Significance
The early evolution of animal nervous systems is poorly understood, but comparative genomics provides a new window into the past. One important controversy is about whether nervous systems evolved just once or independently in different animal lineages. In this work, we explore the history of the gene families most central to nervous system function: ion channels. We track when these gene families expanded in animal evolution and find that these gene families radiated on several occasions and, in some cases, underwent periods of contraction. The multiple origins of these gene families may signify large-scale convergent evolution of nervous system complexity.
Keywords: ancestral genome reconstruction, ASIC/ENaC, Cys-loop receptor, ionotropic glutamate receptor, potassium channel
Abstract
Multicellularity has evolved multiple times, but animals are the only multicellular lineage with nervous systems. This fact implies that the origin of nervous systems was an unlikely event, yet recent comparisons among extant taxa suggest that animal nervous systems may have evolved multiple times independently. Here, we use ancestral gene content reconstruction to track the timing of gene family expansions for the major families of ion-channel proteins that drive nervous system function. We find that animals with nervous systems have broadly similar complements of ion-channel types but that these complements likely evolved independently. We also find that ion-channel gene family evolution has included large loss events, two of which were immediately followed by rounds of duplication. Ctenophores, cnidarians, and bilaterians underwent independent bouts of gene expansion in channel families involved in synaptic transmission and action potential shaping. We suggest that expansions of these family types may represent a genomic signature of expanding nervous system complexity. Ancestral nodes in which nervous systems are currently hypothesized to have originated did not experience large expansions, making it difficult to distinguish among competing hypotheses of nervous system origins and suggesting that the origin of nerves was not attended by an immediate burst of complexity. Rather, the evolution of nervous system complexity appears to resemble a slow fuse in stem animals followed by many independent bouts of gene gain and loss.
Animal nervous systems are complex cellular networks that encode internal states and behavioral output. They achieve this complexity primarily in two ways. First, nervous systems encode information in a wiring scheme whose connections differ in strength and sign (excitatory or inhibitory). The strengths can often change in an activity-dependent fashion (1). Second, nervous systems have a dynamic neural code made up of all-or-none action potentials and subtler graded potentials (2). The shape, timing, and duration of evoked electrical potentials vary greatly among—and even within—neurons and can also be activity-dependent. These two types of complex signaling, respectively, among and within cells are the fundamental work of nervous systems (1), and they are made possible by the great variety of ion channel proteins expressed in neurons.
Recent studies have found that most ion channels and proteins involved in the formation of synapses are ancient, having originated long before the advent of nervous systems or even of animal multicellularity (3–7). However, the nature of the first animals and of the cells from which nervous systems evolved are not well understood, although many theories exist (8–11), and little is known about the genomic events that facilitated the rise of complex nervous systems. New information about animal phylogeny has demanded a return to these old questions concerning the nature of the first animals and the evolutionary history of nervous systems (12–15).
This new information concerns the placement of the ctenophores, or comb jellies. Recent studies place ctenophores as the sister group to all other metazoans, a surprising finding given that ctenophores are complex predators with fairly sophisticated nervous systems (15). In contrast, sponges, which traditionally were considered to be the sister group of the remaining animals (16), and placozoans do not have nervous systems (but see refs. 17 and 18). Recent genomic analyses have found that ctenophores are lacking many nervous system and muscle-associated genes, suggesting independent origins of these structures in ctenophores (15, 19, 20). Conversely, the genomic presence and expression of certain developmental genes involved in nervous system differentiation (13, 14) and genes expressed in the synapse (13, 21) indicate deep similarities between ctenophore nervous systems and others. These findings have revived the debate about whether animal nervous systems have one or more origins. Although it is clear that there has been some degree of homoplasy and/or secondary simplifications or losses, the nature and timing of these events remains debatable (refs. 14, 20, and 21 are excellent reviews on this subject).
Many studies have addressed the origin of animal nervous systems by using comparative physiological, developmental, or morphological evidence (22–24). We used a different technique: ancestral gene content reconstruction. This approach has been used to explore the origin of multicellularity (25), the evolution of prokaryotic metabolism (26), and the expansion of G-protein–coupled receptors in animals (27). Gene duplication has long been known to be a major source of novelty and complexity (28), and many of the families we analyzed play few known roles outside of nervous systems. We therefore hypothesized that the elaboration of nervous systems coincided with an expansion of the ion-channel families that are expressed there. We used two methods (27, 29) to reconstruct the ancestral copy number for a variety of ion channel families and tracked the evolution of gene duplications across the animal and fungal tree. The evolution of some of these families have been studied by other groups (15, 27, 30–32), but here we combine current methods of ancestral genome content reconstruction with dense sampling of early branching species and gene families to search for patterns of gene duplication that might illuminate the early history of nervous systems.
Results
Large-Scale Patterns of Gene Gain and Loss.
We used a custom bioinformatics pipeline (SI Appendix, SI Methods) to collect and annotate predicted proteins from 16 ion channel families (Table 1) for 41 broadly sampled opisthokonts (the group that includes animals, fungi, and related protists) and an apusozoan outgroup. The ion-channel families we analyzed play diverse roles in nervous systems (Table 1). Some families, such as the voltage-gated families, are almost solely associated with nervous system function in animals, whereas others, such as P2X receptors, play more diverse roles, with only some isoforms being expressed in nervous systems. This dataset was then used to infer ancestral genome content and the timing of gene duplications using EvolMap (27).
Table 1.
Ion channel families used in this study
| Abbreviation | Full names | Function |
| Ano | Anoctamin, Ca2+ activated Cl− | Smooth muscle, excitability |
| ASC | Epithelial (ENaC), acid sensing channel (ASIC) | Osmoregulation, synaptic transmission |
| CNG/HCN | Cyc. nucleotide gated | Sensory transduction, heart |
| Cav | Voltage-gated Ca+ channel | AP, muscle contraction, secretion |
| ClC | Voltage-gated Cl− channel | Muscle membrane potential, kidney |
| GIC | Glutamate receptor (iGluR) | Synaptic transmission |
| LIC | Ligand-gated, Cys-loop receptor | Synaptic transmission |
| Kv | Voltage-gated K+ channel | AP, membrane potential regulation |
| Nav | Voltage-gated Na+ channel | AP propagation |
| Leak | Sodium leak (NALCN), yeast calcium channel (Cch1) | Regulation of excitability |
| P2X | Purinurgic receptor | Vascular tone, swelling |
| PCC | Polycystine, Mucolipin | Sensory transduction, kidney |
| RyR | Ryanodine receptor, IP3 receptor | Intracellular, muscle contraction |
| Slo | Voltage and ligand-gated K+ | AP, resting potential |
| TPC | Two-pore channel | Intracellular, NAADP signaling |
| TRP | Transient receptor potential | Sensory transduction |
The channels play a variety of roles. Some are almost exclusively associated with nervous system function, whereas others have additional roles outside the nervous system.
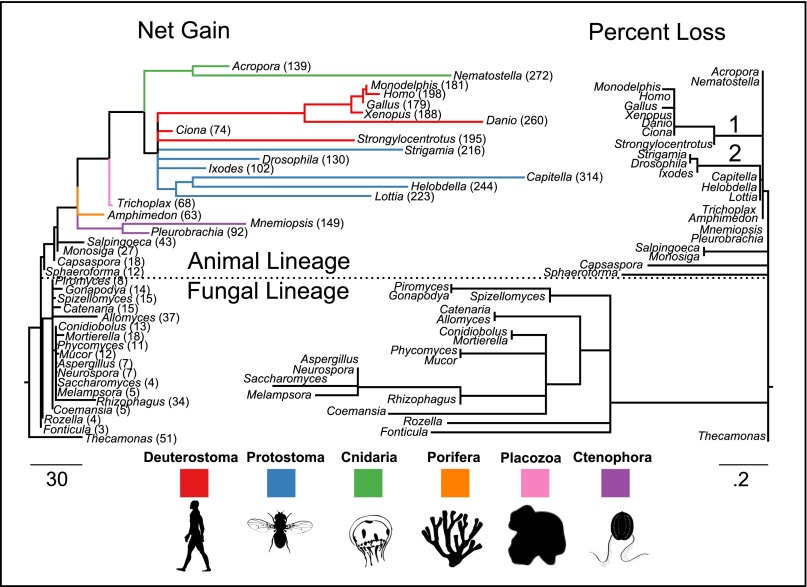
Consistent with previous literature (4–6, 15), we found that these gene families are ancient, with all but two [Cys-loop receptor (LIC) and acid sensing channel (ASC)] being found in the most recent common ancestor (MRCA) of the taxa examined here. Only the ASC family was found to be metazoan-specific. We then pooled all of the families together and plotted net gains and percent losses on the species tree (Fig. 1). The animal lineage has been dominated by gains and the fungal lineage by losses. These patterns are not without exception, however: Two major loss events occurred in the common ancestors of deuterostomes and ecdysozoans (Fig. 1 and SI Appendix, Fig. S1). Both loss events occurred just before major gene family expansions. We also found that the peripheral branches (near the tips) were especially enriched for gene duplications, suggesting multiple independent rounds of gene duplication among the taxa examined.
Fig. 1.
Gain and loss of ion-channel families in opisthokont evolution. The two trees have identical topologies. The branch lengths of the tree on the left are the net gain (gains minus losses), and the branch lengths of the tree on the right represent percent loss (losses minus gains as a percentage of parent copy number). Total numbers of ion channels in each taxon are shown on the left-hand tree. Two branches in animals that had large loss events are labeled: the common ancestors of deuterostomes (1) and of ecdysozoans (2).
Convergent Evolution of Gene Content in Animal with Nervous Systems.
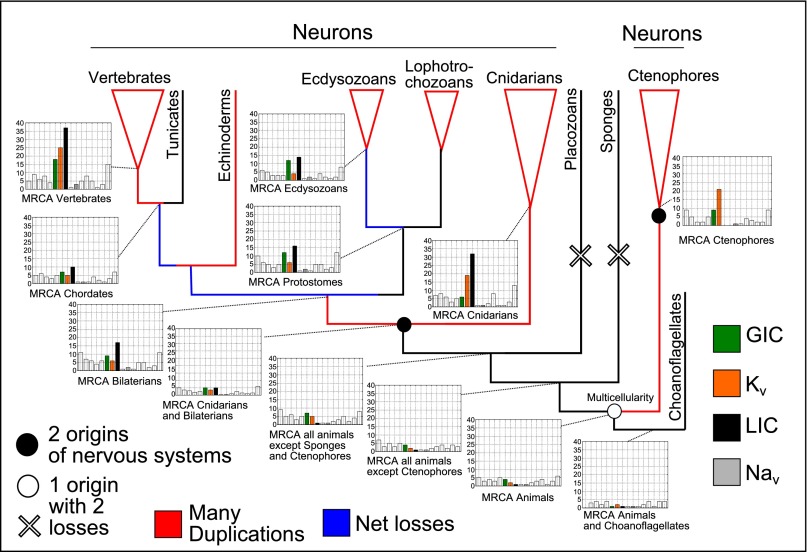
To tease apart the role of the different gene families in these broad-scale patterns, we inferred ancestral gene content and the phylogenetic pattern of gain and loss for each of the 16 ion channel families separately. Counts for key internal nodes are shown on the animal subtree in Fig. 2. We observed large expansions of the LIC, glutamate-gated channel (GIC), and voltage-gated potassium channel (Kv) families at several places on the tree. These gene-family expansions happened independently in the branches leading to the MRCAs of bilaterians, vertebrates, and cnidarians. The vertebrate gene family expansions occurred after the loss event in the branch leading to the MRCA of deuterostomes (Figs. 1 and 2; SI Appendix, Fig. S1). The loss events involved reductions in several families, with the largest families, such as LIC, having the largest losses (SI Appendix, Fig. S1). The branch that gave rise to the MRCA of ctenophores underwent an expansion resembling the expansions in bilaterians and cnidarians, but the LIC family was lost in ctenophores. No expansions were seen in the branches leading to the MRCA of cnidarians plus bilaterians or to the MRCA of animals—two places where nervous systems have been hypothesized to have evolved (12–15, 20).
Fig. 2.
Ion-channel genome content of internal nodes on the animal phylogeny. Ion-channel families that underwent large expansions are colored green (GIC), burnt orange (Kv), and black (LIC). Voltage-gated sodium channels, which drive action potentials but did not experience duplication events on the same scale, are colored gray. All other families are left blank but are shown for comparison. Branches with many duplications are colored red, and those with net losses are colored blue. Two hypotheses about nervous system origins from the literature are also shown. Open symbols show one hypothesis, which posits one origin (open circle) in the common ancestor of animals and two losses (open cross) in placozoans and sponges (13, 14). Filled circles show an alternative hypothesis that nervous systems have two origins—one in the common ancestor of cnidarians and bilaterians and one in the ctenophore lineage (12, 13, 15, 20). Neither hypothesis corresponds with nodes that have large duplication events.
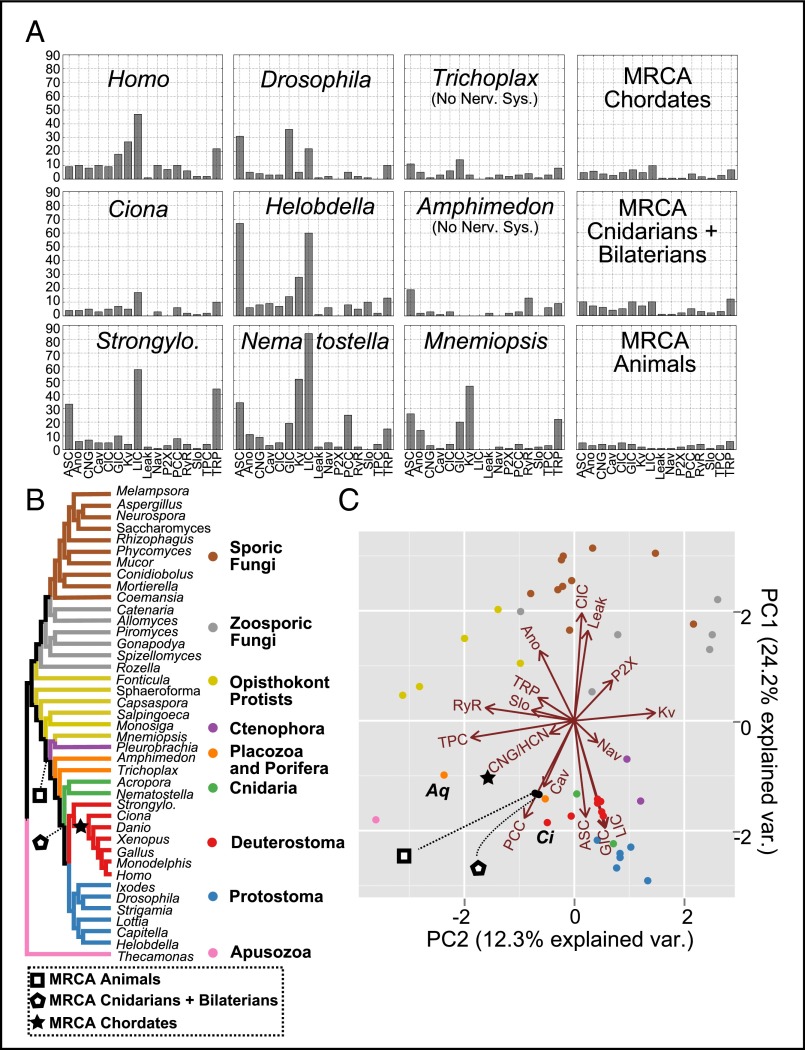
Ecdysozoans and lophotrocozoans also had large expansions of LIC, GIC, and Kv channels, but also had huge expansions of the ASC family (Fig. 3A). These expansions happened mostly in the terminal lineages leading to each species (Fig. 1). Fig. 3A shows ion-channel family counts from representative species from each major lineage represented in Fig. 2. All taxa with nervous systems, with the notable exception of the tunicate Ciona, were enriched for similar gene families. The two taxa without nervous systems, Trichoplax and Amphimedon, had smaller ion-channel complements. The MRCAs of chordates, cnidarians plus bilaterians, and animals each had ion-channel complements that resembled extant animals without nervous systems more than animals with nervous systems.
Fig. 3.
(A) Channel counts of extant species and ancestral species. (B) Species tree showing the relationships of extant taxa and the location of key ancestral nodes. (C) PCA of normalized ion channel gene contents for all tips and three ancestral nodes. Proximity in the space of the two PCs indicates similar gene contents. Loadings of the ion-channel families are shown as vectors in the two axes. The size and direction of the loading vector indicates its correlation with the two components. Loading arrows point to regions where that gene family is in high relative abundance. Labeled species are Amphimedon (Aq) and Ciona (Ci).
To visualize the genomic complements for all channels at all tips, we used principal components analysis (PCA) to reduce the 16 original dimensions (counts for each ion channel family) into the first two principal components (PCs) (Fig. 3C). PCA transforms high-dimensional data into new variables, the PCs, which are linear combinations of the original variables and are ordered by how much of the variance they explain. Proximity in the space of the first two PCs therefore represents similar gene content distributions. Fig. 3C shows the normalized gene contents plotted in the space of the first two PCs and the loadings of each gene on these two axes. We also plot the ion-channel loadings, which show how the abundance of each channel family correlates with the PCs. Thus, dots that cluster near arrows represent genomes with a high relative content of that ion-channel type.
We found that the genome contents of the major lineages were distinguished from each other on the PCA (Fig. 3C). The first PC primarily distinguished fungi—which were dominated by the Leak (Cch1) and ClC families and had lost most of the other types—from animals—which mostly had all of the gene families. Fungi with a swimming zoospore, however, tended to have more channel types, including Cav channels (33). The second PC separated genomes with a higher content of the Ca2+ channel families RyR and two-pore channel (TPC) from those that had more voltage-gated types, primarily Kv. Most genomes that were dominated by Ca2+ channels were from protists.
Animal genomes have a relatively high proportion of synaptic (GIC, LIC, and ASC) and voltage-gated channel types (Nav and Kv). These are the gene families in our dataset most closely associated with nervous system function. Genomes of animals with nervous systems clustered together to the exclusion of the two animals lacking nervous systems: the sponge Amphimedon and the placozoan Trichoplax. These two animals clustered closer to protists due to a larger proportion of Ca2+ channels. The tunicate Ciona was again an interesting exception. Ciona branched from the deuterostome lineage after the major loss event and before the major bout of gene duplication in the ancestor of vertebrates (Figs. 1 and 2). It therefore clustered closer to protists and animals without nervous systems.
The MRCAs of chordates, cnidarians plus bilaterians, and all animals grouped more closely to sponges, placozoans, and protists than to any extant animal with a nervous system, including their immediate descendents (Fig. 3C). This grouping suggests independent gene family expansions of the ion channels that were enriched in extant animals with nervous systems. From the three ancestral genome contents represented by the symbols in the lower left quadrant of Fig. 3C, which is characterized by a relatively high proportion of calcium channels (TPC and Cavs), the ctenophores, cnidarians, and bilaterians independently evolved similar genome contents that caused their genomes to cluster in the lower right quadrant, which is characterized by a high proportion of synaptic ion channel (ASC, GIC, and LIC) and voltage-gated (Kv and Nav) types (Fig. 3C).
We note that our results require accurate reconstruction of gene family histories, and such reconstructions are often difficult over vast evolutionary distances. The best methods to do so are a matter of some debate (34). We relied on two fairly recent methods (27, 29) and found that the patterns of gain and loss described above are not sensitive to the method of ancestral genome reconstruction (SI Appendix, SI Text and Figs. S2 and S3). Our results are also consistent with other studies that have found independent gene family expansions in some of the families studied here (31, 32). Intriguingly, the patterns we found extend to other proteins associated with neural signaling, such as G-protein–coupled receptors, but are not found in proteins that are not strongly associated with neural function (SI Appendix, SI Text and Figs. S4 and S5).
Discussion
We have shown that the major lineages of animals with nervous systems have acquired similar ion-channel complements via convergent gene-family expansions. The gene families that underwent the greatest expansions were two synaptic ion channel types, the LICs and the GICs, as well as ASCs, and Kv. The LIC family was lost in ctenophores, however. Recent evidence suggests that ASCs play a role in synaptic transmission and associative learning (35). Moroz et al. (15) suggested that these genes are key neurotransmitter receptors in ctenophores. Perhaps ASCs fill some of the roles that LICs do in other organisms. Early branching lineages such as ctenophores may therefore be good model systems to explore these understudied channels. The independent bouts of gene-family expansion complement previous studies that have shown convergent evolution of voltage-gated sodium channels between cnidarians and bilaterians (6, 30), of sodium leak channels in many bilaterian lineages (36), of Erg-family potassium channel biophysics in cnidarians and bilaterians (37), and of ligand specificity in LIC channels between protostomes and deuterostomes (32).
Curiously, the major expansions we observed did not occur on any of the nodes where nervous systems are currently hypothesized to have evolved (Fig. 2). Rather, they occurred much later in the common ancestors of vertebrates, bilaterians, cnidarians, and ctenophores and also within the individual lineages of protostomes (Figs. 1–3). The animal stem lineage—from the MRCA of all animals to the MRCA of cnidarians plus bilaterians—experienced very little change in ion-channel genome content, and this content did not differ substantially from the unicellular ancestor of animals and choanoflagellates (Fig. 2). The simplest explanation for this pattern is that nervous systems originated early, were very rudimentary for a long period, and then convergently evolved in complexity by relying on duplications of similar channel types. Another explanation is that stem animals used nervous-system–associated genes in proto-nervous tissues to mediate simple behaviors—as do Trichoplax and the phototactic larvae of sponges (8, 18, 38)—and extant nervous systems were derived independently from these excitable, but nonneural, tissue types (20, 39). Regardless of which scenario is true, our findings suggest a very large role for convergence in extant animal nervous systems. A particularly striking feature of this convergence is the similarity between extant taxa in the relative abundances of the different ion-channel families that underwent the largest expansions (Figs. 2 and 3).
A large repertoire of synaptic channels may have helped nervous systems encode more complex behaviors by facilitating neuronal connections of differing strengths, sign, and context-dependent activity. Kv channels shape action potentials and spike trains, so an expansion of this family may have enabled a dynamic electrical code. This combination of electrical and network complexity is a hallmark of complex nervous systems. Gene-family expansions of channel types associated with these two types of complexity may therefore be a genomic signature for increasing nervous system complexity—a signature that we found to occur at several places in the animal phylogeny.
Our findings may help to explain the distribution of nervous system complexity across the animal tree. In an early attempt to synthesize comparative electrophysiological data and evolutionary theory, Bishop (2) remarked of the phylogenetic distribution of nervous system characteristics, that animals “seem to have available most of the tricks of functioning that any of them employ. Some other factor than availability determines the overall pattern.” This observation, although true, contrasts with the fact that animals with nervous systems do not form a monophyletic group (12, 13, 15), and neither do animals with highly complex, centralized nervous systems (20). We suggest that the ancient origin and independent expansions of the ion-channel types explored here has helped determine this seemingly contradictory pattern. The ancient origins help explain why nervous systems use similar genes in similar roles, and the independent expansions explain why, for instance, neurons and circuits in vertebrates, protostomes, and nonbilaterian invertebrates have such different morphologies (1, 39).
Ion-channel gene expansion has not been monotonic throughout animal evolution. There were two major loss events in the ancestors of deuterostomes and ecdysozoans (Fig. 1 and SI Appendix, Fig. S1). The deuterostome loss events caused the MRCA of chordates and the extant animal Ciona to seem to “revert” to more protist-like genomes (Figs. 2 and 3). The sessile lifestyle and simplified adult brain of ascidians (40), such as Ciona, may be a result or cause of this loss of ion channels. Both loss events were immediately followed by bouts of gene expansions, suggesting the possibility of genomic revolutions where loss events “clear the deck” for a period of increasing complexity and perhaps innovation.
The evolution of animal nervous systems is therefore more complex than has been appreciated. Our results suggest repeated bouts of elaboration and simplification of nervous systems that correlated with expansions and contractions of ion channels (Figs. 1–3) and GPCRs (SI Appendix, Figs. S4 and S5). The shifts in ion-channel gene content are largely captured by the second PC of Fig. 3C, meaning that animal genomes have fluctuated between a higher relative content of Ca2+-channels (TPC, RyR, and Cav) and a higher relative content of other voltage-gated types (Nav and Kv) and synaptic channels (LIC, GIC, and ASC). A switch from Ca2+-based intracellular signaling, which all eukaryotes use, to complex electrical signaling between cells has long been understood as a key animal innovation (4, 41). Our results suggest that this deemphasis of intracellular Ca2+ signaling was not a single evolutionary transition, but that these two types of signaling represent alternate stable states of animal complexity.
Our results are consistent with recent evidence that striated muscle evolved independently in multiple lineages (19) and that ctenophores lack many neurotransmitters associated with vertebrate nervous systems (15). Some studies have already begun to biophysically characterize the expansions of Kv channels and Nav channels in cnidarians and relate them to their homologs in vertebrates, which evolved convergently (30, 31, 37). Investigators have found striking similarities between these channels and those of vertebrates, despite their independent origins. Further study of the biophysical details of genome evolution in animals will help clarify the parallel origins of nervous system functions.
Methods
A more detailed description of the workflow and methods is given in the SI Appendix, SI Methods.
Protein sequences.
Protein sequences were collected from proteomes obtained from the Joint Genome Institute MycoCosm (genome.jgi.doe.gov/programs/fungi/index.jsf), the Origins of Multicellularity Database (www.broadinstitute.org/annotation/genome/multicellularity_project/MultiHome.html), Ensembl (www.ensembl.org/index.html), the Ctenophore genome project websites provided by the Baxevanis and Moroz laboratories (Mnemiopsis Genome Project Portal, research.nhgri.nih.gov/mnemiopsis/; the Pleurobrachia Genome, neurobase.rc.ufl.edu/pleurobrachia), and the Matz laboratory website (matzLABDATA, www.bio.utexas.edu/research/matz_lab/matzlab/Data.html) (42). Details for each proteome used are available in SI Appendix, Table S1. Only proteomes that had protein-locus information were used to avoid redundancy, and only the longest isoform was used for each gene. These sequences were then hand-annotated and filtered to create a conservative estimate for the number of genes for each species.
Analysis.
We used the programs EvolMap (27) and Notung (29) to reconstruct ancestral gene copy number. EvolMap relies on pairwise alignment scores to reconstruct gene duplication history, whereas Notung uses parsimonious gene tree/species tree reconciliation. For gene tree inference, Mafft (43) was used to align protein sequences, and RAxML (44) was used to estimate maximum-likelihood gene trees. The species tree was constructed from the literature (12, 13, 45, 46). A more detailed explanation of the choice of species tree is given in SI Appendix, SI Methods.
PCA was performed in R by using the package prcomp (47). Before PCA was performed, the matrices of gene-family counts for all tips and three interior nodes were normalized by the total gene count for each genome and then centered and scaled by the mean and SD for each column. We decided not to use phylogenetically corrected PCA (48) because this method estimates ancestral states using Brownian motion, whereas we wished to calculate ancestral copy number from protein sequences and plot these values directly on the PCA. The PCA is therefore only interpreted only as a visualization tool, and not as a means to calculate correlations between variates or anything else of that nature.
Program Availability.
All data files are available via Dryad (datadryad.org/; DOI: 10.5061/dryad.60mm2), and all scripts used for the analysis are available on Github (https://github.com/bliebeskind). Commit numbers and repository information can be found in SI Appendix, Table S3. Deposited data include information on the custom programs that were used to parse the output from EvolMap and Notung. The programs used for parsing relied heavily on the python packages pandas, dendropy, and BioPython (49–51).
Supplementary Material
Acknowledgments
We thank Genevieve Smith and Siavash Mirarab for their input on this analysis and Casey W. Dunn for his helpful advice on the manuscript. We also thank the Origins of Multicellularity project; the 1000 Fungal Genomes project; and the Baxevanis, Moroz, and Matz laboratories for making their genomic data freely available. This work was supported by National Science Foundation Cooperative Agreement DBI-0939454.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in Dryad, datadryad.org (DOI: 10.5061/dryad.60mm2).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1501195112/-/DCSupplemental.
References
- 1.Bullock TH, Horridge AG. Structure and Function in the Nervous Systems of Invertebrates. 1st Ed W. H. Freeman; San Francisco: 1965. [Google Scholar]
- 2.Bishop GH. Natural history of the nerve impulse. Physiol Rev. 1956;36(3):376–399. doi: 10.1152/physrev.1956.36.3.376. [DOI] [PubMed] [Google Scholar]
- 3.Burkhardt P, et al. Primordial neurosecretory apparatus identified in the choanoflagellate Monosiga brevicollis. Proc Natl Acad Sci USA. 2011;108(37):15264–15269. doi: 10.1073/pnas.1106189108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai X, Clapham DE. Ancestral Ca2+ signaling machinery in early animal and fungal evolution. Mol Biol Evol. 2012;29(1):91–100. doi: 10.1093/molbev/msr149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chiu J, DeSalle R, Lam HM, Meisel L, Coruzzi G. Molecular evolution of glutamate receptors: A primitive signaling mechanism that existed before plants and animals diverged. Mol Biol Evol. 1999;16(6):826–838. doi: 10.1093/oxfordjournals.molbev.a026167. [DOI] [PubMed] [Google Scholar]
- 6.Liebeskind BJ, Hillis DM, Zakon HH. Evolution of sodium channels predates the origin of nervous systems in animals. Proc Natl Acad Sci USA. 2011;108(22):9154–9159. doi: 10.1073/pnas.1106363108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakarya O, et al. A post-synaptic scaffold at the origin of the animal kingdom. PLoS ONE. 2007;2(6):e506. doi: 10.1371/journal.pone.0000506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jékely G, et al. Mechanism of phototaxis in marine zooplankton. Nature. 2008;456(7220):395–399. doi: 10.1038/nature07590. [DOI] [PubMed] [Google Scholar]
- 9.Nielsen C. Six major steps in animal evolution: Are we derived sponge larvae? Evol Dev. 2008;10(2):241–257. doi: 10.1111/j.1525-142X.2008.00231.x. [DOI] [PubMed] [Google Scholar]
- 10.Pantin CFA. Croonian Lecture: The elementary nervous system. Proc R Soc Lond Ser B - Biol Sci. 1952;140(899):147–168. doi: 10.1098/rspb.1952.0052. [DOI] [PubMed] [Google Scholar]
- 11.Passano LM. Primitive nervous systems. Proc Natl Acad Sci USA. 1963;50(2):306–313. doi: 10.1073/pnas.50.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunn CW, et al. Broad phylogenomic sampling improves resolution of the animal tree of life. Nature. 2008;452(7188):745–749. doi: 10.1038/nature06614. [DOI] [PubMed] [Google Scholar]
- 13.Ryan JF, et al. NISC Comparative Sequencing Program The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science. 2013;342(6164):1242592. doi: 10.1126/science.1242592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan JF. Did the ctenophore nervous system evolve independently? Zoology (Jena) 2014;117(4):225–226. doi: 10.1016/j.zool.2014.06.001. [DOI] [PubMed] [Google Scholar]
- 15.Moroz LL, et al. The ctenophore genome and the evolutionary origins of neural systems. Nature. 2014;510(7503):109–114. doi: 10.1038/nature13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Philippe H, et al. Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19(8):706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- 17.Leys SP, Mackie GO, Meech RW. Impulse conduction in a sponge. J Exp Biol. 1999;202(Pt 9):1139–1150. doi: 10.1242/jeb.202.9.1139. [DOI] [PubMed] [Google Scholar]
- 18.Smith CL, et al. Novel cell types, neurosecretory cells, and body plan of the early-diverging metazoan Trichoplax adhaerens. Curr Biol. 2014;24(14):1565–1572. doi: 10.1016/j.cub.2014.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinmetz PRH, et al. Independent evolution of striated muscles in cnidarians and bilaterians. Nature. 2012;487(7406):231–234. doi: 10.1038/nature11180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moroz LL. On the independent origins of complex brains and neurons. Brain Behav Evol. 2009;74(3):177–190. doi: 10.1159/000258665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marlow H, Arendt D. Evolution: ctenophore genomes and the origin of neurons. Curr Biol. 2014;24(16):R757–R761. doi: 10.1016/j.cub.2014.06.057. [DOI] [PubMed] [Google Scholar]
- 22.Arendt D, Denes AS, Jékely G, Tessmar-Raible K. The evolution of nervous system centralization. Philos Trans R Soc Lond B Biol Sci. 2008;363(1496):1523–1528. doi: 10.1098/rstb.2007.2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Holland LZ, et al. Evolution of bilaterian central nervous systems: a single origin? Evodevo. 2013;4(1):27. doi: 10.1186/2041-9139-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Watanabe H, Fujisawa T, Holstein TW. Cnidarians and the evolutionary origin of the nervous system. Dev Growth Differ. 2009;51(3):167–183. doi: 10.1111/j.1440-169X.2009.01103.x. [DOI] [PubMed] [Google Scholar]
- 25.Richter DJ, King N. The genomic and cellular foundations of animal origins. Annu Rev Genet. 2013;47(1):509–537. doi: 10.1146/annurev-genet-111212-133456. [DOI] [PubMed] [Google Scholar]
- 26.Boussau B, Karlberg EO, Frank AC, Legault B-A, Andersson SGE. Computational inference of scenarios for α-proteobacterial genome evolution. Proc Natl Acad Sci USA. 2004;101(26):9722–9727. doi: 10.1073/pnas.0400975101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakarya O, Kosik KS, Oakley TH. Reconstructing ancestral genome content based on symmetrical best alignments and Dollo parsimony. Bioinformatics. 2008;24(5):606–612. doi: 10.1093/bioinformatics/btn005. [DOI] [PubMed] [Google Scholar]
- 28.Ohno S. Evolution by Gene Duplication. Allen & Unwin; Springer; London: 1970. [Google Scholar]
- 29.Chen K, Durand D, Farach-Colton M. NOTUNG: A program for dating gene duplications and optimizing gene family trees. J Comput Biol. 2000;7(3-4):429–447. doi: 10.1089/106652700750050871. [DOI] [PubMed] [Google Scholar]
- 30.Gur Barzilai M, et al. Convergent evolution of sodium ion selectivity in metazoan neuronal signaling. Cell Reports. 2012;2(2):242–248. doi: 10.1016/j.celrep.2012.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jegla T, et al. Expanded functional diversity of shaker K(+) channels in cnidarians is driven by gene expansion. PLoS ONE. 2012;7(12):e51366. doi: 10.1371/journal.pone.0051366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kehoe J, et al. Aplysia cys-loop glutamate-gated chloride channels reveal convergent evolution of ligand specificity. J Mol Evol. 2009;69(2):125–141. doi: 10.1007/s00239-009-9256-z. [DOI] [PubMed] [Google Scholar]
- 33.Liebeskind BJ, Hillis DM, Zakon HH. Independent acquisition of sodium selectivity in bacterial and animal sodium channels. Curr Biol. 2013;23(21):R948–R949. doi: 10.1016/j.cub.2013.09.025. [DOI] [PubMed] [Google Scholar]
- 34.Hahn MW. Bias in phylogenetic tree reconciliation methods: Implications for vertebrate genome evolution. Genome Biol. 2007;8(7):R141. doi: 10.1186/gb-2007-8-7-r141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wemmie JA, et al. The acid-activated ion channel ASIC contributes to synaptic plasticity, learning, and memory. Neuron. 2002;34(3):463–477. doi: 10.1016/s0896-6273(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 36.Senatore A, Monteil A, van Minnen J, Smit AB, Spafford JD. NALCN ion channels have alternative selectivity filters resembling calcium channels or sodium channels. PLoS ONE. 2013;8(1):e55088. doi: 10.1371/journal.pone.0055088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinson AS, et al. Functional evolution of Erg potassium channel gating reveals an ancient origin for IKr. Proc Natl Acad Sci USA. 2014;111(15):5712–5717. doi: 10.1073/pnas.1321716111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leys SP, Degnan BM. Cytological basis of photoresponsive behavior in a sponge larva. Biol Bull. 2001;201(3):323–338. doi: 10.2307/1543611. [DOI] [PubMed] [Google Scholar]
- 39.MacKie GO. The elementary nervous system revisited. Am Zool. 1990;30(4):907–920. [Google Scholar]
- 40.Mackie GO, Burighel P. The nervous system in adult tunicates: Current research directions. Can J Zool. 2005;83(1):151–183. [Google Scholar]
- 41.Hille B. The Sharpey-Schafer Lecture. Ionic channels: Evolutionary origins and modern roles. Q J Exp Physiol. 1989;74(6):785–804. doi: 10.1113/expphysiol.1989.sp003349. [DOI] [PubMed] [Google Scholar]
- 42.Moya A, et al. Whole transcriptome analysis of the coral Acropora millepora reveals complex responses to CO₂-driven acidification during the initiation of calcification. Mol Ecol. 2012;21(10):2440–2454. doi: 10.1111/j.1365-294X.2012.05554.x. [DOI] [PubMed] [Google Scholar]
- 43.Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: Improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33(2):511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22(21):2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Torruella G, et al. Phylogenetic relationships within the Opisthokonta based on phylogenomic analyses of conserved single-copy protein domains. Mol Biol Evol. 2012;29(2):531–544. doi: 10.1093/molbev/msr185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.James TY, et al. Reconstructing the early evolution of fungi using a six-gene phylogeny. Nature. 2006;443(7113):818–822. doi: 10.1038/nature05110. [DOI] [PubMed] [Google Scholar]
- 47.R Development Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna: 2008. Available at www.R-project.org. [Google Scholar]
- 48.Revell LJ. Size-correction and principal components for interspecific comparative studies. Evolution. 2009;63(12):3258–3268. doi: 10.1111/j.1558-5646.2009.00804.x. [DOI] [PubMed] [Google Scholar]
- 49.Cock PJA, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25(11):1422–1423. doi: 10.1093/bioinformatics/btp163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McKinney W. Python for Data Analysis. O’Reilly; Beijing: 2013. [Google Scholar]
- 51.Sukumaran J, Holder MT. DendroPy: A Python library for phylogenetic computing. Bioinformatics. 2010;26(12):1569–1571. doi: 10.1093/bioinformatics/btq228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.