Significance
Reduced sleep duration is a hallmark of modern-day society and is increasingly associated with medical conditions, such as diabetes, obesity, metabolic syndrome, and cardiovascular disease. Here we present data from a rat model and human clinical study of chronic sleep restriction, both revealing that two metabolites in blood, oxalic acid and diacylglycerol 36:3, are quantitatively depleted under sleep-restricted conditions and restored after recovery sleep. Our findings also reveal a significant overall shift in lipid metabolism, with higher levels of phospholipids in both species and evidence of a systemic oxidative environment. This work provides a potential link between the known pathologies of reduced sleep duration and metabolic dysfunction.
Keywords: metabolomics, lipidomics, sleep restriction, mass spectrometry, sleep deprivation
Abstract
Sleep is an essential biological process that is thought to have a critical role in metabolic regulation. In humans, reduced sleep duration has been associated with risk for metabolic disorders, including weight gain, diabetes, obesity, and cardiovascular disease. However, our understanding of the molecular mechanisms underlying effects of sleep loss is only in its nascent stages. In this study we used rat and human models to simulate modern-day conditions of restricted sleep and addressed cross-species consequences via comprehensive metabolite profiling. Serum from sleep-restricted rats was analyzed using polar and nonpolar methods in two independent datasets (n = 10 per study, 3,380 measured features, 407 identified). A total of 38 features were changed across independent experiments, with the majority classified as lipids (18 from 28 identified). In a parallel human study, 92 metabolites were identified as potentially significant, with the majority also classified as lipids (32 of 37 identified). Intriguingly, two metabolites, oxalic acid and diacylglycerol 36:3, were robustly and quantitatively reduced in both species following sleep restriction, and recovered to near baseline levels after sleep restriction (P < 0.05, false-discovery rate < 0.2). Elevated phospholipids were also noted after sleep restriction in both species, as well as metabolites associated with an oxidizing environment. In addition, polar metabolites reflective of neurotransmitters, vitamin B3, and gut metabolism were elevated in sleep-restricted humans. These results are consistent with induction of peroxisome proliferator-activated receptors and disruptions of the circadian clock. The findings provide a potential link between known pathologies of reduced sleep duration and metabolic dysfunction, and potential biomarkers for sleep loss.
Sleep duration has decreased in modern society, with potential implications for increased incidence of disease. Chronic restriction of sleep duration results in weight gain (1, 2) and has been linked to common clinical phenotypes, such as obesity (3, 4), as well as metabolic (5) and cardiovascular (6) diseases. The mechanisms underlying such effects of sleep restriction are not understood, but an intriguing possibility is that sleep loss alters metabolic function, which could impact many physiological systems. Indeed, links between sleep loss and metabolism have been identified, with sleep-deprived humans displaying perturbations in carbohydrate metabolism and endocrine function (7) and circadian metabolic processes (8). Recent work suggests that sleep drives metabolite clearance within the brain (9) and thus may act as a reparative process at the metabolic level. However, the systemic factors that drive metabolic perturbations during sleep loss and the basis of downstream pathophysiology are largely unknown.
The impact of sleep restriction on circadian biology is particularly relevant given the recent demonstration of circadian oscillation of metabolites in humans (10–13). Further investigations have determined that fatty acids and lipids constitute a major portion of these cycling metabolites (11, 12). Specific lipids, such as phosphocholine 18:0/18:1 that are regulated in a diurnal fashion, are themselves specific regulators of hepatic feeding metabolism (14). Gene-expression profiling indicates that oscillations of clock, oxidative stress, and metabolic genes are perturbed by acute total sleep deprivation (8).
Motivated by this emergent connection between sleep debt and metabolism, we sought to elucidate a common signature of perturbed metabolism in response to sleep restriction via comprehensive metabolomic analysis of serum from both rats and humans. Notably, sleep restriction, in contrast to acute total sleep deprivation, more closely represents real-world situations and is a condition experienced by millions of people on a daily basis (15, 16). In addition, some studies have reported differences in behavioral, sleep homeostatic, and physiological responses to these different types of sleep disturbances (reviewed in ref. 17). Thus, we subjected rats and humans to chronic sleep restriction protocols over a 5-d period. We used a completely independent rat dataset to detect and validate a set of robust markers, and followed this approach with validation in a human dataset.
Results and Discussion
Metabolomic Analysis of Sleep-Restricted Rats.
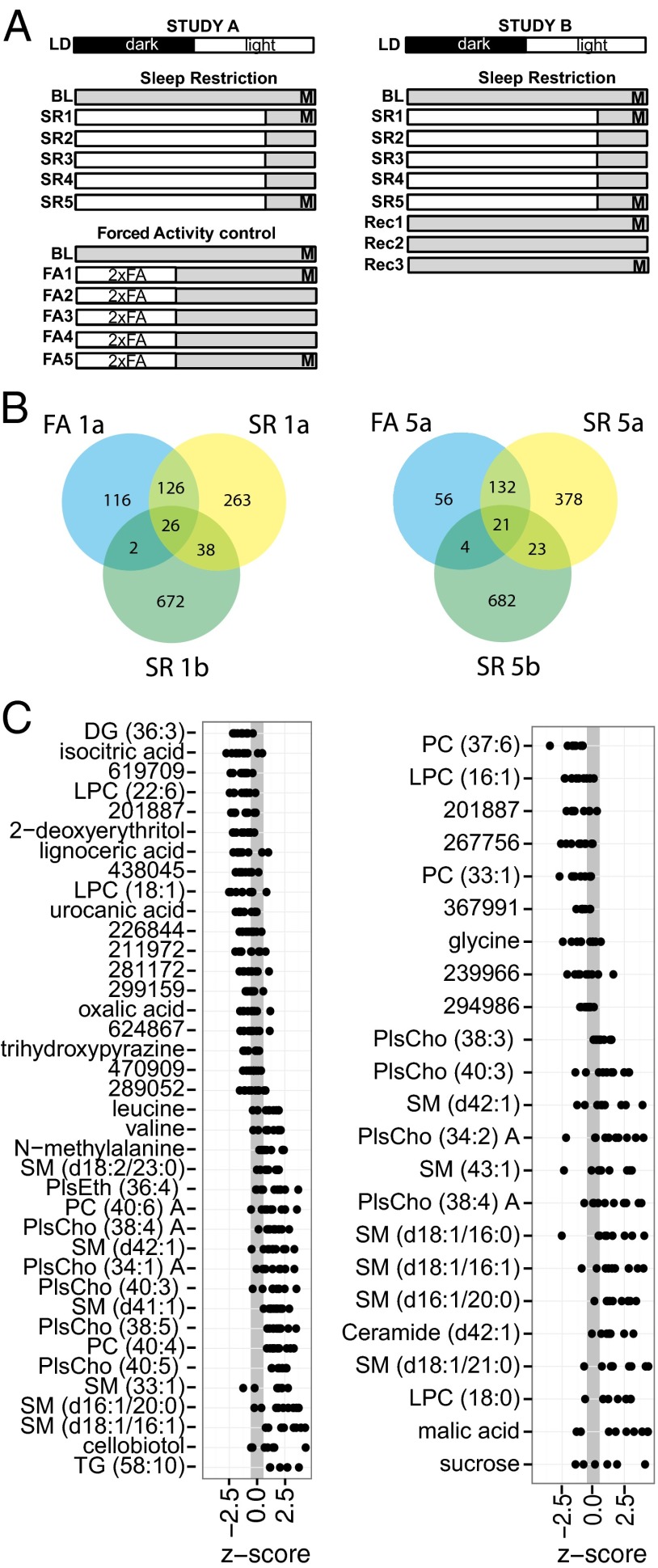
To determine the extent to which systemic metabolism is impacted by sleep restriction, we measured the comprehensive serum metabolic profiles of rats undergoing a sleep restriction protocol (Fig. 1A; see Supporting Information for details on the protocol). Blood samples were taken at baseline and after 1 and 5 d of sleep restriction (SR1 and SR5, respectively) in two separate studies. Study A included a concurrent forced-activity (FA) control group subjected to FA during the waking period (with blood samples at baseline, FA1a and FA5a). These rats had total activity levels equivalent to those of SR rats, but were allowed to sleep, providing a control for the effects of activity induced by the SR protocol. Study B included samples drawn from two recovery time points at day 1 and 3 post-SR (Rec1b and Rec3b). Metabolic profiles consisted of both polar and nonpolar metabolite measurements using a combination of GC-qTOF-MS, as well as hydrophilic interaction liquid chromatography (HILIC) and reverse-phase LC-qTOF-MS measurements. Specific details of metabolomics measurements are described below and in Supporting Information. A representative principal components analysis scores plot from the multivariate analysis of all metabolite measurements is provided as Fig. S1. As described in Supporting Information, body weight and food intake did not account for changes in metabolite levels.
Fig. 1.
Rat serum metabolite biomarker study design. (A) Animals in study A (Left) were subjected to SR by FA for 5 d (SR1–SR5) and a FA control group was subjected to FA for half the time at double speed (2xFA) for 5 d (FA1–FA5). White bars indicate periods of forced activity. Blood samples for metabolomics (M) were taken just before the end of the light phase on day 0 (baseline), day 1 (acute SR), and day 5 (chronic SR). Animals in study B (Right) were similarly SR, followed by 3 d of unrestricted recovery sleep (Rec1–Rec3). Samples were taken as in study A and at Rec1 (acute recovery), and Rec3 (extended recovery). (B) Overlap in metabolites noted as significantly different between acute SR and BL time points (Left) and chronic SR and BL time points (Right). FA denotes a forced activity control from study A. Suffix “a” or “b” denotes study A or study B, respectively. (C) Z-score plots of metabolites (ordered by mean change) that were significantly different between SR and BL time points in study A. A positive value denotes an elevated level under SR conditions. Numeric identifiers indicate unidentified metabolites, and details of their retention index and quantified m/z values are available in Table S1.
Reproducible Metabolite Markers of SR in Rats.
A total of 3,380 unique metabolite species were detected across the two independent studies of rat serum. Of these species, 407 were identified. To compare the change in metabolic profile as a function of SR, univariate comparisons with multiple testing correction were performed between baseline (BL) and SR1 time points to assess acute effects, and between BL and SR5 time points to assess chronic effects. All features were significant at P < 0.01, false-discovery rate (FDR) ≤ 0.05 when both studies were considered together. In addition, to ensure that the response was robust and not highly influenced by one of the two studies (18), each measurement from an individual study was required to meet thresholds of P < 0.1 and FDR < 0.2. Results from the combined data showing P < 0.01 (Table S2) were compared with the equivalent analysis of the control FA group, and the extent of overlap between the groups is detailed in Fig. 1B. Under acute SR, a total of 26 features were reproducibly observed to be attributable to FA, and so were removed from consideration. Of the 38 that were unique to SR (Fig. 1C, Left), 28 were identified. The majority of these metabolites (18 of 28) were lipids. Reduced nonlipid metabolites included: isocitrate from the Krebs cycle/lipogenesis; urocanic acid, a product of histidine catabolism; oxalic acid from glycolate metabolism; and 2-deoxyerythritol and trihydroxypyrazine, two nonmammalian metabolites that may be of dietary or gut microbial origin. Nonlipid metabolites that were increased included leucine, valine, N-methylalanine, and cellobiotol.
Following 5 d of chronic SR, 44 features were significantly altered across studies A and B: 23 were unique to the SR condition (Fig. 1C, Right), and 18 were named, of which 15 were lipids. Glycine was reduced and the Krebs cycle intermediate malate and sucrose were elevated.
A strength of these studies is the total replication of both experimental and analytical procedures for the first 6 d in studies A and B. With this replication, we can be confident that the common markers observed are resistant to experimental variations. There was significant variation between the two studies when examined in aggregate. For example, 64 metabolites were found in both studies to be similarly changed under acute SR, and 44 under chronic SR (Fig. 1B), whereas the number of metabolites significantly different from controls across both studies was ∼400–740 in the acute case and 550–725 in the chronic case. This difference may result from variation in the environmental and sampling procedures or in the analytical analysis. Although a limitation of this study, this caveat generally applies to “omics”-type experiments and reinforces the need for complete experimental replication, as done here.
Temporal Dynamics of SR-Specific Metabolic Alterations.
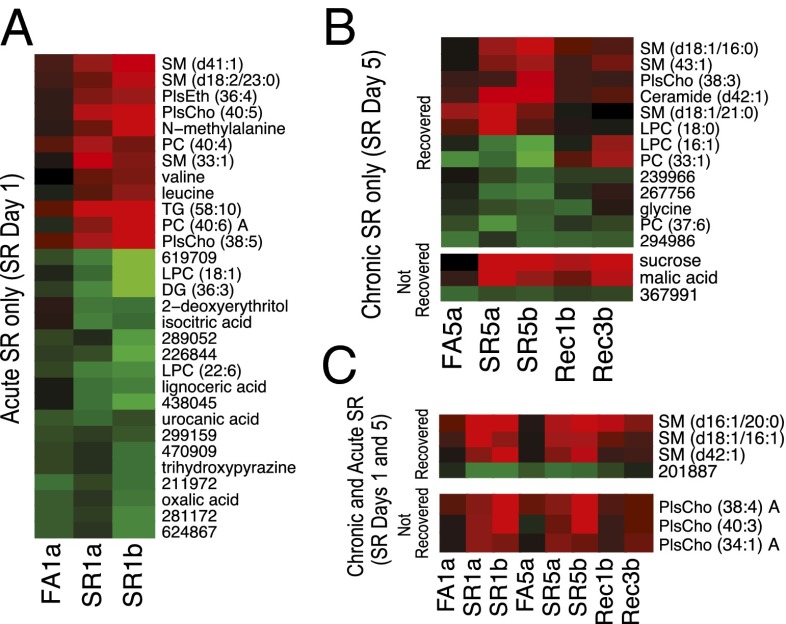
To better understand the potential roles of these metabolites as markers of sleep debt, we examined the temporal relationship between metabolites significantly altered under SR conditions (Fig. 2). Measurements were z-scored and normalized to the BL samples. Fig. 2A depicts the metabolites altered only under acute SR conditions from both studies A and B, as well as the day 1 FA group. Metabolite features were ordered using hierarchical cluster analysis. Fig. 2B highlights metabolites altered under chronic SR at day 5, and is divided into those metabolites that normalized to BL levels by recovery day 3 (Fig. 2B, Upper) and those that did not recover (Fig. 2B, Lower). We have not interpreted the metabolic results from recovery day 1 because they may represent intermediate transitional measurements between more stable physiological states. Sucrose and malate are two notable metabolites that did not recover, both possibly related to energy metabolism, although sucrose levels are likely controlled by intestinal absorption. Fig. S2 shows the fraction of each lipid class found to be significant as a function of all lipids measured in that class, and demonstrates that phosphatidylcholines (PCs), lysophosphatidylcholines (LPCs), choline plasmalogen (PlsCho), and sphingomyelin (SM) species are significantly altered equivalently between acute and chronic situations. In contrast, polar metabolites were primarily altered following acute SR, with only glycine specifically altered following chronic SR. We also identified those compounds that were altered across both acute and chronic conditions, all of which were phospholipids (Fig. 2C). Intriguingly, three SM species recovered, and three PlsCho species did not recover based on our statistical thresholds (P < 0.05, FDR < 0.2 compared with baseline). Although the thresholds are somewhat arbitrary, as demonstrated by visual similarity between the recovered and nonrecovered groups, the clustering of SM and PlsCho species suggests a regulated process in which specific lipid classes/species are preferentially returned to non-SR levels.
Fig. 2.
Metabolites perturbed by SR in the rat model. (A) Metabolite features altered only under acute SR. Each data point represents the mean of 10 measurements across each group for each feature; FA1, forced activity control day 1; SR1, sleep restriction day 1. Suffix “a” or “b” denotes study A or study B, respectively. Red indicates elevation and green indicates reduction compared with BL. (B) Metabolites altered only under chronic SR are divided into those that recovered (Upper) and those that remained altered (Lower) following 3-d recovery sleep. Labels as in Fig. 1, with recovery (Rec) time points at days 1 and 3 after SR. (C) Metabolites altered under both acute and chronic SR are divided into those that recovered (Upper) and those that remained perturbed (Lower).
Altered Metabolic Phenotype in Humans Under SR.
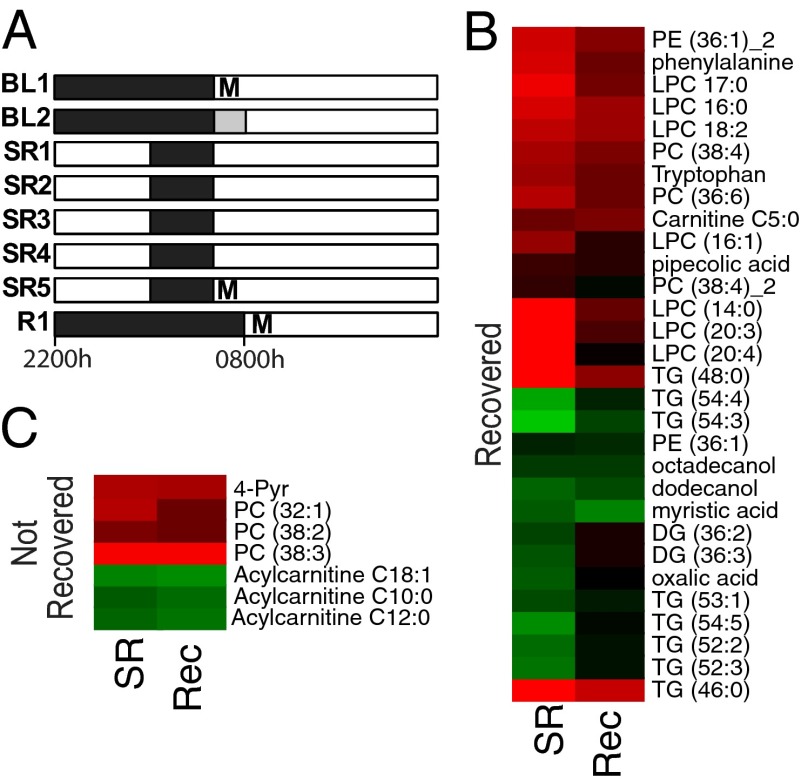
Recent work indicates that total sleep deprivation induces a measurable perturbation to the blood metabolome (19); however, little is known about the metabolic effects of SR. We analyzed the metabolome in blood samples drawn from 10 subjects sampled according to the SR protocol in Fig. 3A, including a sample at BL, following 5 d of SR, which included 4 h of sleep opportunity (SR) each night, and following a night of recovery sleep (Rec). A similar filter was applied (P < 0.1, FDR < 0.2) as with the rat data (Dataset S1) because our goal was to validate the rat results in a human population. Comparison of the human BL and SR samples revealed 92 features that were altered, of which 37 were identified (Fig. 3B; unidentified metabolites not shown for clarity). Of the identified species, 32 were lipid- or fatty acid-related compounds. With regard to polar species, the amino acids tryptophan (Trp), phenylalanine (Phe), as well as pipecolic acid and 1-Methyl-4-pyridone-5-carboxamide (4-Pyr) were elevated, but oxalic acid was reduced.
Fig. 3.
Experimental protocol and metabolomics of human subjects. (A) Protocol for human SR study consisting of two BL nights followed by five nights of SR and one night recovery (Rec) sleep. Blood samples for metabolomics (M) were collected in the morning as indicated. Black bars, sleep periods; white bars, wake periods; gray bar, 0800 hours to 1000 hours additional TIB sleep at BL2 in one protocol. (B and C) Heatmaps of metabolites significantly different between BL and SR divided into those that recovered (B) and those that remained perturbed (C). Each data point represents the mean of measurements from 10 individuals across each group for each feature.
As with the rat study, the majority of metabolites (30 identified and 46 unknown) recovered to pre-SR levels, whereas 7 identified and 11 unknown did not recover over the period measured. Aside from 4-Pyr, the nonrecovered metabolites were either PCs (32:1, 38:2, 38:3), or acylcarnitines (C10:0, C12:0, and C18:1). The differences between baseline (presleep loss) and recovery (postsleep loss) could indicate incomplete recovery.
Polar Metabolites Reflective of Neurotransmitter, Vitamin B3, and Gut Microflora Metabolism Are Altered by SR in Humans.
Both Trp and Phe were elevated in human subjects during SR. Trp is a major precursor for serotonin and melatonin and is elevated under acute total sleep deprivation (19). Intriguingly, Trp is also the precursor to liver synthesis of nicotinic acid (vitamin B3), which interconverts with the amide form, nicotinamide. In support of an effect on this pathway, we observed elevated levels of 4-Pyr, a primary end metabolite of nicotinamide. Phe is also a precursor to several neurotransmitters, including dopamine, norepinephrine, and epinephrine via tyrosine. An increase in Phe might partly reflect increases in catecholamines in response to sleep loss (20). In humans, acute total sleep deprivation in the laboratory increases levels of stress hormones, such as catecholamines and cortisol (e.g., refs. 20 and 21). However, many laboratory studies have reported that SR, the type of sleep loss we used in our study, does not produce significant effects on cortisol levels (e.g., ref. 22). Altogether, it is unlikely that effects of SR in the present study are explained by globally elevated stress hormones, although we cannot entirely exclude this possibility.
Finally, gut metabolism also likely plays a role in the observed metabolic profile (23). For example pipecolic acid is a metabolite of lysine unique to gut microflora and was elevated after SR.
Relevance of Circadian Control to the SR Metabolome.
Gene-expression profiling has established that there is a complex, but defined, interaction between the circadian system and sleep homeostasis (8) in humans. A comparison of the metabolites perturbed in our study with studies of metabolites demonstrating circadian oscillations supports this relationship. Lipidomics analysis has elucidated a set of lipids that oscillate across biological subtypes defined by stratification by lipid composition (11). From this group of rhythmic metabolites, we found eight overlapping species of significance, including LPC 16:0; triacylglycerols (TGs) (52:2, 52:3, TG 54:3, 54:4, 54:5), and diacylglycerols (DGs) (36:2, 36:3). Other groups have reported a mixture of polar and nonpolar metabolites that cycle in a circadian manner, and consistently found that the proportion of lipids is elevated. For example, Dallman et al. (12) found that 33 of 40 lipid species were regulated in a circadian manner, of which carnitine C12:0 overlaps with our study. Similarly, Ang et al. (13) found 25 of 34 to be lipid species, with lysoPCs (18:2, 20:3) acylcarnitine (12:0 and 18:0) and Phe overlapping with our results.
Although we did not examine the expression of clock genes in the rat protocol, we have previously found that a 20-h forced desynchrony protocol does not alter the period of the body temperature rhythm (24). This finding is consistent with work indicating that FA does not affect the clock in the suprachiasmatic nucleus (25). We cannot exclude effects on peripheral clocks, and note that some of the metabolic phenotypes could arise from a loss of synchrony between central and peripheral clocks. Indeed, circadian asynchrony could be one of the consequences of SR and merits future investigation. In support of this idea, 1 wk of 5.70 h per night SR protocol was shown to affect rhythms of the human blood transcriptome (8).
Comparison with Metabolic Profile of Acute Total Sleep Deprivation in Humans.
Similar to a recent study using acute total sleep deprivation by Davies et al. (19), the majority of metabolites altered in our chronic SR study were lipid- or fatty acid-related species (38 of 41 vs. 32 of 37 in the current study). Twelve of these were common between the two studies, including PCs (32:1, 36:6, 38:4, 38:2, 38:3), lysoPCs (14:0, 16:1, 17:0), and acylcarnitines (C5:0, C10:0, C12:0). All of the above-mentioned metabolites were elevated in the acute sleep deprivation study. In our study, SR led to increases in all of these metabolites except acylcarnitines C10:0 and C12:0, which were reduced. Because these findings might reflect the energetic process of fatty acid oxidation, the discrepancy may be a function of the difference in protocol (1 d vs. 5 d and total sleep deprivation vs. chronic SR), leading to a difference in relative energy requirements. In both studies, Trp was also elevated. Investigation of the human plasma metabolome suggests that amino acid metabolism is particularly perturbed as a function of SR (26). The study of Davies et al. (19) identified metabolites that increased but found none that decreased under total sleep deprivation. Here we have identified decreases in a number metabolites as a function of SR, which were not part of the analytical platform used in the Davies et al. study (19). These metabolites included TGs, DGs, fatty alcohols, myristic acid, a phosphatidylethanolamine (PE) species and oxalic acid.
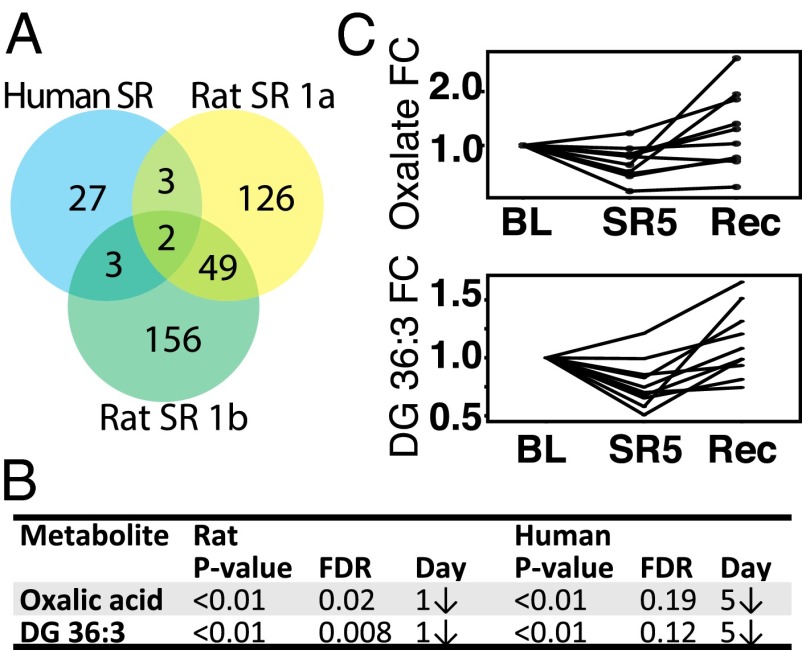
Oxalic Acid and DG 36:3 Are Putative Translational Markers of Sleep Debt.
To examine the feasibility of using metabolite markers to assess sleep debt across species, we compared the list of significantly altered metabolites in rats and humans (Fig. 4). Two identified species were common across rat and humans: oxalic acid and DG 36:3 (Fig. 4 A and B). These metabolites are from highly disparate metabolic pathways yet behave in the same manner, and thus may provide a robust measure of sleep debt (Fig. 4C and Fig. S3). Bootstrapped hierarchical cluster analysis of the entire set of measured metabolites (Dataset S2) indicates a significant branch that includes oxalate, as well hypoxanthine, medium-chain acylcarnitines (C6, C10, C12, C12:1), and arachidonic acid (Fig. S4). Although these metabolites do not share obvious metabolic pathways, they may be linked by cellular function, such as peroxisomal processing, as described below.
Fig. 4.
Oxalate and DG 36:3 reflect cross-species sleep loss. (A) Venn diagram indicating two identified metabolites common to both rat and human studies. (B) Table of statistical values for rat and human datasets. (C) Fold change (FC) in oxalate (Upper) and DG 36:3 (Lower) levels from human serum. Measurement in each subject is linked by a solid line.
The primary sources of blood oxalate are diet-derived plant sources, vitamin C (ascorbate) degradation, and endogenous synthesis pathways in erythrocytes and liver (27). Endogenous oxalate has conventionally been considered an end product of mammalian metabolism and primarily studied in the context of kidney stone formation. Recent evidence suggests that blood levels are also influenced by gut microbiota, with over a dozen oxalate-degrading gut bacterial species identified (28). Although dietary oxalate with high bioavailability increases plasma and urine levels (29), excreted oxalate is minimally dependent on diet in an epidemiological cohort (30). Based on these data, as well as the highly regulated nature of presample fasting and diet for our rat and human studies, we postulate that the observed reduction in blood oxalate is a result of either reduced synthesis or increased gut microbiota processing and is not of dietary origin. Endogenous substrates for oxalate synthesis include glycolate in peroxisomes and hydroxyproline. Several days of total sleep deprivation do not impact hydroxyproline levels in rats (31), nor did we observe differences in this study, and thus we posit that any alterations in endogenous synthesis are the result of peroxisomal glycolate perturbations. This idea is consistent with the other evidence for oxidative stress and peroxisomal activation described below. Another possible mechanism for reduced oxalate levels in blood is via increased urinary clearance. Acute total sleep deprivation increases diuresis (32), and oxalate is readily filtered from plasma (33).
With regard to DG 36:3, it is not clear why this particular species (along with DG 36:2 in humans) but not other DGs is altered in response to SR. This metabolite is also regulated in a circadian fashion in humans (11), and as such may be an indicator of both circadian time and sleep debt.
Energetic and Lipid Metabolism Is Impacted Differentially by SR Across Species.
Systemic metabolic disruption as a function of SR has been reflected in model systems, such as in rats experimentally manipulated to sleep less, either through mechanical stimulation (34) or through lesions in the ventrolateral preoptic area (35). These studies consistently find that energy expenditure increases without a concomitant change in food intake (34, 35). Furthermore, plasma levels of glucose, insulin, and leptin are reduced (34). In contrast, humans undergoing sleep restriction increase caloric intake (1, 2, 36–38) and show alterations in energy expenditure (39–41). Despite the differences in caloric intake, it is clear that sleeps loss affects metabolic function across species.
Specific metabolic changes as a result of sleep deprivation are difficult if not impossible to decouple from metabolic demands of the organism under increased wakefulness. In our rat study, the effects of activity—which may account for much of the effect of wake per se—were controlled by including a FA group. Although a number of metabolites were common to SR and FA groups, we focused on those specific to SR. For the human study, physical activity levels in our subjects were limited. Studies in humans show that energy expenditure (resting metabolic rate and so forth) contributes very little to metabolic changes during sleep restriction, and that energy intake is the largest contributor to metabolic homeostasis (42). Thus, we also attempted to separate the metabolic effects from the sleep effects by enforcing a 10- to 12-h fasting period before metabolomic sampling, such that observed changes could be attributed to sleep/sleep restriction effects.
The metabolites altered in our study are reflective of energetic changes, particularly the overrepresentation of lipid species. Across both species, phospholipids (PC, LPC, PE, SM) were the most elevated as a function of SR (20 of 25 in rats and 14 of 15 in humans) (Fig. S2). This finding raises the possibility that there is a common source for the elevated phospholipids, such as membrane breakdown or release from circulating lipoprotein particles. Elevated LPCs in blood are possibly derived from secreted phospholipase A2 acting on lipoprotein PC, lecithin–cholesterol–acyl-transferase in liver acting on LDL or HDL, or endothelial phospholipase A2 acting on HDL. In turn, LPCs may act in a signaling capacity to stimulate PKC, NF-κB, or COX-2 (43). Consistent with this observation, sleep deprivation increases murine cerebral cortex NF-κB (44). Additionally, COX-2 products, such as prostaglandin D2, are sleep-promoting in rat cerebrospinal fluid (CSF) (45), which may support the idea that sleep-promoting molecules are elevated during sleep loss. Our data suggest a possible mechanism for sleep deprivation-induced increases in NF-κB and prostaglandins. Our results further indicate a significant clustering relationship between oxalate and arachidonic acid (Fig. S4), which is the substrate for COX-2, although we note that arachidonic acid was not significantly changed by our criteria.
Seven plasmalogen species were elevated in the rat under acute or chronic SR conditions, indicative of oxidative stress. This finding is consistent with several studies suggesting that the function of sleep is restorative and involves metabolite clearance from the brain (9), and that sleep restores antioxidant balance in peripheral tissues (46). Although plasmalogens were not significantly changed in the human study, transcript studies have shown that pathways involved in oxidative stress and reactive oxygen species are significantly induced in blood upon sleep loss (8). The lack of detectable lipid oxidation signal in humans may be a result of the increased capacity of human metabolism to regulate oxidative stress, or greater heterogeneity inherent in human datasets.
In contrast, the human data showed a marked reduction in six TG species (>52 carbon chains) and two DG species (36 carbon chains), whereas the rat data demonstrated a reduction in a single DG species and elevation of TG 58:10. Reduction in total blood TGs of humans has been observed previously using a similar protocol (47). Of note, the two TG species elevated in humans were relatively short (≤48 carbons, average chain length ≤16 carbons). This finding implies a possible carbon shift from longer- to shorter-chain TGs. The combination of an oxidative environment, β-oxidation of long-chain fatty acids as implied by TAG shortening, as well as the observed association of acylcarnitines with oxalate (Fig. S4), suggests a distinct role for peroxisomes, possibly via peroxisome proliferator-activated receptors (PPARs).
Oxidative Stress and the Potential Role of PPARs.
Mounting evidence points to sleep loss inducing an oxidative metabolic state (8, 48). One mechanism may be via the induction of PPARs, which play a critical role in lipid and glucose metabolism. Bezafibrate, an antihyperlipidemic PPAR-α agonist, has been shown in a mouse model to affect both BL sleep, with increased EEG δ-power, and recovery sleep following sleep restriction, with a reduced rebound in EEG δ-power (49). As a result, the authors suggested that activation of PPAR-α may provide a protective effect against sleep restriction. In the same model, SR was shown to elevate brain mRNA levels of PPAR-α (50). Oleoylethanolamide, a potent activator of PPAR-α, is also elevated in CSF, and to a lesser extent in serum of sleep-deprived humans (51). Although oleoylethanolamide was not directly measured in our study, a number of metabolite changes in the human data, including the reduced oxalate levels, can be explained by a mechanism that invokes changes in PPAR-α and, more generally, in peroxisomes.
The peroxisome is the central site of fatty acid alcohol processing (52), which generates either diacylphospholipids or ether-linked phospholipids. Induction of peroxisome biogenesis is consistent with the observed decrease in both fatty alcohols (humans) and an increase in phospholipid species (humans and rats), particularly plasmalogens. Furthermore, the reduced levels of long-chain TGs and elevation in shorter-chain TGs observed in humans may reflect peroxisomal β-oxidation processes that are preferential for longer-chain fatty acids. Reduction in both oxalate (humans and rats) and glycine (rats, SR5) might be explained by depletion of glyoxylate, which is a common precursor to both substances in peroxisomes. Finally, the direct relationship between oxalate, hypoxanthine, and medium-chain acylcarnitines (C6, C10, C12, C12:1) in cluster analysis (Fig. S4) may also be explained by peroxisomal processes. Direct conversion of oxalate to hypoxanthine has been observed in extreme oxidative environments (53), although it is not clear if this occurs in vivo.
Conclusion.
The complex effects of sleep loss on metabolism regulatory pathways are increasingly recognized as relevant systemic perturbations. Our cross-species analysis of serum from both rats and humans provides evidence for a common architecture driving such metabolic perturbations. Our comprehensive metabolite profiling results present a picture of an oxidative environment impacting lipid metabolism and processing, albeit with some interspecies variations. The identification of two metabolites, oxalic acid and DG 36:3, which change with SR in rats and humans, provides potential biomarkers for sleep loss. We find that metabolism of neurotransmitters and gut-derived metabolites are also affected by sleep loss, underscoring the system-wide impact of SR.
Methods
Rat Study Protocol.
In two separate studies (Fig. 1A), adult male Sprague–Dawley rats (Harlan) were SR by FA 20 h per day for 5 d and were allowed to rest during the last 4 h of the light phase (n = 10 in all groups). To differentiate the effects of FA from SR, the second study included a FA control group in which animals walked the same distance as SR animals in the dark phase, but had sufficient time to sleep. For an overview of the two studies, see Fig. 1A; for details on the methodology, see Supporting Information. The experiment was approved by the Ethical Committee of Animal Experiments of the University of Groningen.
Human Study Protocol.
Ten healthy subjects, aged 22–50 y (27.5 ± 5.6 y; five females), participated in one of two controlled SR experimental protocols consisting of two BL nights of 10- or 12-h time-in-bed (TIB) per night (2200 hours to 0800 hours/1000 hours) followed by five nights of SR of 4-h TIB per night (0400 hours to 0800 hours) and one night of 12-h TIB recovery (R) sleep (2200 hours to 1000 hours). After 10–12 h of fasting, blood samples for metabolomics were collected in the morning after the first night of BL sleep, after the fifth night of SR, and after the night of recovery sleep. To control for morning-evening effects, sampling occurred during the 2 h following waketime (between 0800 hours and 1000 hours on BL1 and SR5 and between 1000 hours and 1200 hours on R1). Table S3 contains subject demographic data and clinical parameters. The protocols were approved by the Institutional Review Board of the University of Pennsylvania. For all subjects, written informed consent was obtained according to the principles expressed in the Declaration of Helsinki before entry; all subjects received compensation for participation.
Mass Spectrometry.
Small polar molecules were profiled using GC-qTOF-MS technique and HILIC-ESI-MS. Lipids were profiled using reverse-phase ESI-qTOF-MS. The experiments were performed in the West Coast Metabolomics core facility at the University of California, Davis, CA. Further details are provided as Supporting Information.
Statistical Analysis of the Data.
Statistical analyses were performed on the normalized mass spectral data. In each case, comparisons were done by t test using multiple test correction (1,000 permutations, unpaired for animal and paired for human data). Mass spectral features were judged significant if P < 0.1 and FDR < 0.2 in each study, such that the pooled result would yield P < 0.01 (Table S2). Metabolite Set Enrichment Analysis was performed on metabolites from rats and humans (Table S4).
Supplementary Material
Acknowledgments
For human work, we thank the subjects who participated in the experiments and the staff of the Division of Sleep and Chronobiology, who helped acquire the metabolomic data. This research was supported by the Department of the Navy, Office of Naval Research Award N00014-11-1-0361 (to N.G.); National Aeronautics and Space Administration Grant NNX14AN49G (to N.G.); the National Space Biomedical Research Institute through the National Aeronautics and Space Administration NCC 9-58 (to D.F.D.); National Institutes of Health Grant R01 NR004281 (to D.F.D.); Clinical and Translational Research Center Grant UL1TR000003; Defense Advanced Research Projects Agency and the US Army Research Office Grant W911NF1010093 (to T.A.). A. Sehgal is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417432112/-/DCSupplemental.
References
- 1.Spaeth AM, Dinges DF, Goel N. Effects of experimental sleep restriction on weight gain, caloric intake, and meal timing in healthy adults. Sleep. 2013;36(7):981–990. doi: 10.5665/sleep.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markwald RR, et al. Impact of insufficient sleep on total daily energy expenditure, food intake, and weight gain. Proc Natl Acad Sci USA. 2013;110(14):5695–5700. doi: 10.1073/pnas.1216951110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh M, Drake CL, Roehrs T, Hudgel DW, Roth T. The association between obesity and short sleep duration: A population-based study. J Clin Sleep Med. 2005;1(4):357–363. [PubMed] [Google Scholar]
- 4.Di Milia L, Vandelanotte C, Duncan MJ. The association between short sleep and obesity after controlling for demographic, lifestyle, work and health related factors. Sleep Med. 2013;14(4):319–323. doi: 10.1016/j.sleep.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 5.Schmid SM, Hallschmid M, Schultes B. The metabolic burden of sleep loss. Lancet Diabetes Endocrinol. 2014;3(1):52–62. doi: 10.1016/S2213-8587(14)70012-9. [DOI] [PubMed] [Google Scholar]
- 6.Ayas NT, et al. A prospective study of sleep duration and coronary heart disease in women. Arch Intern Med. 2003;163(2):205–209. doi: 10.1001/archinte.163.2.205. [DOI] [PubMed] [Google Scholar]
- 7.Spiegel K, Leproult R, Van Cauter E. Impact of sleep debt on metabolic and endocrine function. Lancet. 1999;354(9188):1435–1439. doi: 10.1016/S0140-6736(99)01376-8. [DOI] [PubMed] [Google Scholar]
- 8.Möller-Levet CS, et al. Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc Natl Acad Sci USA. 2013;110(12):E1132–E1141. doi: 10.1073/pnas.1217154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xie L, et al. Sleep drives metabolite clearance from the adult brain. Science. 2013;342(6156):373–377. doi: 10.1126/science.1241224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasukawa T, et al. Human blood metabolite timetable indicates internal body time. Proc Natl Acad Sci USA. 2012;109(37):15036–15041. doi: 10.1073/pnas.1207768109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chua EC-P, et al. Extensive diversity in circadian regulation of plasma lipids and evidence for different circadian metabolic phenotypes in humans. Proc Natl Acad Sci USA. 2013;110(35):14468–14473. doi: 10.1073/pnas.1222647110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallmann R, Viola AU, Tarokh L, Cajochen C, Brown SA. The human circadian metabolome. Proc Natl Acad Sci USA. 2012;109(7):2625–2629. doi: 10.1073/pnas.1114410109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ang JE, et al. Identification of human plasma metabolites exhibiting time-of-day variation using an untargeted liquid chromatography-mass spectrometry metabolomic approach. Chronobiol Int. 2012;29(7):868–881. doi: 10.3109/07420528.2012.699122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, et al. A diurnal serum lipid integrates hepatic lipogenesis and peripheral fatty acid use. Nature. 2013;502(7472):550–554. doi: 10.1038/nature12710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goel N, Rao H, Durmer JS, Dinges DF. Neurocognitive consequences of sleep deprivation. Semin Neurol. 2009;29(4):320–339. doi: 10.1055/s-0029-1237117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banks S, Dinges DF. Behavioral and physiological consequences of sleep restriction. J Clin Sleep Med. 2007;3(5):519–528. [PMC free article] [PubMed] [Google Scholar]
- 17.Goel N. Genetics of sleep timing, duration and homeostasis in humans. Sleep Med Clin. 2011;6(2):171–182. doi: 10.1016/j.jsmc.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood J, Freemantle N, King M, Nazareth I. Trap of trends to statistical significance: Likelihood of near significant P value becoming more significant with extra data. BMJ. 2014;348:g2215. doi: 10.1136/bmj.g2215. [DOI] [PubMed] [Google Scholar]
- 19.Davies SK, et al. Effect of sleep deprivation on the human metabolome. Proc Natl Acad Sci USA. 2014;111(29):10761–10766. doi: 10.1073/pnas.1402663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meerlo P, Sgoifo A, Suchecki D. Restricted and disrupted sleep: Effects on autonomic function, neuroendocrine stress systems and stress responsivity. Sleep Med Rev. 2008;12(3):197–210. doi: 10.1016/j.smrv.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 21.Minkel J, et al. Sleep deprivation potentiates HPA axis stress reactivity in healthy adults. Health Psychol. 2014;33(11):1430–1434. doi: 10.1037/a0034219. [DOI] [PubMed] [Google Scholar]
- 22.Pejovic S, et al. Effects of recovery sleep after one work week of mild sleep restriction on interleukin-6 and cortisol secretion and daytime sleepiness and performance. Am J Physiol Endocrinol Metab. 2013;305(7):E890–E896. doi: 10.1152/ajpendo.00301.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wikoff WR, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106(10):3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strijkstra AM, Meerlo P, Beersma DG. Forced desynchrony of circadian rhythms of body temperature and activity in rats. Chronobiol Int. 1999;16(4):431–440. doi: 10.3109/07420529908998718. [DOI] [PubMed] [Google Scholar]
- 25.Al-Safadi S, et al. Stress-induced changes in the expression of the clock protein PERIOD1 in the rat limbic forebrain and hypothalamus: Role of stress type, time of day, and predictability. PLoS ONE. 2014;9(10):e111166. doi: 10.1371/journal.pone.0111166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bell LN, et al. Effects of sleep restriction on the human plasma metabolome. Physiol Behav. 2013;122:25–31. doi: 10.1016/j.physbeh.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marengo SR, Romani AMP. Oxalate in renal stone disease: The terminal metabolite that just won’t go away. Nat Clin Pract Nephrol. 2008;4(7):368–377. doi: 10.1038/ncpneph0845. [DOI] [PubMed] [Google Scholar]
- 28.Miller AW, Dearing D. The metabolic and ecological interactions of oxalate-degrading bacteria in the mammalian gut. Pathogens. 2013;2(4):636–652. doi: 10.3390/pathogens2040636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holmes RP, Ambrosius WT, Assimos DG. Dietary oxalate loads and renal oxalate handling. J Urol. 2005;174(3):943–947, discussion 947. doi: 10.1097/01.ju.0000169476.85935.e2. [DOI] [PubMed] [Google Scholar]
- 30.Taylor EN, Curhan GC. Determinants of 24-hour urinary oxalate excretion. Clin J Am Soc Nephrol. 2008;3(5):1453–1460. doi: 10.2215/CJN.01410308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Landis CA, Whitney JD. Effects of 72 hours sleep deprivation on wound healing in the rat. Res Nurs Health. 1997;20(3):259–267. doi: 10.1002/(sici)1098-240x(199706)20:3<259::aid-nur8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 32.Kamperis K, Hagstroem S, Radvanska E, Rittig S, Djurhuus JC. Excess diuresis and natriuresis during acute sleep deprivation in healthy adults. Am J Physiol Renal Physiol. 2010;299(2):F404–F411. doi: 10.1152/ajprenal.00126.2010. [DOI] [PubMed] [Google Scholar]
- 33.Kasidas GP, Rose GA. Measurement of plasma oxalate in healthy subjects and in patients with chronic renal failure using immobilised oxalate oxidase. 1986;154(1):49–58. doi: 10.1016/0009-8981(86)90087-2. [DOI] [PubMed] [Google Scholar]
- 34.Barf RP, et al. Metabolic consequences of chronic sleep restriction in rats: Changes in body weight regulation and energy expenditure. Physiol Behav. 2012;107(3):322–328. doi: 10.1016/j.physbeh.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Vetrivelan R, Fuller PM, Yokota S, Lu J, Saper CB. Metabolic effects of chronic sleep restriction in rats. 2012;35(11):1511–1520. doi: 10.5665/sleep.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.St-Onge M-P, et al. Short sleep duration increases energy intakes but does not change energy expenditure in normal-weight individuals. Am J Clin Nutr. 2011;94(2):410–416. doi: 10.3945/ajcn.111.013904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nedeltcheva AV, et al. Sleep curtailment is accompanied by increased intake of calories from snacks. Am J Clin Nutr. 2009;89(1):126–133. doi: 10.3945/ajcn.2008.26574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brondel L, Romer MA, Nougues PM, Touyarou P, Davenne D. Acute partial sleep deprivation increases food intake in healthy men. Am J Clin Nutr. 2010;91(6):1550–1559. doi: 10.3945/ajcn.2009.28523. [DOI] [PubMed] [Google Scholar]
- 39.Shechter A, Rising R, Albu JB, St-Onge M-P. Experimental sleep curtailment causes wake-dependent increases in 24-h energy expenditure as measured by whole-room indirect calorimetry. Am J Clin Nutr. 2013;98(6):1433–1439. doi: 10.3945/ajcn.113.069427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buxton OM, et al. Adverse metabolic consequences in humans of prolonged sleep restriction combined with circadian disruption. Sci Transl Med. 2012;4(129):29ra43. doi: 10.1126/scitranslmed.3003200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benedict C, et al. Acute sleep deprivation reduces energy expenditure in healthy men. Am J Clin Nutr. 2011;93(6):1229–1236. doi: 10.3945/ajcn.110.006460. [DOI] [PubMed] [Google Scholar]
- 42.Spaeth AM, Dinges DF, Goel N. Sex and race differences in caloric intake during sleep restriction in healthy adults. Am J Clin Nutr. 2014;100(2):559–566. doi: 10.3945/ajcn.114.086579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sevastou I, Kaffe E, Mouratis M-A, Aidinis V. Lysoglycerophospholipids in chronic inflammatory disorders: The PLA(2)/LPC and ATX/LPA axes. Biochim Biophys Acta. 2013;1831(1):42–60. doi: 10.1016/j.bbalip.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 44.Chen Z, Gardi J, Kushikata T, Fang J, Krueger JM. Nuclear factor-kappaB-like activity increases in murine cerebral cortex after sleep deprivation. Am J Physiol. 1999;276(6 Pt 2):R1812–R1818. doi: 10.1152/ajpregu.1999.276.6.R1812. [DOI] [PubMed] [Google Scholar]
- 45.Ram A, et al. CSF levels of prostaglandins, especially the level of prostaglandin D2, are correlated with increasing propensity towards sleep in rats. Brain Res. 1997;751(1):81–89. doi: 10.1016/s0006-8993(96)01401-1. [DOI] [PubMed] [Google Scholar]
- 46.Everson CA, Laatsch CD, Hogg N. Antioxidant defense responses to sleep loss and sleep recovery. Am J Physiol Regul Integr Comp Physiol. 2005;288(2):R374–R383. doi: 10.1152/ajpregu.00565.2004. [DOI] [PubMed] [Google Scholar]
- 47.Reynolds AC, et al. Impact of five nights of sleep restriction on glucose metabolism, leptin and testosterone in young adult men. PLoS ONE. 2012;7(7):e41218. doi: 10.1371/journal.pone.0041218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vollert C, et al. Exercise prevents sleep deprivation-associated anxiety-like behavior in rats: Potential role of oxidative stress mechanisms. Behav Brain Res. 2011;224(2):233–240. doi: 10.1016/j.bbr.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Chikahisa S, Tominaga K, Kawai T, Kitaoka K, Oishi K. Bezafibrate, a peroxisome proliferator-activated receptors agonist, decreases body temperature and enhances electroencephalogram delta-oscillation during sleep in mice. Endocrinology. 2008;149(10):5262–5271. doi: 10.1210/en.2008-0285. [DOI] [PubMed] [Google Scholar]
- 50.Chikahisa S, Shimizu N, Shiuchi T, Séi H. Ketone body metabolism and sleep homeostasis in mice. Neuropharmacology. 2014;79:399–404. doi: 10.1016/j.neuropharm.2013.12.009. [DOI] [PubMed] [Google Scholar]
- 51.Koethe D, et al. Sleep deprivation increases oleoylethanolamide in human cerebrospinal fluid. J Neural Transm. 2009;116(3):301–305. doi: 10.1007/s00702-008-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lodhi IJ, Semenkovich CF. Peroxisomes: A nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19(3):380–392. doi: 10.1016/j.cmet.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holian J, Garrison WM. Mechanism and stoicheiometry in the radiolytic oxidation of purines and aminopurines in aqueous solution. Chem Commun (Camb) 1967:676–677. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.