Significance
High-throughput sequencing of B-cell immunoglobulin receptors is providing unprecedented insight into adaptive immunity. A key step in analyzing these data involves assignment of the germline variable (V), diversity (D), and joining (J) gene-segment alleles that comprise each immunoglobulin sequence by matching them against a database of known V(D)J alleles. However, this process will fail for sequences that use previously undetected alleles, whose frequency in the population is unclear. Here we describe TIgGER, a computational method that significantly improves V(D)J allele assignments by first determining the complete set of gene segments carried by a subject, including novel alleles. The application of TIgGER identifies a surprisingly high frequency of novel alleles, highlighting the critical need for this approach.
Keywords: next-generation sequencing, B-cell repertoire, adaptive immunity, somatic hypermutation, variable gene segment
Abstract
Individual variation in germline and expressed B-cell immunoglobulin (Ig) repertoires has been associated with aging, disease susceptibility, and differential response to infection and vaccination. Repertoire properties can now be studied at large-scale through next-generation sequencing of rearranged Ig genes. Accurate analysis of these repertoire-sequencing (Rep-Seq) data requires identifying the germline variable (V), diversity (D), and joining (J) gene segments used by each Ig sequence. Current V(D)J assignment methods work by aligning sequences to a database of known germline V(D)J segment alleles. However, existing databases are likely to be incomplete and novel polymorphisms are hard to differentiate from the frequent occurrence of somatic hypermutations in Ig sequences. Here we develop a Tool for Ig Genotype Elucidation via Rep-Seq (TIgGER). TIgGER analyzes mutation patterns in Rep-Seq data to identify novel V segment alleles, and also constructs a personalized germline database containing the specific set of alleles carried by a subject. This information is then used to improve the initial V segment assignments from existing tools, like IMGT/HighV-QUEST. The application of TIgGER to Rep-Seq data from seven subjects identified 11 novel V segment alleles, including at least one in every subject examined. These novel alleles constituted 13% of the total number of unique alleles in these subjects, and impacted 3% of V(D)J segment assignments. These results reinforce the highly polymorphic nature of human Ig V genes, and suggest that many novel alleles remain to be discovered. The integration of TIgGER into Rep-Seq processing pipelines will increase the accuracy of V segment assignments, thus improving B-cell repertoire analyses.
The production by B cells of immunoglobulin (Ig) proteins, which are expressed on the cell surface as B-cell receptors and secreted by subsets of B cells as antibodies, is a key component of the adaptive immune system in humans. Through their specific binding to an enormously diverse range of foreign bodies, Ig proteins are able to elicit further immunological response and provide protection. These proteins are assembled in B cells from two pairs of polypeptide chains, termed heavy and light. The antigen-binding portions of these genes are created through the somatic recombination of gene segments, termed variable (V), diversity (D), and joining (J). During the recombination process, one each of the ∼46 V, 23 D, and 6 J gene segments (1) recombine to make the antigen-binding region of the heavy chain; the light chain is created by a similar process, although involving one of two different loci (λ and κ) containing V and J genes only. Over three million different Ig sequences can be created through this V(D)J recombinatorial process alone (2). The potential diversity of these sequences is further expanded to the order of trillions (2) when combined with the random insertion and deletion of nucleotides at the gene segment junctions and with somatic hypermutation (SHM), the latter of which introduces nucleotide changes at a rate of of 10−3 per base pair per division (3, 4).
Variations in a subject’s germline gene segment alleles and expressed repertoire (i.e., the collection of different Igs circulating in that subject) have been associated with various aspects of immune system and health status. Previous studies have, for example: revealed the association of certain germline genotypes with susceptibility to such diseases as rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and multiple sclerosis (MS) (5, 6); found correlations of age with a reduced Ig clonal diversity and less intense response to immune challenge (7, 8); found overly expanded clones in cases of lymphoma (9, 10); and discovered convergent Ig evolution across subjects in response to certain immune challenges (11, 12). These repertoire-sequencing (Rep-Seq) studies have benefitted from improvements in sequencing technologies, which allow for the generation of millions of reads per run (13). Previously, the 454 platform (Roche) was preferred because of its unique ability to generate reads long enough to span the V(D)J rearrangement, although now the MiSeq platform (Illumina) is able to generate paired-end reads also long enough to span the V(D)J rearrangement (13). Rep-Seq use is growing rapidly, even spurring the creation of commercial start-ups to provide researchers and clinical laboratories with sequencing and analysis services (14).
Analysis of Rep-Seq data depends critically on the determination of the germline V, D, and J alleles used by each of the Ig sequences, and several methods exist to perform the task of V(D)J germline assignment (2, 15–17). All of these methods essentially involve alignment of sample sequences to a database of germline alleles of all known V, D, and J gene segments. The IMGT (18) database of germline Ig alleles is the most widely used, and the National Center for Biotechnology Information refers to IMGT for its reference genome. However, recent studies have discovered the presence of numerous V segments and alleles not reported in any published databases (6, 19–21), as well as a several novel D and J alleles (19). Some of these V alleles have been incorporated into the IMGT database or alternative databases of germline alleles [such as VBASE2 (22) or the UNSWIg human heavy chain repertoire (23)]. The completeness of the germline V(D)J database may greatly influence downstream analysis results, including clinically relevant decision processes (24), as unreported alleles can skew estimated segment distributions and because novel polymorphisms will appear as recurrent somatic mutations.
No automated methods exist for detection of novel V(D)J alleles, and most current Rep-Seq studies simply assume that the current databases are complete. One strategy to search for potential Ig polymorphisms involves identifying, from among the least mutated sequences of each clonal group, V genes that have a high frequency of mutation to a single nucleotide at a given position (10% if the nucleotide is a classical SHM hotspot, 5% otherwise), use a wide variety of D and J alleles, and can be ruled out as not having resulted from a PCR chimera (19). Although application of such filtering-based methods have successfully identified novel alleles (19), the resulting predictions require manual curation, and their sensitivity and specificity have not been evaluated. It is unclear whether this approach can distinguish polymorphic positions from SHM hot-spots, which can be mutated in >40% of sequences (25, 26). Here we present a Tool for Immunoglobulin Genotype Elucidation via Rep-Seq (TIgGER). TIgGER includes a sensitive algorithm for the identification of novel V segment alleles, as well as an Ig genotype-determination step, which it uses to correct germline allele assignments from existing V(D)J assignment tools, like IMGT/HighV-QUEST. Application of TIgGER to Rep-Seq data from seven subjects identified 11 new alleles, demonstrating the importance of incorporating novel allele detection into analysis pipelines. TIgGER is available at clip.med.yale.edu/tigger.
Results
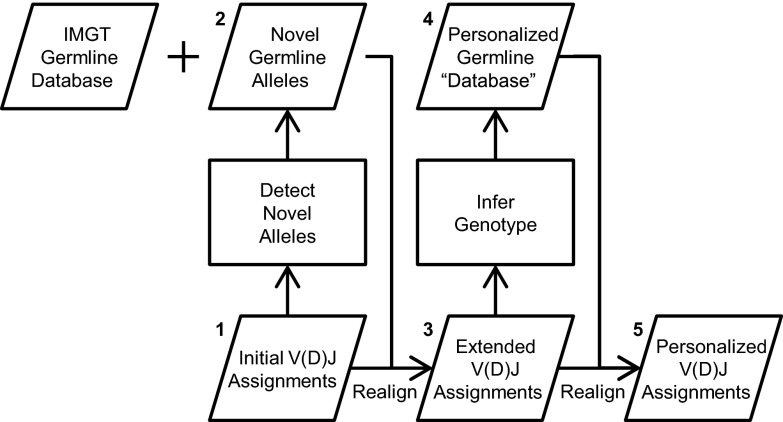
The TIgGER workflow consists of five steps (Fig. 1):
Fig. 1.
Overview of the TIgGER workflow. IMGT/HighV-QUEST is used to determine initial V(D)J assignments (step 1). TIgGER uses these initial gene segment assignments to analyze mutation patterns and detect a putative set of novel alleles (step 2). The germline gene segment database is then extended by adding these novel alleles to improve the initial V(D)J assignments (step 3). The extended V(D)J assignments are then analyzed to determine the genotype of a subject, and generate their personalized germline database (step 4). A final set of V allele assignments is then made (step 5).
Initial V(D)J assignments.
First, existing software is applied to determine initial V(D)J allele assignments for each sequence in the dataset. Throughout this report, IMGT/HighV-QUEST (27) is used for this step.
Novel allele detection.
Second, mutations are determined by comparing each sequence with its initial V(D)J assignments, and novel V alleles are detected based on analysis of these mutation patterns.
Extended V(D)J assignments.
Third, the V allele assigned to each Ig sequence is reassigned by realigning each sequence to the set of novel V alleles as well as the known database germline alleles. Sequences that better align to novel germline alleles will be reassigned from their initial assignments.
Inferred subject-specific genotype.
Fourth, frequencies of allele assignments among sequences aligning to each gene are calculated and used to determine which alleles are actually part of a subject’s genotype. These will serve as the subject’s personalized germline database.
Personalized V allele assignments.
Fifth, sequences that had best aligned to one or more alleles not in the subject’s personalized germline database are realigned to reassign them to V alleles from that set.
Many Nucleotide Positions Are Mutated at High Frequency.
High-throughput sequencing of the Ig heavy chain was carried out from blood and tissue samples of seven subjects (Table 1). Three subjects (PGP1, hu420143, and 420IV) were part of an influenza vaccination study (28), and four subjects (M2, M3, M4, and M5) were part of a study on multiple sclerosis (29). One subject (PGP1) was sequenced both on the 454 GS FLX and Illumina MiSeq platforms. As described in Methods, these sequencing data were processed using pRESTO (30) (clip.med.yale.edu/presto) to arrive at high-quality Ig sequences, which were then submitted to IMGT/HighV-QUEST for initial germline V(D)J gene segment identification. This processing resulted in an average of ∼140,000 sequences per dataset (Table 1).
Table 1.
High-throughput sequencing datasets used in this study
| Source | Disease state | Subject | Technology | Raw reads | Processed reads | Reads assigned 2+ V alleles* | Alleles per V gene (mean)* | Reads used for allele detection | Reads used for genotyping |
| (25) | Healthy | PGP1 | 454 | 117,188 | 70,722 | 9,042 | 4.1 | 14,833 | 43,752 |
| (25) | Healthy | hu420143 | 454 | 178,584 | 76,901 | 16,395 | 4.3 | 35,767 | 27,222 |
| (25) | Healthy | 420IV | 454 | 398,517 | 243,043 | 31,401 | 4.4 | 41,374 | 175,477 |
| (23) | Healthy | PGP1 | MiSeq | 3,851,658 | 110,053 | 47,828 | 4.3 | 31,397 | 53,670 |
| (26) | MS | M2 | MiSeq | 7,691,509 | 121,742 | 51,236 | 4.5 | 27,224 | 47,818 |
| (26) | MS | M3 | MiSeq | 3,641,633 | 103,189 | 30,386 | 4.5 | 22,737 | 4,633 |
| (26) | MS | M4 | MiSeq | 3,714,152 | 137,936 | 49,095 | 4.5 | 27,399 | 6,783 |
| (26) | MS | M5 | MiSeq | 10,917,517 | 277,913 | 123,860 | 4.5 | 51,072 | 16,638 |
| Average across all datasets | 3,813,845 | 142,687 | 44,905 | 4.4 | 31,475 | 46,999 | |||
As assigned by IMGT/HighV-QUEST.
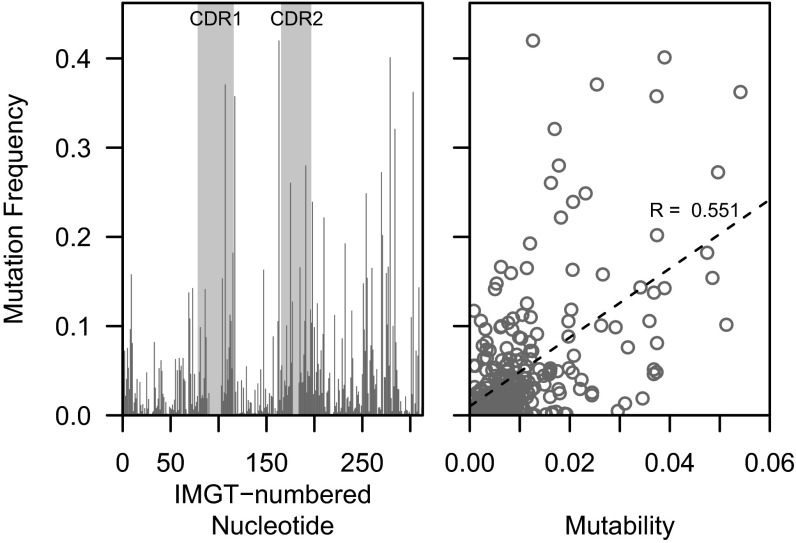
Ig sequences that are derived from novel alleles are assigned to the most similar allele contained in the germline repertoire database. The polymorphic positions are thus interpreted as mutations in the sequence, and will appear to have a high overall mutation frequency (19). Across all sets of sequences assigned to a particular Ig heavy chain variable (IGHV) allele, position-by-position analysis of mutation frequency identified thousands of highly mutated positions. In fact, among alleles occurring at least 100 times in one of the three 454 datasets, 1,206 nucleotide positions were found to be mutated at a frequency of >30%. Although many of these positions occurred at well-known hot-spot motifs, others did not. A typical example is provided by IGHV1-2*02 (Fig. 2, Left). In this case, six positions were mutated in >30% of the sequences. Four of these positions occurred at classic WRC/GYW mutation hotspots, and there was a mild overall correlation between the predicted mutability [according to the S5F targeting model (25)] and the observed mutation frequency (R = 0.551) (Fig. 2, Right). Thus, intrinsic biases in SHM targeting may explain some of these frequently mutated positions, whereas others may represent polymorphisms because of the presence of a novel IGHV allele in the subject. There is no obvious way to draw thresholds on the mutation frequency or position mutability to differentiate these two possibilities.
Fig. 2.
Mutation frequencies of IGHV positions. The mutation frequency of each IMGT-numbered nucleotide position was determined for sequences that best aligned to IGHV1-2*02 in subject hu410143. Somatic mutations were determined through comparison with the germline sequence reported by IMGT/HighV-QUEST, and sequences that were assigned to multiple alleles including IGHV1-2*02 were included in the analysis. (Left) The mutation frequency plotted as a function of IMGT-numbered nucleotide position. (Right) The mutation frequency plotted as a function of predicted mutability under the S5F targeting model.
Polymorphisms Exhibit a Distinct Pattern of Mutation Accumulation.
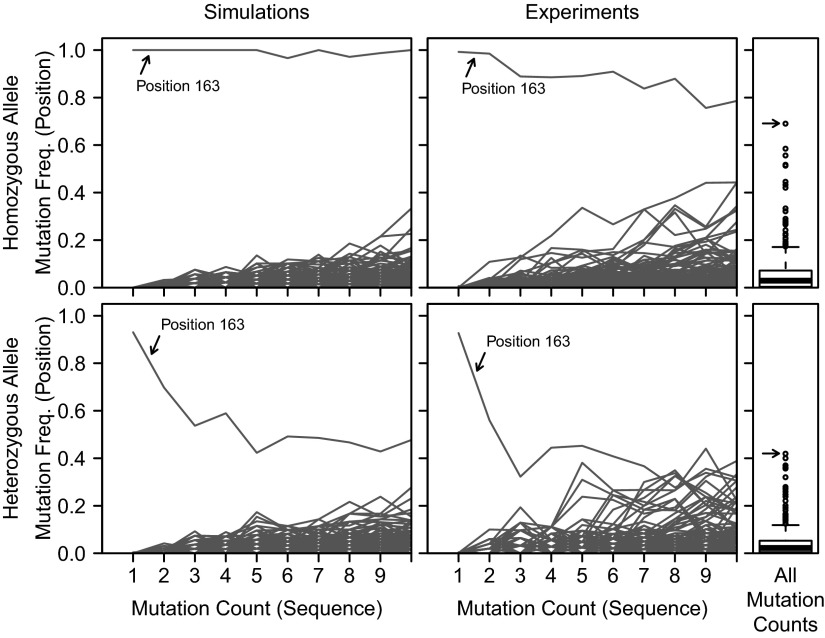
Although many nucleotide positions exhibited a high frequency of mutations, we reasoned that the pattern of mutation accumulation at polymorphic positions would be distinct from that found at other positions. Specifically, we hypothesized that nonpolymorphic positions would accumulate mutations at a frequency proportional to sequence-wide mutation counts, whereas polymorphic positions would exhibit a negative correlation. To investigate this hypothesis, a single base change was introduced into an existing IGHV allele to simulate a novel allele. Then, a computer simulation was used to sequentially introduce point mutations and generate a repertoire that reproduced the number of sequences and mutation count distribution observed in the data from a specified subject (Methods). Mutations were identified by comparing these simulated sequences to the existing IGHV allele (i.e., assuming no knowledge of the polymorphism). When all simulated sequences contained the polymorphism (i.e., the subject was homozygous for the novel allele), then the pattern of mutation accumulation at polymorphic positions was clearly distinct from other positions (Fig. 3, Upper Left). Polymorphic positions exhibited an extremely high mutation frequency (almost 100%) that was practically independent of the sequence-wide mutation count, although there was a small negative slope, as expected, because there is a low probability that this position could obtain a mutation that reverts the sequence to the existing IGHV germline. In contrast, the mutation frequency at nonpolymorphic positions was near zero when sequence-wide mutation counts were low, and was positively correlated with sequence-wide mutation counts.
Fig. 3.
Mutation patterns for polymorphic positions in IGHV sequences. The pattern of mutation accumulation in IGHV1-2*02 and a hypothetical unknown allele of IGHV1-2*02 (containing a polymorphism at position 163) was simulated as described in Methods. The fraction of sequences carrying a mutation at each IMGT position (lines) was then determined for groups of sequences sharing the same total IGHV mutation count, assuming that the subject was homozygous (Upper Left) or heterozygous (Lower Left) for the unknown allele. Equal allele use was assumed for the heterozygous case. The same analysis was performed on experimentally observed sequences that aligned to IGHV1-2*02 from subjects M5 (Upper Center) and hu420143 (Lower Center). For these experimental data, the fraction of sequences carrying a mutation at each IMGT position (points), irrespective of total IGHV mutation count, was also analyzed (Right). IMGT position 163 is indicated by an arrow in all panels.
The mutation pattern of polymorphic positions changed dramatically when the subject was assumed to be heterozygous for the novel allele (i.e., when one allele of the IGHV segment is known and the other contains a polymorphism). Although nonpolymorphic positions behaved similarly to the homozygous case, the polymorphic positions exhibited an unexpected pattern (Fig. 3, Lower Left). The mutation frequency of the polymorphic position was relatively high when the overall mutation count of the sequence was low (one to two mutations per sequence), but this frequency quickly fell to levels that were indistinguishable from other positions. This pattern results from the fact that the proportion of sequences derived from each of the two V segment alleles (novel and existing) changes dramatically depending on the mutation count per sequence. In general, the frequency of sequences with varying mutation counts (per sequence) follows a bimodal distribution for unsorted B cells (Fig. S1). Many sequences (derived from naive or IgM cells) have no mutations, or perhaps a small number because of sequencing errors, whereas many other sequences (derived from class-switched sequences) are generally highly mutated. Thus, when all IGHV alleles are known, there are few sequences expected in the range of one to five mutations. Consequently, when a novel allele exists, it creates sequences that appear to carry a single “mutation” (or a low number of mutations depending on how many polymorphisms are present), and such sequences will be highly overrepresented in these groups (Fig. S2). Overall, these results show that polymorphic positions exhibit distinct patterns of mutation accumulation, and that these patterns differ depending on whether the novel allele is homozygous or heterozygous in the subject being analyzed.
To determine whether the predicted patterns for V segment polymorphisms could be found in experimental data, mutation accumulation plots were generated for every germline IGHV allele for each dataset in Table 1. Manual inspection of these plots found that although most nucleotide positions tended to exhibit higher mutation frequencies as sequence-wide mutation counts increased, as predicted for nonpolymorphic positions, several potential polymorphisms were also identified (Fig. 3, Center). For example, position 163 in sequences aligning to IGHV1-2*02 in subject M5 appeared to be a homozygous polymorphism (Fig. 3, Upper Center), whereas this same polymorphism in subject hu420143 appeared to be heterozygous (Fig. 3, Lower Center). Interestingly, the putative homozygous polymorphism was the most frequently mutated position (Fig. 3, Upper Right), suggesting that a simple analysis of mutation frequency may have uncovered this allele in subject M5. However, in subject hu420143, where the polymorphism appears to be heterozygous, there were many other positions that had a similar mutation frequency (Fig. 3, Lower Right). This finding suggests that identifying polymorphisms based on the pattern of mutation accumulation (rather than overall mutation frequency) should have increased sensitivity for detecting novel alleles, particularly when they are heterozygous.
Automated Method to Detect Novel Alleles from V(D)J-Rearranged Samples.
Leveraging the distinct pattern of mutation accumulation exhibited by polymorphic positions, a regression-based method was developed to automate the process of recognizing single nucleotide polymorphisms, which could then be combined to predict novel IGHV alleles. For each IMGT-numbered position in each existing germline IGHV allele, polymorphisms were detected by regressing the mutation frequency of the specific position against the mutation count of the entire V segment (complete details are provided in Methods). Nucleotide positions with y-intercepts above 0.125 (as determined by a Student’s t test with P < 0.05) were considered potentially polymorphic, with the specific polymorphism defined by the most commonly mutated-to nucleotide at that position. Nonpolymorphic positions are expected to have a y-intercept of zero, and the threshold of 0.125 was chosen for polymorphisms to detect heterozygous alleles that may be expressed at low frequency. Manual inspection of the data suggested that this was a reasonable threshold, and the simulation results below confirm this choice.
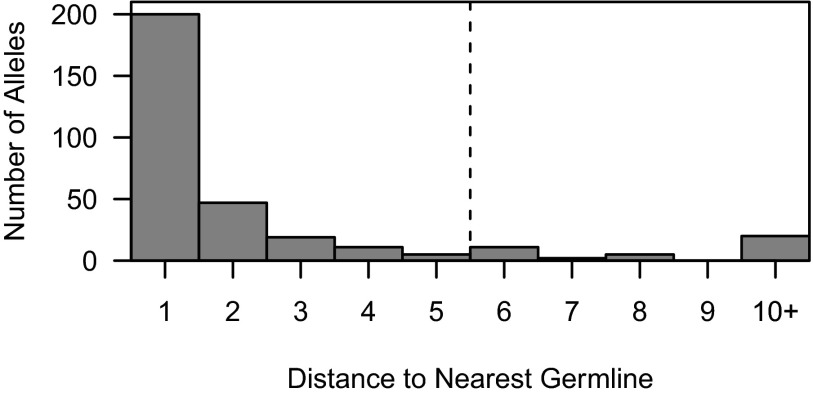
The apparent mutation count in germline sequences using novel IGHV alleles is determined by the most closely related known allele. This is because sequences using the novel allele will get mapped to these existing alleles, and any polymorphisms will be interpreted as mutations. Thus, if the novel allele contains a single nucleotide difference relative to an existing allele, then all of the germline sequences using the novel allele will appear to have one mutation (at the polymorphic position). If the novel allele contains multiple polymorphisms, then few sequences will be found with mutation counts below the number of polymorphisms. In this case, the lower bound on the mutation count for regression analysis should be set to the number of polymorphisms differentiating the novel allele from its most closely related known allele. We detected this lower bound by analyzing the observed number of sequences at mutation counts between one and five. If the number of sequences at any of these counts was found to be a statistical outlier (i.e., higher than expected; see Methods), then the lower bound of the regression was set to be the count at the smallest such outlier. The range of one to five was chosen because 88% of known IGHV alleles are related to another allele that is less than six nucleotides away (Fig. 4), and we thus expect that most novel alleles will be similarly related to at least one known allele. Additionally, the 16 new IGHV alleles discovered in a study of Papua New Guineans differed by at most four nucleotides from known alleles (20). The regression also used an upper bound of 10 for the mutation count, because accumulation patterns deviate from linear after this point, and outliers at mutation counts greater than five were not considered so that the regression would have sufficient data. Overall, the proposed method was designed to detect polymorphisms in novel alleles that are related to known alleles by five or fewer nucleotide changes, which we expect will constitute nearly 90% of novel alleles, as seen in Fig. 4.
Fig. 4.
Distances between known alleles. For each IGHV germline allele sequence in the IMGT database, the Hamming distance to every other germline allele was calculated to determine the nearest allele. Gaps and degenerate alleles were excluded from the distance calculation, and pairs of alleles with distance zero were excluded altogether.
To construct the novel IGHV alleles implied by the individual polymorphisms identified in the regression-based approach, potential germline versions of the novel allele were generated by introducing every combination of polymorphisms into the best-aligning germline allele. The entire dataset was then searched to identify perfect matches for each of the proposed novel alleles. To rule out the possibility that the observed pattern was caused by a large clonal expansion, only novel alleles for which the most prominent J gene-junction length combination accounted for 15% or less of these perfect-match sequences were retained. An example of these steps (y-intercept detection, mutated-to nucleotide frequency, and J gene/junction length distribution) for a given position in a given allele can be seen in Fig. S3, in addition to the nucleotide sequence of a proposed novel allele.
Once the set of novel IGHV alleles were determined, the initial V(D)J assignments provided by IMGT/HighV-QUEST for each sequence were re-evaluated to test if one of the novel alleles provided a better alignment. If one of the novel alleles had a Hamming distance lower than the existing germline V assignment, then the existing assignment was replaced by the novel allele with the minimum distance. In the case of ties, all of the alleles were included in the assignment.
Automated Method Detects Novel IGHV Alleles from Experimental Samples.
Eleven putative novel alleles (Table 2) were identified by applying the regression-based approach and filtering (described above) to the Ig sequencing data from seven subjects listed in Table 1. These alleles were missing from the IMGT database, and were also not present in VBASE2 (22) or the UNSWIg human heavy chain repertoire (23). Excluded from Table 2 are six alleles that were sequencing artifacts (further discussed in SI Text), and which were eventually removed by the genotyping process described later. Of the novel alleles listed in Table 2, nine differed by a single position from the nearest known allele, one allele differed by two positions, and another by three positions. Interestingly, every subject examined contained at least one novel IGHV allele, suggesting that existing germline segment databases are largely incomplete, as previously suspected (19–21).
Table 2.
Novel IGHV alleles identified by TIgGER
| No. | Nearest allele | Polymorphic site(s) | Subject(s) | Technology | Perfect matches |
| 1† | 1-2*02 | T163C | hu420143 | 454 | 629 |
| M5 | MiSeq | 736 | |||
| M4 | MiSeq | 283 | |||
| 2 | 1-8*02 | G234T | PGP1 | 454 | 647 |
| PGP1 | MiSeq | 453 | |||
| 3 | 1-18*01 | T111C | M2 | MiSeq | 1560 |
| 4 | 3-11*03 | T13G | 420IV | 454 | 866 |
| 5 | 3-11*03 | C300T | M2 | MiSeq | 101 |
| 6 | 3-20*01 | C307T | PGP1 | 454 | 187 |
| PGP1 | MiSeq | 85 | |||
| 7 | 1-69*06 | C191T | M3 | MiSeq | 284 |
| 8†† | 2-70*01 | T164G | PGP1 | 454 | 220 |
| 9 | 3-64*05 | A210C, G265C | M2 | MiSeq | 251 |
| 10 | 3-43*01 | A112G, C222T, A286G | 420IV | 454 | 192 |
| 11 | 3-9*01 | C296T | PGP1 | 454 | 128 |
| PGP1 | MiSeq | 202 |
Inferred novel alleles are listed in order of prevalence. For each novel allele, the nearest IGHV allele from IMGT is listed, along with the sites that differ between the two. (The first letter represents the database nucleotide, the number is the IMGT-numbered nucleotide position, and the second letter represents the novel allele’s nucleotide at that position). Allele 8 could not be detected in the PGP1 MiSeq data as the allele-differentiating position is in the primer area. Excluded from this table are six alleles that were predicted by TIgGER but are believed to be artifacts (see SI Text for further discussion).
The same sequence is observed if the polymorphic site T299C is introduced into IGHV1-2*05.
The same sequence is observed if the polymorphic site G2A is introduced into IGHV2-70*11.
Several observations in the data support the validity of these predicted novel alleles. First, in all cases, hundreds of unique sequences (i.e., differing by CDR3 or J segment) carrying an unmutated form of the novel IGHV allele were observed (Table 2). Second, for the subject where data were available from multiple sequencing platforms (PGP1), the same set of novel alleles was predicted in both platforms. In this comparison, it is important to note that one of the novel alleles could not be detected in the MiSeq data because the position required to differentiate it from IGHV2-70*11 (IMGT position 2) was part of the V primer used for amplification (and thus masked by pRESTO). Third, the novel allele which differs from IGHV1-2*02 at position 163 was discovered independently in three subjects (hu420143, M4, and M5). Subject M5 appears to be homozygous for the novel allele, whereas subjects hu420143 and M4 appear to be heterozygous (Table S1). Finally, four of the predicted novel alleles (numbered 3, 4, 9, and 11 in Table 2) were subsequently added to the IMGT database after our analysis was complete (as IGHV1-18*04, IGHV3-11*05, IGHV3-64D*06, and IGHV3-9*03, respectively). The addition of these alleles to the IMGT database was the result of genomic DNA sequenced as a part of two studies unrelated to our own (20, 21). Taken together, these features strongly suggest that the sequence predictions made by TIgGER represent true novel alleles.
IGHV Alleles Are Detected with High Sensitivity.
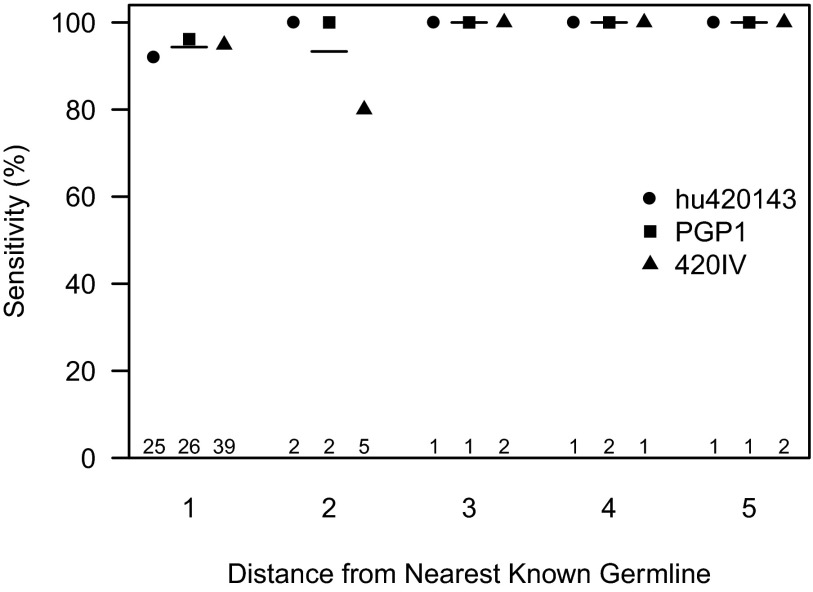
Performance was quantified by removing known alleles from the IMGT germline database, and testing whether our method could recover the “novel” allele. Only alleles assigned to at least 500 Ig sequences, and for which our method did not predict a novel allele in the full dataset, were included in the analysis. Additionally, we ensured that the next-best aligning germline did not differ from the initial alignment beyond the third framework (structural) region of the Ig. This analysis was carried out using the 454 datasets, as in some cases the primer positions for the MiSeq data overlap with the start of the V sequence and prevent accurate germline identification. As shown in Fig. 5, novel alleles with a single polymorphism were detected with 95% sensitivity, as were novel alleles with up to five polymorphisms. Most novel alleles are expected to be found in this range (Fig. 4), and no novel alleles with more than five polymorphisms were detected, as expected based on the design of the method. For the three subjects analyzed (hu420143, PGP1, and 420IV) the false-discovery rate (new alleles called incorrectly divided by all new alleles called) was zero. Thus, the proposed method exhibited excellent performance, and should be effective at identifying most novel alleles.
Fig. 5.
Sensitivity of polymorphism detection method. For each allele assignment given to at least 500 sequences in a subject, matching sequences were all reassigned to another allele of that gene. The TIgGER polymorphism detection method was then applied, to test whether the positions required to recreate the artificially excluded known allele could be detected. This analysis was performed for all alleles in samples derived from the 454 sequencing platform, excluding those in which TIgGER had previously identified polymorphisms. Horizontal bars indicate mean sensitivity across the three subjects tested. For each subject, the number of alleles falling into each distance group is indicated along the bottom.
Novel IGHV Alleles Appear at High Frequency.
Although IGHV genes can be duplicated in some subjects, with IGHV1-69 being a frequent example (as reviewed in ref. 5), it is expected that each subject carries either one or two alleles of most genes. However, IMGT/HighV-QUEST assigned an average of 4.3 alleles per gene across the three subjects sequenced by 454, as shown in Table 1. Thus, many of these initial V allele assignments are likely to be incorrect. This problem results in part from the difficulty in identifying the specific allele for highly mutated sequences, and IMGT/HighV-QUEST often assigns multiple potential alleles in such cases. Indeed, 13–21% of sequences were assigned multiple alleles in the three subjects sequenced by 454, as shown in Table 1. We propose that many of these assignments could be corrected by analyzing the global repertoire properties of each subject to identify a subject-specific genotype (i.e., the set of IGHV alleles carried by the subject), which could then be used to constrain the potential allele assignments. Improved IGHV assignments would allow us to better determine the prevalence of novel alleles in the population.
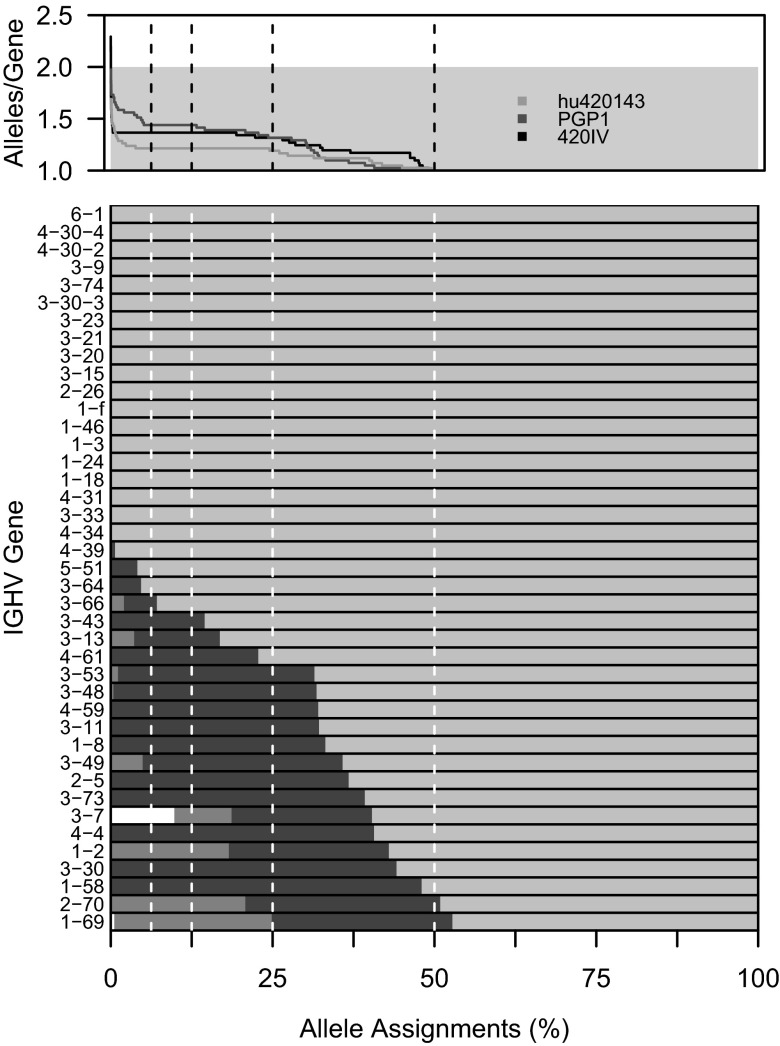
Existing approaches for inferring Ig genotypes are based on identifying the set of alleles that appear above a specified frequency (6, 19). This frequency is calculated using the set of unmutated sequences, as existing V(D)J segment identification tools are most accurate at identifying and aligning these sequences to their germline sequences (2). Indeed, we found that limiting our calculation to unmutated sequences led to average numbers of alleles per gene of 2.0 across the 454 datasets. A key issue is how to determine the appropriate frequency for inclusion in the genotype. Using the set of IGHV genes that were observed in more than 0.01% of sequences, we found that even a very low allele frequency cut-off (6.25%, or 1 of 16, of unmutated sequences assigned to the IGHV gene) reduced the total number of alleles per subject from 80 to 55 (Fig. 6). The allele count continued to decrease as this threshold for inclusion was increased, but appeared to stabilize between minimum frequencies of inclusion of 6.25% and 12.5%, at which point there was an average of 1.3 alleles per IGHV gene (Fig. 6). Based on this observation, we defined the subject-specific genotype to include all alleles whose frequency was 12.5% or higher. Application of this approach to the seven subjects in Table 1 found that each subject had a unique genotype, a result similar to that found in ref. 6. Most (63%) IGHV genes were homozygous, and duplications of IGHV1-69 or IGHV2-70 were found in three cases (Table S1). Duplications of these genes have also been observed in other studies (21). Significantly, 11 of the novel alleles appeared in the final subject-specific genotypes. Collectively, the genotypes contained 82 unique IGHV alleles, 11 of which (13%) were novel alleles. This high frequency of novel alleles suggests that current germline databases are incomplete, and that many more IGHV alleles remain to be discovered.
Fig. 6.
The influence of allele assignment frequency cut-offs on IGHV genotype zygocity. TIgGER was used to determine subject-specific IGHV genotypes using different values for the allele assignment frequency cut-off (i.e., fraction of assignments to a gene segment that are required to be composed of a single allele to be included in the genotype). For each of the three 454 datasets, the number of alleles included in the inferred IGHV genotype is shown as a function of the allele assignment frequency cut-off (Upper). For PGP1, the distribution of allele assignments is shown for each gene included in the inferred genotype (Lower). For each bar, the lightest gray represents the most common allele, darkest gray the second most common, medium gray the third, and white for all others.
Subject-Specific Genotype Improves Allele Assignments.
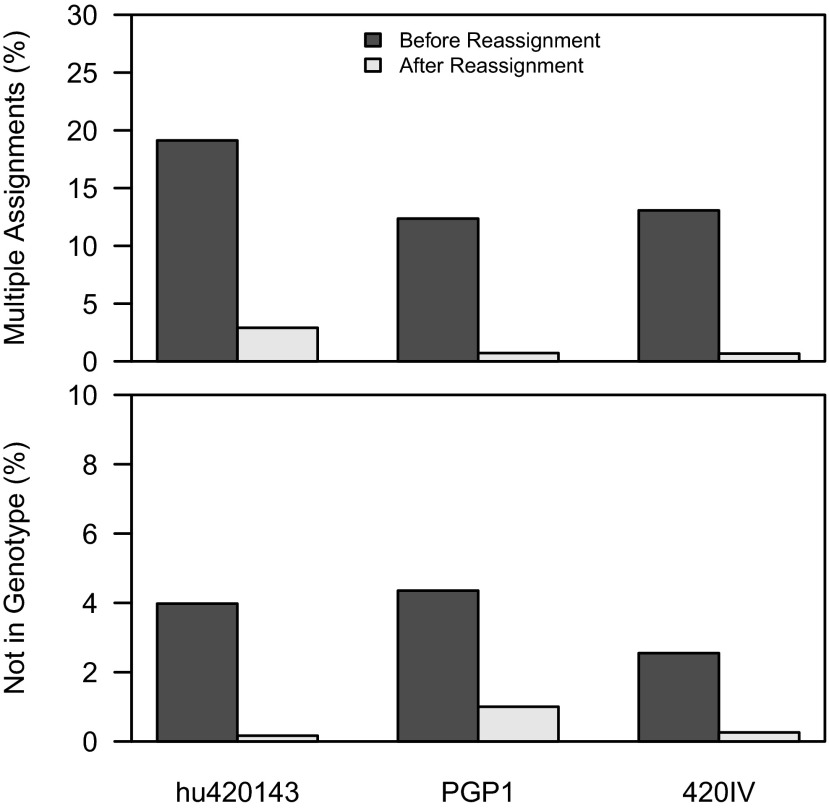
On average, 19% of Ig sequences included IMGT/HighV-QUEST assignments to V segments that were not included in the subject-specific genotype. Moreover, in 4% of sequences, none of the V assignments were included in the genotype. Among the latter, virtually all (93%) of the sequences were mutated, and they tended to carry a higher mutation load (Fig. S4). These sequences also tended to be assigned to multiple V segments, reflecting the difficulty in choosing the V segment. Thus, these initial assignments provided by existing germline determination software are likely to be incorrect. To address this problem, we reassigned these sequences to the best matching segment that was included in the subject-specific genotype. By removing V assignments not in the personalized germline database, the number of Ig sequences assigned to multiple IGHV alleles was reduced by 92% (Fig. 7, Upper). In addition, the number of low-confidence assignments was reduced by a similar amount (90%) (Fig. 7, Lower). This rereassignment was most likely to impact sequences that were closely related to multiple germline segments (Fig. S5), and the rate was consistent with the previously estimated error rate for allele assignment methods (∼5%) (2). Following these personalized allele assignments, an average of 4,835 sequences per dataset (∼3%; range 1,316–14,389, or about 1–5%) were assigned to novel alleles. Overall, this process produced high-quality assignments for 98% of sequences.
Fig. 7.
IGHV genotyping greatly improves allele assignments. The percentage of sequences with multiple IGHV allele assignments before and after genotype-based allele reassignment was determined for the three subjects sequenced by 454 (Upper). The percentage of sequences which could not be assigned to genotype alleles before and after genotype-based allele reassignment for the same three subjects was also determined (Lower).
Discussion
Adaptive immune responses are a critical component of the human defense against infection, but overactivation can also lead to pathology. Genetic polymorphisms in immune-related genes have been implicated in several diseases, including SLE, MS, and RA (31–33). The MHC locus is highly polymorphic, and is often one of the strongest signals in genome wide association studies (34). Subjects also differ greatly in the set of V(D)J segment alleles that they carry (6, 19, 21) and some V segment alleles have been associated with disease (including SLE, MS, RA, and type 1 diabetes, as summarized in ref. 5). Along with the underlying genetics of the Ig locus, properties of the expressed B-cell Ig repertoire, such as diversity, have been associated with disease and clinical status (5–10). A critical step in identifying these linkages is the accurate determination of the set of germline V(D)J segment alleles carried by a subject.
We have created TIgGER, an automated method for identifying novel Ig V segment alleles based on the analysis of mutation patterns in Rep-Seq data. Existing methods for Ig polymorphism detection are based on the identification of positions that are perceived as mutated in an overwhelming number of sequences (19). Although such approaches may detect novel alleles that are homozygous, polymorphisms in heterozygous alleles can easily be missed. Despite the simplicity of these approaches, few existing Rep-Seq studies include any kind of novel allele detection step in their analysis pipeline. These studies implicitly assume that the germline allele databases used for V(D)J assignment (most often IMGT) are complete. Our results clearly show that this assumption is faulty, and many subjects are likely to carry novel V segment alleles. Of the seven subjects analyzed in this study, all carried at least one novel V segment allele. In fact, these novel IGHV alleles accounted for 13% of the unique alleles determined to be in the genotypes of those subjects. Although the ethnicity of most of the subjects was unknown, PGP1 and hu420143 represent subjects of European ancestry; thus, it appears that novel alleles exist even among the most-studied ethnic group. Clearly, we have just begun to identify the extent of V(D)J segment diversity in the human population, and polymorphism detection should be a standard part of Rep-Seq analysis pipelines.
During the course of this study, four of the novel alleles detected by TIgGER were added to the IMGT database as a result of two independent studies, which identified these alleles from DNA (20, 21). IMGT will not include the additional seven novel V segment alleles identified by TIgGER because they do not meet the current requirement that new alleles must have been amplified from the genomic germline region and include the full sequence. Given the large number of Rep-Seq studies being carried out on mRNA, these strict requirements mean that many novel alleles will not be represented in IMGT. This represents a significant challenge for Rep-Seq analysis with clinical implications (24), which could be avoided by adopting a classification system to indicate alleles with varying levels of underlying evidence, similar to what has been proposed (23). We are currently working to submit the novel alleles predicted by TIgGER to the UNSWIg human heavy chain repertoire (23), and they will also be hosted on our website (clip.med.yale.edu/tigger).
Along with identifying novel alleles carried by a subject, the accuracy of Rep-Seq analysis depends on determining which alleles are not carried by a subject. Existing V(D)J segment assignment tools, such as IMGT/HighV-QUEST (15), SoDA2 (2), iHMMune-Align (16), and IgBLAST (17), operate on a sequence-by-sequence basis. A consequence of this approach is that many different alleles for each gene can be assigned in a single subject. Indeed, in the seven subjects analyzed in this study, an average of 4.4 V segment alleles per gene was assigned by IMGT/HighV-QUEST. Because most genes are expected to be present at single copy, this average should be well below two for most subjects. To correct these allele assignments, we inferred each subject’s IGHV genotype based on a simple threshold frequency for inclusion. This personalized database was then used to reassign alleles. This approach had two important consequences. First, Ig sequences that were assigned to V segments not included in the genotype could be reassigned to a V segment in the genotype. We also found that most Ig sequences that were initially assigned to multiple alleles by IMGT/HighV-QUEST were reassigned to a single allele. Indeed, using this method we were able to confidently reassign single alleles to 92% of sequences with multiple allele assignments, and the ∼4% of sequences whose allele assignments were not in the inferred genotype was reduced to ∼0.5%. Ideally, the same method used to generate the initial V(D)J assignments would be applied to carry out the reassignment process using the personalized germline database. Unfortunately, IMGT/HighV-QUEST does not currently allow for the use of a custom germline database.
In designing Rep-Seq analysis pipelines, it is important to carefully consider the order of the analysis steps. We have determined that detection of polymorphisms must be done before genotype inference. If the order of these steps is reversed, no unmutated Ig sequences assigned to novel alleles will be found (and thus the allele will be wrongfully excluded from the genotype). However, carrying out polymorphism detection first leads to many sequences with multiple germline allele assignments during this step. Such sequences cannot be excluded from the analysis. If sequences with multiple allele assignments are excluded, the approach will be unable to detect polymorphic nucleotides in positions that differentiate known alleles. TIgGER identified just such a case. Ig sequences using the novel allele IGHV1-2*02_T163C were initially assigned to both IGHV1-2*02 and IGHV1-2*05 by IMGT/HighV-QUEST. These two known alleles differ from each other in two positions, and the novel allele differs from each of the known alleles by one position.
TIgGER has high sensitivity for identifying novel IGHV alleles that differ from known alleles by five or fewer single nucleotide polymorphisms. Because most known IGHV alleles are within five mutations of another known allele, this approach should detect most novel alleles. However, ∼10% of known alleles are six or more nucleotides from their closest known allele. Thus, it will be important to develop methods that can find more distantly related novel alleles. Alleles that differ by insertions or deletions are also not addressed by TIgGER. The sensitivity of TIgGER is dependent on having a large number of sequences with relatively low mutation frequency (≤10 mutations per sequence); although this is reasonable for many studies, it would not be the case for studies that focus only on mature, highly mutated populations. In such cases, one option is to include a collection of less mutated “background” populations as part of the experimental design. Alternatively (or additionally), other methods might also be developed to work on these highly mutated subsets. Although we have focused on applying TIgGER to IGHV allele detection, this approach should also be applicable to allele detection of any other Ig gene segment. Once TIgGER has identified novel alleles, the V(D)J segments of all sequences are reassigned in case they use one of these new segments. Whereas any of the existing V(D)J assignment approaches can be used for this task (2, 15–17), this study used a method based on simple Hamming distance because the implementation of current V(D)J assignment methods does not allow for easy incorporation of a modified germline database. It should be straightforward to extend these methods for future analysis.
TIgGER identified 11 novel IGHV alleles in just seven subjects using moderately deep Rep-Seq data. The fact that so many novel alleles were identified in so few subjects implies that existing gene segment allele databases are substantially incomplete, and much remains to be discovered about these highly polymorphic genes. We expect that applying TIgGER to Rep-Seq data from other subjects will identify additional novel alleles, and germline segment databases will enter a phase of rapid expansion. We strongly recommend that Rep-Seq studies include polymorphism detection as part of their standard analysis pipeline.
Methods
Data Collection and Preprocessing.
Samples coincide with those used by a previous study (25), and were originally collected and sequenced as part of two other studies (28, 29). Sequencing results were preprocessed to remove low-quality reads, annotate and mask primers, assemble paired-end reads, and remove duplicate sequences using the Repertoire Sequencing Toolkit (pRESTO) (30) (clip.med.yale.edu/presto) as previously described (25). Sequences were then assigned to germline IGHV gene segment alleles, based on alignments to a database of known IGHV alleles, using IMGT/HighV-QUEST (27). The 454 datasets were analyzed with IMGT/HighV-QUEST in January 2012; the MiSeq datasets from subjects M2, M3, M4, and M5 in February 2013; and the MiSeq data from PGP1 in September 2013. The alignment-based nucleotide numbering scheme of IMGT was used throughout the analyses (18). Finally, when sequences were grouped by V allele assignment for analysis, all sequences assigned to multiple alleles by IMGT/HighV-QUEST were considered as part of each allele group independently. For example, a single sequence assigned to both IGHV1-2*02 and IGHV1-2*05 was included in the analysis of sequences aligning to IGHV1-2*02 as well as the analysis of sequences aligning to IGHV1-2*05.
Simulation of Polymorphic Sequences.
Using the experimental data from subject hu420143, Sm was defined as the number of sequences with an IGHV mutation count of m. For each m ∈ {0,1,…,10}, simulations were used to generate Sm sequences, each carrying m mutations, based on the method described in previous work (35) with the “S5F” SHM targeting and substitution models (25). To investigate the mutation pattern in a homozygous polymorphic allele, the starting sequence for the simulations was a novel allele (IGHV1-2*02_T163C) that was created by introducing a single nucleotide substitution (T → C at position 163) into a known germline IGHV gene segment (IGHV1-2*02). Next, 1,887 simulated sequences were randomly selected without replacement to match the total number of sequences aligning to IGHV1-2 with m ≤ 10 in subject hu420143. To investigate the mutation pattern in a heterozygous gene consisting of one known allele and one polymorphic allele, an additional set of Sm sequences was generated for m ∈ {0,1,…,10} using IGHV1-2*02 as the starting sequence for the simulation. Then, 1,887 sequences were randomly selected without replacement from the combined IGHV1-2*02 and IGHV1-2*02_T163C simulations to match the total number of sequences aligning to IGHV1-2 with m ≤ 10 in subject hu420143. This sampling scheme assumed that the known and polymorphic alleles are expressed at equal frequency. For both the homozygous and heterozygous cases, the location of mutations was determined by comparing the simulated sequences with the known germline (IGHV1-2*02).
Polymorphism Detection Method.
For each IMGT-numbered position in each germline IGHV allele, polymorphisms were detected by regressing the mutation frequency at the specific position against the mutation count of the entire V segment. Specifically, all sequences assigned to a given germline allele were first binned into groups based on the number of mutations per sequence (1, 2, 3, …, 10), and the mutation frequency (number of times mutated/number of times sequenced) of each position was calculated for each bin. A linear model was then fit to these mutation frequencies versus the number of mutations per sequence using a least-squares objective function. Nucleotide positions with y-intercepts above 0.125 (as determined by a Student’s t test with P < 0.05) were considered potentially polymorphic. The specific polymorphism was defined by the most commonly mutated-to nucleotide at that position. To account for alleles that might contain multiple polymorphic positions, the range of mutations per sequence to be included was determined by testing each bin to see whether the number of sequences included in the bins carrying two, three, four, or five mutations per sequence was an outlier (i.e., more than 1.5-times the interquartile range greater than the third quartile of the number of sequences in the bins carrying between 1 and 10 mutations). If any such bin was found to be a significant outlier, then all bins with fewer IGHV mutations per sequence were excluded from the regression.
Supplementary Material
Acknowledgments
We thank Jason Vander Heiden and Namita Gupta for their helpful discussions and help in testing the Tool for Ig Genotype Elucidation via Repertoire Sequencing (TIgGER). This work was supported by National Institutes of Health Grant R01AI104739; D.G.-M. was supported by National Institutes of Health Grant T15LM07056 from the National Library of Medicine.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. S.D.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417683112/-/DCSupplemental.
References
- 1.Lefranc MP. Nomenclature of the human immunoglobulin heavy (IGH) genes. Exp Clin Immunogenet. 2001;18(2):100–116. doi: 10.1159/000049189. [DOI] [PubMed] [Google Scholar]
- 2.Munshaw S, Kepler TB. SoDA2: A hidden Markov model approach for identification of immunoglobulin rearrangements. Bioinformatics. 2010;26(7):867–872. doi: 10.1093/bioinformatics/btq056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 4.Papavasiliou FN, Schatz DG. Somatic hypermutation of immunoglobulin genes: Merging mechanisms for genetic diversity. Cell. 2002;109(Suppl):S35–S44. doi: 10.1016/s0092-8674(02)00706-7. [DOI] [PubMed] [Google Scholar]
- 5.Watson CT, Breden F. The immunoglobulin heavy chain locus: Genetic variation, missing data, and implications for human disease. Genes Immun. 2012;13(5):363–373. doi: 10.1038/gene.2012.12. [DOI] [PubMed] [Google Scholar]
- 6.Kidd MJ, et al. The inference of phased haplotypes for the immunoglobulin H chain V region gene loci by analysis of VDJ gene rearrangements. J Immunol. 2012;188(3):1333–1340. doi: 10.4049/jimmunol.1102097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyd SD, Liu Y, Wang C, Martin V, Dunn-Walters DK. Human lymphocyte repertoires in ageing. Curr Opin Immunol. 2013;25(4):511–515. doi: 10.1016/j.coi.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gibson KL, et al. B-cell diversity decreases in old age and is correlated with poor health status. Aging Cell. 2009;8(1):18–25. doi: 10.1111/j.1474-9726.2008.00443.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zuckerman NS, et al. Ig gene diversification and selection in follicular lymphoma, diffuse large B cell lymphoma and primary central nervous system lymphoma revealed by lineage tree and mutation analyses. Int Immunol. 2010;22(11):875–887. doi: 10.1093/intimm/dxq441. [DOI] [PubMed] [Google Scholar]
- 10.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson KJL, et al. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell Host Microbe. 2014;16(1):105–114. doi: 10.1016/j.chom.2014.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parameswaran P, et al. Convergent antibody signatures in human dengue. Cell Host Microbe. 2013;13(6):691–700. doi: 10.1016/j.chom.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benichou J, Ben-Hamo R, Louzoun Y, Efroni S. Rep-Seq: Uncovering the immunological repertoire through next-generation sequencing. Immunology. 2012;135(3):183–191. doi: 10.1111/j.1365-2567.2011.03527.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eisenstein M. Personalized, sequencing-based immune profiling spurs startups. Nat Biotechnol. 2013;31(3):184–186. doi: 10.1038/nbt0313-184b. [DOI] [PubMed] [Google Scholar]
- 15.Alamyar E, Giudicelli V, Duroux P, Lefranc M-P. 2010. IMGT/HighV-QUEST: A high-throughput system and web portal for the analysis of rearranged nucleotide sequences of antigen receptors. High-throughput version of IMGT/V-QUEST. 11thJournées Ouvertes en Biologie, Informatique et Mathématiques September 7–9, 2010, Montpellier, France. Available at www.imgt.org/IMGTindex/IMGTHighV-QUEST.html. Accessed January 22, 2015.
- 16.Gaëta BA, et al. iHMMune-align: Hidden Markov model-based alignment and identification of germline genes in rearranged immunoglobulin gene sequences. Bioinformatics. 2007;23(13):1580–1587. doi: 10.1093/bioinformatics/btm147. [DOI] [PubMed] [Google Scholar]
- 17.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: An immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41(Web Server issue):W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lefranc M-P, et al. IMGT unique numbering for immunoglobulin and T cell receptor variable domains and Ig superfamily V-like domains. Dev Comp Immunol. 2003;27(1):55–77. doi: 10.1016/s0145-305x(02)00039-3. [DOI] [PubMed] [Google Scholar]
- 19.Boyd SD, et al. Individual variation in the germline Ig gene repertoire inferred from variable region gene rearrangements. J Immunol. 2010;184(12):6986–6992. doi: 10.4049/jimmunol.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, et al. Genomic screening by 454 pyrosequencing identifies a new human IGHV gene and sixteen other new IGHV allelic variants. Immunogenetics. 2011;63(5):259–265. doi: 10.1007/s00251-010-0510-8. [DOI] [PubMed] [Google Scholar]
- 21.Watson CT, et al. Complete haplotype sequence of the human immunoglobulin heavy-chain variable, diversity, and joining genes and characterization of allelic and copy-number variation. Am J Hum Genet. 2013;92(4):530–546. doi: 10.1016/j.ajhg.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Retter I, Althaus HH, Münch R, Müller W. VBASE2, an integrative V gene database. Nucleic Acids Res. 2005;33(Database issue):D671–D674. doi: 10.1093/nar/gki088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Jackson KJL, Sewell WA, Collins AM. Many human immunoglobulin heavy-chain IGHV gene polymorphisms have been reported in error. Immunol Cell Biol. 2008;86(2):111–115. doi: 10.1038/sj.icb.7100144. [DOI] [PubMed] [Google Scholar]
- 24.Xochelli A, et al. Immunoglobulin heavy variable (IGHV) genes and alleles: New entities, new names and implications for research and prognostication in chronic lymphocytic leukaemia. Immunogenetics. 2015;67(1):61–66. doi: 10.1007/s00251-014-0812-3. [DOI] [PubMed] [Google Scholar]
- 25.Yaari G, et al. Models of somatic hypermutation targeting and substitution based on synonymous mutations from high-throughput immunoglobulin sequencing data. Front Immunol. 2013;4:358. doi: 10.3389/fimmu.2013.00358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohm-Laursen L, Barington T. Analysis of 6912 unselected somatic hypermutations in human VDJ rearrangements reveals lack of strand specificity and correlation between phase II substitution rates and distance to the nearest 3′ activation-induced cytidine deaminase target. J Immunol. 2007;178(7):4322–4334. doi: 10.4049/jimmunol.178.7.4322. [DOI] [PubMed] [Google Scholar]
- 27.Brochet X, Lefranc M-P, Giudicelli V. IMGT/V-QUEST: The highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36(Web Server issue):W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laserson U, et al. High-resolution antibody dynamics of vaccine-induced immune responses. Proc Natl Acad Sci USA. 2014;111(13):4928–4933. doi: 10.1073/pnas.1323862111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stern JN, et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci Transl Med. 2014;6(248):ra107. doi: 10.1126/scitranslmed.3008879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vander Heiden JA, et al. pRESTO: A toolkit for processing high-throughput sequencing raw reads of lymphocyte receptor repertoires. Bioinformatics. 2014;30(13):1930–1932. doi: 10.1093/bioinformatics/btu138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beecham AH, et al. International Multiple Sclerosis Genetics Consortium (IMSGC) Wellcome Trust Case Control Consortium 2 (WTCCC2) International IBD Genetics Consortium (IIBDGC) Analysis of immune-related loci identifies 48 new susceptibility variants for multiple sclerosis. Nat Genet. 2013;45(11):1353–1360. doi: 10.1038/ng.2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kochi Y, Suzuki A, Yamamoto K. Genetic basis of rheumatoid arthritis: A current review. Biochem Biophys Res Commun. 2014;452(2):254–262. doi: 10.1016/j.bbrc.2014.07.085. [DOI] [PubMed] [Google Scholar]
- 33.Cui Y, Sheng Y, Zhang X. Genetic susceptibility to SLE: Recent progress from GWAS. J Autoimmun. 2013;41:25–33. doi: 10.1016/j.jaut.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 34.de Bakker PIW, Raychaudhuri S. Interrogating the major histocompatibility complex with high-throughput genomics. Hum Mol Genet. 2012;21(R1):R29–R36. doi: 10.1093/hmg/dds384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uduman M, et al. Detecting selection in immunoglobulin sequences. Nucleic Acids Res. 2011;39(Web Server issue) Suppl 2:W499–W504. doi: 10.1093/nar/gkr413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.