Significance
G-protein signaling contributes to nearly all aspects of cell physiology. In order for signaling to occur, the G-protein αβγ heterotrimer must be assembled from its individual subunits. The G-protein βγ subcomplex does not assemble spontaneously, but requires the protein-folding chaperone cytosolic chaperonin containing TCP-1 (CCT) and its cochaperone phosducin-like protein 1 (PhLP1). To understand the molecular mechanism underlying this process, we determined the structures of two key intermediates in Gβγ assembly, Gβ-CCT and PhLP1–Gβ-CCT, using cryo-electron microscopy and chemical cross-linking. These structures reveal the molecular basis for chaperone-mediated Gβγ assembly that can be exploited to design novel therapeutics to regulate G-protein signaling.
Keywords: G-protein, chaperonin, phosducin-like protein, electron cryo-microscopy, cross-linking
Abstract
G-protein signaling depends on the ability of the individual subunits of the G-protein heterotrimer to assemble into a functional complex. Formation of the G-protein βγ (Gβγ) dimer is particularly challenging because it is an obligate dimer in which the individual subunits are unstable on their own. Recent studies have revealed an intricate chaperone system that brings Gβ and Gγ together. This system includes cytosolic chaperonin containing TCP-1 (CCT; also called TRiC) and its cochaperone phosducin-like protein 1 (PhLP1). Two key intermediates in the Gβγ assembly process, the Gβ-CCT and the PhLP1–Gβ-CCT complexes, were isolated and analyzed by a hybrid structural approach using cryo-electron microscopy, chemical cross-linking coupled with mass spectrometry, and unnatural amino acid cross-linking. The structures show that Gβ interacts with CCT in a near-native state through interactions of the Gγ-binding region of Gβ with the CCTγ subunit. PhLP1 binding stabilizes the Gβ fold, disrupting interactions with CCT and releasing a PhLP1–Gβ dimer for assembly with Gγ. This view provides unique insight into the interplay between CCT and a cochaperone to orchestrate the folding of a protein substrate.
Cells detect and respond to a myriad of extracellular signals via G-protein signaling pathways. G proteins form complexes consisting of Gα, Gβ, and Gγ subunits that play a key role in propagating signals between activated receptors and downstream effectors (1). To perform this role, the G-protein heterotrimer must be assembled from its nascent polypeptides. A critical step in this process is the formation of the Gβγ dimer (2). Gβγ is an obligate dimer in which the individual subunits cannot fold into a stable structure on their own, but require the molecular chaperone cytosolic chaperonin containing tailless complex polypeptide 1 (CCT; also called TRiC) and its cochaperone phosducin-like protein 1 (PhLP1) (3, 4).
CCT is a member of the group II chaperonin family found in eukaryotes. It is a large protein-folding machine, made up of eight homologous subunits that assemble to form a double-ring structure, with each ring encompassing a central cavity. Nascent polypeptides and denatured proteins bind these cavities and are thereby sequestered from the other proteins in the cytosol and protected from aggregation (5). Each of the eight CCT subunits is an ATPase, and the binding and hydrolysis of ATP generates a conformational change in CCT that encapsulates the protein and assists in its folding (6–8). An important class of CCT substrates is WD40 repeat proteins that form β-propeller structures (9). One of these WD40 substrates is Gβ, which forms a seven-bladed β-propeller (10) with the assistance of CCT (3). However, unlike other CCT substrates, Gβ cannot achieve its native structure and release from CCT on its own, but requires the help of PhLP1 (3, 4). PhLP1 triggers the release of Gβ from CCT, allowing Gβ to interact with Gγ and form the Gβγ dimer (3, 4, 11).
Given their vital roles in Gβγ assembly, it is important to understand at the molecular level how CCT and PhLP1 mediate Gβ folding. To achieve this objective, we have isolated two key intermediates in the Gβγ assembly process, the Gβ-CCT complex and the PhLP1–Gβ-CCT complex, and examined their structures by cryo-electron microscopy (cryo-EM), site-specific chemical cross-linking using unnatural amino acids, and chemical cross-linking coupled with mass-spectrometric identification of the cross-links (XL-MS). This analysis reveals a complex molecular mechanism by which CCT and PhLP1 fold Gβ and assist in Gβγ assembly.
Results
Cryo-EM Structure of the Gβ-CCT Complex.
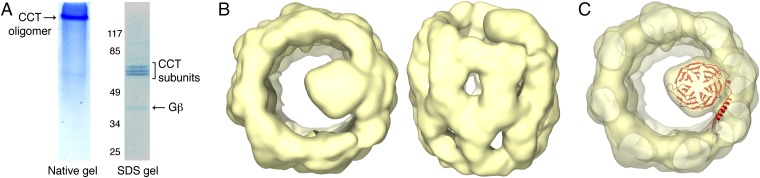
To isolate the Gβ-CCT complex for structural studies, we expressed human Gβ1 in insect cells and purified the complex between Gβ and the endogenous insect CCT using an affinity capture approach. The purified product was analyzed by native and SDS gel electrophoresis (Fig. 1A), which revealed bands of ∼95% purity migrating at the correct molecular weights for CCT subunits and Gβ. The presence of CCT subunits and Gβ in the complex was confirmed by immunoblotting (Fig. S1). The complex was purified in the absence of exogenous ATP and is thus in a nucleotide-free conformation.
Fig. 1.
Cryo-EM structure of the Gβ-CCT complex. (A) Electrophoretic analysis of the purified Gβ-CCT complex by native gel and SDS/PAGE. (B) Top and side views of the reconstructed Gβ-CCT complex at 19-Å resolution. (C) Docking of Gβ into the Gβ-CCT cryo-EM structure. The atomic structure of Gβ [red; Protein Data Bank (PDB) ID code 2TRC (19)] was docked into the mass attributable to Gβ in the cryo-EM reconstruction. The orientation of Gβ was chosen only to show the fit in the cryo-EM mass.
A cryo-EM analysis of the purified Gβ-CCT was performed to determine its molecular structure. Solutions containing Gβ-CCT were vitrified and imaged by cryo-EM. Particle classification of a total of 23,288 particles yielded 17,521 homogenous particles that were used to generate a 19-Å 3D reconstruction of the Gβ-CCT complex without imposing any symmetry throughout the reconstruction procedure (Fig. S2). The reconstruction revealed the typical double-ring structure of CCT in an open conformation with an additional cylindrical mass sitting inside one ring that could be attributed to Gβ (Fig. 1B). This mass was positioned to one side of the ring, interacting with the apical region of one or two CCT subunits. The size of the mass was slightly larger than the atomic structure of Gβ in the Gβγ dimer (10), suggesting that Gβ had reached a near-native β-propeller structure within the CCT folding cavity (Fig. 1C). However, the compact cylindrical shape of the Gβ β-propeller made it difficult to rotationally orient Gβ in the cryo-EM electron density. As a result, we performed two different cross-linking analyses to determine the orientation of Gβ within the CCT folding cavity and to identify the CCT subunits that contact Gβ.
Unnatural Amino Acid Cross-Linking of Gβ to CCT.
Using amber stop codon suppression methods (12), we introduced the unnatural amino acid p-benzoyl-l-phenylalanine (BzF) photo-cross-linker at different positions within the Gβ cDNA sequence (Fig. S3) and transfected these variants into HEK-293T cells along with the suppressor tRNA and BzF tRNA synthetase. Cells were subsequently incubated with BzF, and cell extracts were cross-linked with UV light. Gβ was immunoprecipitated and immunoblotted for Gβ and CCT subunits to detect Gβ photo-cross-links. When activated, BzF creates a high-energy intermediate that reacts rapidly with atoms within an ∼3-Å radius (13). Thus, any cross-links between Gβ and CCT subunits would indicate regions of close contact.
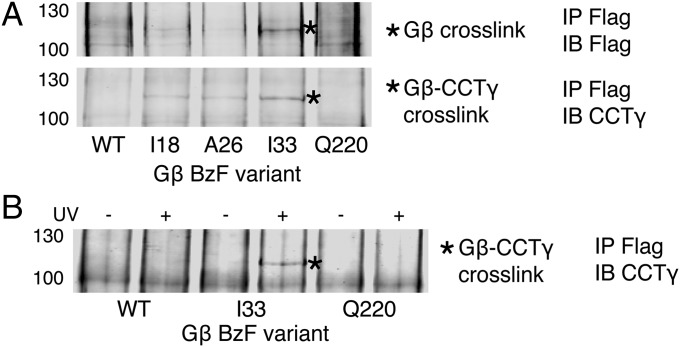
Of 175 Gβ variants tested, only 3 with BzF at positions I18, A26, and I33 showed a unique banding pattern in Gβ immunoblots (Fig. 2A). These variants had a band at 115 kDa not found with WT Gβ. The variants were subsequently immunoblotted for each of the eight CCT subunits (Fig. S3), and the same 115-kDa band was detected only in the CCTγ blot (Fig. 2A). The band was clearly visible with the I33 variant, but weak bands were also observed with the I18 and A26 variants. These results indicate that the N-terminal α-helical region of Gβ, which is not part of the Gβ β-propeller, makes contacts with the CCTγ subunit. Interestingly, each of the three residues I18, A26, and I33 are involved in hydrophobic contacts with Gγ as part of the coiled-coil interaction between Gβ and Gγ in the Gβγ structure (10).
Fig. 2.
BzF cross-linking of Gβ. (A) Flag-Gβ and CCTγ immunoblots of cross-linked Flag-Gβ immunoprecipitates. Cells transfected with Flag-Gβ variants incorporating BzF at the indicated residues were lysed, and extracts were treated with UV light, immunoprecipitated for Flag-Gβ, and blotted for Flag or CCTγ. A unique band with mobility of 115 kDa (asterisk) is observed with the I18, A26, and I33 variants, but not with the WT or Q220 controls. (B) CCTγ immunoblot of Flag-Gβ immunoprecipitates showing the UV dependence of the 115-kDa cross-linked band with the Gβ I33 variant.
XL-MS Analysis of the Gβ-CCT Complex.
The BzF cross-linking established contacts between the Gβ N-terminal α-helix and CCTγ, but gave no information about the orientation and interactions of the β-propeller region. To address these issues, we used XL-MS, a method that has been used to characterize CCT/substrate complexes (14). The Gβ-CCT complex was treated with disuccinimidyl suberate (DSS), which cross-links adjacent lysine residues. The complex was digested with protease, and the masses of the resulting peptides were measured by tandem mass spectrometry (MS/MS). The cross-linked peptides were identified by using the xQuest software (15).
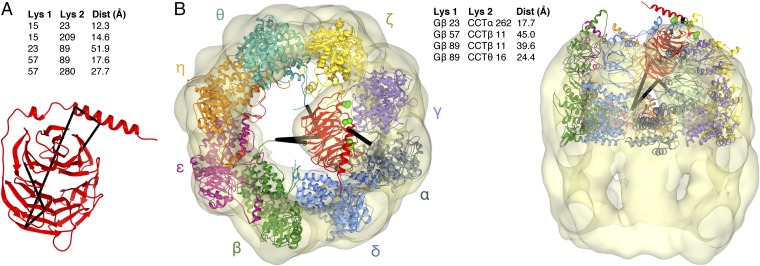
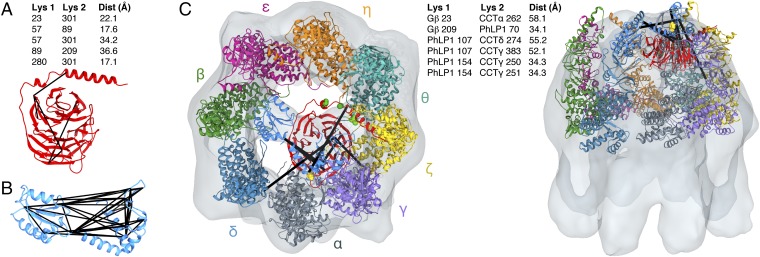
XL-MS identified five intramolecular cross-links within Gβ (Fig. 3A). These cross-links were used to assess the folded state of Gβ in the CCT cavity. The distance between the Cα carbons of the cross-linked lysines, taking into account normal movement within the protein backbone, has an ∼30-Å cutoff (16). When mapped on the native structure of Gβ found in the Gβγ dimer (10), four of the intramolecular cross-links were within this distance (Fig. 3A), whereas one of the cross-links (K23-K89) was not, indicating a deviation from the native structure. These results suggest that Gβ in the CCT folding cavity has achieved a near-native state; nevertheless, the β-propeller is not in the stable conformation found in the Gβγ dimer.
Fig. 3.
XL-MS of the Gβ-CCT complex. (A) Intramolecular Gβ cross-links (black lines) are mapped on the atomic structure of Gβ. The distances between the Cα atoms of the cross-linked lysines in the structure are indicated. (B) The atomic structures of Gβ and CCT [PDB ID code 2XSM (20)] are docked into the cryo-EM electron density envelope to minimize the distance of the intermolecular Gβ-CCT cross-links (black lines). The flexible CCTβ and CCTθ N termini are extended toward Gβ. Gβ I18, A26, and I33 are shown as green spheres. The distances between the Cα atoms of the cross-linked lysines are indicated. In the docking, the helical extensions of the CCT apical domains sometimes extend above the EM electron density because of missing density caused by mobility in these regions.
The XL-MS analysis identified a number of intermolecular cross-links between CCT subunits (Dataset S1) that were consistent with the arrangement of the subunits in the CCT ring proposed from previous XL-MS studies (16, 17). In addition, there were four cross-links between Gβ and CCT (Fig. 3B). These cross-links were used to dock the atomic structure of the Gβ β-propeller into the cryo-EM density in an orientation that minimized the cross-link distances. The docking placed the N-terminal α-helix of Gβ at the top of the CCT cavity and blade 1 of the β-propeller at the bottom of the cavity, orienting the β-propeller on its side with its central axis nearly perpendicular to the central axis of the CCT toroid (Fig. 3B). The BzF cross-linking (Fig. 2) further constrained the docking by requiring that the Gγ-binding surface of Gβ make contact with the CCTγ subunit, leaving the PhLP1-binding surface of Gβ facing the center of the CCT folding cavity.
In this structural model, the distance between the Cα atoms of Gβ K23 and CCTα K262 is well within the 30 Å required for cross-linking to occur (Fig. 3B). However, for the other cross-links, conformational flexibility in the N termini of CCTβ and CCTθ as well as in blade 1 of the Gβ β-propeller is needed to bring the lysines within cross-linking distance. Extension of the CCTθ N terminus brings CCTθ K16 within cross-linking distance of Gβ K89 in blade 1 in its native position in the Gβ β-propeller (Fig. 3B), whereas extension of the CCTβ N terminus and flexibility in Gβ blade 1 are both necessary for CCTβ K11 to cross-link with Gβ K57 and K89. Interestingly, blade 1 mobility could also explain the intramolecular cross-link of K89 to K23 in the Gβ α-helix. Blade 1 is the most N-terminal blade of the β-propeller and must make contact with blade 7, the most C-terminal blade, to close the β-propeller. The XL-MS data suggest that in the Gβ-CCT complex blade 1 is not fixed in the Gβ β-propeller and the β-propeller is not completely closed.
Cryo-EM Structure of the PhLP1–Gβ-CCT Complex.
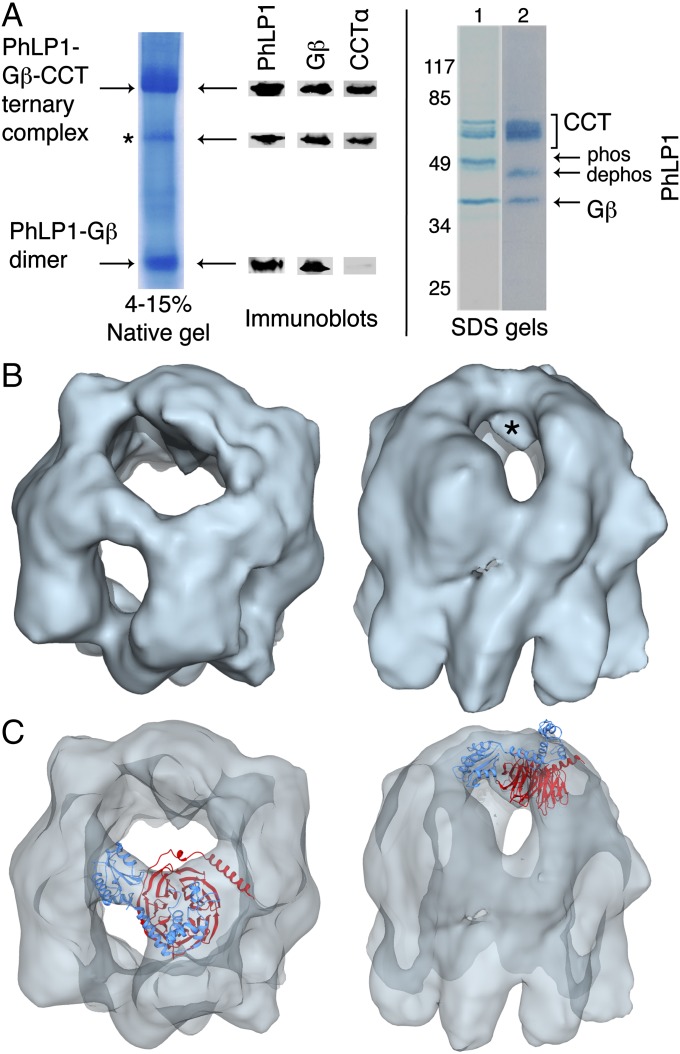
We went on to perform a similar structural analysis of the PhLP1–Gβ-CCT ternary complex. We coexpressed human Gβ1 and PhLP1 in insect cells and purified the ternary complex with endogenous insect CCT using a tandem affinity capture approach. An SDS gel of the purified complex showed very pure bands corresponding to CCT subunits, PhLP1 and Gβ (Fig. 4A) as confirmed by mass spectrometry (MS; Fig. S1). Addition of Gγ to the complex resulted in the formation of Gβγ dimers, indicating that the complex was biologically active (Fig. S4). Immunoblots from the native gel showed that the complex was only moderately stable with some dissociated PhLP1–Gβ as well as the ternary complex (Fig. 4A). This instability is consistent with the observation that PhLP1 enhances the release of Gβ from CCT (3). Accordingly, the cryo-EM analysis revealed several subpopulations of particles that could be sorted by particle classification. From 22,724 particles examined, 11,634 homogeneous PhLP1–Gβ-CCT particles were used to generate an 18.5-Å 3D reconstruction of the complex without imposing any symmetry throughout the reconstruction procedure (Fig. S5). The structure resembles that of the PhLP1–CCT complex (18), with a mass spanning the CCT folding cavity, interacting with subunits on both sides of the cavity (Fig. 4B). There is an additional mass within the folding cavity, not seen in the PhLP1–CCT structure, which is attributable to Gβ (Fig. 4B, asterisk). Docking of an atomic model of the PhLP1–Gβ dimer based on the phosducin (Pdc)–Gβγ complex (19) into the reconstruction allowed only one orientation, with Gβ in the mass inside the CCT folding cavity and PhLP1 above Gβ in the mass spanning the cavity (Fig. 4C).
Fig. 4.
Cryo-EM structure of the PhLP1–Gβ-CCT complex. (A) Electrophoretic analysis of the purified PhLP1–Gβ-CCT complex by native gel and SDS/PAGE without (lane 1) and with (lane 2) alkaline phosphatase treatment. Increased mobility of PhLP1 after alkaline phosphatase indicates that PhLP1 is phosphorylated at S18–20 (3). Native gel bands were excised and immunoblotted for PhLP1, Gβ1, and CCTα. The minor band marked with an asterisk appears to be a CCT subcomplex containing PhLP1, Gβ, and CCTα. (B) Top and side views of the reconstructed PhLP1–Gβ-CCT complex at 18.5-Å resolution. The asterisk identifies the mass attributed to Gβ. (C) Docking of PhLP1 (light blue) and Gβ (red) into the PhLP1–Gβ-CCT cryo-EM structure. The model of the PhLP1–Gβ dimer based on the Pdc–Gβγ structure [PDB ID code 2TRC (19)] was docked into the masses attributable to PhLP1 and Gβ in the cryo-EM reconstruction. Top and side views are shown.
Unnatural Amino Acid Cross-Linking of PhLP1 to CCT.
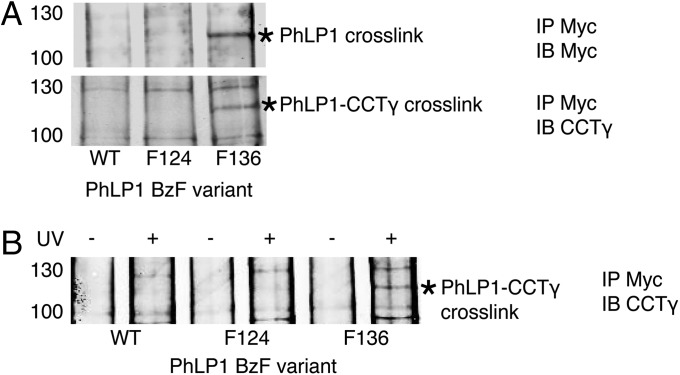
Site-specific BzF cross-linking was again used to determine the CCT subunits that interact with PhLP1 and to orient PhLP1 on CCT. BzF was inserted at six positions in the PhLP1 sequence. When BzF was substituted for F136 of PhLP1, a strong cross-link was made between PhLP1 and CCTγ, whereas BzF at the other five positions did not cross-link (Fig. 5 and Fig. S6). In addition, none of the other CCT subunits made cross-links with BzF at F136 (Fig. S6). Interestingly, F136 is next to a stretch of negatively charged amino acids between PhLP1 residues 131 and 135 that has been shown by mutagenesis to be important in PhLP1 binding to CCT (18). The F136 BzF cross-linking confirms that this region of the PhLP1 N-terminal domain makes close contact with CCT and places the N-terminal domain of PhLP1 in close contact with CCTγ, as was also observed with Gβ (Fig. 2). These findings support the docking model of Fig. 4C that positions the N-terminal domain of PhLP1 over Gβ in the CCT cavity to occupy its binding site on Gβ.
Fig. 5.
BzF cross-linking of PhLP1. (A) PhLP1-Myc and CCTγ immunoblots of cross-linked PhLP1-Myc immunoprecipitates. Cells transfected with PhLP1-Myc variants incorporating BzF at the indicated residues were lysed, and extracts were treated with UV-light, immunoprecipitated for PhLP1-Myc, and blotted for Myc or CCTγ. A unique band with a mobility of 120 kDa (asterisk) was observed in both blots with the F136 variant, but not with the WT or F124 controls. (B) CCTγ immunoblot of PhLP1-Myc immunoprecipitates showing the UV dependence of the 120-kDa cross-linked band with the F136 variant.
XL-MS Analysis of the PhLP1–Gβ-CCT Complex.
To assess the folding of Gβ in the PhLP1–Gβ-CCT ternary complex and to further examine the orientation of PhLP1 and Gβ on CCT, an XL-MS analysis of the ternary complex was performed. A total of five intramolecular Gβ cross-links were identified in the ternary complex (Fig. 6A). One of the cross-links was also found in the Gβ-CCT complex, whereas the four others were unique. Of the five, three cross-links were within the 30-Å distance constraint for DSS in the native Gβγ structure, whereas the other two were slightly longer (Fig. 6A). These cross-links show that Gβ in the ternary complex has achieved a more native structure than that seen in the Gβ-CCT complex. For example, the Gβ intramolecular cross-link observed in the Gβ-CCT complex between K23 of the α-helix and K89 of blade 1 that required significant deviations away from the β-propeller structure was not found in the presence of PhLP1. Interestingly, PhLP1 makes several contacts with both blades 1 and 7 of Gβ (19), and these contacts may close the β-propeller, decreasing the mobility of blade 1 and making long-range cross-links with K23 impossible.
Fig. 6.
XL-MS cross-linking of the PhLP1–Gβ-CCT complex. (A) Intramolecular Gβ cross-links (black lines) are mapped on the atomic structure of Gβ. The distances between the Cα atoms of the cross-linked lysines are indicated. (B) Intramolecular PhLP1 cross-links (black lines) are mapped on the structural model of PhLP1. See Dataset S2 for the cross-link distances in the model. (C) The PhLP1–Gβ structure and CCT structure are docked into the cryo-EM electron density envelope to minimize the distance of the intermolecular PhLP1, Gβ, and CCT cross-links (black lines). Gβ I18, A26, and I33 are shown as green spheres. PhLP F136 is shown as a yellow sphere. The distances between the Cα atoms of the cross-linked lysines are indicated. Two cross-links (PhLP1 70–CCTα 129 and Gβ 209–CCTη 108) at the ring interface were omitted from the docking analysis because to form they would require dissociation of the CCT complex.
An examination of the PhLP1 intramolecular cross-links also provides useful information about the structure of PhLP1 in the PhLP1–Gβ-CCT complex. A total of 38 PhLP1 intramolecular cross-links were found, which was many more than with Gβ. This increase can be attributed to more lysines in PhLP1 (22 compared with 10 for Gβ) and greater flexibility of the PhLP1 N-terminal domain. The cross-links were mapped onto the structural model of PhLP1, which showed 21 of the 38 PhLP1 intramolecular cross-links within the 30-Å cross-linking limit (Fig. 6B and Dataset S2). Of the other 17 cross-links, 15 involved residues from highly flexible regions of the N-terminal domain (19). Thus, these cross-links support the argument that the PhLP1 structure in the PhLP1–Gβ-CCT complex is similar to that of Pdc in the Pdc–Gβγ complex, indicating that in the ternary complex, PhLP1 adopts a conformation that is competent for Gβ binding.
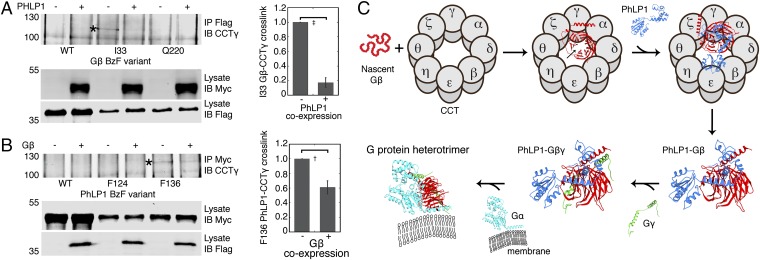
XL-MS also identified several intermolecular cross-links involving PhLP1 or Gβ (Fig. 6C). These cross-links were used to guide the docking of PhLP1 and Gβ into the cryo-EM mass and create an accurate structural model of the PhLP1–Gβ-CCT complex (Fig. 6C). To create the model, PhLP1 and Gβ were held in their binding orientation relative to each other and fit into the cryo-EM mass while minimizing the XL-MS cross-link distances. The model suggests two significant movements of Gβ in the folding cavity upon PhLP1 binding. First, the β-propeller has rotated up in the cavity so that its central axis is more parallel to that of the CCT toroid. This rotation moves Gβ K57 and K89 of blade 1 away from the bottom of the cavity, but keeps Gβ K23 near the top of the cavity—possibly explaining why cross-links between blade 1 and the bottom of the CCT cavity were not observed in the ternary complex but that the cross-link between K23 and CCTα K262 at the top of the cavity was retained. Second, the β-propeller has rotated counter-clockwise in the cavity, displacing Gβ I33 from its binding site on CCTγ. Disrupting this contact may contribute to PhLP1-mediated release of Gβ from CCT (Fig. 7).
Fig. 7.
PhLP1 mediates release of Gβ from CCT. (A) Effects of PhLP1 on Gβ I33 BzF cross-linking to CCTγ. CCTγ immunoblots of Flag-Gβ immunoprecipitates from UV-light–treated extracts of cells cotransfected with Flag-Gβ BzF variants with and without PhLP1. An asterisk indicates the cross-linked band. The graph shows the quantification of the cross-linked band from three experiments. ‡P < 0.01. (B) Effects of Gβ on PhLP1 F136 BzF cross-linking to CCTγ. CCTγ immunoblots of PhLP1-Myc immunoprecipitates from UV-light–treated extracts of cells cotransfected with PhLP1-Myc BzF variants with and without Gβ. An asterisk indicates the cross-linked band. The graph shows the quantification of the cross-linked band from four experiments. †P < 0.05. (C) Schematic diagram of the mechanism of G-protein heterotrimer assembly. In the Gβ-CCT complex, the Gβ β-propeller is shown in an open conformation with a gap between blades 1 and 7 (arrow). Upon PhLP1 binding, contacts between PhLP1 and blades 1 and 7 close the Gβ β-propeller and disrupt interactions of Gβ with CCT. The PhLP1–Gβ complex is released from CCT, and Gβγ assembly is able to proceed. See text for a more detailed description of the mechanism.
With regard to the position of PhLP1, the three cross-links between the N-terminal domain of PhLP1 and CCTγ confirm that the N-terminal domain of PhLP1 is oriented toward CCTγ. All of the intermolecular cross-links have distances beyond the 30-Å limit, but two involve PhLP1 K107 that is found in a large unstructured loop of the N-terminal domain of PhLP1 (19), and four involve residues (CCTα 262, CCTγ K250 and K251, and CCTδ 274) in the flexible helical extensions of the CCT subunits (20). Thus, when conformational flexibility is taken into account, the cross-links fit the proposed model of the PhLP1–Gβ-CCT complex.
The model places both the PhLP1 N-terminal domain and Gβ near CCTα and CCTγ in the folding cavity and orients Gβ inside the cavity with PhLP1 above the cavity. The cross-link between PhLP1 K70 and Gβ K209 supports this orientation and provides additional evidence that PhLP1 is bound to Gβ in the ternary complex. The location of PhLP1, near the tips of the CCT apical domains and spanning the folding cavity, is very similar to that of PhLP1 in the PhLP1–CCT cryo-EM structure (18). Hence, when PhLP1 occupies its binding sites on CCT, it is positioned to also bind Gβ, and little conformational energy is lost in the process.
PhLP1 Mediates the Release of Gβ from CCT.
PhLP1 has been shown biochemically to release Gβ from CCT, and this release has been proposed to be important for Gβγ assembly (3). Consistent with this notion, the native gel analysis showed that the purified PhLP1–Gβ-CCT complex contained a significant amount of PhLP1–Gβ dimer not associated with CCT (Fig. 4A). To better understand the structural basis for PhLP1-mediated release of Gβ from CCT, we measured the effects of PhLP1 on BzF cross-linking of Gβ to CCTγ. Coexpression of PhLP1 with Gβ containing BzF at residue 33 resulted in an 80% decrease in the cross-linking of Gβ to CCTγ (Fig. 7A). In the reciprocal experiment, coexpression of Gβ with PhLP1 containing BzF at residue 136 caused a 40% decrease in the cross-linking of PhLP1 to CCTγ (Fig. 7B). Together, these results confirm that when PhLP1 interacts with Gβ on CCT, Gβ changes its conformation in the CCT folding cavity, and contacts between Gβ and CCT are lost.
Discussion
The structures of the Gβ-CCT and PhLP1–Gβ-CCT complexes provide insight into the molecular mechanism of CCT and PhLP1-mediated Gβ folding and Gβγ assembly. Based on the structures, a detailed mechanism of Gβγ assembly can be proposed (scheme in Fig. 7C). The Gβ-CCT structure shows Gβ in a globular shape in the CCT folding cavity with slightly expanded dimensions compared with the atomic structure of the Gβ β-propeller (Fig. 1C). This structure is more compact than those of β-actin and α- and β-tubulin, which have more extended shapes in the folding cavity (20–22). Despite its globular form, the XL-MS analysis identified several cross-links that could only be made if blade 1 of Gβ were flexible (Fig. 3). Instability in blade 1 could result from an inability of blade 1 to interact effectively with blade 7 to close the β-propeller.
The BzF unnatural amino acid cross-linking revealed a close association between the N-terminal region of Gβ and CCTγ (Fig. 2). This same region of Gβ makes a coiled-coil interaction with Gγ in the native Gβγ dimer. Hence, Gγ cannot access its binding site while Gβ is bound to CCTγ. This observation explains previous biochemical data showing that Gγ does not associate with Gβ bound to CCT (3). The inability of Gγ to access its binding site, coupled with the inability of Gβ to form a stable β-propeller in the absence of Gγ, creates a situation in which Gβ cannot efficiently release from CCT and assemble with Gγ. Thus, Gβ remains bound to CCT, and the Gβγ assembly process is stalled in the absence of PhLP1.
PhLP1 binding to Gβ in the CCT folding cavity allows Gβγ assembly to proceed (3). Analysis of the PhLP1–Gβ-CCT structure shows how PhLP1 binding contributes to the folding of Gβ and its release from CCT. When PhLP1 occupies its binding site near the tips of the CCT apical domains, its N-terminal domain is in contact with CCTγ and is poised to bind Gβ (Fig. 6C). Interactions of PhLP1 with blades 1 and 7 of Gβ (19) appear to close the β-propeller. This stabilization disrupts interactions between blade 1 and the N termini of CCTβ and θ, allowing Gβ to rotate up toward the top of the folding cavity. In addition, PhLP1 binding appears to cause another rotation in Gβ that moves I33 away from its binding site on CCTγ. These changes decrease the contacts between Gβ and CCT and promote the release of the PhLP1–Gβ dimer from the complex (Figs. 4A and 7A). Once released, the Gγ-binding site on Gβ is completely available for binding. Hence, Gγ can associate with Gβ, forming the Gβγ dimer and stabilizing the Gβ β-propeller.
PhLP1 has been recently shown by mouse gene targeting to be absolutely required for assembly of Gβγ dimers in vivo (11). This structural analysis of chaperone-bound intermediates in Gβγ assembly demonstrates why PhLP1 plays such an important role. This knowledge can be used to create therapeutics to regulate Gβγ assembly and thereby treat some of the many diseases linked to defects in G-protein signaling.
Materials and Methods
Purification of Gβ-CCT and PhLP1–Gβ-CCT Complexes.
High Five insect cells were coinfected with a baculovirus construct of human Gβ1 with a C-terminal HPC4 tag with or without a similar construct of human PhLP1 with a C-terminal His6 tag. The Gβ-CCT and PhLP1–Gβ-CCT complexes, with endogenous insect CCT, were affinity-purified from cell extracts, and the composition was confirmed by immunoblotting and MS.
EM and Image Processing.
For cryo-EM, aliquots of the Gβ-CCT or PhLP1–Gβ-CCT complexes were applied to glow-discharged Quantifoil 1.2-μm holey carbon grids, blotted, and fast-frozen in liquid ethane. Low-dose images (<10e- A-2) of the complexes were taken on a FEI Tecnai G2 FEG200 electron microscope at 200 kV by using a Gatan side-entry cryo-holder with a nominal magnification of 62,000× and 1.8- to 3.0-μm underfocus. Images were classified by using reference-free methods (23), and the selected averages were used to build a reference volume using common lines (24), which was subsequently used for the 3D reconstruction procedure (25). Visualization and docking of the atomic structures were carried out by using Chimera (26).
Unnatural Amino Acid Cross-Linking.
The amber codon (TAG) was introduced at positions in Gβ1 or PhLP1 cDNAs in the pcDNA3.1 vector by using site-directed mutagenesis. The Gβ or PhLP1 expression vectors were transfected into cells along with vectors containing the amber suppressor tRNA and the BzF tRNA synthetase (12). BzF was added and allowed to incorporate. Cell lysates were exposed to UV light to initiate cross-linking. Gβ or PhLP1 was immunoprecipitated and immunoblotted for each of the CCT subunits to identify cross-links with CCT.
Chemical Cross-Linking Coupled with MS.
The chemical cross-linking and MS identification of the cross-links followed described protocols (27). Briefly, purified Gβ-CCT or PhLP1–Gβ-CCT complexes were cross-linked with a 50% (mol/mol) mixture of heavy isotope-labeled DSS and then digested with protease. Cross-linked peptides were analyzed by MS/MS on an Orbitrap Velos Pro mass spectrometer and identified in the mass spectra by using the xQuest software (15). See SI Materials and Methods for additional experimental details.
Supplementary Material
Acknowledgments
We thank Caleb Stowell and Devon Blake for their technical assistance. This work was supported by National Institutes of Health Grants GM078550 and EY012287 (to B.M.W.) and Grants BFU2013-44202 from the Spanish Ministry of Economy and Innovation and S2013/MIT-2807 from the Madrid Regional Government (to J.M.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419595112/-/DCSupplemental.
References
- 1.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85(4):1159–1204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 2.Willardson BM, Tracy CM. Chaperone-mediated assembly of G protein complexes. Subcell Biochem. 2012;63:131–153. doi: 10.1007/978-94-007-4765-4_8. [DOI] [PubMed] [Google Scholar]
- 3.Lukov GL, et al. Mechanism of assembly of G protein betagamma subunits by protein kinase CK2-phosphorylated phosducin-like protein and the cytosolic chaperonin complex. J Biol Chem. 2006;281(31):22261–22274. doi: 10.1074/jbc.M601590200. [DOI] [PubMed] [Google Scholar]
- 4.Lukov GL, Hu T, McLaughlin JN, Hamm HE, Willardson BM. Phosducin-like protein acts as a molecular chaperone for G protein betagamma dimer assembly. EMBO J. 2005;24(11):1965–1975. doi: 10.1038/sj.emboj.7600673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yébenes H, Mesa P, Muñoz IG, Montoya G, Valpuesta JM. Chaperonins: Two rings for folding. Trends Biochem Sci. 2011;36(8):424–432. doi: 10.1016/j.tibs.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 6.Dekker C, et al. The crystal structure of yeast CCT reveals intrinsic asymmetry of eukaryotic cytosolic chaperonins. EMBO J. 2011;30(15):3078–3090. doi: 10.1038/emboj.2011.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Douglas NR, et al. Dual action of ATP hydrolysis couples lid closure to substrate release into the group II chaperonin chamber. Cell. 2011;144(2):240–252. doi: 10.1016/j.cell.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang J, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463(7279):379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valpuesta JM, Martín-Benito J, Gómez-Puertas P, Carrascosa JL, Willison KR. Structure and function of a protein folding machine: The eukaryotic cytosolic chaperonin CCT. FEBS Lett. 2002;529(1):11–16. doi: 10.1016/s0014-5793(02)03180-0. [DOI] [PubMed] [Google Scholar]
- 10.Sondek J, Bohm A, Lambright DG, Hamm HE, Sigler PB. Crystal structure of a G-protein beta gamma dimer at 2.1A resolution. Nature. 1996;379(6563):369–374. doi: 10.1038/379369a0. [DOI] [PubMed] [Google Scholar]
- 11.Lai CW, et al. Phosducin-like protein 1 is essential for G-protein assembly and signaling in retinal rod photoreceptors. J Neurosci. 2013;33(18):7941–7951. doi: 10.1523/JNEUROSCI.5001-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ye S, et al. Site-specific incorporation of keto amino acids into functional G protein-coupled receptors using unnatural amino acid mutagenesis. J Biol Chem. 2008;283(3):1525–1533. doi: 10.1074/jbc.M707355200. [DOI] [PubMed] [Google Scholar]
- 13.Sato S, et al. Crystallographic study of a site-specifically cross-linked protein complex with a genetically incorporated photoreactive amino acid. Biochemistry. 2011;50(2):250–257. doi: 10.1021/bi1016183. [DOI] [PubMed] [Google Scholar]
- 14.Herzog F, et al. Structural probing of a protein phosphatase 2A network by chemical cross-linking and mass spectrometry. Science. 2012;337(6100):1348–1352. doi: 10.1126/science.1221483. [DOI] [PubMed] [Google Scholar]
- 15.Rinner O, et al. Identification of cross-linked peptides from large sequence databases. Nat Methods. 2008;5(4):315–318. doi: 10.1038/nmeth.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leitner A, et al. The molecular architecture of the eukaryotic chaperonin TRiC/CCT. Structure. 2012;20(5):814–825. doi: 10.1016/j.str.2012.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kalisman N, Adams CM, Levitt M. Subunit order of eukaryotic TRiC/CCT chaperonin by cross-linking, mass spectrometry, and combinatorial homology modeling. Proc Natl Acad Sci USA. 2012;109(8):2884–2889. doi: 10.1073/pnas.1119472109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martín-Benito J, et al. Structure of the complex between the cytosolic chaperonin CCT and phosducin-like protein. Proc Natl Acad Sci USA. 2004;101(50):17410–17415. doi: 10.1073/pnas.0405070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaudet R, Bohm A, Sigler PB. Crystal structure at 2.4 angstroms resolution of the complex of transducin betagamma and its regulator, phosducin. Cell. 1996;87(3):577–588. doi: 10.1016/s0092-8674(00)81376-8. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz IG, et al. Crystal structure of the open conformation of the mammalian chaperonin CCT in complex with tubulin. Nat Struct Mol Biol. 2011;18(1):14–19. doi: 10.1038/nsmb.1971. [DOI] [PubMed] [Google Scholar]
- 21.Llorca O, et al. Eukaryotic chaperonin CCT stabilizes actin and tubulin folding intermediates in open quasi-native conformations. EMBO J. 2000;19(22):5971–5979. doi: 10.1093/emboj/19.22.5971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villebeck L, Moparthi SB, Lindgren M, Hammarström P, Jonsson BH. Domain-specific chaperone-induced expansion is required for beta-actin folding: A comparison of beta-actin conformations upon interactions with GroEL and tail-less complex polypeptide 1 ring complex (TRiC) Biochemistry. 2007;46(44):12639–12647. doi: 10.1021/bi700658n. [DOI] [PubMed] [Google Scholar]
- 23.Scheres SH, et al. Maximum-likelihood multi-reference refinement for electron microscopy images. J Mol Biol. 2005;348(1):139–149. doi: 10.1016/j.jmb.2005.02.031. [DOI] [PubMed] [Google Scholar]
- 24.Ludtke SJ, Baldwin PR, Chiu W. EMAN: Semiautomated software for high-resolution single-particle reconstructions. J Struct Biol. 1999;128(1):82–97. doi: 10.1006/jsbi.1999.4174. [DOI] [PubMed] [Google Scholar]
- 25.Frank J, et al. SPIDER and WEB: Processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116(1):190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 26.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 27.Leitner A, Walzthoeni T, Aebersold R. Lysine-specific chemical cross-linking of protein complexes and identification of cross-linking sites using LC-MS/MS and the xQuest/xProphet software pipeline. Nat Protoc. 2014;9(1):120–137. doi: 10.1038/nprot.2013.168. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.