Significance
Most sexually dimorphic features of Drosophila melanogaster are specified by the action of sex-specific transcription factors encoded by the doublesex (dsx) gene. Evolutionary changes in such sexually dimorphic features are often a result of changes in the cis-regulatory sequences of the DSX target genes. When a particular target gene is directly regulated by DSX in multiple tissues, evolutionarily conflicting constraints may be generated. The research we present here reveals that such conflict can be solved by deploying different cis-regulatory modules (sharing the DSX-binding site) for different tissues. This mechanism could also apply to other transcription factors.
Keywords: sex determination, transcription factors, DNA binding site, evolution
Abstract
“Regulatory evolution,” that is, changes in a gene’s expression pattern through changes at its regulatory sequence, rather than changes at the coding sequence of the gene or changes of the upstream transcription factors, has been increasingly recognized as a pervasive evolution mechanism. Many somatic sexually dimorphic features of Drosophila melanogaster are the results of gene expression regulated by the doublesex (dsx) gene, which encodes sex-specific transcription factors (DSXF in females and DSXM in males). Rapid changes in such sexually dimorphic features are likely a result of changes at the regulatory sequence of the target genes. We focused on the Flavin-containing monooxygenase-2 (Fmo-2) gene, a likely direct dsx target, to elucidate how sexually dimorphic expression and its evolution are brought about. We found that dsx is deployed to regulate the Fmo-2 transcription both in the midgut and in fat body cells of the spermatheca (a female-specific tissue), through a canonical DSX-binding site in the Fmo-2 regulatory sequence. In the melanogaster group, Fmo-2 transcription in the midgut has evolved rapidly, in contrast to the conserved spermathecal transcription. We identified two cis-regulatory modules (CRM-p and CRM-d) that direct sexually monomorphic or dimorphic Fmo-2 transcription, respectively, in the midguts of these species. Changes of Fmo-2 transcription in the midgut from sexually dimorphic to sexually monomorphic in some species are caused by the loss of CRM-d function, but not the loss of the canonical DSX-binding site. Thus, conferring transcriptional regulation on a CRM level allows the regulation to evolve rapidly in one tissue while evading evolutionary constraints posed by other tissues.
How sex-specific characteristics are generated during development, what roles they have in the biology of an organism, and how they have evolved are fundamental questions in biology. In Drosophila melanogaster, the molecular mechanisms leading to the sex-specific and temporally and spatially restricted deployment of the transcription factors encoded by the terminal genes in the sex-determination regulatory hierarchy, doublesex (dsx) and fruitless (fru), have been well documented (1–4). In addition, although it has been established that the FRUM and DSXM and DSXF proteins control nearly all aspects of somatic sexual differentiation, including anatomical and behavioral differences, much less is known about the direct target genes through which these transcription factors act (2, 3, 5–11).
It has been increasingly recognized that evolutionary significant changes in gene expression are often a result of mutations in the regulatory sequences of genes, rather than in their coding sequences (12). Furthermore, it has been noted that evolutionary changes are most likely in the cis-acting regulatory elements of the downstream targets of transcription factors, rather than in the genes encoding the transcription factors (13, 14). This finding is likely because changes at the transcription factors or changes at the coding sequence of the target gene could both have deleterious pleiotropic effects (15). Indeed, it was recently shown that both dsx and fru genes are very conserved in their expression and function among Drosophila species (16). Therefore, although closely related Drosophila species often differ in their sexual dimorphisms, it is likely that such rapidly evolved traits reflect changes in the expression of downstream targets of dsx and fru in these species (17). Thus, knowing the genes that are direct targets of dsx and fru would greatly facilitate studies on the evolution of sex. However, to date there are only a few validated direct gene targets for dsx: Yolk Proteins (18, 19), bric-à-brac 1 (20), and desatF (also known as Fad2) (21). No direct target gene has been validated for fru, although the binding sequences for different Fru isoforms were recently identified (22, 23). The lack of known direct targets of dsx and fru places obstacles in our path to understanding the biology and evolution of sex.
To fulfill this need, we previously identified potential direct dsx target genes through a genome-wide screen for DSX binding sites (24). Characterization of these candidate genes should facilitate our understanding of the molecular mechanisms and evolution of the differences between males and females. As an example, here we characterized one such candidate target gene, flavin-containing monooxygenase 2 (Fmo-2), a top-ranking dsx direct target from our screen.
Flavin-containing monooxygenases (Fmos) are present across all phyla of eukaryotes and support diverse biological functions (25–28). Sexually dimorphic expression of the Fmos is quite often observed in vertebrates. In rabbits, Fmo-2 is expressed at higher levels in several female tissues, including esophagus, nasal mucosa, and kidney (29). In mice, hepatic expression of Fmo1 and Fmo3 are higher in females and are under the regulation of sex steroids (30). The male-specific repression of Fmo-3 expression in the livers of certain Mus species allows the male-biased production of trimethylamine in the urine, which serves as an olfactory cue for species-specific behavior (31). Although little is known about the in vivo function or the transcriptional regulation of Fmo-2, the Drosophila Fmo-2 has been characterized in vitro for its molecular properties (32), which suggest it likely functions as a terminal gene of the sex-determination pathway.
We found that D. melanogaster Fmo-2 expression in the midgut, specifically at the posterior part of the cardia and a small anterior section of the ventriculus that adjoins the cardia (together denoted CVJ, for cardia ventriculus junction), is sexually dimorphic. The manifestation of this sexual dimorphism requires both dsx function and the DSX-binding site at the Fmo-2 gene. In addition, dsx also acts through this same DSX-binding site to activate the Fmo-2 transcription in the fat body cells of a female-specific tissue, the spermatheca. We found that expression of Fmo-2 in the midgut has evolved rapidly, and that DNA sequence changes upstream of the Fmo-2 gene appear sufficient to account for differences in the expression between a number of species in the melanogaster group. In contrast, the expression of Fmo-2 in the fat body cells encapsulating the spermatheca appears to be very conserved, therefore placing an evolution constraint on the DSX-binding site. We identified two cis-regulatory modules (CRMs) in the Fmo-2 regulatory sequence. The more proximally (relative to Fmo-2) located CRM (denoted CRM-p) directs sexually monomorphic Fmo-2 transcription across the entire anterior midgut. The more distally located CRM (denoted CRM-d) contains a DSX-binding site and directs sexually dimorphic Fmo-2 transcription at the CVJ. Combinations of changes at these two CRMs produce diverse Fmo-2 expression patterns in the midgut across species in the melanogaster group. Most notably, the reversion of Fmo-2 expression in some species from sexually dimorphic to sexually monomorphic is caused by the loss of CRM-d function but not the loss of the DSX-binding site in CRM-d. Thus, integrating regulatory information through a CRM, but not just a mere DSX-binding site, to control sex-specific gene transcription in a specific tissue is one mechanism to get around evolutionary constraints posed by other tissues in the case of pleiotropic sex-specific transcriptional regulation.
Results
The Expression of Fmo-2 Is Sexually Dimorphic at the Junction of the Posterior Cardia and Anterior Ventriculus in the Midgut of D. melanogaster.
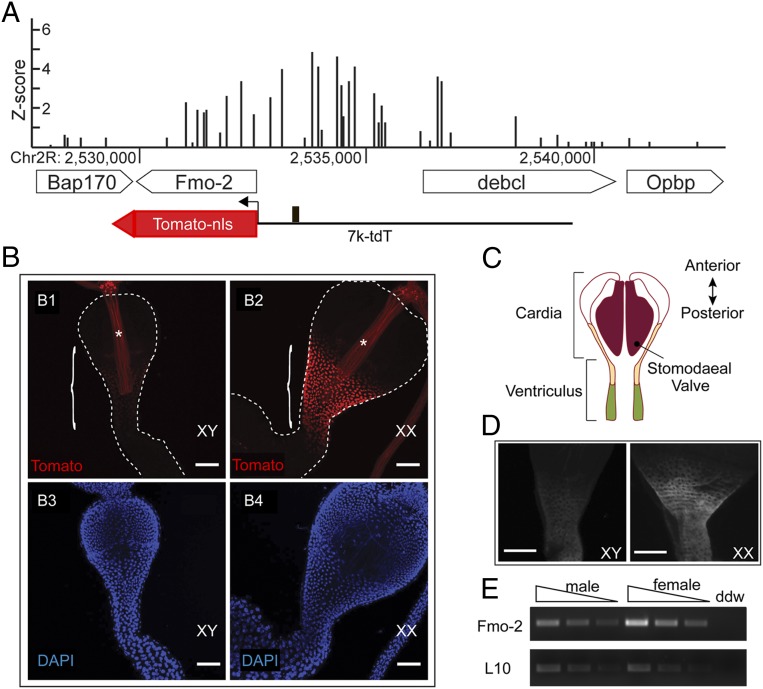
The Fmo-2 gene was among the highest ranking candidates identified by our previous genome-wide screen for direct dsx targets: the region 5′ to the Fmo-2 gene was consistently bound by DSX in vivo and this region also contains a perfect match to the 13-bp optimal DSX-binding site that is bound by DSX in vitro (Fig. 1A and SI Appendix, Fig. S1) (24).
Fig. 1.
Fmo-2 expression in the cardia and anterior ventriculus is sexually dimorphic in D. melanogaster. (A) The in vivo DSX binding peak at the Fmo-2 locus measured by DamID (24) and the Tomato reporter (7k-tdT) driven by the 7-kb genomic sequence, from −6994 to −61 relative to the translation initiation site of the Fmo-2 gene. The z-score indicates the difference in the levels of methylation at each GATC site between the wild type DSX_Dam and its non–DNA-binding mutant control samples (24). The solid box indicates the position of the consensus DSX binding site (locating at ch2R: 2533346–2533358 of the genome). (B) Fmo-2 Tomato reporter expression at the CVJ is much higher in females (B2) than in males (B1). The region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. Autofluorescence from the inner lining of the digestive tract is indicated by the asterisk. The dashed lines demark the borders of the samples. The DAPI staining of the nuclear DNA is also shown for reference (B3 and B4). (Scale bars, 50 μm.) (C) A schematic drawing of a cross-section of the anterior midgut. The orange region indicates where Fmo-2 is highly expressed in females. The muscular stomodaeal valve is indicated in dark red. The green shaded region indicates where in the ventriculus the Tomato reporter is not detected. (D) Representative images of the whole-mount staining of the anterior midgut of male (Left, XY) and female (Right, XX) flies with anti–FMO-2 (gray). (Scale bars, 50 μm.) (E) Quantitative RT-PCR using RNA of dissected cardia samples from male and female flies for Fmo-2 (Upper) and ribosomal RNA L10 (as a control, Lower). A serial dilution for each sample was used as template to ensure the PCR is within linear range.
To identify the tissues in which dsx might regulate Fmo-2, we first examined the temporal and spatial patterns of expression of the Fmo-2 gene. For this purpose, we created a transgenic reporter in which a 7-kb genomic sequence immediately upstream of the transcriptional start site of the Fmo-2 gene was linked to a cassette encoding a nuclear red fluorescent protein, tdTomato-nls (Fig. 1A). The 7-kb genomic sequence contains the entire intergenic region between the divergent transcriptional start sites for Fmo-2 and debcl, as well as the first intron of the debcl gene. Flies carrying the transgene (7k-tdT) were examined in larval and adult stages and Tomato fluorescence was observed in the renal system, the digestive system, and fat body cells of both larvae and adults, and in the spermatheca of adult females. These findings are consistent with transcript profiling of Fmo-2 in FlyAtlas (33), suggesting that the reporter recapitulates the endogenous Fmo-2 expression pattern.
When we compared the reporter expression in tissues present in both adult males and females, we only observed sexually dimorphic levels of Tomato fluorescence in part (CVJ) of the midgut (Fig. 1B). Specifically, in females there was a high level of Tomato fluorescence at the posterior cardia (R0 section) (34) and adjoining anterior ventriculus (R1a and maybe R1b sections) (34) (illustrated by the orange color in Fig. 1C), whereas fluorescence was barely detectable in males in this tissue (Fig. 1B). In addition, we observed Tomato fluorescence in the spermatheca, a tissue that only present in the females.
To confirm that the female-biased Fmo-2 reporter expression at the CVJ indeed reflects the endogenous gene expression pattern, the FMO-2 protein level in adults was examined by antibody staining against FMO-2 (32). We detected a much higher level of FMO-2 immuno-reactivity at the CVJ in females than in males, consistent with what was observed for the 7k-tdT reporter expression (Fig. 1D). Furthermore, quantitative RT-PCR from dissected CVJs indicated that Fmo-2 transcript levels were higher in females than males (Fig. 1E). In contrast, debcl, the gene that shares the 5′ intergenic region with Fmo-2, did not show sex-biased expression in the same RT-PCR assay.
Together, our results show that expression of Fmo-2 is sexually dimorphic in the CVJ and that this dimorphism is likely generated at the level of Fmo-2 transcription. We also conclude that expression of the 7k-tdT transgene faithfully recapitulates transcription of the endogenous Fmo-2 gene.
Fmo-2 Transcription Is Directly Regulated by DSX Through an Optimal DSX-Binding Site.
If Fmo-2 is directly regulated by dsx, Fmo-2–expressing cells should also express dsx. Thus, we examined the simultaneous expression of 7k-tdT and dsxGAL4(Δ2), a GAL4 knocked into the dsx locus that reflects the transcription of dsx (35), in the CVJ of females. We found that all CVJ cells that were positive for 7k-tdT were also positive for dsxGAL4(Δ 2), driving a nuclear GFP (SI Appendix, Fig. S2). In addition, there were GFP+ cells that were not tdT+, suggesting that factors in addition to dsx are required for Fmo-2 expression in some cells of this tissue.
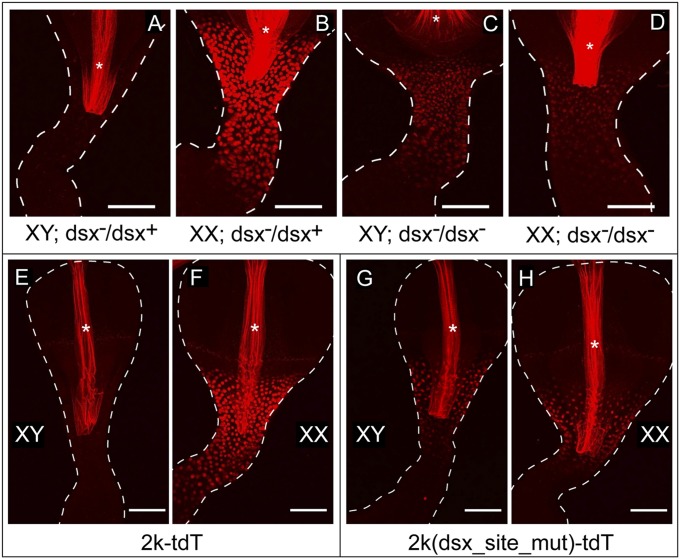
We then examined the effects of dsx mutations on the expression of the 7k-tdT reporter in chromosomal males and females. The male-specific DSXM and the female-specific DSXF proteins are encoded by sex-specific mRNAs produced by alternative splicing of a primary dsx transcript common to both sexes. Both DSX proteins are zinc-finger transcription factors with identical DNA-binding domains, but different C termini that interact differently with cofactors, and thus can have different effects on the transcription of target genes (36–38). We found that in XY;dsx−/dsx− flies, the 7k-tdT reporter expression strongly increased compared with their XY;dsx−/+ siblings. On the other hand, the 7k-tdT reporter expression significantly decreased in the XX;dsx−/dsx− flies in comparison with their XX;dsx−/+ siblings (Fig. 2 A–D). Thus, in contrast to the sexually dimorphic expression observed in wild-type flies, XX and XY flies lacking dsx function exhibited monomorphic Fmo-2 reporter expression. Similar results were observed at the FMO-2 protein level as detected by antibody staining (SI Appendix, Fig. S3). Therefore, the sexually dimorphic Fmo-2 expression at the CVJ is controlled by dsx.
Fig. 2.
Fmo-2 transcription is regulated by dsx directly through the DSX-binding site. (A–D) The 7k-tdT reporter expression in dsx−/dsx+ male (A) and female (B) flies, as well as in XY, dsx−/dsx− (C) and XX, dsx−/dsx− (D) flies. The dsx− alleles used here are dsx683-7058 and dsx1649-9625. Similar results were observed with another dsx allele combination (dsx1/dsxM+R13). (E–G) The expression of 2k-tdT in males (E) or females (F), and the expression of 2k(dsx_site_mut)-tdT, in which the DSX-binding site was mutated from GCAACATTGTTGC to GgggggggggggC, in males (G) or females (H). The region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. Autofluorescence from the inner lining of the digestive tract is indicated by the asterisk. (Scale bars, 50 μm.)
These results also revealed that each of the sex-specific DSX isoforms plays an active role in regulating Fmo-2 transcription: DSXF activates, whereas DSXM represses it. To further test this conclusion, a dominant dsx allele, dsxM, which produces only the male isoform of DSX regardless of chromosomal sex (39), was introduced into XX flies to examine its effect on the Fmo-2 expression. We found that the level of 7k-tdT fluorescence, which was already reduced to an intermediate level in XX;dsx−/dsx− flies, was further reduced in XX;dsx−/dsxM flies to a barely detectable level that was similar to that seen in XY;dsx−/+ flies (SI Appendix, Fig. S3), confirming the repressive role of DSXM on Fmo-2 expression.
To further delimit the genomic region that controls female-biased Fmo-2 expression in the CVJ, a reporter driven by a smaller 2-kb fragment immediately upstream of the Fmo-2 gene was constructed (named 2k-tdT). This reporter also showed female-biased expression at the CVJ (Figs. 2 E and F), suggesting that regulatory sequences within this 2-kb region are sufficient to account for the sex-biased and tissue-specific Fmo-2 expression in the gut. Within this 2-kb sequence, there is a perfect match for the optimal 13-bp DSX-binding site. We introduced mutations at this 13-bp sequence to test if it was required for the sexually dimorphic expression of 2k-tdT. The expression of the mutated reporter [named 2k(dsx_site_mut)-tdT] at the CVJ was comparable between males and females (Fig. 2 G and H), and at a level that was intermediate of those seen for the original 2k-tdT in males and females (Figs. 2 E and F). These results, in combination with the observations that DSX binds both to the Fmo-2 promoter region in vivo (Fig. 1A) and to the 13-bp sequence in vitro (SI Appendix, Fig. S1), demonstrate that DSX directly regulates the sexually dimorphic Fmo-2 transcription at the CVJ through this 13-bp DSX-binding site.
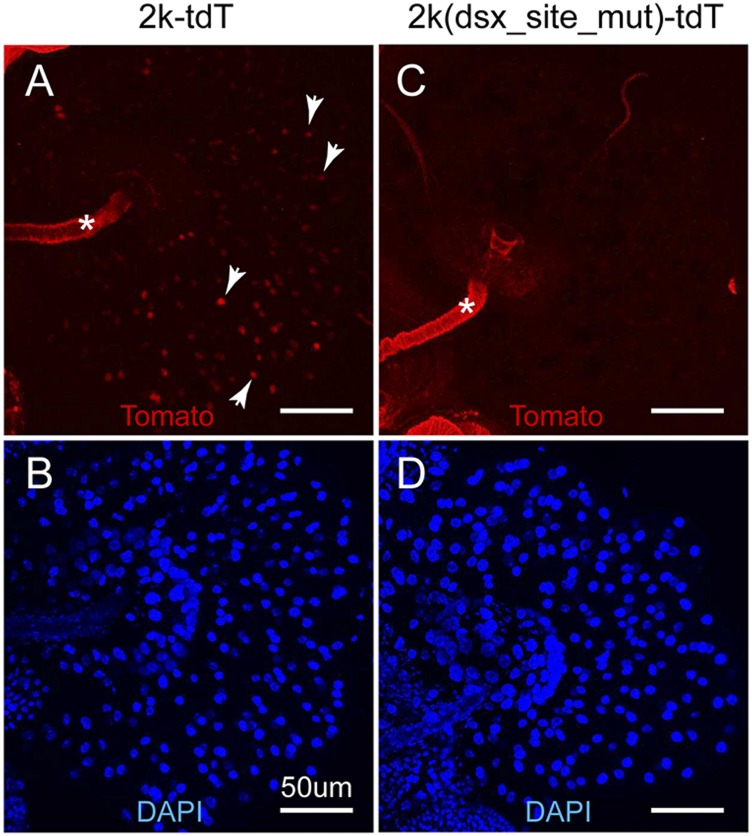
We also found that similar to the 7kb-tdT reporter, the 2k-tdT reporter is also expressed in the spermatheca (Fig. 3). The fat body cells that encapsulate the spermatheca show high level of reporter expression. Mutation of the DSX-binding site in the D. melanogaster 2k-tdT reporter reduced the expression in these fat body cells to barely detectable levels (Fig. 3), suggesting that the DSX-binding site is required for the Fmo-2 transcriptional activation in the fat body cells of the spermatheca. However, we were not able to conclude confidently whether other fat body cells in the adult also show sex-specific expression of Fmo-2, because of the reporter expression being highly variable among fat body cells from different part of the body of the same fly.
Fig. 3.
The DSX-binding site is required for the Fmo-2 transcriptional activation in the fat body cells of the spermatheca. The expression of the reporters 2k-tdT (A) and 2k(dsk_site_mut)-tdT (C), in the spermatheca were examined. Examples of the nuclear Tomato red fluorescence are indicated by the arrowheads. The DAPI staining showed the nuclei of these samples (B and D). Samples in A and C were processed in parallel and imaged with the same confocal settings. Autofluorescence is indicated by the asterisk.
Presence of the DSX-Binding Site in the Fmo-2 Regulatory Region Does Not Necessarily Confer Sexually Dimorphic Expression in the Midgut.
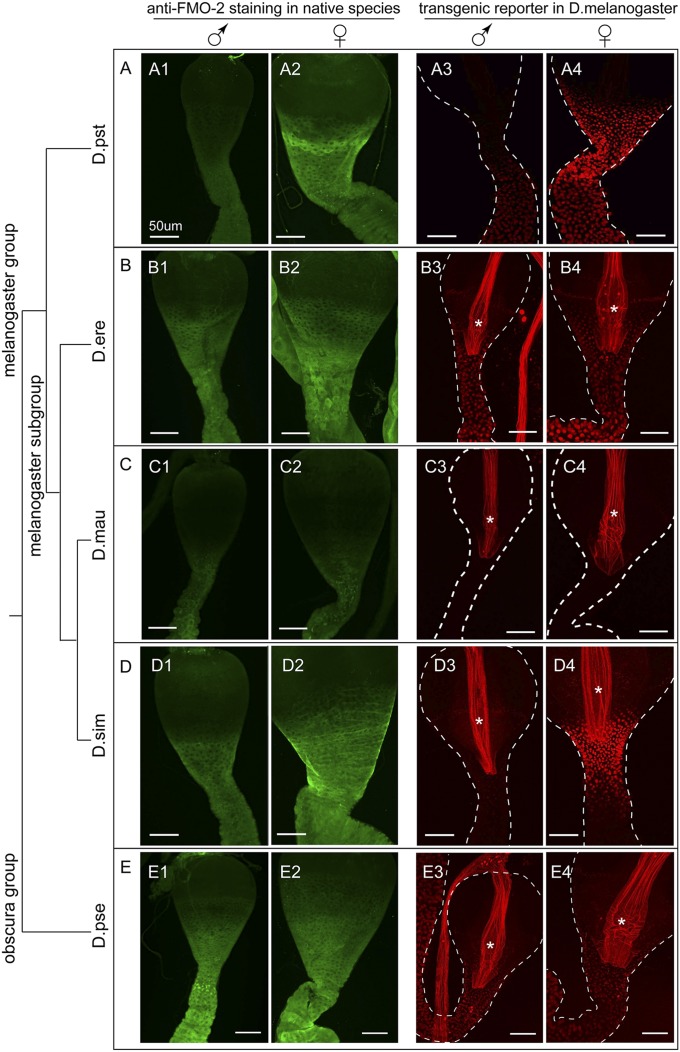
We previously found that the Fmo-2 gene of all five sequenced species in the melanogaster subgroup contained a sequence matching at least 12 bp of the 13-bp optimal DSX-binding site (24). To test whether the conservation of the DSX-binding site at the Fmo-2 gene correlates with conservation of sexually dimorphic Fmo-2 expression, we examined the Fmo-2 expression in six species from the melanogaster subgroup, as well as three species in the melanogaster group but not in the melanogaster subgroup (Fig. 4 and SI Appendix, Fig. S5). All these species have a sequence within the 5′ regulatory region of the Fmo-2 gene that matches at least 12 bp of the 13-bp optimal DSX binding site (SI Appendix, Fig. S5). As a control, we also included one species from the obscura group (Drosophila pseudoobscura), whose Fmo-2 gene does not have an as well-matched DSX-binding sequence (Fig. 4 and SI Appendix, Fig. S5).
Fig. 4.
Changes in the regulatory sequence appear sufficient to account for the diverse Fmo-2 expression in closely related species. (A) The FMO-2 protein level in the midgut of the male (A1) and female (A2) D. pseudotakahashii (D.pst) was examined by immunofluorescence using an anti–FMO-2 antibody. Male (A3) and female (A4) D. melanogaster transgenic flies carrying the Tomato reporter driven by the 5′ regulatory sequence from the D.pst Fmo-2 gene were examined for reporter expression in the midgut. (B) Similar to A, except that the species is D. erecta (D.ere). (C) Similar to A except that the species is D. Mauritiana (D.mau). (D) Similar to A except that the species is D. simulans (D.sim). (E) Similar to A except that the species is D. pseudoobscura (D.pse). The phylogenetic tree for these species is on the left (drawing not to the scale with respect to time). The region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. Autofluorescence from the inner lining of the digestive tract is indicated by the asterisk.
Using immunofluorescence staining, we focused exclusively on determining whether the FMO-2 protein level was sexually dimorphic in each species, because the staining intensity difference between species could be in part because of the different affinities of the antibody for the FMO-2 proteins of these species. To our surprise, despite the conservation of a well-matched DSX-binding sequence in the 5′ regulatory region, the expression pattern for the FMO-2 protein in the midgut is very diverse among the melanogaster group species (Fig. 4 and SI Appendix, Fig. S5). In five species (D. melanogaster, Drosophila pseudotakahashii, Drosophila santomea, Drosophila yakuba, and Drosophila simulans), we observed significant sexually dimorphic staining at the CVJ. Two species (Drosophila erecta, Drosophila teissieri) showed a monomorphic staining throughout the CVJ and the ventriculus. Three species (Drosophila prostipennis, Drosophila mauritiana, and Drosophila eugracilis) showed such weak staining at both the CVJ and the ventriculus that the difference between males and females could not be determined. The weak staining in the latter three species is not likely a result of either the loss of the Fmo-2 gene in the genome or to the antibody not recognizing the FMO-2 protein in these species, because we can detect strong staining in a group of corpus cardiacum/hypocerebral ganglion neurons in the same flies. The obscura group species (D. pseudoobscura) showed monomorphic staining at both the CVJ and the ventriculus (Fig. 4 and SI Appendix, Fig. S5). Thus, the presence of the DSX-binding site does not necessarily confer sexually dimorphic expression in the midgut.
Changes in cis-Regulatory Sequences Appear Sufficient to Account for the Rapid Evolution of Fmo-2 Expression in the Midgut Across Closely Related Species.
The differences in the Fmo-2 expression pattern in these species could be a result of: (i) changes in its cis-regulatory sequences; (ii) changes in the expression/properties of its up-stream transcription factors; (iii) posttranscriptional changes affecting the detectable levels of the FMO-2 proteins; or (iv) a combination of these changes. Under the first scenario, we would expect that the regulatory sequence for the Fmo-2 gene, when moved to another species, would be able to direct an expression pattern similar to what it directs in its native species. We generated Tomato reporters driven by the 5′ genomic sequences of the Fmo-2 genes from D. pseudotakahashii, D. erecta, D. mauritiana, D. simulans, and D. pseudoobscura. These transgenes were integrated into the same attP site in the D. melanogaster genome and their expression analyzed in males and females. We found that the midgut expression pattern for each of the reporters in D. melanogaster closely resembled the patterns of anti–FMO-2 staining in the respective native species (Fig. 4). The reporter driven by the D. pseudotakahashii sequence showed highly sexually dimorphic expression at both the CVJ and the ventriculus that was quite similar to the antibody staining in its native species (Fig. 4A). Both reporters driven by D. erecta (Fig. 4B) and D. pseudoobscura (Fig. 4E) sequences showed a sexually monomorphic expression in both the CVJ and the ventriculus, with the expression in the ventriculus being stronger. These are close to the patterns of antibody staining in their respective native species. The reporter driven by the D. mauritiana (Fig. 4C) sequence showed no detectable expression at either the CVJ or the ventriculus, similar to the very low intensity of antibody staining in its native species. The reporter driven by the D. simulans sequence showed a sexually dimorphic expression at the CVJ, closely resembling the antibody staining in D. simulans (Fig. 4D). Thus, the differences in Fmo-2 midgut expression are likely caused by changes within the cis-regulatory sequences of the Fmo-2 gene.
On the other hand, we found that the D. erecta and the D. mauritiana reporters both showed expression in the fat body cells of the spermatheca (SI Appendix, Fig. S6), although they showed sexually monomorphic expression or no expression in the midgut. These results suggested that: (i) the transcriptional regulation of Fmo-2 by dsx in the spermatheca is very conserved; and (ii) the presence of a conserved DSX-binding site in the Fmo-2 gene is likely because of it being required for the conserved transcriptional activation in the spermatheca.
Two CRMs Control the Transcription of Fmo-2 in the Midgut.
Next we addressed how alterations in the cis-acting Fmo-2 regulatory sequences might lead to the observed changes in Fmo-2 expression in the midgut. We focused on D. melanogaster and D. erecta because the reporters driven by the Fmo-2 genomic sequences from these two species showed very different expression patterns: the D. melanogaster sequence confers high, sexually dimorphic expression in the CVJ, and very low expression in the ventriculus (hence, whether ventricular expression is sexually dimorphic could not be assessed), whereas the D. erecta reporter is not expressed in a sexually dimorphic pattern although it is expressed in both the CVJ and the ventriculus, with the expression in the ventriculus being much stronger than in the CVJ. To delimit the locations and properties of the sequences governing Fmo-2 expression in the midgut, we swapped sequences between the reporters driven by the 2-kb genomic sequence immediately upstream of the D. melanogaster Fmo-2 coding sequence and the 2.2-kb orthologous sequence from D. erecta (Fig. 5A).
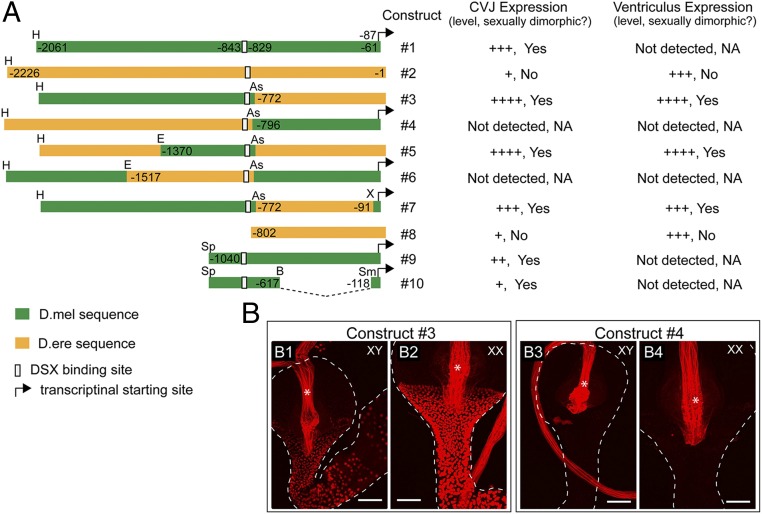
Fig. 5.
Two CRMs control the Fmo-2 transcription in the CVJ and anterior midgut (including the CVJ and the ventriculus), respectively. (A) Diagram of the regulatory sequences that are used to drive the tdTomato reporter in the constructs 2k-tdT (#1) and ere-tdT (#2), and the derivative constructs with sequences swapped between the two. The letters denote the restriction site used for the sequence swapping: As, AseI; B, BbvCI; E, EcoRV; H, HindIII; Sp, SphI; Sm for SmaI; X, XhoI. The position of each restriction site (relative to the translation initiation site of the Fmo-2 gene) is indicated. The translation initiation site was used as a reference for both species because the Fmo-2 transcriptional start site has not been mapped for D. erecta. The expression of the each construct is summarized on the right. The level of reporter activity is indicated by the number of “+”. NA indicates that the expression is too low to determine whether it is sexually dimorphic. (B) Examples of the sequence-swapped reporter expression in D.mel transgenic flies. (Left) Construct #3 (B1, male; B2, female); (Right) construct #4 (B3, male; B4, female). The region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. Autofluorescence from the inner lining of the digestive tract is indicated by the asterisk. (Scale bars, 50 μm.)
For the sake of clarity, we first discuss the effects of these chimeric constructs on expression in the CVJ. Examination of the expression of these reporters showed that the combination of the distal part (∼1.2 kb) of the D. melanogaster sequence (with respect to the Fmo-2 transcriptional start site) and the proximal part (∼0.8 kb) of the D. erecta sequence drove a D. melanogaster-like pattern (although at a higher level) of reporter expression in the CVJ (high, sexually dimorphic) (Fig. 5A, construct #3, and Fig. 5B). Notably, this distal sequence contains the DSX-binding site (Fig. 5A). Conversely, the combination of the distal part of the D. erecta sequence (∼1.4 kb) and the proximal part of the D. melanogaster sequence (∼0.8 kb) drove a D. erecta-like pattern of expression in the CVJ (no detectable expression) (Fig. 5A, construct #4, and Fig. 5 B3 and B4). These results suggested that there must be at least one functional CRM in the distal portion of the D. melanogaster sequence that specifies sexually dimorphic expression in the CVJ. In support of this hypothesis, a smaller (∼0.6 kb) substitution of D. melanogaster sequence into the D. erecta sequence is sufficient to produce a reporter with a high, sexually dimorphic expression pattern in the CVJ (Fig. 5A, construct #5). Conversely, when the D. erecta counterpart of the 0.6-kb sequence was swapped into the reporter driven by the D. melanogaster sequence, there was no detectable reporter expression in the CVJ of either sex (Fig. 5A, construct #6). In addition, two deletion constructs derived from the D. melanogaster sequence, one with 1 kb of the distal sequence removed and the other one with additional proximal sequence removed (Fig. 5A, constructs #9 and #10), can both drive sexually dimorphic reporter expression in the CVJ, further demonstrating that the D. melanogaster sequence around the DSX binding site contains a CRM (named CRM-d) that regulates Fmo-2 transcription in the CVJ.
Observation of ventricular expression in the sequence swapping experiments above (constructs #3, #4, #5, and #6) suggested that there exists another CRM (denoted CRM-p) in the 0.8-kb region at the proximal end of the D. erecta Fmo-2 sequence that is required for activation of Fmo-2 transcription in the ventriculus (Fig. 5A). This CRM must be deleted or nonfunctional in the orthologous D. melanogaster genomic sequence. Thus, in constructs that contain this D. erecta sequence (constructs #2, #3, #5), we observed high levels of expression in the ventriculus that was either sexually monomorphic or sexually dimorphic (the sexually dimorphic aspect of this expression is discussed below), whereas in the constructs that contain the orthologous D. melanogaster counterpart (constructs #1, #4, #6), we observed little reporter expression in the ventriculus (Fig. 5A).
We subsequently realized that the D. erecta Fmo-2 genomic sequence in the above reporter constructs includes extra sequence from the 5′ UTR of the D. erecta Fmo-2 transcript (because of the different annotations for these two species in FlyBase). However, we found that addition of the extra 5′ UTR sequence into the 2k-tdT reporter did not change the reporter expression pattern. On the other hand, when a 682-bp sequence immediately upstream of the TATA box of the D. erecta Fmo-2 gene, was swapped into the 2k-tdT reporter, the resulting reporter (construct #7) showed a high level of expression in both the CVJ and the ventriculus in females (Fig. 5A). This expression pattern is very similar to that of the reporter construct #3, suggesting that CRM-p is within this 682-bp sequence. Finally, when the distal sequence (including the DSX-binding site) was removed from the D. erecta reporter, the resulting construct #8 retained a sexually monomorphic expression in the ventriculus, and at a lower level, in the CVJ, similar to that in the parental construct (Fig. 5A). This result further demonstrates that the proximal part of D. erecta sequence contains a functional CRM that activates Fmo-2 transcription in the ventriculus (and to a lower level, in the CVJ) in a sexually monomorphic manner.
Thus, the absence of CRM-p in D. melanogaster leads to the lack of Fmo-2 transcription in the ventriculus, and the absence of CRM-d in D. erecta leads to the lack of sexually dimorphic Fmo-2 expression.
Changes Outside of the DSX-Binding Site Impair the Function of CRM-d in D. erecta.
We tried to identify the sequence critical for Fmo-2 expression in the CVJ, as this might suggest the possible causes for the loss of CRM-d function in D. erecta and other species. The reporter driven by the proximal 1 kb (from −1040 to −61) of the D. melanogaster sequence (construct #9, denoted as 1k-tdT) has a CVJ expression pattern that is very similar to, albeit slightly lower than, that of the 2k-tdT reporter (Fig. 5A), suggesting that most of the sequence comprising CRM-d is retained in this 1-kb region. The swapping experiments above (constructs #5, #6) also suggested that the D. melanogaster sequence between −1370 and −795 is important for the CVJ expression. Thus, CRM-d is likely located within the 246-bp sequence between −1040 and −795. Indeed, when this 246-bp sequence in the 2k-tdT reporter was replaced with the orthologous sequence from D. erecta, the resulting reporter [CRM-d(m->e)] showed little expression at the CVJ (Fig. 5A). The latter result suggested that the sequence changes within this 246-bp region are sufficient to cause the loss of CRM-d function in D. erecta.
There are many changes scattering across the orthologous D. erecta sequence compared with this 246-bp D. melanogaster sequence (Fig. 6B). First, the putative DSX-binding site in the D. erecta Fmo-2 sequence differs in 1 bp from the optimal DSX-binding site, and there are also changes immediately adjacent to the DSX-binding site (Fig. 6B). To test whether these particular base substitutions within and near the DSX-binding site are responsible for the absence of sexually dimorphic expression in D. erecta, they were mutated in the D. melanogaster 2k-tdT construct to those of D. erecta. The resulting reporter [denoted dsxsite(m->e)] was expressed in a sexually dimorphic pattern similar to that of its parental (D. melanogaster) construct (Fig. 6A), suggesting that other changes in the 246-bp sequence but not in the DSX-binding site or its immediate flanking sequence impair CRM-d function in D. erecta.
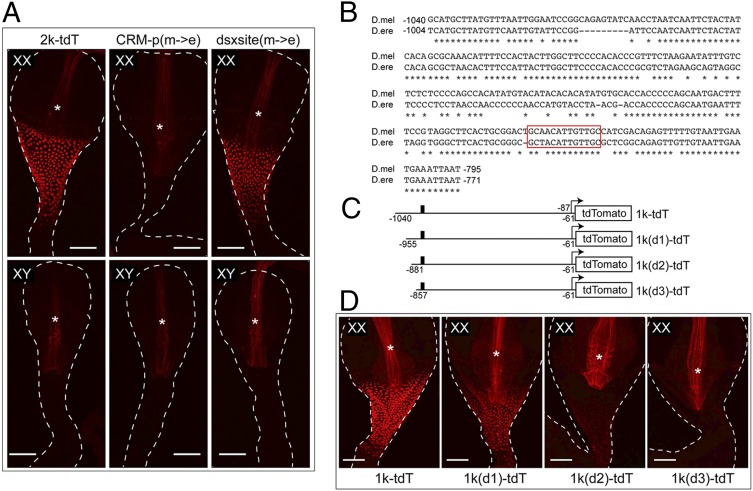
Fig. 6.
Changes in other positions instead of the DSX-binding site impaired the function of CRM-d in D. erecta. (A) Mutations in other positions but not the DSX-binding site eliminated the function of CRM-d in D.ere. Two mutant reporters, named CRM-d(m->e) and dsxsite(m->e), were derived from 2k-tdT. In the reporter CRM-d(m->e), a 246-bp sequence (from −1040 to −795) containing the putative CRM-d was replaced with the orthologous D.ere sequence. In the reporter dsxsite(m->e), only the DSX-binding site (located between −842 and −830) and the flanking sequence (ACTGCAACATTGTTGCCA, the DSX-binding site underlined) were changed to the D.ere sequence (gc-GCtACATTGTTGCgc). Confocal images show the expression of the 2k-tdT and the two mutated reporters in females (Upper) or in males (Lower). The reporter expression in the female CVJ was lost in CRM-d(m->e) but not in dsxsite(m->e). (B) Alignment of the 246-bp D. mel Fmo-2 regulatory sequence containing the putative CRM-d, and the orthologous D.ere sequence. The conserved nucleotides are indicated by the asterisk underneath them. The DNA positions relative to the translation start site are indicated. The DSX-binding sites are red-boxed. (C) Diagram of the 1k-tdT, 1k(d1)-tdT, 1k(d2)-tdT and 1k(d3)-tdT reporters. The DSX-binding site is indicated by the filled box. (D) The expression of the reporters from C in females. None of these constructs has detectable reporter expression in the male samples. For A and D, the region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. Autofluorescence from the inner lining of the digestive tract is indicated by the asterisk. (Scale bars, 50 μm.)
To further localize the sequences that are important for Fmo-2 expression at the CVJ, we made progressive deletions from the 5′ end of the 1k-tdT reporter, each deletion removing some additional sequence that has changed in D. erecta (Fig. 6C). We found that each of the deletions caused an additional decrease in the expression level of the reporter, but the sexually dimorphic expression pattern was preserved until the deletion that removed most of the sequence upstream of the DSX binding site, for which there was no longer detectable reporter expression in either sex (Fig. 6D). These results suggest two possibilities: (i) multiple transcription factors contribute to the tissue-specific activation of Fmo-2 transcription at the CVJ, and that the loss of function for CRM-d in D. erecta is a result of cumulative changes at the binding sites for these transcriptional factors; or (ii) alternatively, there might be gain of repressors binding in the D. erecta orthologous sequence of CRM-d that suppress the binding/action of these activating transcription factors.
Beside the DSX-Binding Site, Other Sequences in CRM-d Are Required for Sexually Dimorphic Fmo-2 Transcription in the Ventriculus.
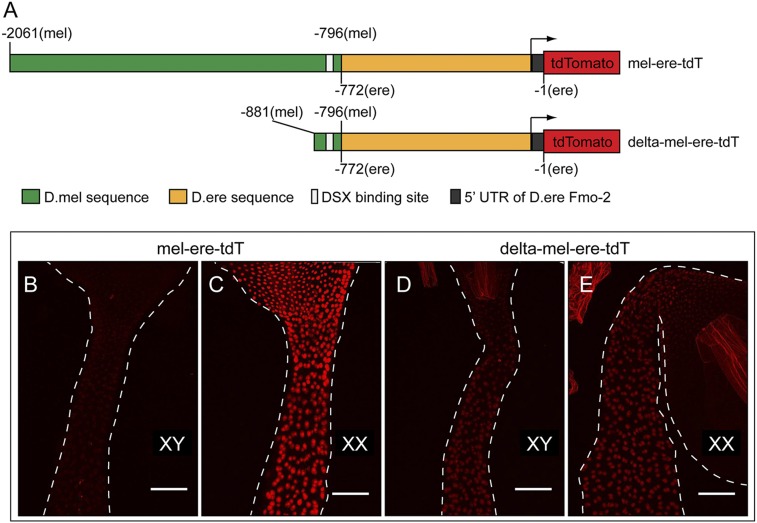
In the sequence-swapping experiments described above, we observed an unexpected combinatorial effect of the distal CRM-d (mel) and proximal CRM-p (ere) in regulating reporter transcription in the ventriculus. The distal CRM-d (mel) itself does not activate the reporter in the ventriculus when together in cis with the proximal (mel) sequence (Fig. 5A, construct #1). However, when together in cis with the proximal (ere) CRM-p (constructs #3, #5 and #7), we observed high levels of sexually dimorphic expression in the ventriculus (Fig. 5A). As noted above, the D. erecta sequence contains an intact DSX-binding site yet directs sexually monomorphic expression in the ventriculus. In addition, the positions of the DSX-binding sites (in relationship to CRM-p) in the native D. erecta sequence (Fig. 5A, construct #2) and the chimeric constructs (Fig. 5A, #3, #5 and #7) are identical, with only 4-bp substitutions in the spacing sequence. Thus, it is possible that a functional CRM-d, not just the DSX-binding site, is required for sexually dimorphic transcriptional activation by CRM-p in the ventriculus. To test this hypothesis, we revisited construct #3, henceforth referred to as “mel-ere-tdT.” We deleted most sequences upstream of the DSX-binding site in this reporter construct to create a new construct (named delta-mel-ere-tdT) that has a DSX-binding site but not a functional CRM-d linked to the 5′ end of the CRM-p sequence (Fig. 7A). This new deletion construct and the parental mel-ere-tdT construct both contain the same DSX-binding site with the same spacing sequence relative to CRM-p. The same deletion in a purely D. melanogaster reporter almost completely abolishes the CRM-d function (d2 deletion) (Fig. 6 C and D). The expression of the delta-mel-ere-tdT reporter in the ventriculus is at similar levels in males and females, which is drastically different from that of the parental mel-ere-tdT reporter, but very similar to that of the D. erecta reporter (Fig. 7 B–E). Thus, the sexually dimorphic Fmo-2 transcription in the ventriculus also requires a functional CRM-d, and the presence of just a DSX-binding site is insufficient to confer regulation by dsx.
Fig. 7.
The presence of the DSX-binding site itself is insufficient to confer the sex-specific regulation of Fmo-2 by dsx in the ventriculus. (A) Diagram of the reporter mel-ere-tdT (construct #3, Fig. 4A) and its derived reporter delta-mel-ere-tdT. The DNA positions relative to the translation start site are indicated. (B–F) The reporter activity at the anterior midgut was analyzed for mel-ere-tdT in males (B) and females (C), and for delta-mel-ere-tdT in males (D) and females (E). The region where the reporter is active is bracketed. Male and female samples were imaged with the same confocal settings. (Scale bars, 50 μm.)
We noted that the expression pattern directed by the combination of CRM-d (mel) and CRM-p (ere) is very similar to the reporter expression driven by the D. pseudotakahashii Fmo-2 regulatory sequence. Therefore, it is very likely that in those sibling species whose Fmo-2 expression is sexually dimorphic throughout the CVJ and the ventriculus, the Fmo-2 regulatory sequence is composed of these two CRMs.
Discussion
We have shown that dsx is deployed to regulate Fmo-2 in more than one tissue: the sexually dimorphic transcription of Fmo-2 in the midgut is directly activated by DSXF and repressed by DSXM through a canonical DSX-binding site; the transcriptional activation of Fmo-2 in the female-specific spermatheca also requires the action of DSXF through the same canonical binding site. We further established Fmo-2 transcription in the midgut is controlled through two CRMs, and changes within these two CRMs—but not at the canonical DSX-binding site—caused the diverse midgut Fmo-2 expression patterns while maintaining the conserved spermathecal expression found in the melanogaster group. Thus, these CRMs form the foundation for the rapid evolution of Fmo-2 transcriptional regulation in the midgut. Below we discuss some implications of these findings.
Rapid Change of the Fmo-2 Regulatory Sequences Drive the Diversified Fmo-2 Expression in Drosophila Species.
We have shown that the midgut expression pattern of Fmo-2 evolved rapidly in the melanogaster group. The rapid changes in Fmo-2 expression are likely because of changes in the Fmo-2 regulatory sequences, as the regulatory sequences of the Fmo-2 gene from all species tested were able to drive expression of a reporter in D. melanogaster in a pattern similar to the Fmo-2 expression pattern in the respective native species. This finding is consistent with the general notion that evolutionarily significant changes are most likely in the cis-acting regulatory elements of the downstream targets of the transcription factors, rather than in the genes encoding the transcription factors (12–14), which appears to be especially true for sexually dimorphic gene expression (40).
We found two CRMs (CRM-d and CRM-p) controlling Fmo-2 expression in the CVJ and the ventriculus in the melanogaster group. CRM-d contains a DSX-binding site and can direct sexually dimorphic expression in the CVJ, whereas CRM-p directs sexually monomorphic expression in both the CVJ and the ventriculus. Because we could not find a homologous sequence for CRM-d in out-group species, such as D. pseudoobscura, we believe that CRM-d arose de novo in the common ancestor of the melanogaster group. On the other hand, we did find homology for the CRM-p containing sequence in the D. pseudoobscura Fmo-2 regulatory sequence, which can direct a monomorphic reporter expression in both the CVJ and the ventriculus in D. melanogaster. Therefore, we believe that the D. pseudoobscura sequence contains a functional CRM-p and that CRM-p represents a more ancestral CRM (arose before the branching of the obscura group and the melanogaster group). However, because of the rapid evolution of the Fmo-2 regulatory sequence, we could not determine more precisely when CRM-p arose.
We propose that changes at these two CRMs lead to the diversified patterns of Fmo-2 expression in the melanogaster group (summarized in Fig. 8). The gain of a functional CRM-d in the common ancestor of the melanogaster group had two consequences: (i) sexually dimorphic activation of Fmo-2 transcription in the CVJ; and (ii) adding a layer of sex-specific regulation on Fmo-2 transcription (originally directed by CRM-p) in the ventriculus. Further modifications of these two CRMs in descendent species resulted in diverse patterns of Fmo-2 expression. There was only one event of a gain of sexually dimorphic regulation, but several events of loss of sexually dimorphic regulation in the melanogaster group. This finding is consistent with the “losses are easy, gains are harder” principle of regulatory evolution (12), possibly because of a functional CRM-d requires gain of multiple transcription factor binding sites in an appropriate configuration, whereas losing of the CRM-d function might require just one change.
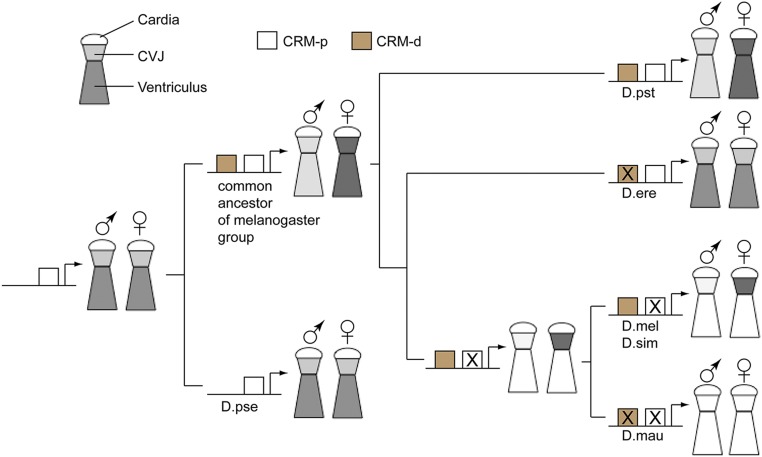
Fig. 8.
A model for the evolution of Fmo-2 transcriptional regulation at the midgut. The common ancestor for the melanogaster group and the out-group species started with the proximal CRM-p in the Fmo-2 regulatory sequence; therefore the expression is monomorphic. This monomorphic transcription was retained in the out-group species (e.g., D.pse). The gain of distal CRM-d (indicated by the filled box) in the common ancestor of the melanogaster group led to sexually dimorphic expression in both the CVJ and the ventriculus, which was retained by some descendent species (e.g., D.pst). Some species (e.g., D.ere) lose the function (indicated by the cross) of CRM-d while maintaining CRM-p intact, causing Fmo-2 transcription to revert to the more ancestral monomorphic pattern in both the CVJ and the ventriculus. The common ancestor of D.mau, Dmel, and D.sim lose the function of CRM-p while CRM-d was intact; therefore, Fmo-2 transcription was highly sexually dimorphic in the CVJ but was at a minimal level in the ventriculus. This feature was maintained in some descendent species (e.g., D.mel and D.sim). D.mau may have further lost the function of CRM-d, thus the Fmo-2 transcription is very low in both the CVJ and the ventriculus of both sexes. The intensity of the gray shade symbolizes the level of transcription in each segment of the anterior midgut.
Insight into the Tissue- and Sex-Specific Gene Regulation.
There are two mechanisms that could affect the tissue- and sex-specific gene expression directly regulated by dsx. The first mechanism is through the changes in the dsx expression pattern (4, 41). The other mechanism, maybe more commonly, is through the changes at the cis-regulatory sequences of the dsx target genes. For the latter mechanism, previous studies on individual genes that are the direct regulatory targets of DSX have revealed how tissue- and sex-specific information is integrated in the transcriptional regulation of these target genes. For the Yolk Protein genes, the fat body- and female-specific expression is brought about through enhancers containing multiple identified protein-binding sites (42, 43). The binding of the DSX proteins provides sex-specific regulation on top of the fat body-specific transcription established by the other proteins. In the case of the bric-à-brac 1 gene, the HOX protein Abdominal-B binds to multiple sites in the bric-à-brac 1 regulatory sequence to produce tissue-specific transcriptional activation, which is further modified sex-specifically by the direct binding of DSXF in females or DSXM in males to the bric-à-brac 1 regulatory sequence (20). Thus, a general feature for the transcriptional regulation of direct dsx target genes is tissue-specific transcription, established by the action of one or more tissue-specific transcription factors, in combination with DSX-binding sites recruiting the direct binding of DSXF and DSXM to achieve sex-specificity. We have shown that the transcriptional regulation of Fmo-2 in the CVJ follows this theme.
However, the sex-specific regulation of Fmo-2 transcription in the ventriculus appears to be more complex. Ventricular-specific transcriptional activation is established through CRM-p; however, it requires a functional CRM-d, not just a DSX-binding site, to confer regulation by DSX, although CRM-d itself does not activate Fmo-2 transcription in the ventriculus. The activation may require other factors binding in CRM-d either to secure DSX to its binding site or to bridge the interaction between the two CRMs. Regardless, the tissue- and sex-specific Fmo-2 transcription in the ventriculus is the result of the integration of regulation through two CRMs.
Overcoming the Evolutionary Constraints on the Regulatory Sequence in Pleiotropic Transcriptional Regulation.
Evidence has accumulated in recent years that “regulatory evolution” is the most pervasive mechanism underlying the morphological changes during evolution (12). Under this theory, changes in the expression pattern of the genes that govern the development of a particular feature, through changes at the regulatory sequence rather than changes at the coding sequence of the gene or changes of the upstream transcription factors, lead to changes of that feature. In addition, the cis-regulatory elements of a particular gene are often organized as modules and the gain or loss of the trait could be a result of the independent changes of these modules (12, 44). Regulatory evolution provides great flexibility and minimizes deleterious pleiotropic effects (12, 15).
It is conceivable that the regulation of a direct target gene by a particular transcription factor is pleiotropic, meaning that the gene is under the transcriptional regulation of this transcription factor in multiple tissues. In some of these tissues the regulation might be more resistant to change, consequently placing an evolutionary constraint on the changes in other tissues. This theory leads to the question of how such evolutionary constraints on the pleiotropic transcriptional regulation are overcome. Under the regulatory evolution viewpoint, this conflict could be resolved by independent changes of different regulatory modules. However, very few real examples have been shown to demonstrate that this is the case. One reason might be the difficulty of clearly identifying the in vivo binding sites of that transcription factor within the tissue-specific regulatory modules of the target gene. Generally, transcription factor binding sites are short (5–8 bp) and imprecise, thus the sequence for a given transcription factor binding site occurs very often in the genome just by chance. Therefore, many of these consensus matches are not bona fide binding sites and have no effect on transcription (45).
The transcriptional regulation by dsx provides a great paradigm to address how evolutionary constraints on pleiotropic transcriptional regulation are overcome. First, the optimal DSX-binding site is exceptional in that it is a 13-bp palindrome, and thus rarely occurs in the genome just by chance. Second, sex-specific features evolve very rapidly (46), and such rapidly evolved traits are likely a result of changes in the expression of downstream targets of dsx (17).
Unfortunately, the previous identified direct dsx targets are all regulated by dsx in a single tissue (Yolk Protein genes in the fat body, desat-F gene in the oenocytes, and bric-à-brac 1 gene in the abdominal cuticle). Here we found that dsx directly regulated Fmo-2 in two different tissues, the midgut and the spermatheca. The dsx control of Fmo-2 transcription in the spermatheca appears to be very conserved, whereas the dsx regulation of Fmo-2 transcription in the midgut has evolved rapidly. Therefore, the Fmo-2 transcriptional regulation by dsx provides a great opportunity to investigate how such a constraint shapes the evolution of dsx regulation. By maintaining the DSX-binding site (and possibly other sequence) required for the Fmo-2 transcription in the spermatheca, and changing other sequence critical for CRM-d function, Fmo-2 transcriptional regulation in the midgut could evolve without a pleiotropic effect. Thus, we provide an example at a single transcription-factor binding-site resolution to demonstrate how a constraint posed by pleiotropic transcriptional regulation is evaded during evolution.
Experimental Procedures
Reporter Constructs and Transgenic Fly Production.
DNA sequences used to drive the tdTomato-nls (47) reporter were PCR-amplified from genomic DNA of the indicated species, and cloned into site-specific transformation vectors carrying the tdTomato-nls reporter. Additional information on the reporter construction, PCR primers, and examination of the reporter expression are provided in SI Appendix, Experimental Procedures and Table S1. All of the reporter constructs were integrated at the attP40 site.
Drosophila Species.
Fly stocks for other species (SI Appendix, Table S2) were obtained from the University of California, San Diego stock center.
Fly Stocks.
The dsx alleles used were: dsx1, dsxM+R13 (47), dsxM (39), dsx683-7058 and dsx1649-9625 (48), dsxGAL4(Δ2) (35). A wild type w+ allele on the paternal X chromosome was used to distinguish XY, dsx−/dsx− from XX, dsx−/dsx−, or XY, dsx−/+ from XX, dsx−/dsxM.
Whole-Mount Antibody Staining.
The midgut was dissected from age-matched flies (5-d-old, unless otherwise specified) and stained with anti–FMO-2 antibody (32) diluted 1:300, followed by secondary antibody Alexa Fluor 488 (or 568) goat anti-rabbit IgG (Invitrogen) diluted 1:1,000 as described previously (49) with modifications (SI Appendix, Experimental Procedures). The confocal images were quantified for the intensity of the antibody staining using ImageJ (see SI Appendix, Fig. S5 for more details on methods).
Quantitative RT-PCR.
Total RNA from 20 cardias (dissected in PBS on ice, attached with a section of ventriculus about the same length of the cardia) was isolated using TRIzol (Life Technologies). First-strand cDNA was synthesized with a SuperScript III first strand synthesis system (Life Technologies), and used as templates for PCR reactions (see SI Appendix, Experimental Procedures for detail PCR conditions and primers).
Supplementary Material
Acknowledgments
We thank Dr. Michael Scharf for the anti–FMO-2 antibody; and Troy Shirangi and members of the B.S.B. laboratory for helpful discussions and suggestions. This work was supported by the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501192112/-/DCSupplemental.
References
- 1.Salz HK. Sex determination in insects: A binary decision based on alternative splicing. Curr Opin Genet Dev. 2011;21(4):395–400. doi: 10.1016/j.gde.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Christiansen AE, Keisman EL, Ahmad SM, Baker BS. Sex comes in from the cold: The integration of sex and pattern. Trends Genet. 2002;18(10):510–516. doi: 10.1016/s0168-9525(02)02769-5. [DOI] [PubMed] [Google Scholar]
- 3.Camara N, Whitworth C, Van Doren M. The creation of sexual dimorphism in the Drosophila soma. Curr Top Dev Biol. 2008;83:65–107. doi: 10.1016/S0070-2153(08)00403-1. [DOI] [PubMed] [Google Scholar]
- 4.Robinett CC, Vaughan AG, Knapp JM, Baker BS. Sex and the single cell. II. There is a time and place for sex. PLoS Biol. 2010;8(5):e1000365. doi: 10.1371/journal.pbio.1000365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoli DS, Fan P, Fraser EJ, Shah NM. Neural control of sexually dimorphic behaviors. Curr Opin Neurobiol. 2013;23(3):330–338. doi: 10.1016/j.conb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Manoli DS, Meissner GW, Baker BS. Blueprints for behavior: Genetic specification of neural circuitry for innate behaviors. Trends Neurosci. 2006;29(8):444–451. doi: 10.1016/j.tins.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto D. The neural and genetic substrates of sexual behavior in Drosophila. Adv Genet. 2007;59:39–66. doi: 10.1016/S0065-2660(07)59002-4. [DOI] [PubMed] [Google Scholar]
- 8.Pavlou HJ, Goodwin SF. Courtship behavior in Drosophila melanogaster: Towards a ‘courtship connectome’. Curr Opin Neurobiol. 2013;23(1):76–83. doi: 10.1016/j.conb.2012.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dauwalder B. The roles of fruitless and doublesex in the control of male courtship. Int Rev Neurobiol. 2011;99:87–105. doi: 10.1016/B978-0-12-387003-2.00004-5. [DOI] [PubMed] [Google Scholar]
- 10.Siwicki KK, Kravitz EA. Fruitless, doublesex and the genetics of social behavior in Drosophila melanogaster. Curr Opin Neurobiol. 2009;19(2):200–206. doi: 10.1016/j.conb.2009.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Billeter JC, Rideout EJ, Dornan AJ, Goodwin SF. Control of male sexual behavior in Drosophila by the sex determination pathway. Curr Biol. 2006;16(17):R766–R776. doi: 10.1016/j.cub.2006.08.025. [DOI] [PubMed] [Google Scholar]
- 12.Prud’homme B, Gompel N, Carroll SB. Emerging principles of regulatory evolution. Proc Natl Acad Sci USA. 2007;104(Suppl 1):8605–8612. doi: 10.1073/pnas.0700488104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tautz D. Evolution of transcriptional regulation. Curr Opin Genet Dev. 2000;10(5):575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 14.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134(1):25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 15.Stern DL. Evolutionary developmental biology and the problem of variation. Evolution. 2000;54(4):1079–1091. doi: 10.1111/j.0014-3820.2000.tb00544.x. [DOI] [PubMed] [Google Scholar]
- 16.Cande J, Stern DL, Morita T, Prud’homme B, Gompel N. Looking under the lamp post: Neither fruitless nor doublesex has evolved to generate divergent male courtship in Drosophila. Cell Reports. 2014;8(2):363–370. doi: 10.1016/j.celrep.2014.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nat Rev Genet. 2009;10(11):797–804. doi: 10.1038/nrg2687. [DOI] [PubMed] [Google Scholar]
- 18.Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10(9):2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes Dev. 1993;7(1):42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- 20.Williams TM, et al. The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell. 2008;134(4):610–623. doi: 10.1016/j.cell.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shirangi TR, Dufour HD, Williams TM, Carroll SB. Rapid evolution of sex pheromone-producing enzyme expression in Drosophila. PLoS Biol. 2009;7(8):e1000168. doi: 10.1371/journal.pbio.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalton JE, et al. Male-specific Fruitless isoforms have different regulatory roles conferred by distinct zinc finger DNA binding domains. BMC Genomics. 2013;14:659. doi: 10.1186/1471-2164-14-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neville MC, et al. Male-specific fruitless isoforms target neurodevelopmental genes to specify a sexually dimorphic nervous system. Curr Biol. 2014;24(3):229–241. doi: 10.1016/j.cub.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo SD, Shi GW, Baker BS. Direct targets of the D. melanogaster DSXF protein and the evolution of sexual development. Development. 2011;138(13):2761–2771. doi: 10.1242/dev.065227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Suh JK, Robertus JD. Role of yeast flavin-containing monooxygenase in maintenance of thiol-disulfide redox potential. Methods Enzymol. 2002;348:113–121. doi: 10.1016/s0076-6879(02)48631-4. [DOI] [PubMed] [Google Scholar]
- 26.Stepanova AN, et al. The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell. 2011;23(11):3961–3973. doi: 10.1105/tpc.111.088047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phillips IR, Shephard EA. Flavin-containing monooxygenases: Mutations, disease and drug response. Trends Pharmacol Sci. 2008;29(6):294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 28.Motika MS, Zhang J, Cashman JR. Flavin-containing monooxygenase 3 and human disease. Expert Opin Drug Metab Toxicol. 2007;3(6):831–845. doi: 10.1517/17425255.3.6.831. [DOI] [PubMed] [Google Scholar]
- 29.Shehin-Johnson SE, Williams DE, Larsen-Su S, Stresser DM, Hines RN. Tissue-specific expression of flavin-containing monooxygenase (FMO) forms 1 and 2 in the rabbit. J Pharmacol Exp Ther. 1995;272(3):1293–1299. [PubMed] [Google Scholar]
- 30.Falls JG, Ryu DY, Cao Y, Levi PE, Hodgson E. Regulation of mouse liver flavin-containing monooxygenases 1 and 3 by sex steroids. Arch Biochem Biophys. 1997;342(2):212–223. doi: 10.1006/abbi.1997.9965. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, et al. Synchronous evolution of an odor biosynthesis pathway and behavioral response. Curr Biol. 2013;23(1):11–20. doi: 10.1016/j.cub.2012.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scharf ME, Scharf DW, Bennett GW, Pittendrigh BR. Catalytic activity and expression of two flavin-containing monooxygenases from Drosophila melanogaster. Arch Insect Biochem Physiol. 2004;57(1):28–39. doi: 10.1002/arch.20012. [DOI] [PubMed] [Google Scholar]
- 33.Chintapalli VR, Wang J, Dow JAT. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat Genet. 2007;39(6):715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 34.Buchon N, et al. Morphological and molecular characterization of adult midgut compartmentalization in Drosophila. Cell Reports. 2013;3(5):1725–1738. doi: 10.1016/j.celrep.2013.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Pan Y, Robinett CC, Baker BS. Turning males on: Activation of male courtship behavior in Drosophila melanogaster. PLoS ONE. 2011;6(6):e21144. doi: 10.1371/journal.pone.0021144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56(6):997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 37.Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc finger related DNA binding domain. EMBO J. 1993;12(2):527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garrett-Engele CM, et al. intersex, a gene required for female sexual development in Drosophila, is expressed in both sexes and functions together with doublesex to regulate terminal differentiation. Development. 2002;129(20):4661–4675. doi: 10.1242/dev.129.20.4661. [DOI] [PubMed] [Google Scholar]
- 39.Baker BS, Ridge KA. Sex and the single cell. I. On the action of major loci affecting sex determination in Drosophila melanogaster. Genetics. 1980;94(2):383–423. doi: 10.1093/genetics/94.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meiklejohn CD, Coolon JD, Hartl DL, Wittkopp PJ. The roles of cis- and trans-regulation in the evolution of regulatory incompatibilities and sexually dimorphic gene expression. Genome Res. 2014;24(1):84–95. doi: 10.1101/gr.156414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tanaka K, Barmina O, Sanders LE, Arbeitman MN, Kopp A. Evolution of sex-specific traits through changes in HOX-dependent doublesex expression. PLoS Biol. 2011;9(8):e1001131. doi: 10.1371/journal.pbio.1001131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An W, Wensink PC. Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J. 1995;14(6):1221–1230. doi: 10.1002/j.1460-2075.1995.tb07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li H, Baker BS. hermaphrodite and doublesex function both dependently and independently to control various aspects of sexual differentiation in Drosophila. Development. 1998;125(14):2641–2651. doi: 10.1242/dev.125.14.2641. [DOI] [PubMed] [Google Scholar]
- 44.Prud’homme B, et al. Repeated morphological evolution through cis-regulatory changes in a pleiotropic gene. Nature. 2006;440(7087):1050–1053. doi: 10.1038/nature04597. [DOI] [PubMed] [Google Scholar]
- 45.Wray GA, et al. The evolution of transcriptional regulation in eukaryotes. Mol Biol Evol. 2003;20(9):1377–1419. doi: 10.1093/molbev/msg140. [DOI] [PubMed] [Google Scholar]
- 46.Darwin C. The Descent of Man and Selection in Relation to Sex. John Murray; London: 1871. [Google Scholar]
- 47.Mellert DJ, Knapp JM, Manoli DS, Meissner GW, Baker BS. Midline crossing by gustatory receptor neuron axons is regulated by fruitless, doublesex and the Roundabout receptors. Development. 2010;137(2):323–332. doi: 10.1242/dev.045047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pan Y, Baker BS. Genetic identification and separation of innate and experience-dependent courtship behaviors in Drosophila. Cell. 2014;156(1-2):236–248. doi: 10.1016/j.cell.2013.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Patel NH. 1994 Imaging Neuronal subset and other cell type in whole mount Drosophila embryos and larvae using antibody probes. Drosophila melanogaster: Practical Uses in Cell Biology, Methods in Cell Biology, eds Goldstein LSB, Fyrberg E (Academic, New York), Vol 44, pp 446–487. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.