and at the plug in G2.
Lemon Capsid Exhibits Heterogeneity.
To improve the map resolution of the lemon-shaped virus, a larger dataset of 30 tomograms was collected and reconstructed, yielding a total of 2,500 subtomograms of the virus particles. We noted visually that the length and the width of the virions vary. A histogram of the capsid width for all of the particles reveals a range of ∼360–390 Å (SI Appendix, Fig. S3), indicating intrinsic variance in the capsid dimensions.
We then split the subtomogram particles into four classes based on their widest diameter: class I, 362 ± 4.5 Å (n = 353); class II, 371 ± 4.5 Å (n = 388); class III, 380 ± 4.5 Å (n = 377); and class IV, 389 ± 4.5 Å (n = 312). The rest of the subtomogram particles did not fall into any of these four classes and were not subject to any further analysis. Subtomogram averaging within each width class was performed (details are described in Materials and Methods and SI Appendix, Figs. S4–S6). The final maps for different width classes show that the lemon body width steadily becomes larger from class I to IV, as indicated by the radial coloring (SI Appendix, Fig. S7). The final map of the class IV average (SI Appendix, Fig. S7D), which has the widest capsid diameter, appears to be the shortest in length compared with the other three classes (SI Appendix, Fig. S7 A–C). This diameter and length correlation mimics a ripe lemon: when the two ends are pushed, the body gets shorter but at the same time wider. The lipid modification of the major capsid protein may contribute to the elasticity of the lemon capsid shell.
We have measured the interior volumes for the four width classes, which are 32.25 × 106 Å3 for class I; 28.44 × 106 Å3 for class II; 37.66 × 106 Å3 for class III; and 38.48 × 106 Å3 for class IV. The inner volumes are approximately consistent for the four classes, indicating the encapsulation of the same size of DNA. We then measured the inner surface area of the capsid shell for the four width classes, which are 712.9 × 103 Å2 for class I; 610.4 × 103 Å2 for class II; 692.2 × 103 Å2 for class III; and 703.1 × 103 Å2 for class IV. The inner surface areas tend to be very consistent except for class II, indicating that the same amounts of capsid proteins were used in the four classes.
The resolution of the final map in each width diameter class was evaluated by calculating the Fourier Shell Correlations (FSCs) from independent refinements (SI Appendix, Fig. S8). For each of the four classes, the final map resolutions of the whole lemon virus are around 57–58 Å. However, when computing the resolutions in the tail part only, all four classes showed improved resolutions to 47–49 Å, suggesting that the tail structure of the lemon virus is relatively more constant than that of the capsid protein in the lemon body. This result prompted us to further explore the tail structure for improvements in the resolution.
Organization of the Tail and the Interior Genome.
To interrogate the structure of the tail, we masked away the lemon capsid with a Gaussian soft mask to avoid sharp edges and focused alignment on the tail end for all available subtomograms (details are described in Materials and Methods and SI Appendix, Figs. S9 and S10). FSCs calculated from each pair of independent refinements without imposing symmetry revealed a resolution of ∼51 Å for the virus particle, but a much better resolution, ∼28 Å, for the tail alone (Fig. 1 B and C, and SI Appendix, Fig. S11). After applying sixfold symmetry (Fig. 1 D and E), the resolution of the tail part further improved to ∼20 Å (SI Appendix, Fig. S11). Such improvement confirms our original hypothesis that the tail part of the lemon virus is relatively uniform regardless of the width of the lemon body.
From the final averaged map of all 2,444 subtomograms, more details of the organization of the tail hub and spikes were revealed (Fig. 1 and Movie S3). The six tail spikes are organized in a horizontal fashion (Fig. 1 B, D, and G1). The central tail hub, to which the six spikes are attached, is hexagonal and sits at the end of the capsid (Fig. 1G1). Inside the tail hub, a small cavity ∼15 Å in height and ∼30 Å in width is seen (Fig. 1G1). Between the tail hub and the capsid, there is another central cavity (Fig. 1G3) about 50 Å in both width and height. These two cavities are separated by a density (Fig. 1G2) that could serve as a valve to control the genome transport through the tail. We also examined the spacing of the layers inside the capsid from our subtomographic average map. The spacing between the two layers in the subtomogram average map was shown to be ∼50 Å, which possibly indicated two layers of typical 25-Å DNA genome spacing.
Dissociation Analyses Show That His1 Virion Is Extremely Stable.
Highly purified virions were treated with different proteases, detergents, solvents, and chaotropic agents as well as a variety of pH and temperature conditions. All treatments were performed at low salinity, except for freezing and thawing that was at high salinity (SI Appendix, Table S1). After each treatment, viral infectivity was determined and the dissociation products were analyzed by rate zonal centrifugation. DNA and protein content were determined for the gradient fractions, and the appearance of the dissociation products was assayed by negative-stain transmission electron microscopy (SI Appendix, Table S1).
Remarkably, His1 remained highly infectious even under extremely harsh conditions (SI Appendix, Table S1). Some treatments inactivated the virus (SI Appendix, Table S1), but during the rate zonal centrifugation no difference was observed in the sedimentation rate between the treated and untreated particles. Using protease treatments, no major digestion of the MCP VP21 was observed, suggesting that it is protease-resistant (SI Appendix, Fig. S12A). However, the amount of the minor protein VP26 decreased upon the proteinase K digestion (SI Appendix, Fig. S12A).
Most of the conditions that inactivated the virus resulted in the same end products: genome release and transformation of the lemon-shaped particle into a tube (Figs. 2 and 3). These conditions included strong alkaline treatment, addition of the nonionic detergents Triton X-100 (TX-100) or Nonidet P-40 (Nonidet P-40), incubation at 80 °C, and boiling. Interestingly, even extremely low concentrations of TX-100 [0.1% (vol/vol)] or Nonidet P-40 [0.01% (vol/vol)] induced the genome release with concomitant loss of infectivity (SI Appendix, Fig. S13A). Both detergents also solubilized approximately half of VP27 (Nonidet P-40 in Fig. 2B), and higher concentrations of the detergents resulted in partial solubilization of both forms of VP21 (Nonidet P-40 in SI Appendix, Fig. S12B). However, even high Nonidet P-40 concentrations [6% (vol/vol)] were not sufficient to completely solubilize VP21 as individual molecules, as empty particles could be observed after such treatments (SI Appendix, Fig. S14 A and B). Furthermore, with higher Nonidet P-40 concentrations sedimentation properties of the particles varied (SI Appendix, Figs. S12B and S14 A and B). Major structural changes or rearrangements upon DNA release were irreversible. His1 lost its infectivity between 70 and 80 °C at low salinity (SI Appendix, Fig. S13 B and C). In contrast to the detergent and boiling treatments, incubation at 80 °C at low salinity produced two light-scattering zones in a gradient after rate zonal centrifugation: a minor one containing tubes with attached DNA and a major one containing empty tubes only (Fig. 2). In addition, nonionic detergent octyl-β-d-glucopyranoside released the genome, but the empty tube-like particles were more variable compared with the TX-100 or Nonidet P-40 treatments (SI Appendix, Fig. S15A). Also, chloroform treatment resulted in the release of the genome, but the empty particles were pleomorphic with variable sedimentation behaviors (SI Appendix, Fig. S15 B and C).
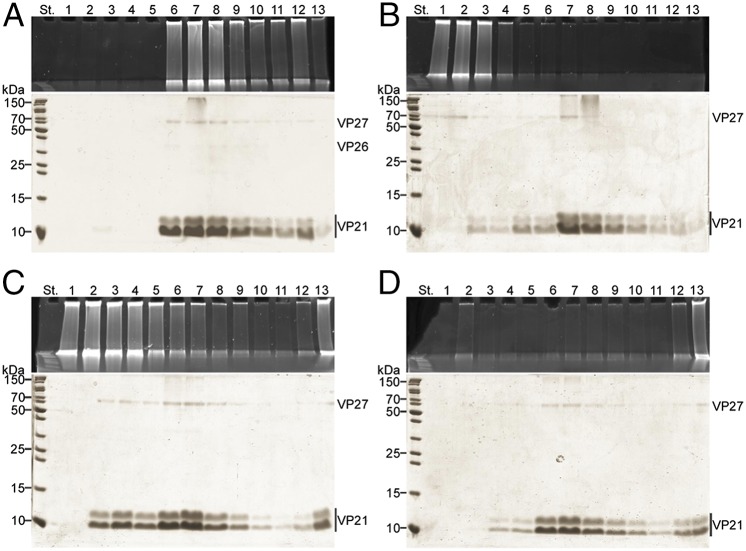
Fig. 2.
Production of empty tubes of His1. The virions and dissociation products were sedimented by rate zonal centrifugation, and the gradient fractions were analyzed for DNA and proteins by gel electrophoresis. Upper represents EtBr and Lower represents Coomassie blue staining of the SDS/PAGE gels (for nucleic acids and proteins, respectively). Numbers at the top indicate gradient fractions 1–13 starting from the surface of the gradient. Numbers on the left indicate the molecular mass markers. Centrifugation time was three times longer in B and C compared with the one used in A (Materials and Methods). (A) Untreated 2× purified virions. (B) Virions treated with 0.01% (vol/vol) Nonidet P-40 at low salinity at room temperature for 90 min. (C) Virions incubated at 80 °C at low salinity for 5 min. (D) Virions boiled at low salinity for 10 min.
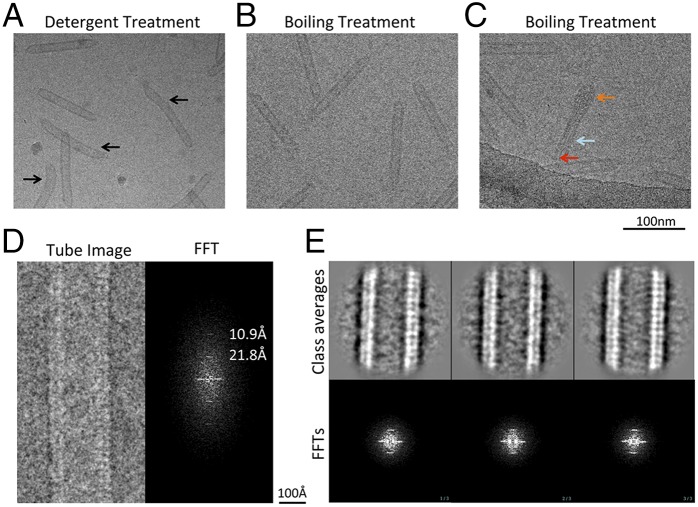
Fig. 3.
Transformation of the lemons into empty tubes. (A) A typical micrograph of the empty tubes produced with detergent treatment. Some of the tubes were bent, as indicated by the black arrow. (B) A typical micrograph of the empty tubes produced with a boiling treatment. (C) An intermediate stage of a lemon (orange arrow) transforming into a tube (cyan arrow) with genome being ejected (red arrow). (D) A representative micrograph of one tube with its Fourier transformation on the right. Layer lines with periodicity of 21.8 and 10.9 Å are labeled. (E) Reference-free 2D classification and averaging with three class averages with clear repeats on the Top and the corresponding power spectra on the Bottom.
Empty tube-like particles were also observed after the 1.5% (vol/vol) HCl treatment at 75 °C (SI Appendix, Fig. S15D) that has previously been observed to dissociate the lipid-modified, higher-molecular-mass form of VP21, most probably changing it into the lower-molecular-mass form (21). These tubes were highly aggregative, and we could also observe thinner rod-like particles (SI Appendix, Fig. S15D). This result indicates that the lower-molecular-mass form of VP21 is able to form tube-like particles on its own. The only treatment tested that was observed to quantitatively solubilize both forms of VP21 was incubation with SDS (SI Appendix, Table S1).
Visualization of the Empty Tube.
The empty tubes produced by both the detergent and boiling treatments were imaged using cryo-EM. The untreated particles were lemon shaped (Fig. 1A) whereas the detergent treatment resulted in empty bent tubes (Fig. 3A). However, heat treatment induced empty tubes (Fig. 3B) that were straighter than the detergent-treated ones. We also observed an intermediate stage (Fig. 3C) with one end still retaining the lemon shape and the other end forming the empty tube. DNA strands that are being released from the capsid can also be seen in the middle of the tube. Capturing this intermediate state enables the direct visualization of the transforming process and provides confirmatory evidence for the tube-triggering experiments.
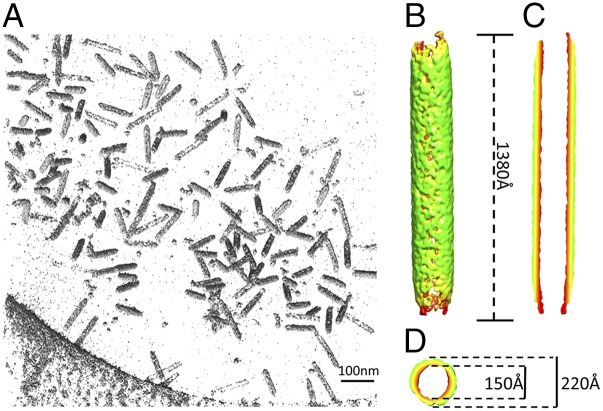
Tubes produced by the boiling treatment were chosen for further characterization due to their rigid appearance and approximately constant length and width. A long and straight tube showed periodicity at 21.8 and 10.9 Å evidenced in its corresponding power spectrum (Fig. 3D). The tubular segments were then computationally segmented into square boxes treated as individual particles. Two-dimensional class averages show clear periodicity in agreement with the raw micrographs and their power spectra (Fig. 3E). Although the short-range order can be clearly determined at 21.8 Å, long-range order remains ambiguous. Cryo-ET was then used to visualize the empty tubes in three dimensions. Five tilt series of the empty tubes embedded in vitreous ice were collected (Movie S4) and reconstructions of the tilt series yielded 3D volumes containing the densities of the tubes (Fig. 4A and Movie S5). Approximately 200 tube subtomograms were extracted and computationally classified and averaged using the all-vs.-all algorithm. The averaged map from the major group of 54 subtomograms revealed the dimensions of the empty tube (Fig. 4B): length, ∼1,380 Å; outer diameter, ∼220 Å; inner diameter, ∼150 Å (Fig. 4 C and D). This demonstrates the drastic transformation from an ∼900-Å–long and ∼400-Å–wide lemon shape into an ∼1,380-Å–long and ∼220-Å–wide tube upon the genome release.
Fig. 4.
Tomographic analysis of the empty tubes. (A) A representative volume rendering of a reconstructed tomogram. (B) The symmetry-free average of the tubes generated from 54 subtomograms using model-free all-vs.-all alignment method. (C) Central section from the side and (D) the top of the averaged map in B.
Discussion
The ability to survive under environmental extremes has stimulated great interest in investigating special properties of archaeal viruses. With advances in genomic and biochemical analyses, classification of archaeal viruses has been possible (4, 20, 35). However, the exceptional virion architectures of many archaeal viruses pose challenges to a comprehensive structural understanding, and, consequently, the resolved structures of unusual archaeal virion morphotypes are limited.
Although the lemon-shape capsid exhibits structure variation, we undertook an image-processing strategy to perform posttomographic averaging of different parts of the virion that had less structure heterogeneity among particles. We achieved a significantly improved subtomogram average of His1 with features resolved at the tail. To assess these features, we applied sixfold symmetry to the map, which further improved the resolution of the tail to ∼20 Å (Fig. 1). Comparison of His1 tail organization to existing structures of tailed bacterial and eukaryotic viruses reveals similar tail structures, such as P-SSP7 (36) and PBCV-1 (37); thus the lemon-shaped His1 and tailed bacteriophages may have similar adsorption and genome ejection mechanisms. We propose that in archaeal virus His1, as in tailed bacteriophages, the tail plays a key role not only in facilitating and securing the virus to its host, but also in regulating the translocation of its genetic material into the host. These processes are controlled by the interaction between the spike proteins and host receptors and the coordination between the tail hub proteins and the plug. However, His1 is a nonlytic virus in contrast to tailed bacteriophages and thus likely uses different mechanisms for virion assembly and release (21).
The organization of the genome injection apparatus has been widely studied in tailed bacteriophages. In the case of Epsilon 15 (38) and P22 (39, 40), the tail spikes are perpendicular to the surface of the capsid, whereas the tail fibers of P-SSP7 (36) extend along the capsid surface in the mature virion and change into a horizontal extension after the capsid encounters the bacterial surface to signal genome release. In this study of the lemon-shaped virus His1, the tail spikes extend horizontally from the central hub in the mature virion. In addition to the tail hub and spikes, another internal feature between the tail hub and capsid was also observed. A cavity and a plug separate the genome from the exterior milieu. During genome transfer, the potential conformational changes of the tail spikes may open up the plug (Fig. 1G2) between the small cavity in the tail hub and central cavity, as in the case of P-SSP7 (36). The newly formed larger cavity may function as a channel-like structure that provides room for the ejection of the DNA. The cavity and plug organization may be conserved as has been described in the Paramecium bursaria chlorella virus (PBCV-1) (37).
For several tailed bacteriophages, it has been reported that the genome ejection is a two-step process: pressure inside the virion starts the ejection, but other factors are needed to finish it (41–43). His1 is so far the only archaeal virus in which genome ejection has been studied. Single-molecule experiments and visualization of the ejection by total internal reflection fluorescence microscopy showed that the DNA ejection from His1 virions in vitro is unidirectional and randomly paused, indicating that host cell factors are required to finish the ejection (44).
Dissociation data (SI Appendix, Table S1) illustrate the extremely stable nature of the His1 virion. The MCP VP21 has a theoretical low isoelectric point, 4.14, which is typical for halophilic organisms or their viruses and most likely contributes to the protein stability and solubility in hypersaline environments (45). Protein VP21 has been shown to have two forms in the virion—native and lipid modified—and it is mainly composed of hydrophobic amino acids forming two domains (21). It might be the hydrophobic nature and lipid modification that provide such strong interactions, making it difficult to dissociate the particle. The only major dissociation products observed were an empty tube-like particle and the released genome. To our knowledge, no other virus particle has been described as undergoing such a dramatic rearrangement upon genome release. Importantly, this transformation was achieved without detectable changes in the protein composition (Fig. 2C), indicating that the capsid subunits are able to switch from one arrangement to another. Lemon-shaped archaeal viruses have also previously been reported to be able to transform their capsids, as Acidianus two-tailed virus forms long tails at both ends of the lemon body outside the host cells (17, 46). Thus, this lemon architecture seemingly allows the virion to adopt several conformations. The His1 genome, however, seems to have quite a strong interaction with the capsid, as some empty tubes still had bound DNA after incubation at 80 °C. Negatively stained EM images of the treated His1 virions also revealed that under certain conditions empty tubes had variable length and thickness (SI Appendix, Fig. S15). In addition, empty pleomorphic particles could be observed by negative-staining EM after the chloroform treatment (SI Appendix, Fig. S15). Interestingly, these resembled empty membrane vesicles reported for pleomorphic, membrane-containing archaeal viruses (47) although His1 virions have no lipid membrane. Thus, the lipid modification of the His1 MCP may facilitate such transformation. Another lemon-shaped virus, SSV1, has also been reported to be very stable under harsh conditions such as 6 M urea. However, SSV1 is also very sensitive to organic solvents and alkaline conditions that induce the genome release leaving an empty, lemon-shaped shell (48).
In our biochemical dissociation experiments, the lemon-tube transition seems to happen simultaneously with the release of the genome DNA. It is tempting to speculate that a similar mechanism may be used when the virus is infecting its host and ejecting its genome. Combined with our structural observations, we hypothesize that during the infection the tail spikes of His1 recognize the receptors on the cell wall of its archaeal host, the process of which orients the tail hub perpendicularly to the host cell wall and initiates the genome injection. Furthermore, upon genome release, the lemon-shaped capsid transforms into an empty tube to facilitate the process of the genome release. A confirmation of this hypothesized infection mechanism in vivo will certainly require future experiments. Interestingly, tailless icosahedral bacteriophage PRD1 has been shown to have a unique packaging vertex and form a tube-like structure from its inner membrane to deliver the genome across the cell envelope (49, 50). As tailed bacteriophages also use their tube-like tails to eject the genome, tube-resembling devices may work as a common genome-injection needle among different virus morphotypes.
Materials and Methods
Production and Purification of His1 Virion.
H. hispanica strain ATCC 33960 was used to propagate His1 (22, 51). Cultures were grown aerobically at 37 °C in the modified growth media (MGM) (52). Broth, solid, and top-layer MGM contained 23, 20, and 18% (wt/vol) artificial salt water (SW), respectively. Five grams of peptone (Oxoid) and 1 g of Bacto yeast extract (Difco Laboratories) were added per liter. Solid and top-layer media contained 14 and 4 g of Bacto agar (Difco Laboratories) per liter, respectively. The 30% (wt/vol) stock solution of SW contains 240 g NaCl, 30 g MgCl2·6H2O, 35 g MgSO4·7H2O, 7 g KCl, 5 mL of 1 M CaCl2·2H2O, and 80 mL of 1 M Tris⋅HCl (pH 7.2) per liter (www.haloarchaea.com).
His1 virus stocks were prepared as previously described, using confluent plates (53). These were used to infect H. hispanica cultures with a multiplicity of infection of 10 followed by incubation at 37 °C overnight. The overnight cultures were treated with DNase I (50 µg/mL) for 1 h at 37 °C. Virus particles were purified from the culture supernatants as previously described (21), and the concentrated “2× purified virus” was further purified in a linear 5–20% sucrose gradient in His1 buffer [500 mM NaCl, 35 mM MgCl2, 10 mM KCl, 1 mM CaCl2, and 50 mM Tris⋅HCl (pH 7.2)] (Sorvall TH641, 35,000 × g, 40 min at 15 °C). The light-scattering virus zone was collected, concentrated, and washed with His1 buffer or 10-fold–diluted His1 buffer [50 mM NaCl, 3.5 mM MgCl2, 1 mM KCl, 0.1 mM CaCl2, and 50 mM Tris⋅HCl (pH 7.2)] using ultrafiltration. The resulting material was designated as “3× purified” virus.
Virus Inactivation Experiments.
Sensitivity of His1 to different conditions was tested using 2× purified virus material. For detergent sensitivity, virus particles were treated with 0.01–1% (vol/vol) TX-100 or Nonidet P-40 in 10-fold–diluted His1 buffer for 90 min at room temperature. For temperature sensitivity, virus particles were incubated at 4–80 °C in 10-fold–diluted His1 buffer for 5 min and at 80 °C for 5–90 min. After incubations, infectivity was determined using the plaque assay.
Virion Dissociation.
All dissociation experiments were carried out with 2× purified virus particles. Treatments included (i) proteases, (ii) pH, (iii) detergents, (iv) temperature, (v) chaotropic agents, (vi) solvents, and (vii) combinations of these (SI Appendix, Table S1). Incubations were carried out either in His1 buffer (high salinity) or in 10-fold–diluted His1 buffer (low salinity). After each treatment, infectivity was determined using the plaque assay, and dissociation products were analyzed in a linear 5–20% sucrose gradient in His1 buffer by rate zonal centrifugation (Beckman SW50.1, 40,000 × g, 25–65 min at 15 °C or TH641, 35,000 × g, 40–120 min at 15 °C). The virus amounts used in the reactions were A260 = 1.85 (SW50.1) or A260 = 4.50 (TH641), except that in SDS and ethanol treatments A260 = 2.50 (SW50.1) and A260 = 3.70 (SW50.1) were used. After centrifugation, gradients were fractionated and the fractions were analyzed by SDS/PAGE with 4 and 16% (wt/vol) acrylamide concentrations in the stacking and separation gels, respectively (54) and by staining with ethidium bromide for DNA and Coomassie blue for proteins.
Dissociation products were visualized by negative-staining transmission electron microscopy. Treated particles were allowed to adsorb on grids for 1 min before staining with 3% (wt/vol) uranyl acetate (pH 4.5) for 1 min. The micrographs were taken with a JEOL 1200EX electron microscope operating at 80 kV in the Electron Microscopy Unit of the Institute of Biotechnology, University of Helsinki.
For cryo-EM, empty virion tubes were prepared using the following dissociations: (i) treatment with 0.01% (vol/vol) Nonidet P-40 in 10-fold–diluted His1 buffer for 90 min at room temperature, (ii) boiling in 10-fold–diluted His1 buffer for 10 min, and (iii) incubation at 80 °C in 10-fold–diluted His1 buffer for 5 min. After the treatment, the specimens were purified in a linear 5–20% sucrose gradient in His1 buffer by rate zonal centrifugation (TH641, 35,000 × g, 40–120 min at 15 °C). The virus amount used in the reactions was A260 = 4.50. After centrifugation, the light-scattering zones were collected and concentrated and washed with 10-fold–diluted His1 buffer using ultrafiltration as described above.
Cryo-ET Data Acquisition.
Aliquots of 2.5 μL of 3× purified His1 virions were applied to 200-mesh R2/2 Quantifoil grids in a Vitrobot MarkIII (FEI) blotted for 2 s and flash-frozen in liquid ethane. The frozen grids were then imaged on a 200-kV JEM2200FS electron microscope (JEOL) equipped with in-column energy filter with a 20-eV slit width. To collect tilt series, 3× purified His1 virions were mixed with 60 Å gold before vitrification. Microscope magnification of 25 K× corresponding to 4.52 Å/pixel sampling on a Gatan 4K×4K CCD was used to collect each tilt series from −65° to +65° with 2° step size with total dose of ∼100e−⋅Å-2. The intended defocus was set to 5 μm in SerialEM. The 3D reconstruction for each tilt series was performed in IMOD software package tracking with gold fiducial markers (55).
All-vs.-All Classification and Refinements Within Each Width Class.
The initial stage of processing was performed with 100 subtomograms computationally extracted from one tomogram. As a first-step, the lemon-shaped subtomograms were aligned to a featureless cylindrical density so that the long axis of the virions were all aligned to the z axis of the cylinder model. All-vs.-all cross-correlation classification was performed, as described previously (34). The two degrees of freedom considered in the alignment were flipping of the two ends and rotational alignment along the long axis (SI Appendix, Fig. S1). Through this iterative approach, the 100 subtomograms were classified into different groups and averaged without a starting model or imposing any symmetry.
A larger dataset of 30 tomograms was collected subsequently in three different microscope sessions. The intended defocus in SerialEM was set to 1 μm to push the first zero cutoff of the contrast transfer function to ∼20 Å. A total of ∼2,500 subtomograms were extracted and aligned to the initial all-vs.-all map. Quantitatively, we measured the diameter of the capsid for all of the subtomogram particles by radially plotting the densities of the central section and recording the highest peak, which corresponds to the capsid shell (SI Appendix, Fig. S3) (56). The particles were separated based on the widest width of the lemon body (SI Appendix, Fig. S3). For each width class, 100 particles were randomly selected for all-vs.-all classification, and the major group average from the all-vs.-all was used as a starting model for further refinement (SI Appendix, Figs. S4 and S5). The independently refined maps for the three subsets were used to calculate the FSCs (SI Appendix, Figs. S6 and S8). The final map for each width class was obtained by combining the refined maps of the three subsets.
Refinement of the Tail Structure by Combining All Data.
For the refinement of the tail structure, all subtomograms were also split into three subsets. The lemon capsid was masked away using a soft Gaussian mask to avoid artifacts introduced by sharp edges, and only the tail was included for further independent refinement within each subset (SI Appendix, Fig. S9). The resulting rotations and translations from the alignment were applied to the raw particles for averaging. Through the iterative process, the final average map was combined using all 2,444 particles without imposing symmetry (SI Appendix, Fig. S10). Sixfold symmetry was then applied to the final average. Resolution assessment was done by calculating the FSC against each pair of the three independently reconstructed maps (SI Appendix, Fig. S11).
Cryo-EM Imaging and Tomographic Analysis of the Empty Tubes.
The frozen grids of the empty tubes were prepared in the same way as described for the virions and imaged in a 200-kV JEM2010F electron microscope (JEOL) with same imaging conditions without energy filtering. Approximately 750 long nonoverlapping helices from ∼86 CCD frames were boxed out and then cut into short square boxes (200 by 200 pixels) with 80% overlap between successive boxes using e2helixboxer.py in EMAN2 (57). Each square box was later treated as a single particle image for 2D reference-free classification and averaged using e2refine2d.py.
The boiling-induced empty tubes were also mixed with 6 nm gold and flash-frozen on 200-mesh R2/2 Quantifoil grids. Five tilt series of the frozen grids were collected and reconstructed to yield tomograms in the same way as described for the virion. About 200 subtomograms of the empty tubes were computationally extracted from the five tomograms and then classified and averaged using the all-vs.-all algorithm. One major group containing 54 subtomograms generated the final averaged map.
Supplementary Material
Acknowledgments
The authors thank Htet Khant, Päivi Hannuksela, and Helin Veskiväli for excellent technical assistance and Dr. David Prangishvili of Institut Pasteur and Dr. Juha T. Huiskonen of Wellcome Trust Centre for Human Genetics for their helpful comments on our manuscript. This work was supported by the NIH (Grant P41GM103832); the Robert Welch Foundation (Grant Q1242) (to W.C.); and the Academy of Finland (Academy Professor Funding Grants 255342 and 256518 to D.H.B.). We thank University of Helsinki and Academy of Finland (Grants 271413 and 272853) for supporting the European Union European Strategy Forum on Research Infrastructures (ESFRI) Instruct Centre for Virus Purification (ICVIR) used in this study.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the Electron Microscopy Data Bank (EMDB) under accession no. EMD-6220 for the initial all-vs.-all His1 virion map; EMD-6221 for refined symmetry-free His1 virion map; EMD-6222 for the sixfold symmetrized refined His1 virion map; and EMD-6223 for the all-vs.-all empty tube map.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1425008112/-/DCSupplemental.
References
- 1.Abrescia NGA, Bamford DH, Grimes JM, Stuart DI. Structure unifies the viral universe. Annu Rev Biochem. 2012;81:795–822. doi: 10.1146/annurev-biochem-060910-095130. [DOI] [PubMed] [Google Scholar]
- 2.Bamford DH. Do viruses form lineages across different domains of life? Res Microbiol. 2003;154(4):231–236. doi: 10.1016/S0923-2508(03)00065-2. [DOI] [PubMed] [Google Scholar]
- 3.Benson SD, Bamford JK, Bamford DH, Burnett RM. Does common architecture reveal a viral lineage spanning all three domains of life? Mol Cell. 2004;16(5):673–685. doi: 10.1016/j.molcel.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 4.Pina M, Bize A, Forterre P, Prangishvili D. The archeoviruses. FEMS Microbiol Rev. 2011;35(6):1035–1054. doi: 10.1111/j.1574-6976.2011.00280.x. [DOI] [PubMed] [Google Scholar]
- 5.Tourna M, et al. Nitrososphaera viennensis, an ammonia oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108(20):8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437(7058):543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 7.DeLong EF, Pace NR. Environmental diversity of bacteria and archaea. Syst Biol. 2001;50(4):470–478. [PubMed] [Google Scholar]
- 8.López-Bueno A, et al. High diversity of the viral community from an Antarctic lake. Science. 2009;326(5954):858–861. doi: 10.1126/science.1179287. [DOI] [PubMed] [Google Scholar]
- 9.Oren A, Bratbak G, Heldal M. Occurrence of virus-like particles in the Dead Sea. Extremophiles. 1997;1(3):143–149. doi: 10.1007/s007920050027. [DOI] [PubMed] [Google Scholar]
- 10.Rachel R, et al. Remarkable morphological diversity of viruses and virus-like particles in hot terrestrial environments. Arch Virol. 2002;147(12):2419–2429. doi: 10.1007/s00705-002-0895-2. [DOI] [PubMed] [Google Scholar]
- 11.Redder P, et al. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol. 2009;11(11):2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 12.Rice G, et al. Viruses from extreme thermal environments. Proc Natl Acad Sci USA. 2001;98(23):13341–13345. doi: 10.1073/pnas.231170198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos F, et al. Metagenomic approach to the study of halophages: The environmental halophage 1. Environ Microbiol. 2007;9(7):1711–1723. doi: 10.1111/j.1462-2920.2007.01289.x. [DOI] [PubMed] [Google Scholar]
- 14.Sime-Ngando T, et al. Diversity of virus-host systems in hypersaline Lake Retba, Senegal. Environ Microbiol. 2011;13(8):1956–1972. doi: 10.1111/j.1462-2920.2010.02323.x. [DOI] [PubMed] [Google Scholar]
- 15.Borrel G, et al. Unexpected and novel putative viruses in the sediments of a deep-dark permanently anoxic freshwater habitat. ISME J. 2012;6(11):2119–2127. doi: 10.1038/ismej.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiang X, et al. Sulfolobus tengchongensis spindle-shaped virus STSV1: Virus-host interactions and genomic features. J Virol. 2005;79(14):8677–8686. doi: 10.1128/JVI.79.14.8677-8686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prangishvili D, et al. Structural and genomic properties of the hyperthermophilic archaeal virus ATV with an extracellular stage of the reproductive cycle. J Mol Biol. 2006;359(5):1203–1216. doi: 10.1016/j.jmb.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 18.Martin A, et al. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J. 1984;3(9):2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krupovic M, Quemin ER, Bamford DH, Forterre P, Prangishvili D. Unification of the globally distributed spindle-shaped viruses of the Archaea. J Virol. 2014;88(4):2354–2358. doi: 10.1128/JVI.02941-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy: Ninth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press; London: 2012. p. 1327. [Google Scholar]
- 21.Pietilä MK, Atanasova NS, Oksanen HM, Bamford DH. Modified coat protein forms the flexible spindle-shaped virion of haloarchaeal virus His1. Environ Microbiol. 2013;15(6):1674–1686. doi: 10.1111/1462-2920.12030. [DOI] [PubMed] [Google Scholar]
- 22.Bath C, Dyall-Smith ML. His1, an archaeal virus of the Fuselloviridae family that infects Haloarcula hispanica. J Virol. 1998;72(11):9392–9395. doi: 10.1128/jvi.72.11.9392-9395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bath C, Cukalac T, Porter K, Dyall-Smith ML. His1 and His2 are distantly related, spindle-shaped haloviruses belonging to the novel virus group, Salterprovirus. Virology. 2006;350(1):228–239. doi: 10.1016/j.virol.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 24.Gorlas A, Koonin EV, Bienvenu N, Prieur D, Geslin C. TPV1, the first virus isolated from the hyperthermophilic genus Thermococcus. Environ Microbiol. 2012;14(2):503–516. doi: 10.1111/j.1462-2920.2011.02662.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schleper C, Kubo K, Zillig W. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: Demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci USA. 1992;89(16):7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serre MC, Letzelter C, Garel JR, Duguet M. Cleavage properties of an archaeal site-specific recombinase, the SSV1 integrase. J Biol Chem. 2002;277(19):16758–16767. doi: 10.1074/jbc.M200707200. [DOI] [PubMed] [Google Scholar]
- 27.Huiskonen JT, et al. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J Virol. 2010;84(10):4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schur FK, et al. 2014. Structure of the immature HIV-1 capsid in intact virus particles at 8.8 Å resolution. Nature 517(7535):505–508.
- 29.Arnold HP, et al. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology. 2000;267(2):252–266. doi: 10.1006/viro.1999.0105. [DOI] [PubMed] [Google Scholar]
- 30.Häring M, Rachel R, Peng X, Garrett RA, Prangishvili D. Viral diversity in hot springs of Pozzuoli, Italy, and characterization of a unique archaeal virus, Acidianus bottle-shaped virus, from a new family, the Ampullaviridae. J Virol. 2005;79(15):9904–9911. doi: 10.1128/JVI.79.15.9904-9911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vestergaard G, et al. Structure of the acidianus filamentous virus 3 and comparative genomics of related archaeal lipothrixviruses. J Virol. 2008;82(1):371–381. doi: 10.1128/JVI.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bize A, et al. Viruses in acidic geothermal environments of the Kamchatka Peninsula. Res Microbiol. 2008;159(5):358–366. doi: 10.1016/j.resmic.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 33.Stedman KM, DeYoung M, Saha M, Sherman MB, Morais MC. Structural insights into the architecture of the hyperthermophilic Fusellovirus SSV1. Virology. 2015;474:105–109. doi: 10.1016/j.virol.2014.10.014. [DOI] [PubMed] [Google Scholar]
- 34.Schmid MF, Booth CR. Methods for aligning and for averaging 3D volumes with missing data. J Struct Biol. 2008;161(3):243–248. doi: 10.1016/j.jsb.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ackermann HW, Prangishvili D. Prokaryote viruses studied by electron microscopy. Arch Virol. 2012;157(10):1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat Struct Mol Biol. 2010;17(7):830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang X, et al. Three-dimensional structure and function of the Paramecium bursaria chlorella virus capsid. Proc Natl Acad Sci USA. 2011;108(36):14837–14842. doi: 10.1073/pnas.1107847108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang W, et al. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439(7076):612–616. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang J, Weigele P, King J, Chiu W, Jiang W. Cryo-EM asymmetric reconstruction of bacteriophage P22 reveals organization of its DNA packaging and infecting machinery. Structure. 2006;14(6):1073–1082. doi: 10.1016/j.str.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 40.Lander GC, et al. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312(5781):1791–1795. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 41.Panja D, Molineux IJ. Dynamics of bacteriophage genome ejection in vitro and in vivo. Phys Biol. 2010;7(4):045006. doi: 10.1088/1478-3975/7/4/045006. [DOI] [PubMed] [Google Scholar]
- 42.São-José C, de Frutos M, Raspaud E, Santos MA, Tavares P. Pressure built by DNA packing inside virions: Enough to drive DNA ejection in vitro, largely insufficient for delivery into the bacterial cytoplasm. J Mol Biol. 2007;374(2):346–355. doi: 10.1016/j.jmb.2007.09.045. [DOI] [PubMed] [Google Scholar]
- 43.Hu B, Margolin W, Molineux IJ, Liu J. The bacteriophage t7 virion undergoes extensive structural remodeling during infection. Science. 2013;339(6119):576–579. doi: 10.1126/science.1231887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanhijärvi KJ, Ziedaite G, Pietilä MK, Hæggström E, Bamford DH. DNA ejection from an archaeal virus: A single-molecule approach. Biophys J. 2013;104(10):2264–2272. doi: 10.1016/j.bpj.2013.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kiraga J, et al. The relationships between the isoelectric point and length of proteins, taxonomy and ecology of organisms. BMC Genomics. 2007;8:163. doi: 10.1186/1471-2164-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Häring M, et al. Virology: Independent virus development outside a host. Nature. 2005;436(7054):1101–1102. doi: 10.1038/4361101a. [DOI] [PubMed] [Google Scholar]
- 47.Pietilä MK, et al. Virion architecture unifies globally distributed pleolipoviruses infecting halophilic archaea. J Virol. 2012;86(9):5067–5079. doi: 10.1128/JVI.06915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zillig W, et al. Viruses of archaeabacteria. In: Calendar R, editor. The Bacteriophages. Vol 1. Plenum Press; New York: 1988. pp. 517–558. [Google Scholar]
- 49.Hong C, et al. A structural model of the genome packaging process in a membrane-containing double stranded DNA virus. PLoS Biol. 2014;12(12):e1002024. doi: 10.1371/journal.pbio.1002024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peralta B, et al. Mechanism of membranous tunnelling nanotube formation in viral genome delivery. PLoS Biol. 2013;11(9):e1001667. doi: 10.1371/journal.pbio.1001667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Juez G, Rodriquez-Valera F, Ventosa A, Kushner DJ. Haloarcula hispanica spec. nov. and Haloferax gibbonsii spec. nov., two new species of extremely halophilic Archaebacteria. Syst Appl Microbiol. 1986;8(1-2):75–79. [Google Scholar]
- 52.Nuttall SD, Dyall-Smith ML. HF1 and HF2: Novel bacteriophages of halophilic archaea. Virology. 1993;197(2):678–684. doi: 10.1006/viro.1993.1643. [DOI] [PubMed] [Google Scholar]
- 53.Pietilä MK, Roine E, Paulin L, Kalkkinen N, Bamford DH. An ssDNA virus infecting archaea: A new lineage of viruses with a membrane envelope. Mol Microbiol. 2009;72(2):307–319. doi: 10.1111/j.1365-2958.2009.06642.x. [DOI] [PubMed] [Google Scholar]
- 54.Olkkonen VM, Bamford DH. Quantitation of the adsorption and penetration stages of bacteriophage φ 6 infection. Virology. 1989;171(1):229–238. doi: 10.1016/0042-6822(89)90530-8. [DOI] [PubMed] [Google Scholar]
- 55.Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol. 2005;152(1):36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Schmid MF, et al. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol. 2006;364(3):526–535. doi: 10.1016/j.jmb.2006.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157(1):38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.