Significance
Ribosomes are essential intracellular machines composed of proteins and RNA molecules. The DNA sequences [i.e., ribosomal DNA (rDNA)] encoding rRNAs are tandemly repeated and give rise to the nucleolus. The rRNAs are transcribed from two array kinds (the 5S and the 45S arrays). Here we show that variation in the 5S and 45S rDNA arrays is tightly coupled, despite their location on different chromosomes. Our observations suggest that natural selection contributes to maintain balanced rDNA dosage across unlinked rDNA arrays. Finally, we show that bisphenol A can induce parallel loss of rDNA units in 5S and 45S arrays. These observations raise the prospect that human diseases might be traced to disrupted rDNA dosage balance in the genome.
Keywords: nucleolus, ribosome, gene dosage balance, concerted evolution, bisphenol A
Abstract
Tandemly repeated ribosomal DNA (rDNA) arrays are among the most evolutionary dynamic loci of eukaryotic genomes. The loci code for essential cellular components, yet exhibit extensive copy number (CN) variation within and between species. CN might be partly determined by the requirement of dosage balance between the 5S and 45S rDNA arrays. The arrays are nonhomologous, physically unlinked in mammals, and encode functionally interdependent RNA components of the ribosome. Here we show that the 5S and 45S rDNA arrays exhibit concerted CN variation (cCNV). Despite 5S and 45S rDNA elements residing on different chromosomes and lacking sequence similarity, cCNV between these loci is strong, evolutionarily conserved in humans and mice, and manifested across individual genotypes in natural populations and pedigrees. Finally, we observe that bisphenol A induces rapid and parallel modulation of 5S and 45S rDNA CN. Our observations reveal a novel mode of genome variation, indicate that natural selection contributed to the evolution and conservation of cCNV, and support the hypothesis that 5S CN is partly determined by the requirement of dosage balance with the 45S rDNA array. We suggest that human disease variation might be traced to disrupted rDNA dosage balance in the genome.
Repeated gene arrays have provided unique challenges to genetic and genome analyses, and have remained among the most elusive components of eukaryotic genomes (1, 2). Tandemly repeated loci of high copy number (CN) are labile, evolutionary dynamic, often subjected to concerted evolution of DNA sequences, and display abundant CN variation that emerges from high rates of repeat expansion and contraction (1). Moreover, natural selection contributes to the determination of gene CN, shaping rapid gene amplification in cancer, balanced gene loss after whole genome duplication, and optimal gene CN in locally adapted populations (3–5). For example, higher CN of the amylase gene is present in populations with starch-rich diets across organisms as diverse as humans, dogs, and fungi (3, 6, 7). Remarkably, gene CN may also be developmentally amplified in specific tissues to ensure rates of transcription in genes with high transcriptional demands (8, 9). This is the case, for instance, of the chorion genes in Drosophila, which are amplified up to 80 fold in ovarian cells (10).
The ribosomal DNA (rDNA) arrays display substantial CN variation within and between species (2, 11–16). The variation is functionally relevant with rDNA CN polymorphism modifying chromatin states and gene expression across the genome in humans and flies (17–19). In mammals, the rDNA arrays are dispersed across several chromosomes, and encode the four rRNAs that account for more than 60% of all transcription in the cell (20, 21). Transcription of rDNA loci varies with cell and tissue type and is epigenetically regulated with allelic specificity (22, 23). The four rRNAs are indispensable structural and catalytic components of the ribosome. Three of the rRNAs (18S, 5.8S, and 28S) are spliced from a single precursor RNA (45S rRNA) that is encoded by a tandemly repeated locus (hereafter referred to as the 45S rDNA locus) residing on several chromosomes in humans and mice (Fig. 1 A and B). The 45S rDNA arrays display concerted evolution of DNA sequences that increases sequence uniformity among 45S rDNA units within and between arrays (2, 11, 24–27). The fourth rRNA molecule (5S rRNA) is also encoded by a repeated locus that is physically unlinked from the 45S rDNA loci (Fig. 1 A and B) in humans and mice. The 5S and 45S elements do not share segments of homology and are, indeed, transcribed by a different RNA polymerase [the 45S arrays are transcribed by polymerase I (Pol I) whereas the 5S array is transcribed by Pol III]. The 45S arrays give rise to the nucleolus, a nuclear organelle that is the site of ribosome biogenesis (28–31).
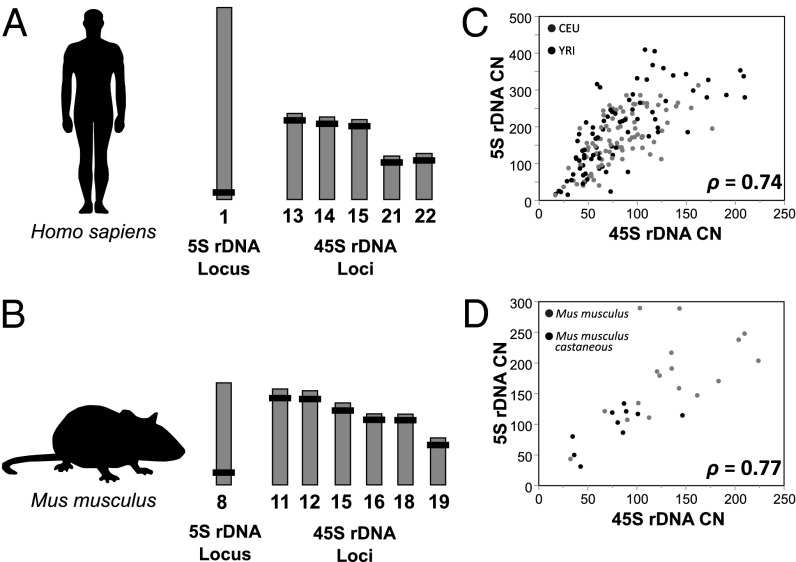
Fig. 1.
cCNV between 5S and 45S rDNA loci residing in different chromosomes. Genomic location of the 5S and 45S rDNA arrays in human (A) and mouse (B). Gray bars represent chromosomes, and black rectangles represent the approximate chromosomal location of the 5S and 45S rDNA loci. 5S (y axis) and 45S (x axis) rDNA CN is highly correlated in human (C) and mouse (D). Gray and black points represent CEU and YRI human populations, and laboratory/inbred and wild mouse isolates, respectively. Spearman correlations reflecting combined populations are reported. The 45S rDNA CN is the average CN of selected segments across the 18S, 5.8S, and 28S components. Correlations are consistent when stratified by population and sex (Table S1).
Here we investigate the relationship between CN variation in the 5S array and the 45S arrays. CN variation in the 5S and 45S DNA elements was ascertained with short-read whole-genome sequence data (19). We applied the methodology to samples from 78 and 90 individuals with African (YRI) and European (CEU) ancestry, respectively (32), and 17 inbred laboratory strains of mice and 10 wild Mus musculus castaneus isolates (33, 34). Human genomic DNA (gDNA) samples originated from whole blood and B cell-derived lymphoblastoid cell lines (LCLs); mouse gDNA was isolated from liver samples. The data revealed a positive association between CN in the 5S and 45S loci and a novel mode of genome variation, which we refer to as concerted CN variation (cCNV). To validate our observations, we experimentally ascertained cCNV across human genotypes with DNA samples isolated from LCL lines and whole blood. The outcome of cCNV is the homogenization of CN between loci on different chromosomes. It operates in the absence of sequence homology and is likely driven by functional requirements to maintain rRNA dosage balance.
Results
First, we developed a computational approach to determine rDNA CN across human and mouse genomes by using whole-genome DNA sequencing (DNA-seq) data (19). By using this method, our CN estimates for a panel of CN variable genes are in close agreement with a previous study (19, 35). We observed more than 10-fold variation in rDNA CN within humans and mice, with remarkable congruence in the lower and upper bounds of variation in 5S (human, minimum 16 and maximum 210; mouse, minimum 32 and maximum 224) and 45S elements (human, minimum 14 and maximum 410; mouse, minimum 31 and maximum 289; Fig. S1). These estimates are within the range expected from experimental analyses (12, 36, 37) and indicate that the rDNA loci are among the most CN variable coding segments of the genome. The 5S and 45S components do not share segments of homology or stretches of sequence similarity; as expected, reads mapped exclusively to one of these components but never to both.
rDNA 5S and 45S Genes Display cCNV Across Genotypes.
Concerted evolution maintains DNA sequence similarity between repeat units of the 45S rDNA array (2, 11, 24–27). The mechanism leads to a uniformity of sequences within a species, but it is not known to influence CN of rDNA arrays. Classical studies have documented rapid rates of expansion and contraction in the 45S rDNA arrays (11). Here, we hypothesize that rDNA loci undergo cCNV across mammalian chromosomes and display a positive correlation in CN among rDNA arrays. To address the issue, we focus on the 5S rDNA locus, which is readily distinguishable and physically unlinked from the 45S rDNA loci in the human (38, 39) and mouse (40–42) genomes (Fig. 1 A and B). In agreement with our hypothesis, we find that CN in the 5S locus is significantly correlated with CN in the 45S rDNA loci in both humans (Spearman correlation ρ = 0.74; P = 1 × 10−30) and in mice (ρ = 0.77; P = 2.4 × 10−6). We have extensively evaluated sources that could potentially confound CN estimates. First, we estimated rDNA CN based on the average read depth across the entire locus, as well as the average read depth across selected segments displaying lowest variances in per-site read depth (Fig. S2). Both analyses revealed robust associations between 45S and 5S rDNA CN. Second, we examined the robustness of the association with the data stratified by population and sex (Table S1). Third, correcting the estimates of CN to mitigate confounding factors caused by pseudogenes had a minor effect on individual estimates (Materials and Methods) (19) (Fig. S3) and did not change our estimates of the strength of cCNV. Finally, we used quantitative PCR (qPCR) to experimentally validate cCNV with human DNA samples obtained from LCLs and whole blood. These analyses confirm the association between CN of the 5S and 45S elements across individual genotypes in these populations (ρ = 0.71, P = 4.58 × 10−9, n = 54 for all individuals; ρ = 0.53, P = 0.006, n = 26 individuals from population with DNA isolated from LCLs; ρ = 0.47, P = 0.013, n = 28 individuals from population with DNA isolated from blood; Fig. S4). The data reveal cCNV between 5S and 45S elements that is manifested across chromosomes and in the absence of sequence homology.
CN of the 5S rDNA Is More Tightly Correlated with Functionally Linked 45S rDNA Segments.
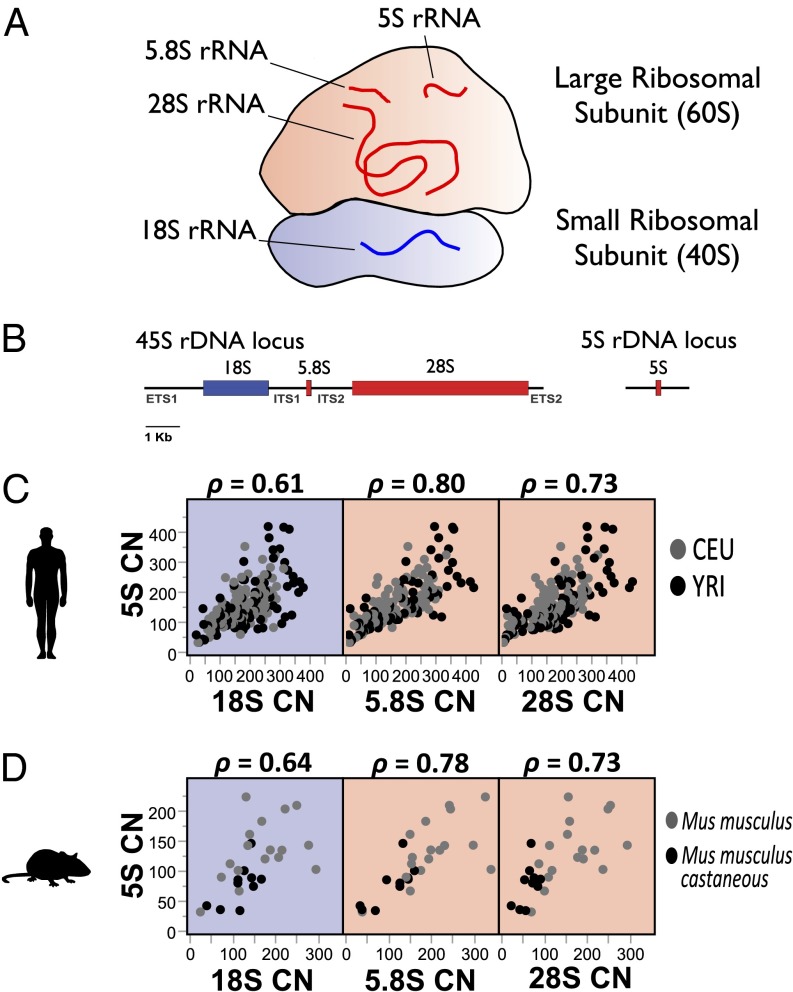
Balanced expression has been unequivocally demonstrated between members of macromolecular complexes (4, 43). Similarly, cCNV between functionally related loci might emerge if balanced gene dosage between the loci is important for cellular fitness. Indeed, the evolutionary conserved associations between CN in the 5S locus and 45S loci suggest that natural selection contributes to the maintenance of balanced CN. The model predicts that stoichiometric demands should be stronger among the 5S, 5.8S, and 28S rRNAs that constitute the RNA portion of the large 60S ribosomal subunit compared with the 18S rRNA, which assembles with the small 40S ribosomal subunit (Fig. 2 A and B). Hence, we examined CN associations between the 5S locus and each of the rRNA coding regions of the 45S rDNA locus (18S, 5.8S, and 28S). In agreement with our predictions, the 5S locus was significantly more highly correlated with the 5.8S and 28S rDNA components than with the 18S rDNA component (P < 0.001 for the equality of correlation coefficients; Fig. 2 C and D). Collectively, our observations are consistent with the expectation that cCNV might have evolved to mitigate gene dosage imbalances between rRNA components of the ribosome.
Fig. 2.
cCNV of rDNA loci is stronger between functionally linked components of the ribosome. (A) Schematic diagram of the ribosome. The large 60S subunit is shown in red, and the small 40S subunit is shown in blue. (B) The 5S, 5.8S, and 28S rRNA molecules assemble with the large subunit, and the 18S rRNA assembles with the small subunit of the ribosome. 5S rDNA CN is more strongly associated with rRNA molecules that comprise the large ribosomal subunit in human (C) and mouse (D). 5S rDNA CN is shown on the y axis, and 18S, 5.8S, and 28S rDNA CNs are depicted on the x axis. Spearman correlations of combined populations are reported.
cCNV Is Manifested Within the Offspring of Human Pedigrees.
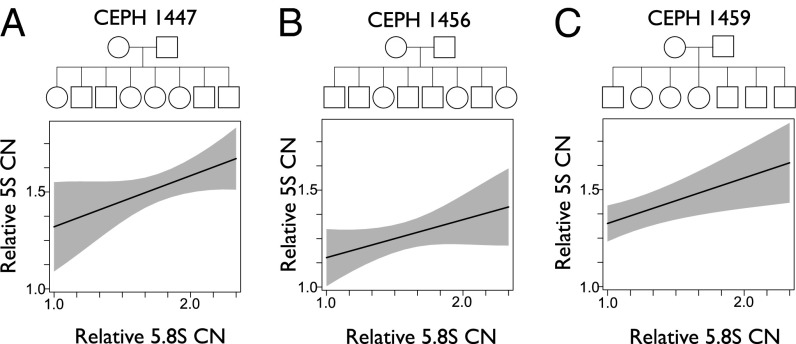
We observe cCNV across genotypes in human and mouse populations. However, cCNV might also operate within sufficiently short time scales and be manifested across individuals in the progeny of a single parental pair. Indeed, rapid amplification and contraction in the 45S rDNA array have been reported in human parent/offspring trios (12) and somatically in Drosophila (17). The 5S locus on human chromosome 1 is unlinked to the 45S arrays and is, therefore, expected to segregate and vary independently in the progeny of a parental pair. On the contrary, our hypothesis of cCNV predicts covariation between the 5S and 45S loci. We experimentally genotyped 5S and 45S CN in three CEU human pedigrees whose terminal branches consist of seven or eight full siblings (Fig. 3). In agreement with our expectations, we observed rDNA expansion and contraction across siblings and the statistically significant association between 5S and 45S elements within pedigrees despite the narrow range of variation (ρ = 0.54, P = 0.002, n = 29 across all data; ρ = 0.50, P = 0.2, n = 9, family 1459; ρ = 0.54, P = 0.1, n = 10, family 1447; ρ = 0.77, P = 0.01, n = 10, family 1456; Fig. 3). The manifestation of cCNV within families suggests that CN of the rDNA loci is not only varying in concert across loci but that CN in 5S and 45S loci is expanding and contracting sufficiently rapid to be manifested across siblings.
Fig. 3.
cCNV between 5S and 45S loci is manifested in human pedigrees. The pedigree and association between the 5S (y axis) and 5.8S (x axis) is reported for each family (A–C). Estimates of CN were divided by the value of the individual with the smallest CN array within each family and component (relative CN). Generalized linear models indicate significant associations between 5S and 5.8S elements (P < 0.01; n = 29). The gray shaded area represents the 95% CI for the linear coefficients within each family.
CN of the 5S rDNA Is Unlinked with Nearby SNP Variation in the 1q42 Segment.
Rapid amplification and contractions in the rDNA loci between generations and the manifestation of cCNV across siblings suggest that rDNA CN evolves rapidly and reversibly. To further address the issue we analyzed SNPs in the HapMap individuals in our sample. Rapid and reversible 5S copy expansions and contractions lead to the expectation that the 5S array will not be statistically associated with proximal SNP variation in the 1q42 segment in which it resides. In agreement with our expectation, we observed that 5S CN variation is unlinked with SNP variation in the 1q42 region. Moreover, estimates of the VST statistic for the 5S rDNA element are close to zero (5S VST = 0.001) and reflects the absence of population differentiation in rDNA CN between CEU and YRI populations (mean 5S CN is 75.2 in YRI vs. 74.3 in CEU populations). On the other hand, we observe that the CEU and YRI populations display substantial population differentiation, with moderately high FST values for SNPs across the whole genome (FST = 0.087) and in the 1q42 segment (FST = 0.091). Principal component analysis of SNP variation in the 1q42 segment shows unequivocal population differentiation. Hence, cCNV results in rapid and reversible expansions and contractions of the 5S rDNA array that uncouples its CN variation from variation in nearby SNPs. The process results in a lack of linkage between CN variation in the 5S element and proximal SNP variation in the neighboring chromosomal segments upstream and downstream of the 5S locus.
Bisphenol A Induces Parallel CN Loss of 5S and 45S rDNA Units.
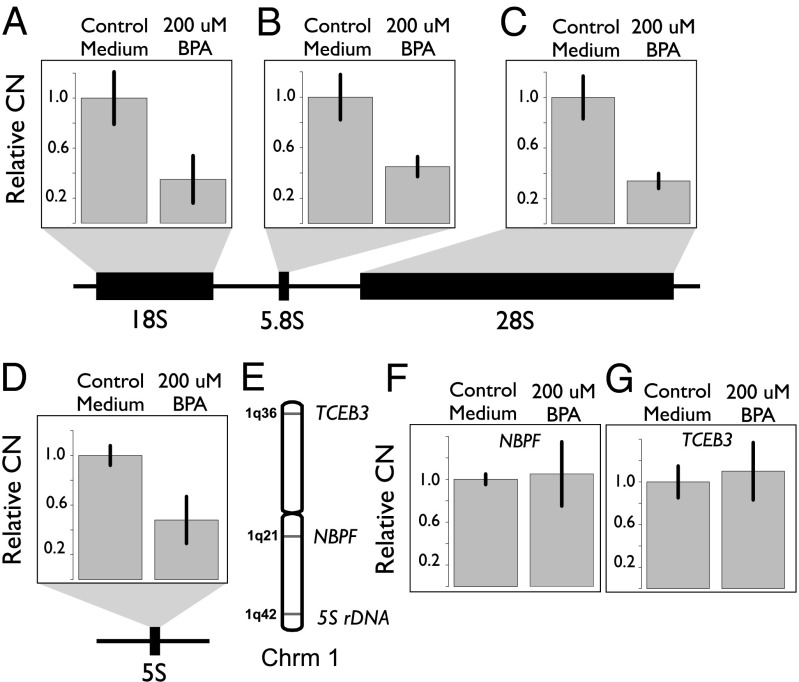
To further address the time scale in which coupling between 5S and 45S elements emerges, we investigated the hypothesis that environmental triggers might rapidly induce concerted CN expansions and contractions in the rDNA loci. In particular, it has recently been suggested that bisphenol A (BPA) disrupts the expression of the genes coding for proteins of the small and large ribosome, and could cause nucleolar stress (44). Disruption in the expression of nucleolar components might cause rDNA instability and trigger concerted modulation of rDNA CN across the 5S and 45S loci. Hence, we treated a human LCL with BPA-containing media (200 µM) for 24 h. We found that components of the rDNA locus were significantly reduced in the BPA treatment relative to the control (Student t test, n = 3, 18S, P = 0.031; 5.8S, P = 0.056; 28S, P = 0.021; Fig. 4 A–C). Furthermore, in agreement with the hypothesis of cCNV, we observed parallel modulation in the 5S rDNA array (5S; P = 0.003; Fig. 4D), whereas the CN of two tandemly repeated CN variable genes NBPF and TCEB3 located on chromosome 1 remained stable between control and BPA treatments (Student t test, n = 3; for both genes, P > 0.62; Fig. 4 E–G). These observations reinforce the hypothesis of cCNV between rDNA loci and indicate that environmental triggers can induce specific, rapid, and parallel rDNA loss in 5S and 45S arrays.
Fig. 4.
Rapid environmental induction of cCNV between the 5S and 45S rDNA arrays. CN of rDNA loci in human cell culture after 24 h growth in control and BPA-containing media. CN of each rDNA component was estimated with qPCR (n = 3; biological replicates). CNs were normalized, with CN of each component set to 1 in the control media (relative CN). Black bars in each graph represent the SD. The CN of the 45S (A–C) and 5S (D) rDNA contracted upon treatment with BPA. Chromosomal location of 5S rDNA array and the TCEB3 and NBPF gene arrays (E). CN of TCEB3 and NBPF, which are CN variable tandemly repeated arrays located on chromosome 1, remained stable upon BPA treatment (F and G).
Discussion
Here we develop and address the hypothesis of cCNV in the genetically unlinked, nonhomologous, but functionally related 5S and 45S rDNA elements. We observe tight coupling between the CN of the rRNA encoding 5S rDNA and 45S rDNA loci in humans and mice populations as well as human pedigrees. Gene dosage balance (43, 45) is a suitable model that fits well with our hypothesis of cCNV. Moreover, cCNV might act together with classical concerted variation in DNA sequences to enhance array uniformity and integrity, thus lessening the contribution from truncated rDNA copies (46), many of which are expected to be transcriptionally active. If balanced rDNA dosage is important to ensure proper ribosome assembly and function, cCNV of the 45S and 5S rDNA loci might emerge and be evolutionarily conserved, despite their transcription through distinct RNA polymerases and absence of sequence homology. This is because the four rRNA molecules are interdependent, contribute to ribosome structure, and make up the catalytic portion of the ribosome. The stoichiometric requirements of the ribosome may be disrupted if 5S and 45S gene dosage were uncoupled. In this model, independent expansion or contraction of 45S or 5S rDNA CN may be detrimental and cause lowered fitness.
Long range interchromosomal associations between loci can emerge through population bottlenecks but also through strong positive selection. In the malaria parasite Plasmodium falciparum, higher CN of the gch1 gene is strongly correlated with a variant of the dhfr gene, which together result in higher resistance to antifolate drugs (47). These genes are located on different chromosomes but participate in the folate biosynthesis pathway. Interchromosomal correlations in gene loci can also emerge from nonrandom gene loss and pseudogene formation following whole-genome duplication. The process is mediated by natural selection to maintain balance in the ribosome and mitigate the fitness costs of imbalance. Accordingly, observations in Arabidopsis indicate that ribosomal protein paralogs are preferentially retained for long periods after the tetraploidization event (48–51). The interpretation is that natural selection disfavored pseudogene formation in ribosomal proteins to maintain balanced dosage among protein members of the ribosome. Indeed, components of highly expressed macromolecular complexes are among the loci that are most sensitive to dosage imbalance (43). Following polyploidization, balanced gene loss operates to maintain equitable dosage among functionally related loci participating in protein complexes. Similarly, studies in yeast have revealed that dosage-sensitive genes are more likely to encode subunits of protein complexes (4, 52), further indicating the strong selective pressure to maintain the stoichiometric interdependence of multiple gene products that form a single functioning macromolecule (43). In humans, dosage imbalance of interacting proteins has been hypothesized to predispose to disease (53).
Cellular mechanisms of CN control and evolutionary processes can explain the emergence of concerted modulation in CN between functionally related loci. An important consideration is that rDNA CN may be remarkably labile, and we expect that random and directed processes could contribute to rDNA CN modulation. In this regard, unusually high rates of rDNA repeat expansion and contraction could facilitate the emergence of cCNV. Mutation rates for CN variation (10−4 to 10−2) (54) are estimated to be four to six orders of magnitude higher than the mutation rate for single nucleotides (10−8) (55). In this model, natural selection at the cellular level could also contribute to the manifestation of cCNV if cells with balanced gene dosage have higher fitness. On the other hand, active mechanisms of developmental and tissue-specific gene amplification and contraction can provided targeted, controlled, and synchronous modulation of rDNA CN (8, 56) as well as impact nucleolar activity in specific developmental stages and tissues (22, 56). In this case, cCNV might emerge through autonomous molecular mechanisms of coordinated gene dosage modulation (56–58). Interestingly, the 45S rDNA repeats make up the core structures of the nucleolus and are transcribed from RNA polymerase I, whereas 5S elements are localized in the periphery of the organelle and are transcribed from RNA polymerase III (59–61). We hypothesize that 45S CN modulation in the nucleolus will drive CN variation in 5S elements at the boundary of the organelle. Evolutionarily, spatial localization of 5S elements in the periphery of the nucleolus might have been driven in part by requirements to coordinate 5S and 45S expression to maintain optimal dosage balance. It is intriguing that, in yeast, the 5S and 45S elements are physically linked and reside in a single rDNA array on chromosome XII despite their transcription from different RNA polymerases (62). All in all, we predict that autonomous cellular mechanisms as well as natural selection in cell populations could contribute to the manifestation of cCNV at the scale of a few cell divisions as well as across individual genotypes in natural populations.
Environmental factors and chemical stressors can drive CN variation (63), including CN of the rDNA locus (14). Indeed, soil bacteria with a greater number of rDNA arrays dominate isolates with fewer rDNA arrays when exposed to herbicide (64). Similarly, numerous studies in plants reveal that rDNA CN is responsive to environmental pressures, such as temperature, humidity, and fire-related stress (65–68). Our data indicate that BPA can induce rapid and parallel 5S and 45S rDNA CN loss in a human cell population. The observation is consistent with evidence that BPA can induce nucleolar stress and affect gene expression of ribosomal genes in Drosophila (44), as well as reports that BPA affects chromosome structure and induce DNA damage (69, 70). Rapid modulation of rDNA CN raises the prospect that miscoordination of rDNA CN in environmentally triggered cCNV could be a new cellular mechanism contributing to disease.
Concerted evolution of DNA sequences operates through gene conversion and unequal recombination, and leads to sequence uniformity between 45S rDNA loci in different arrays (2, 11, 24–26). Importantly, concerted evolution of DNA sequences is limited by the requisite of substantial homology between the sequences involved. On the other hand, cCNV occurs in the absence of sequence homology, between loci in different chromosomes, and is likely driven by functional demands for balanced rDNA expression that emerges during evolution and development. Our results uncover a novel mode of genome variation and raise the question of how widespread cCNV might be across other functionally related loci of high CN and undergoing rapid rates of amplification and contraction. They raise the prospect that human disease phenotypes might be traced to disrupted rDNA dosage balance in the genome.
Materials and Methods
Reference Sequences.
Consensus rDNA locus reference sequences for human (71) and mouse (72) were obtained from GenBank (human, accession no. U13369; mouse, accession no. NR_046233.2). The 5S rDNA sequence was identified in each genome and extracted along with flanking regions from the University of California, Santa Cruz, Genome Browser (73) (human 5S, chr1:228,766,135–228,766,255; mouse 5S, chr8:123,538,584-123,539,104). We used a combination of RNAmmer v1.2 (74), manual sequence alignment between the human and mouse references, and BLAST (75) to annotate the boundaries of each rDNA element, identify conserved segments, and rule out the existence of stretches of sequence similarity between rDNA components. For each species, the complete exon sets were downloaded from the University of California, Santa Cruz browser. We developed a reference single copy exon set as a proxy for background read depth (BRD). To retain putative single copy exons, we applied several filtering metrics. First, exons with significant BLAST hits (E-value < 1 × 10−6) to any other exons were removed to reduce regions with homology to other sequences. Second, exons from the sex chromosomes were removed. Third, we retained only the largest exon of each gene. Finally, only exons ≥ 300 bp were retained.
Short-Read Genome Sequence Data and Read Mapping.
We obtained raw Illumina fastq whole-genome DNA sequence reads from CEU and YRI individuals from the 1000 Genomes Project FTP site (1000genomes.org) (32). Illumina data from laboratory (34) and wild mouse (33) isolates were attained from the National Center for Biotechnology Information sequence read archive (SRA). SRA files were converted to fastq using the “fastq-dump” function in the SRA toolkit v2.3.5–2. All fastq files were quality trimmed by using Trim Galore (www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Residual adapter sequences were removed from reads, and reads were trimmed such that they contained a minimum quality score of 20 at each nucleotide position. Trimmed reads shorter than 50 nt were discarded. Quality trimmed read sets for each sample were mapped against the reference 45S rDNA locus, 5S locus, and single copy exon set by using Bowtie2 v2.0.5 (76). Read mapping was performed unpaired using the “sensitive” preset (command line parameters: –end-to-end -D 15 -R 2 -L 22 -i S,1,1.15). Mapping output was converted to sorted bam format using the samtools (v0.1.18) “view” and “sort” functions, respectively (77).
Estimation of BRD from Single-Copy Exon Panel.
BRD, representing the depth of sequences present in the genome at a single copy, was calculated for each sample and was used as a normalized factor to estimate 45S and 5S rDNA CN. Average per-base BRD was calculated from the sorted bam mapping output file by using the samtools “depth” function (77). Because fewer reads will map to the beginning and ends of reference sequences, we excluded the first and last 108 bp of each exon when calculating BRD. Additionally, for each sample, we excluded read depth values in the upper 5% of the distribution.
5S and 45S rDNA CN Estimation.
We estimated 45S and 5S haploid CN in two ways. First, average read depth of the 45S rDNA locus (ranging from the 18S start site to the 28S stop site) and the 5S rDNA locus was normalized by the BRD. CN is calculated as average read depth divided by BRD. This procedure is detailed in a previous publication (19). Second, average read depth of selected segments of the 45S locus and 5S locus were normalized by the BRD. The first procedure revealed variation in per-base read depth, and nonhomogeneous CN estimates per sites across the 18S and 28S rDNA components. Hence, in the second procedure, we calculated CN variance across sites for nonoverlapping 150-bp windows. We chose a window of 150 bp because it is a similarly sized region to the 5.8S and 5S rDNA loci, both of which show lower variance in read depth and CN across sites. Individual 45S rDNA component CN was calculated from the window with the lowest average variance (Fig. 2 C and D), and a consensus 45S CN was estimated as the average across the three components (Fig. 1 C and D). For the 5.8S and 5S components, the entire region was used as read depth and CN estimates were similar across sites within each locus. Segments with homogeneous CN estimates for each locus are as follows: 18S, 183–327; 5.8S, 1–157 (full sequence); 28S, 114–263; and 5S, 1–122 (full sequence; Fig. S2). Reads that mapped to each coding component of the 5S and 45S rDNA arrays were independently extracted for each sample and mapped against the human reference genome, including all supercontigs that have not been assigned a chromosomal location. As expected, the majority of 45S rDNA reads mapped to the single supercontig that harbors the 45S rDNA locus (GL000220.1; 93.4% mapped to 45S) (19), whereas 5S rDNA reads mapped to chromosome 1q42, which harbors the 5s rDNA locus (90.2%; Fig. S3). For each individual, we used the percentage of reads mapped to the expected chromosomes (chromosome 1 for 5S) and contigs (GL000220.1 for 45S) as a correction factor to obtain best estimates of 5S and 45S CN (19). The correlation between 5S and 45S is robust and remains true and nearly identical when CN of each element is estimated from (i) reads mapping across the whole coding element or from (ii) reads mapping to selected segments of each element, and (iii) regardless of whether the correction for pseudogenes is implemented.
Experimental Validation of cCNV.
Experimental validation of cCNV was performed in a population sample that consists of DNA isolated from LCLs from 15 individuals from CEU origin and 12 individuals from the autism collection, as well as DNA isolated from whole blood of 28 ethnically diverse individuals from the Bioserve repository (Beltsville, MD; Table S2). In addition, validation was also performed in three full pedigrees. DNA derived from B cell-derived LCLs was obtained from the Coriell Cell Repository. Real-time qPCR analyses were carried out with the KAPA SYBR FAST Universal PCR Master Mix (Kapa Biosystems) using 7900HT Fast Real-Time PCR (Applied Biosystems). The reaction was performed using 20 ng of gDNA from each individual and 125 nM of each primer (Table S3). All samples were analyzed three times to check the consistency of the results. The samples were submitted to 40 cycles of 10 s at 95 °C and 30 s at 60 °C. Unspecific amplification was assessed by the dissociation curve, and the results of CN variation were normalized based on delta Ct values from single copy genes.
Analysis of Population Differentiation.
Although there is no evidence for population stratification within CEU or YRI populations (78), these two populations display substantial levels of differentiation from one another. We used HapMap3 data to interrogate SNP variation across the whole genome and in the 1q42 segment of 79 individuals with Northern and Western Europe ancestry (i.e., CEU) and 75 individuals with Western African ancestry (i.e., YRI). The CEU and YRI data were merged into one dataset and analyzed with PLINK software, version 1.07 (79). Multidimensional scaling and principal components analyses displayed evidence of population differentiation across the whole genome and in the 1q42 segment. We calculated FST and VST (80) values to examine population differentiation in the 5S array and 1q42 segment between CEU and YRI (81). The fixation index (Fst) is widely used as a descriptive statistic to measure population differentiation. Fst were calculated by using SNPstats R Bioconductor package (www.bioconductor.org). VST was calculated as (VT − VS)/VT, where VT is the variance in log2 ratios among all unrelated individuals and VS is the population size weighted average variance within each population.
Cell Culture and BPA Treatment.
We obtained an LCL from the Coriell Cell Repository (NA12043). All cells are free from bacterial, fungal, or mycoplasma contamination. Cell were cultivated in T25 flasks under standard conditions with media containing RPMI 1640 enriched with 2 mM l-glutamine and 15% (vol/vol) FBS at 37 °C. For the experimental procedure, cells were seeded at 2 × 105 viable cells per milliliter in 5 mL of medium. After 48 h, three biological replicas were treated with 200 μM of BPA added directly to the culture. Cells were harvested 24 h after the treatment with BPA, and gDNA was isolated using DNeasy Blood and Tissue kit following the method described by the manufacturer (Qiagen). gDNA samples were treated with RNase A, and concentration was estimated with NanoDrop (Thermo Scientific).
Supplementary Material
Acknowledgments
We thank the SEQanswers community for helpful discussions, members of the laboratory of B.L. for constructive comments and discussions, and two anonymous reviewers for their thoughtful comments. The computations in this paper were performed on the Odyssey cluster maintained by the Faculty of Arts and Sciences (FAS) Science Division Research Computing Group at Harvard University. This work was supported by Training Grant T32-HL007118 (NIH), a Smith Family Award for Excellence in Biomedical Research (to B.L.), and an Ellison Medical Foundation New Scholars in Aging Award (to B.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1416878112/-/DCSupplemental.
References
- 1.Finnegan DJ, Rubin GM, Young MW, Hogness DS. Repeated gene families in Drosophila melanogaster. Cold Spring Harb Symp Quant Biol. 1978;42(pt 2):1053–1063. doi: 10.1101/sqb.1978.042.01.106. [DOI] [PubMed] [Google Scholar]
- 2.Coen ES, Thoday JM, Dover G. Rate of turnover of structural variants in the rDNA gene family of Drosophila melanogaster. Nature. 1982;295(5850):564–568. doi: 10.1038/295564a0. [DOI] [PubMed] [Google Scholar]
- 3.Perry GH, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39(10):1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424(6945):194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 5.Fogel S, Welch JW. Tandem gene amplification mediates copper resistance in yeast. Proc Natl Acad Sci USA. 1982;79(17):5342–5346. doi: 10.1073/pnas.79.17.5342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Axelsson E, et al. The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature. 2013;495(7441):360–364. doi: 10.1038/nature11837. [DOI] [PubMed] [Google Scholar]
- 7.Gibbons JG, et al. The evolutionary imprint of domestication on genome variation and function of the filamentous fungus Aspergillus oryzae. Curr Biol. 2012;22(15):1403–1409. doi: 10.1016/j.cub.2012.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claycomb JM, Benasutti M, Bosco G, Fenger DD, Orr-Weaver TL. Gene amplification as a developmental strategy: Isolation of two developmental amplicons in Drosophila. Dev Cell. 2004;6(1):145–155. doi: 10.1016/s1534-5807(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 9.Calvi BR, Lilly MA, Spradling AC. Cell cycle control of chorion gene amplification. Genes Dev. 1998;12(5):734–744. doi: 10.1101/gad.12.5.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Calvi BR, Spradling AC. The nuclear location and chromatin organization of active chorion amplification origins. Chromosoma. 2001;110(3):159–172. doi: 10.1007/s004120100131. [DOI] [PubMed] [Google Scholar]
- 11.Eickbush TH, Eickbush DG. Finely orchestrated movements: Evolution of the ribosomal RNA genes. Genetics. 2007;175(2):477–485. doi: 10.1534/genetics.107.071399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stults DM, Killen MW, Pierce HH, Pierce AJ. Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res. 2008;18(1):13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lyckegaard EM, Clark AG. Ribosomal DNA and Stellate gene copy number variation on the Y chromosome of Drosophila melanogaster. Proc Natl Acad Sci USA. 1989;86(6):1944–1948. doi: 10.1073/pnas.86.6.1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weider LJ, et al. The functional significance of ribosomal (r)DNA variation: Impacts on the evolutionary ecology of organisms. Annu Rev Ecol Evol Syst. 2005;36:219–242. [Google Scholar]
- 15.Prokopowich CD, Gregory TR, Crease TJ. The correlation between rDNA copy number and genome size in eukaryotes. Genome. 2003;46(1):48–50. doi: 10.1139/g02-103. [DOI] [PubMed] [Google Scholar]
- 16.Eagle SH, Crease TJ. Copy number variation of ribosomal DNA and Pokey transposons in natural populations of Daphnia. Mob DNA. 2012;3(1):4. doi: 10.1186/1759-8753-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci USA. 2009;106(42):17829–17834. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paredes S, Branco AT, Hartl DL, Maggert KA, Lemos B. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7(4):e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibbons JG, Branco AT, Yu S, Lemos B. Ribosomal DNA copy number is coupled with gene expression variation and mitochondrial abundance in humans. Nat Commun. 2014;5:4850. doi: 10.1038/ncomms5850. [DOI] [PubMed] [Google Scholar]
- 20.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 21.Moss T, Langlois F, Gagnon-Kugler T, Stefanovsky V. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell Mol Life Sci. 2007;64(1):29–49. doi: 10.1007/s00018-006-6278-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 23.Pontvianne F, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27(14):1545–1550. doi: 10.1101/gad.221648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnheim N, et al. Molecular evidence for genetic exchanges among ribosomal genes on nonhomologous chromosomes in man and apes. Proc Natl Acad Sci USA. 1980;77(12):7323–7327. doi: 10.1073/pnas.77.12.7323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krystal M, D’Eustachio P, Ruddle FH, Arnheim N. Human nucleolus organizers on nonhomologous chromosomes can share the same ribosomal gene variants. Proc Natl Acad Sci USA. 1981;78(9):5744–5748. doi: 10.1073/pnas.78.9.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szostak JW, Wu R. Unequal crossing over in the ribosomal DNA of Saccharomyces cerevisiae. Nature. 1980;284(5755):426–430. doi: 10.1038/284426a0. [DOI] [PubMed] [Google Scholar]
- 27.Ganley AR, Kobayashi T. Highly efficient concerted evolution in the ribosomal DNA repeats: Total rDNA repeat variation revealed by whole-genome shotgun sequence data. Genome Res. 2007;17(2):184–191. doi: 10.1101/gr.5457707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woolford JL, Jr, Baserga SJ. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics. 2013;195(3):643–681. doi: 10.1534/genetics.113.153197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3(3) doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grummt I. The nucleolus—guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122(6):487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 31.Sullivan GJ, et al. Human acrocentric chromosomes with transcriptionally silent nucleolar organizer regions associate with nucleoli. EMBO J. 2001;20(11):2867–2874. doi: 10.1093/emboj/20.11.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abecasis GR, et al. 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halligan DL, et al. Contributions of protein-coding and regulatory change to adaptive molecular evolution in murid rodents. PLoS Genet. 2013;9(12):e1003995. doi: 10.1371/journal.pgen.1003995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keane TM, et al. Mouse genomic variation and its effect on phenotypes and gene regulation. Nature. 2011;477(7364):289–294. doi: 10.1038/nature10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sudmant PH, et al. 1000 Genomes Project Diversity of human copy number variation and multicopy genes. Science. 2010;330(6004):641–646. doi: 10.1126/science.1197005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zentner GE, Saiakhova A, Manaenkov P, Adams MD, Scacheri PC. Integrative genomic analysis of human ribosomal DNA. Nucleic Acids Res. 2011;39(12):4949–4960. doi: 10.1093/nar/gkq1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmickel RD. Quantitation of human ribosomal DNA: Hybridization of human DNA with ribosomal RNA for quantitation and fractionation. Pediatr Res. 1973;7(1):5–12. doi: 10.1203/00006450-197301000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Henderson AS, Warburton D, Atwood KC. Letter: Ribosomal DNA connectives between human acrocentric chromosomes. Nature. 1973;245(5420):95–97. doi: 10.1038/245095b0. [DOI] [PubMed] [Google Scholar]
- 39.Sørensen PD, Frederiksen S. Characterization of human 5S rRNA genes. Nucleic Acids Res. 1991;19(15):4147–4151. doi: 10.1093/nar/19.15.4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurihara Y, Suh DS, Suzuki H, Moriwaki K. Chromosomal locations of Ag-NORs and clusters of ribosomal DNA in laboratory strains of mice. Mamm Genome. 1994;5(4):225–228. doi: 10.1007/BF00360550. [DOI] [PubMed] [Google Scholar]
- 41.Matsuda Y, Chapman VM. Application of fluorescence in situ hybridization in genome analysis of the mouse. Electrophoresis. 1995;16(2):261–272. doi: 10.1002/elps.1150160142. [DOI] [PubMed] [Google Scholar]
- 42.Matsuda Y, et al. Chromosomal mapping of mouse 5S rRNA genes by direct R-banding fluorescence in situ hybridization. Cytogenet Cell Genet. 1994;66(4):246–249. doi: 10.1159/000133704. [DOI] [PubMed] [Google Scholar]
- 43.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109(37):14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Branco AT, Lemos B. High intake of dietary sugar enhances bisphenol A (BPA) disruption and reveals ribosome-mediated pathways of toxicity. Genetics. 2014;197(1):147–157. doi: 10.1534/genetics.114.163170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21(4):219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 46.Caburet S, et al. Human ribosomal RNA gene arrays display a broad range of palindromic structures. Genome Res. 2005;15(8):1079–1085. doi: 10.1101/gr.3970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nair S, et al. Adaptive copy number evolution in malaria parasites. PLoS Genet. 2008;4(10):e1000243. doi: 10.1371/journal.pgen.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99(21):13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16(7):934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casanova-Saez R, Candela H, Micol JL. Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis. Sci Rep. 2014;4:4122. doi: 10.1038/srep04122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rosado A, Raikhel NV. Application of the gene dosage balance hypothesis to auxin-related ribosomal mutants in Arabidopsis. Plant Signal Behav. 2010;5(4):450–452. doi: 10.4161/psb.5.4.11341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semple JI, Vavouri T, Lehner B. A simple principle concerning the robustness of protein complex activity to changes in gene expression. BMC Syst Biol. 2008;2:1. doi: 10.1186/1752-0509-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. 2010;107(20):9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egan CM, Sridhar S, Wigler M, Hall IM. Recurrent DNA copy number variation in the laboratory mouse. Nat Genet. 2007;39(11):1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- 55.Kong A, et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mais C, Scheer U. Molecular architecture of the amplified nucleoli of Xenopus oocytes. J Cell Sci. 2001;114(pt 4):709–718. doi: 10.1242/jcs.114.4.709. [DOI] [PubMed] [Google Scholar]
- 57.Kobayashi T, Heck DJ, Nomura M, Horiuchi T. Expansion and contraction of ribosomal DNA repeats in Saccharomyces cerevisiae: Requirement of replication fork blocking (Fob1) protein and the role of RNA polymerase I. Genes Dev. 1998;12(24):3821–3830. doi: 10.1101/gad.12.24.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Claycomb JM, Orr-Weaver TL. Developmental gene amplification: Insights into DNA replication and gene expression. Trends Genet. 2005;21(3):149–162. doi: 10.1016/j.tig.2005.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Fedoriw AM, Starmer J, Yee D, Magnuson T. Nucleolar association and transcriptional inhibition through 5S rDNA in mammals. PLoS Genet. 2012;8(1):e1002468. doi: 10.1371/journal.pgen.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garcia S, Crhák Khaitová L, Kovařík A. Expression of 5 S rRNA genes linked to 35 S rDNA in plants, their epigenetic modification and regulatory element divergence. BMC Plant Biol. 2012;12:95. doi: 10.1186/1471-2229-12-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Németh A, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6(3):e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petes TD. Yeast ribosomal DNA genes are located on chromosome XII. Proc Natl Acad Sci USA. 1979;76(1):410–414. doi: 10.1073/pnas.76.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang YT, et al. Cigarette smoking increases copy number alterations in nonsmall-cell lung cancer. Proc Natl Acad Sci USA. 2011;108(39):16345–16350. doi: 10.1073/pnas.1102769108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Klappenbach JA, Dunbar JM, Schmidt TM. rRNA operon copy number reflects ecological strategies of bacteria. Appl Environ Microbiol. 2000;66(4):1328–1333. doi: 10.1128/aem.66.4.1328-1333.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bobola MS, Eckert RT, Klein AS. Restriction fragment variation in the nuclear ribosomal DNA repeat unit within and between Picea rubens and Picea mariana. Can J For Res. 1992;22(2):255–263. [Google Scholar]
- 66.Govindaraju DR, Cullis CA. Ribosomal DNA variation among populations of a Pinus rigida Mill. (pitch pine) ecosystem: I. Distribution of copy numbers. Heredity. 1992;69:133–140. [Google Scholar]
- 67.Gupta PK, et al. Polymorphism at rDNA loci in barley and its relation with climatic variables. Theor Appl Genet. 2002;104(2-3):473–481. doi: 10.1007/s001220100690. [DOI] [PubMed] [Google Scholar]
- 68.Sharma S, Beharav A, Balyan HS, Nevo E, Gupta PK. Ribosomal DNA polymorphism and its association with geographical and climatic variables in 27 wild barley populations from Jordan. Plant Sci. 2004;166(2):467–477. [Google Scholar]
- 69.Chapin RE, et al. NTP-CERHR expert panel report on the reproductive and developmental toxicity of bisphenol A. Birth Defects Res B Dev Reprod Toxicol. 2008;83(3):157–395. doi: 10.1002/bdrb.20147. [DOI] [PubMed] [Google Scholar]
- 70.Hunt PA, et al. Bisphenol A alters early oogenesis and follicle formation in the fetal ovary of the rhesus monkey. Proc Natl Acad Sci USA. 2012;109(43):17525–17530. doi: 10.1073/pnas.1207854109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gonzalez IL, Sylvester JE. Complete sequence of the 43-kb human ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics. 1995;27(2):320–328. doi: 10.1006/geno.1995.1049. [DOI] [PubMed] [Google Scholar]
- 72.Grozdanov P, Georgiev O, Karagyozov L. Complete sequence of the 45-kb mouse ribosomal DNA repeat: Analysis of the intergenic spacer. Genomics. 2003;82(6):637–643. doi: 10.1016/s0888-7543(03)00199-x. [DOI] [PubMed] [Google Scholar]
- 73.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagesen K, et al. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35(9):3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 76.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Frazer KA, et al. International HapMap Consortium A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449(7164):851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Purcell S, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Holsinger KE, Weir BS. Genetics in geographically structured populations: Defining, estimating and interpreting F(ST) Nat Rev Genet. 2009;10(9):639–650. doi: 10.1038/nrg2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.