Significance
X-ray fluorescence microscopy provides unparalleled sensitivity for measuring the distribution of trace elements in many-micrometer-thick specimens, whereas ptychography offers a path to the imaging of weakly fluorescing biological ultrastructure at beyond-focusing-optic resolution. We demonstrate here for the first time, to our knowledge, the combination of fluorescence and ptychography for imaging frozen-hydrated specimens at cryogenic temperatures, with excellent structural and chemical preservation. This combined approach will have significant impact on studies of the intracellular localization of nanocomposites with attached therapeutic or diagnostic agents, help elucidate the roles of trace metals in cell development, and further the study of diseases where trace metal misregulation is suspected (including neurodegenerative diseases).
Keywords: ptychography, X-ray fluorescence microscopy, cryogenic biological samples
Abstract
Trace metals play important roles in normal and in disease-causing biological functions. X-ray fluorescence microscopy reveals trace elements with no dependence on binding affinities (unlike with visible light fluorophores) and with improved sensitivity relative to electron probes. However, X-ray fluorescence is not very sensitive for showing the light elements that comprise the majority of cellular material. Here we show that X-ray ptychography can be combined with fluorescence to image both cellular structure and trace element distribution in frozen-hydrated cells at cryogenic temperatures, with high structural and chemical fidelity. Ptychographic reconstruction algorithms deliver phase and absorption contrast images at a resolution beyond that of the illuminating lens or beam size. Using 5.2-keV X-rays, we have obtained sub–30-nm resolution structural images and ∼90-nm–resolution fluorescence images of several elements in frozen-hydrated green algae. This combined approach offers a way to study the role of trace elements in their structural context.
X-ray fluorescence microscopy (XFM) offers unparalleled sensitivity for quantitative mapping of elements, especially trace metals which play a critical role in many biological processes (1–3). It is complementary to light microscopy, which can study some elemental content in live cells (with superresolution techniques possible) but which is more difficult to quantitate because it depends on the binding affinities of fluorophores. However, XFM does not usually show much cellular ultrastructure, because the light elements (such as H, C, N, and O, which are the main constituents of biological materials) have low fluorescence yield (4). At the multi-keV X-ray energies needed to excite most X-ray fluorescence lines of interest, these light elements show little absorption contrast, but phase contrast can be used to image cellular structure (5, 6) and this can be combined with scanned-beam XFM (7–11).
One can also acquire phase-contrast X-ray images with a resolution beyond X-ray lens limits by recording the diffraction pattern from a coherently illuminated, noncrystalline sample in an approach called coherent diffraction imaging (CDI) (12). This approach has been used to image isolated dried cells (13–15), and 3-nm resolution has been achieved when imaging silver nanocubes (16). The traditional CDI approach requires that samples meet a so-called “finite support” (17) requirement with no observable scattering outside of a defined region; although some limited success has been obtained (18, 19), this finite support condition has proven difficult to achieve with single cells surrounded by ice layers. Ptychography (20–22) is a recently realized CDI method [with an older history (23)] that circumvents this isolated cell requirement by instead scanning a limited-size coherent illumination spot across the sample. Ptychography has been used to image freeze-dried diatoms at 30-nm resolution (24) and bacteria at 20-nm resolution (25), and frozen-hydrated yeast at 85-nm resolution (26), whereas ptychographic tomography has been used to image nanoporous glass to 16-nm 3D resolution (27).
The spatial resolution of ptychography can in theory reach the wavelength limit. However, radiation damage limits the achievable resolution in X-ray microscopy of hydrated biological specimens (28). A good approach to reduce beam-induced degradation of the sample is to work with frozen-hydrated biological specimens under cryogenic conditions (29, 30). Cryogenic samples can provide high-fidelity structural (31) and ionic elemental (32–34) preservation, and mitigate the effects of radiation damage (35). As a result, the combination of cryogenic sample conditions with fluorescence and ptychographic X-ray imaging can provide simultaneous views of ultrastructure and elemental compositions of specimens at high resolution.
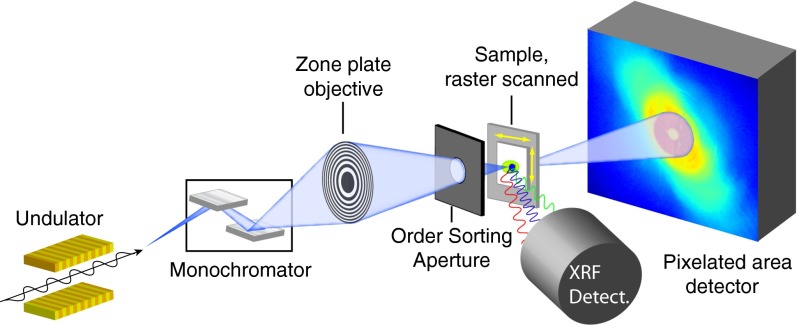
We demonstrate this combination at the Bionanoprobe (34), a hard X-ray fluorescence nanoprobe with cryogenic sample transfer capabilities at beamline 21-ID-D of the Advanced Photon Source at Argonne National Laboratory. Fig. 1 shows the schematic of the experimental layout. A monochromatic X-ray beam at 5.2-keV photon energy is focused by a Fresnel zone plate with 85-nm theoretical Rayleigh resolution onto the sample maintained at a temperature below 110 K in the -torr vacuum environment of the microscope. The fluorescence spectra and far-field diffraction patterns are recorded simultaneously as the sample is raster-scanned, yielding two independent but complementary contrast modes: the fluorescence signal provides quantitative elemental composition from all but the lightest elements, whereas the ptychographic image reconstructed from diffraction patterns shows biological structure at high resolution. The combination of fluorescence and ptychography has been demonstrated for imaging metallic test structures (36), and freeze-dried diatoms (24). We address two challenges in this combination. First, the need for single-mode coherence in traditional CDI would reduce the flux that can be used from partially coherent sources, leading to long imaging times; we have used a multimode reconstruction method (37) to allow us to open up spatially filtering apertures for higher flux (Fig. S1). Second, as the spatial resolution of fluorescence imaging is limited by the beam size, the focused beam needs to be as small as possible (24, 38) (about 80-nm FWHM for the combined probe modes in our experiment; see Fig. S2). This is different from the practice in many other ptychography experiments where a coherent beam spot of –3000 nm is scanned in a spiral pattern with overlaps, yielding a smaller number of diffraction patterns (27, 39, 40). In our case, the smaller beam size combined with the ptychographic overlap requirements leads to a larger dataset for reconstruction; we have used graphical processing units (GPUs) to significantly speed up data processing.
Fig. 1.
Schematic of the experimental layout of combined cryogenic fluorescence and ptychographic imaging. A cryogenic sample is raster-scanned through a focused X-ray beam; at each scan position, an energy-dispersive detector records the X-ray fluorescence spectrum from the sample, whereas a pixelated area detector records the far-field diffraction pattern.
Results
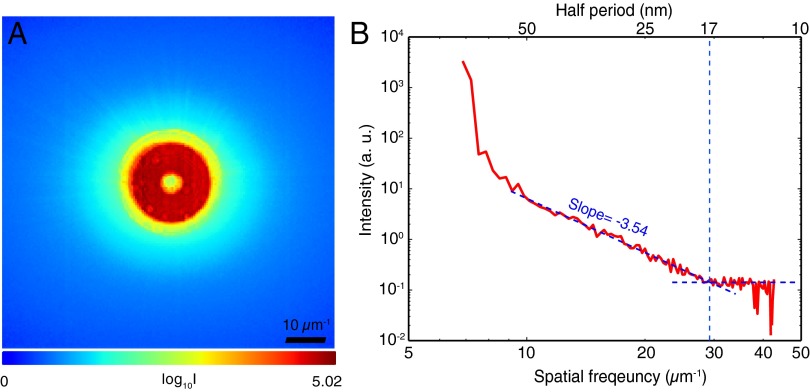
In Fig. 2A, we show the ptychographic data from a frozen-hydrated alga (Ostreococcus sp.), which is the average of far-field diffraction patterns from the scan points within the cell region. The diffraction pattern is dominated by a red annulus, which is the far-field diffraction pattern of a Fresnel zone plate with a central stop. Scattering outside of the annulus is due primarily to diffraction from the alga. Fig. 2B shows the azimuthally averaged power spectrum of this average diffraction pattern after subtracting the average diffraction measured with no sample present; this power spectrum shows that the signal S declines with spatial frequency f in a power-law relationship before reaching a noise floor at high spatial frequencies. This signal decrease is consistent with what is observed in model objects (41), in scanning X-ray microscope images (42, 43), and in other CDI and ptychography experiments (14, 25). The spatial resolution of the ptychography reconstruction cannot exceed that corresponding to the maximum spatial frequency at which photons are distinguishable from noise, which in this case is at 29 μm−1 with a corresponding half-period size of 17 nm.
Fig. 2.
View of the ptychographic data produced by X-ray diffraction from a frozen-hydrated Ostreococcus alga, acquired at 5.2-keV photon energy. (A) The average (logarithmic scale) of 4,000 far-field diffraction pattern recordings from the cell region within the 100 × 100-point scan; the high-intensity red annulus represents the incident beam focused by a Fresnel zone plate with a central stop, whereas the light blue areas represent significant scattering from the cell (the speckles of individual diffraction patterns are not visible in this summed image). By subtracting the diffraction pattern measured with no sample present, and carrying out an azimuthal average, one arrives at B, which is a plot of diffraction from the alga as a function of spatial frequency f. The diffraction intensity signal S decreases as until, at a length scale with a half-period of about 17 nm, it reaches a flat region consistent with spatially uncorrelated noise fluctuations.
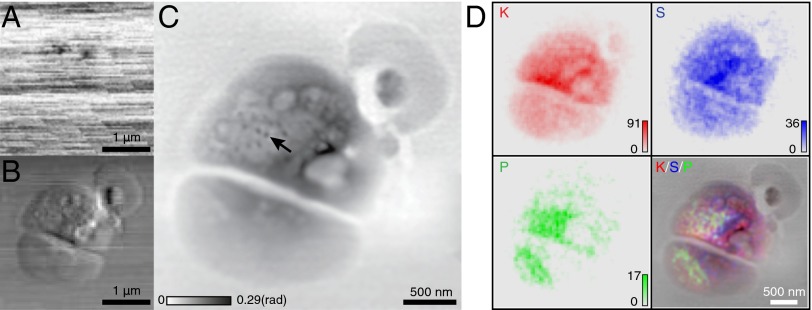
Ptychography allows for the recovery of both the probe and the complex object transmission function (22). This quantifies the X-ray phase shift and absorption due to the object [although phase contrast is much stronger than absorption contrast for hard X-ray imaging of light elements (5, 6)]. The enhanced resolution given by ptychography exceeds the lens resolution limit, unlike that of conventional X-ray phase contrast microscopy. To illustrate this, absorption contrast and horizontal differential phase contrast (DPC) images were extracted from the collected diffraction patterns (22, 44), which are shown in Fig. 3 A and B, respectively. Because light elements are largely transparent to hard X-rays, there are few visible features of the alga in the absorption contrast image, which is dominated by imperfectly normalized incident flux variations. The alga is visible in DPC; however, the resolution of the image is limited by the focused beam size to be no better than the Rayleigh resolution of 85 nm. Ptychography reconstructions were carried out using the extended ptychographic iterative engine (ePIE) algorithm (45), with scan-line to scan-line flux variations (the horizontal lines in Fig. 3A) normalized out by setting the outside-of-cell area to have constant signal values. With a partially coherent source, tradeoffs between coherence degree and total flux of the beam must be considered. We chose to work with higher flux and instead reconstruct several mutually incoherent probe modes along with the object (Fig. S1). The reconstruction details are provided in Materials and Methods. Fig. 3C shows the phase of the cell complex transmission function reconstructed via ptychography. More structural details are shown in this phase image. The dark spots (indicated by an arrow), visible only in the ptychographic image, are suggestive of ribosome-like complexes observed in cryo electron microscopy studies of similar algae (46). The fluorescence maps (Fig. 3D) show the elemental distributions of K, S, and P within the sample, which can be excited with the 5.2-keV beam used in this experiment (beam energies of 10 keV are commonly used in X-ray fluorescence experiments to excite a broader range of elemental signals, but for this first demonstration we emphasized improved contrast in ptychographic images). The presence of potassium (K) within the cell indicates good preservation of membrane integrity in the frozen-hydrated cell preparation. Power spectrum analysis of the fluorescence images shows a rolloff to noise at a half-period of about 90 nm (Fig. S3), which is consistent with the 85-nm calculated Rayleigh resolution of the X-ray probe beam.
Fig. 3.
Images of a frozen-hydrated Ostreococcus alga obtained from 100 × 100-point scan data. (A) Absorption contrast image of the alga, obtained from the total signal recorded on the pixelated area detector at each scan point. (B) Differential phase contrast image in the horizontal direction, obtained by plotting the first moment of the diffraction patterns as a function of position. (C) Phase of the sample complex transmission function reconstructed via ptychography. The arrow points to structures that resemble ribosome-like complexes observed in cryo electron microscopy studies of similar algae (46). (D) X-ray fluorescence maps of the distributions of the elements K, S, and P, along with their color-composite overlay on the ptychographic image C. For the fluorescence images, the numbers of X-ray photons recorded per pixel dwell time are shown as “counts.” The presence of K within the cell suggests good preservation of membrane integrity in the frozen-hydrated sample preparation.
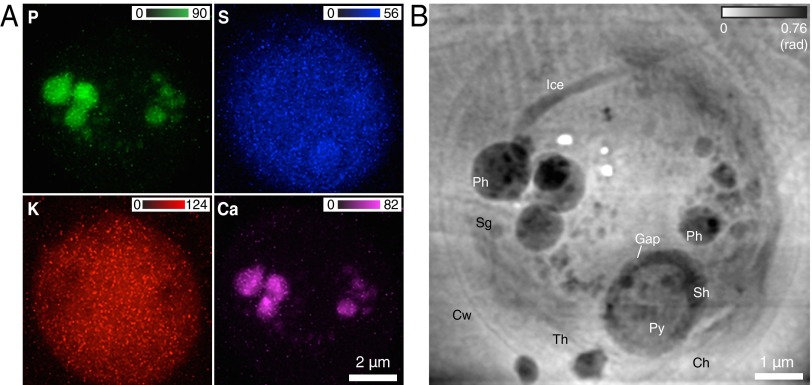
We have also imaged the unicellular green algae Chlamydomonas reinhardtii, which serves as a model organism for molecular genetic studies (47, 48). Fig. 4A shows fluorescence maps of the elements P, S, K, and Ca (where the K map again shows good membrane containment of diffusible ions), whereas Fig. 4B shows the phase of the cell transmission function which is reflective of light elements comprising the main cellular structures. The contrast and visibility of cellular structures in this image is not dissimilar from Chlamydomonas images obtained using soft X-ray cryo microscopy (29). The pyrenoid in the chloroplast, highlighted slightly in the S fluorescence image, can be easily identified in the ptychographic image. A starch sheath with gaps between starch plates is formed at the periphery of the pyrenoid (49, 50). Several electron-dense spherical structures in the ptychographic image show large amounts of P and Ca in fluorescence maps, presumably representing polyphosphate bodies that contain polyphosphate complexed with calcium (51). Structural and elemental features of polyphosphate bodies have been studied by electron microscopy and X-ray microanalysis, respectively (51, 52), whereas both of these are simultaneously revealed in our study.
Fig. 4.
Fluorescence maps and ptychographic image of a frozen-hydrated C. reinhardtii alga obtained from 167 × 151-point scan data. (A) Elemental distributions of P, S, K, and Ca within the cell. (B) Phase image reconstructed via ptychography. A number of organelles can be identified inside the cell wall (Cw): polyphosphate bodies (Ph), pyrenoid (Py), thylakoids (Th), chloroplast (Ch), starch granule (Sg), and starch sheath (Sh). Some ice changes can be seen in the top half of the cell due to a malfunction of a cryogenic component during the second half of the scan (Materials and Methods). One can also see three white spots where, due to glitches in the scan control, the X-ray beam was allowed to dwell for a long time, thus leading to “beam burn,” which has been observed before in frozen-hydrated specimens at cryogenic temperatures (see, e.g., figure 8 of ref. 30).
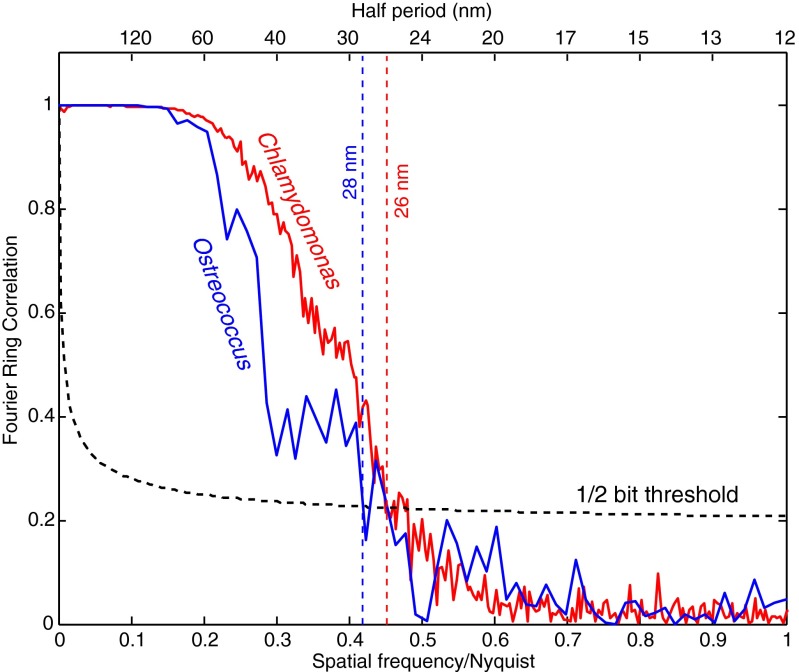
Several methods have been used to estimate the spatial resolution of images obtained from coherent diffraction patterns. For traditional CDI involving a single diffraction pattern, the phase retrieval transfer function (PRTF) (53) [also called the intensity ratio (14)] is a commonly used method to assess resolution by finding the finest periodicity at which phases are consistent in iterative phase retrieval (Fig. S4). However, the diffraction patterns in ptychography have contributions from two unknown but entangled functions (object and probe, with the probe function being stronger and with possible variations between illumination spots), and this complicates PRTF analysis of the reconstructed object (40). Here, we have estimated the spatial resolution of the reconstructed phase image using Fourier ring correlation (FRC) (54, 55), which is a well-established approach to measure resolution in cryo-electron microscopy. Because two independent datasets are needed for FRC analysis, each ptychographic dataset was split into two subsets which were then reconstructed [Fig. S5; this resulted in fewer measurements and less overlap between illumination spots in one reconstruction, which is known to reduce the resolution and fidelity of the reconstructed image (56)]. The FRC was computed between these two independent reconstructed images (details of the FRC analysis are provided in SI Text). Fig. 5 shows the computed FRC as a function of spatial frequency. The 1/2-bit threshold criterion (55) was used to quantitatively estimate the image resolution from the FRC. The intersection between the FRC and 1/2-bit threshold indicates a spatial resolution of 28 nm in the Ostreococcus image and 26 nm in the Chlamydomonas image. This provides a conservative estimate of the spatial resolution achieved in that the images obtained of the full datasets show somewhat greater detail than either of the split-data images, and FRC measurements of full-data reconstructions obtained from different random-phase starts of the ptychographic reconstruction algorithm also suggest higher resolution (SI Text).
Fig. 5.
Resolution estimation of the ptychographic phase images of Ostreococcus (Fig. 3) and Chlamydomonas (Fig. 4) by FRC. In each case, the overlapping beam spot data were divided into two separate sets, with each set reconstructed to yield two images of the same object from independent data. This approach leads to lower resolution images than one obtains by using the full dataset, but it provides a conservative estimate of the spatial resolution. FRC measures the phase correlation of the Fourier transforms of the two images at various spatial frequency ranges, or length scales.
Discussion
The work reported here demonstrates a biological sample imaging method for frozen-hydrated cells by combining cryogenic fluorescence and ptychographic X-ray imaging techniques. By rapidly freezing the sample and maintaining it at cryogenic conditions, its structural and elemental information can be better preserved while radiation damage is minimized. The fluorescence images show some elemental composition within the sample, whereas ptychography visualizes the light-element biological structure at high resolution. By setting the elemental signals in the same context with high-resolution biological structures given by ptychography, the analysis of trace elements can be improved. Although we show here 2D images, both ptychography (39) and fluorescence (57) are compatible with tomographic approaches to nanoscale 3D imaging of biological materials. Compared with “water window” soft X-ray tomography (30, 58, 59), tomography with multi-keV X-rays offers the capability for increased sample thickness, and increased depth of focus for a given spatial resolution, which allows this combined method to image larger cells and tissue sections in 3D. This method will aid the interpretation of studies of the localization of nanoparticles attached to therapeutic agents (1, 60), or the role of metals in cell development (61), and in diseases where trace metal misregulation is implicated as a cause (62). One could of course carry out separate experiments where one first does a fluorescence scan with a finely focused beam, followed by a separate ptychographic imaging experiment with a larger beam spot and fewer illumination points; this would however involve a longer time for the experiment if done in one instrument, or risk of specimen frosting if one instead transferred the cryo sample to a separate instrument. It would also involve a higher radiation dose, because the sample would have to be exposed twice to collect the signals separately.
Materials and Methods
Frozen-Hydrated Sample Preparation.
Ostreococcus algae were purchased from Provasoli-Guillard National Center for Marine Algae and Microbiota at Bigelow Laboratory for Ocean Sciences (cat #CCMP2407). C. reinhardtii [American Type Culture Collection (ATCC) No. 18798] were grown mixotrophically in a tris-acetate-phosphate medium at 296 K on a rotary shaker (100 rpm). Five microliters of fresh cell suspensions were dispersed on Si3N4 windows (200-nm thick, 1.5 × 1.5-mm membrane area) and incubated for 10 min in a humidified Petri dish. The windows were then mounted and plunge frozen in liquid nitrogen cooled liquid ethane using FEI Vitrobot Mark IV plunge freezer following the manufacturer’s instructions with the chamber set at 20 °C, 100% humidity, a blot time of 2 s, a blot offset of 0 mm, and a blot total of 1 to reduce the overlying water layer to about 1-μm thickness. The frozen-hydrated cells on Si3N4 windows were then transferred from liquid ethane to liquid nitrogen and observed under a cryogenic light microscope (Nikon 50i light fluorescent microscope with a N.A. = 0.45 CFI Super Plan Fluor ELWD (extra long working distance) objective, equipped with an Instec CLM77K cryo stage) at −170 °C (Fig. S6). Isolated single cells were chosen for investigation.
Ptychography and Fluorescence Experiments.
Ptychography and fluorescence scanning experiments were conducted at the Bionanoprobe at the 21-ID-D beamline of the Advanced Photon Source, Argonne National Laboratory. A beam energy of keV was used to maximize the partially coherent flux (because coherent flux scales as source brightness divided by ), and ptychographic image contrast. A partially coherent portion of the incident beam was selected by movable slits ∼27 m from the undulator source. The transverse coherence length was governed by the slit width of 50 μm. Longitudinal coherence was determined by the bandwidth of of the double-crystal Si monochromator. The partially coherent beam was focused by a 160-μm-diameter Fresnel zone plate with an outermost zone width of 70 nm. The focused flux was of the order of photons/s. The estimated radiation dose imparted to the specimen with 3-s exposure times per (40 nm)2 pixel was about Gy, which is above the -Gy dose at which atom-to-atom spatial correlations are significantly reduced as observed in X-ray diffraction patterns (63), but well below the dose (which we estimate to be about Gy) used for single metal atom detection in electron microscopy (64). In other experiments using cryo X-ray microscopy, no significant mass loss or redistribution has been observed at Gy doses at 30–100-nm length scales when studying frozen-hydrated specimens at cryogenic temperatures (29, 65). The sample was kept in the Bionanoprobe high-vacuum ( torr) chamber at cryogenic temperature below 110 K. A collimated four-element silicon drift detector (Vortex-ME4, Hitachi High-Technologies Science America) with a total active area of 170 mm2 and maximum solid angle acceptance of about 0.65 steradians was mounted at 90° with regard to the incident X-ray beam to collect fluorescence signals. A Dectris Pilatus 100K photon-counting hybrid pixel array detector was positioned 2.2 m from the sample to record diffraction patterns. Frozen-hydrated samples were raster-scanned using a step size of 40 nm: with positioning and computer overheads, the total scanning time for an Ostreococcus alga (100 × 100 scan grid, 3-s exposure time) was about 10 h, and was about 6.5 h for a Chlamydomonas alga (167 × 151 scan grid, 0.5-s exposure time). One thing to note is that the cold shield (which surrounds the sample to maintain a stable temperature) suffered a mechanical problem during the second half of the Chlamydomonas scan; as a result, the sample chuck suffered a temperature increase to 143 K, leading to some changes in ice condition in the top half of Fig. 4B (scan from the bottom to the top).
Reconstructions.
The diffraction data from Ostreococcus alga were divided into two subsets (and Chlamydomonas into four subsets) because of the memory capacity of the graphical processing unit (Tesla M2050 GPU) used. Each subset was reconstructed using the ePIE algorithm (45) with the multimode modification proposed by Thibault and Menzel (37) with GPU parallel programming. The probe function was set as a superposition of eight probe modes that were individually fully coherent but mutually incoherent. Each reconstruction ran for 500 iterations with the probe updating beginning from the 10th iteration. The reconstructed phases from the subdatasets were set to a matching global phase shift and then recombined into one image. The FRC was computed between two independent ePIE reconstructions from separated datasets (SI Text). Before computing the FRC, the two phase images were aligned with subpixel precision using an efficient image method based on cross-correlation (66) and then were multiplied by a soft-edged mask (Tukey window) to avoid correlation from the image field boundaries. See Nashed et al. (67).
Supplementary Material
Acknowledgments
We thank R. Mak and M. Guizar-Sicairos for valuable discussions, and K. Brister, C. Roehrig, J. VonOsinkski, and M. Bolbat for help during the experiments. We thank NIH National Institute of General Medical Sciences for support of this work under Grant 1R01GM104530. The Bionanoprobe is funded by NIH/National Center for Research Resources High End Instrumentation Grant 1S10RR029272-01 as part of the American Recovery and Reinvestment Act. Use of the Advanced Photon Source, an Office of Science User Facility operated for the US Department of Energy (DOE) Office of Science by Argonne National Laboratory, was supported by the US DOE under Contract DE-AC02-06CH11357.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1413003112/-/DCSupplemental.
References
- 1.Paunesku T, Vogt S, Maser J, Lai B, Woloschak G. X-ray fluorescence microprobe imaging in biology and medicine. J Cell Biochem. 2006;99(6):1489–1502. doi: 10.1002/jcb.21047. [DOI] [PubMed] [Google Scholar]
- 2.Fahrni CJ. Biological applications of X-ray fluorescence microscopy: Exploring the subcellular topography and speciation of transition metals. Curr Opin Chem Biol. 2007;11(2):121–127. doi: 10.1016/j.cbpa.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 3.Majumdar S, et al. Applications of synchrotron μ-XRF to study the distribution of biologically important elements in different environmental matrices: A review. Anal Chim Acta. 2012;755:1–16. doi: 10.1016/j.aca.2012.09.050. [DOI] [PubMed] [Google Scholar]
- 4.Krause M. Atomic radiative and radiationless yields for K and L shells. J Phys Chem Ref Data. 1979;8(2):307–327. [Google Scholar]
- 5.Schmahl G, Rudolph D. In: Proposal for a Phase Contrast X-ray Microscope. Cheng PC, Jan GJ, editors. Springer; Berlin: 1987. pp. 231–238. [Google Scholar]
- 6.Davis TJ, Gao D, Gureyev TE, Stevenson AW, Wilkins SW. Phase-contrast imaging of weakly absorbing materials using hard X-rays. Nature. 1995;373:595–598. [Google Scholar]
- 7.Kaulich B, et al. Diffracting aperture based differential phase contrast for scanning X-ray microscopy. Opt Express. 2002;10(20):1111–1117. doi: 10.1364/oe.10.001111. [DOI] [PubMed] [Google Scholar]
- 8.Hornberger B, et al. Differential phase contrast with a segmented detector in a scanning X-ray microprobe. J Synchrotron Radiat. 2008;15(4):355–362. doi: 10.1107/S0909049508008509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bleuet P, et al. A hard x-ray nanoprobe for scanning and projection nanotomography. Rev Sci Instrum. 2009;80(5):056101. doi: 10.1063/1.3117489. [DOI] [PubMed] [Google Scholar]
- 10.Holzner C, et al. Zernike phase contrast in scanning microscopy with X-rays. Nat Phys. 2010;6(11):883–887. doi: 10.1038/nphys1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kosior E, et al. Combined use of hard X-ray phase contrast imaging and X-ray fluorescence microscopy for sub-cellular metal quantification. J Struct Biol. 2012;177(2):239–247. doi: 10.1016/j.jsb.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Miao J, Charalambous P, Kirz J, Sayre D. An extension of the methods of X-ray crystallography to allow imaging of micron-size non-crystalline specimens. Nature. 1999;400(6742):342–344. [Google Scholar]
- 13.Miao J, et al. Imaging whole Escherichia coli bacteria by using single-particle x-ray diffraction. Proc Natl Acad Sci USA. 2003;100(1):110–112. doi: 10.1073/pnas.232691299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shapiro D, et al. Biological imaging by soft x-ray diffraction microscopy. Proc Natl Acad Sci USA. 2005;102(43):15343–15346. doi: 10.1073/pnas.0503305102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson J, et al. High-resolution x-ray diffraction microscopy of specifically labeled yeast cells. Proc Natl Acad Sci USA. 2010;107(16):7235–7239. doi: 10.1073/pnas.0910874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takahashi Y, et al. High-resolution diffraction microscopy using the plane-wave field of a nearly diffraction limited focused X-ray beam. Phys Rev B. 2009;80:054103. [Google Scholar]
- 17.Fienup JR. Reconstruction of an object from the modulus of its Fourier transform. Opt Lett. 1978;3(1):27–29. doi: 10.1364/ol.3.000027. [DOI] [PubMed] [Google Scholar]
- 18.Huang X, et al. Soft X-ray diffraction microscopy of a frozen hydrated yeast cell. Phys Rev Lett. 2009;103(19):198101. doi: 10.1103/PhysRevLett.103.198101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima E, et al. Cryogenic X-ray diffraction microscopy for biological samples. Phys Rev Lett. 2009;103(19):198102. doi: 10.1103/PhysRevLett.103.198102. [DOI] [PubMed] [Google Scholar]
- 20.Faulkner HML, Rodenburg JM. Movable aperture lensless transmission microscopy: A novel phase retrieval algorithm. Phys Rev Lett. 2004;93(2):023903. doi: 10.1103/PhysRevLett.93.023903. [DOI] [PubMed] [Google Scholar]
- 21.Rodenburg JM, et al. Hard-x-ray lensless imaging of extended objects. Phys Rev Lett. 2007;98(3):034801. doi: 10.1103/PhysRevLett.98.034801. [DOI] [PubMed] [Google Scholar]
- 22.Thibault P, et al. High-resolution scanning x-ray diffraction microscopy. Science. 2008;321(5887):379–382. doi: 10.1126/science.1158573. [DOI] [PubMed] [Google Scholar]
- 23.Hoppe W. Beugung im Inhomogenen Primärstrahlwellenfeld. I. Prinzip einer Phasenmessung. Acta Crystallogr A. 1969;25:495–501. [Google Scholar]
- 24.Vine DJ, et al. Simultaneous X-ray fluorescence and ptychographic microscopy of Cyclotella meneghiniana. Opt Express. 2012;20(16):18287–18296. doi: 10.1364/OE.20.018287. [DOI] [PubMed] [Google Scholar]
- 25.Wilke RN, et al. Hard X-ray imaging of bacterial cells: Nano-diffraction and ptychographic reconstruction. Opt Express. 2012;20(17):19232–19254. doi: 10.1364/OE.20.019232. [DOI] [PubMed] [Google Scholar]
- 26.Lima E, et al. Cryo-scanning X-ray diffraction microscopy of frozen-hydrated yeast. J Microsc. 2012;249(1):1–7. [Google Scholar]
- 27.Holler M, et al. X-ray ptychographic computed tomography at 16 nm isotropic 3D resolution. Sci Rep. 2014;4(3857):1–5. doi: 10.1038/srep03857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kirz J, Jacobsen C, Howells M. Soft X-ray microscopes and their biological applications. Q Rev Biophys. 1995;28(1):33–130. doi: 10.1017/s0033583500003139. [DOI] [PubMed] [Google Scholar]
- 29.Schneider G. Cryo X-ray microscopy with high spatial resolution in amplitude and phase contrast. Ultramicroscopy. 1998;75(2):85–104. doi: 10.1016/s0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- 30.Maser J, et al. Soft X-ray microscopy with a cryo scanning transmission X-ray microscope: I. Instrumentation, imaging and spectroscopy. J Microsc. 2000;197(Pt 1):68–79. doi: 10.1046/j.1365-2818.2000.00630.x. [DOI] [PubMed] [Google Scholar]
- 31.Steinbrecht RA, Zierold K, editors. Cryotechniques in Biological Electron Microscopy. Springer; Berlin: 1987. [Google Scholar]
- 32.Matsuyama S, et al. Elemental mapping of frozen-hydrated cells with cryo-scanning X-ray fluorescence microscopy. XRay Spectrom. 2010;39:260–266. [Google Scholar]
- 33.Bohic S, et al. Biomedical applications of the ESRF synchrotron-based microspectroscopy platform. J Struct Biol. 2012;177(2):248–258. doi: 10.1016/j.jsb.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Chen S, et al. The Bionanoprobe: Hard X-ray fluorescence nanoprobe with cryogenic capabilities. J Synchrotron Radiat. 2014;21(Pt 1):66–75. doi: 10.1107/S1600577513029676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dubochet J, et al. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys. 1988;21(2):129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 36.Schropp A, et al. Hard X-ray nanobeam characterization by coherent diffraction microscopy. Appl Phys Lett. 2010;96:091102. [Google Scholar]
- 37.Thibault P, Menzel A. Reconstructing state mixtures from diffraction measurements. Nature. 2013;494(7435):68–71. doi: 10.1038/nature11806. [DOI] [PubMed] [Google Scholar]
- 38.Schropp A, et al. Hard X-ray scanning microscopy with coherent radiation: Beyond the resolution of conventional X-ray microscopes. Appl Phys Lett. 2012;100:253112. [Google Scholar]
- 39.Dierolf M, et al. Ptychographic X-ray computed tomography at the nanoscale. Nature. 2010;467(7314):436–439. doi: 10.1038/nature09419. [DOI] [PubMed] [Google Scholar]
- 40.Vila-Comamala J, et al. Characterization of high-resolution diffractive X-ray optics by ptychographic coherent diffractive imaging. Opt Express. 2011;19(22):21333–21344. doi: 10.1364/OE.19.021333. [DOI] [PubMed] [Google Scholar]
- 41.Huang X, et al. Signal-to-noise and radiation exposure considerations in conventional and diffraction x-ray microscopy. Opt Express. 2009;17(16):13541–13553. doi: 10.1364/oe.17.013541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jacobsen C, et al. Diffraction-limited imaging in a scanning transmission X-ray microscope. Opt Commun. 1991;86:351–364. [Google Scholar]
- 43.Howells MR, et al. An assessment of the resolution limitation due to radiation-damage in x-ray diffraction microscopy. J Electron Spectrosc Relat Phenom. 2009;170(1-3):4–12. doi: 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thibault P, et al. Contrast mechanisms in scanning transmission X-ray microscopy. Phys Rev A. 2009;80:043813. [Google Scholar]
- 45.Maiden AM, Rodenburg JM. An improved ptychographical phase retrieval algorithm for diffractive imaging. Ultramicroscopy. 2009;109(10):1256–1262. doi: 10.1016/j.ultramic.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 46.Henderson GP, Gan L, Jensen GJ. 3-D ultrastructure of O. tauri: Electron cryotomography of an entire eukaryotic cell. PLoS ONE. 2007;2(8):e749. doi: 10.1371/journal.pone.0000749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harris EH. Chlamydomonas as a model organism. Annu Rev Plant Physiol Plant Mol Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 48.Merchant SS, et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science. 2007;318(5848):245–250. doi: 10.1126/science.1143609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sager R, Palade GE. Structure and development of the chloroplast in Chlamydomonas. I. The normal green cell. J Biophys Biochem Cytol. 1957;3(3):463–488. doi: 10.1083/jcb.3.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rawat M, Henk MC, Lavigne LL, Moroney JV. Chlamydomonas reinhardtii mutants without ribulose-1,5-bisphosphatecarboxylase-oxygenase lack a detectable pyrenoid. Planta. 1996;198:263–270. [Google Scholar]
- 51.Komine Y, Eggink LL, Park H, Hoober JK. Vacuolar granules in Chlamydomonas reinhardtii: Polyphosphate and a 70-kDa polypeptide as major components. Planta. 2000;210(6):897–905. doi: 10.1007/s004250050695. [DOI] [PubMed] [Google Scholar]
- 52.Ruiz FA, Marchesini N, Seufferheld M, Govindjee, Docampo R. The polyphosphate bodies of Chlamydomonas reinhardtii possess a proton-pumping pyrophosphatase and are similar to acidocalcisomes. J Biol Chem. 2001;276(49):46196–46203. doi: 10.1074/jbc.M105268200. [DOI] [PubMed] [Google Scholar]
- 53.Chapman HN, et al. High-resolution ab initio three-dimensional x-ray diffraction microscopy. J Opt Soc Am A Opt Image Sci Vis. 2006;23(5):1179–1200. doi: 10.1364/josaa.23.001179. [DOI] [PubMed] [Google Scholar]
- 54.Saxton WO, Baumeister W. The correlation averaging of a regularly arranged bacterial cell envelope protein. J Microsc. 1982;127(2):127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 55.van Heel M, Schatz M. Fourier shell correlation threshold criteria. J Struct Biol. 2005;151(3):250–262. doi: 10.1016/j.jsb.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 56.Bunk O, et al. Influence of the overlap parameter on the convergence of the ptychographical iterative engine. Ultramicroscopy. 2008;108(5):481–487. doi: 10.1016/j.ultramic.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 57.de Jonge MD, et al. Quantitative 3D elemental microtomography of Cyclotella meneghiniana at 400-nm resolution. Proc Natl Acad Sci USA. 2010;107(36):15676–15680. doi: 10.1073/pnas.1001469107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schneider G, et al. Computed tomography of cryogenic cells. Surf Rev Lett. 2002;9:177–183. [Google Scholar]
- 59.Larabell CA, Le Gros MA. X-ray tomography generates 3-D reconstructions of the yeast, saccharomyces cerevisiae, at 60-nm resolution. Mol Biol Cell. 2004;15(3):957–962. doi: 10.1091/mbc.E03-07-0522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yuan Y, et al. Epidermal growth factor receptor targeted nuclear delivery and high-resolution whole cell X-ray imaging of Fe3O4@TiO2 nanoparticles in cancer cells. ACS Nano. 2013;7(12):10502–10517. doi: 10.1021/nn4033294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim AM, Vogt S, O’Halloran TV, Woodruff TK. Zinc availability regulates exit from meiosis in maturing mammalian oocytes. Nat Chem Biol. 2010;6(9):674–681. doi: 10.1038/nchembio.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ralle M, et al. Wilson disease at a single cell level: Intracellular copper trafficking activates compartment-specific responses in hepatocytes. J Biol Chem. 2010;285(40):30875–30883. doi: 10.1074/jbc.M110.114447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owen RL, Rudiño-Piñera E, Garman EF. Experimental determination of the radiation dose limit for cryocooled protein crystals. Proc Natl Acad Sci USA. 2006;103(13):4912–4917. doi: 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leapman RD. Detecting single atoms of calcium and iron in biological structures by electron energy-loss spectrum-imaging. J Microsc. 2003;210(Pt 1):5–15. doi: 10.1046/j.1365-2818.2003.01173.x. [DOI] [PubMed] [Google Scholar]
- 65.Beetz T, Jacobsen C. Soft X-ray radiation-damage studies in PMMA using a cryo-STXM. J Synchrotron Radiat. 2003;10(Pt 3):280–283. doi: 10.1107/s0909049503003261. [DOI] [PubMed] [Google Scholar]
- 66.Guizar-Sicairos M, Thurman ST, Fienup JR. Efficient subpixel image registration algorithms. Opt Lett. 2008;33(2):156–158. doi: 10.1364/ol.33.000156. [DOI] [PubMed] [Google Scholar]
- 67.Nashed Y, et al. Parallel ptychographic reconstruction. Optics Express. 2014;22(26):32082–32097. doi: 10.1364/OE.22.032082. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.