Significance
It has been known for decades that purified small subunits of the ribosome can interconvert between active and inactive conformations in experiments performed under simplified conditions, but the physiological relevance of this switch has remained unclear. We probed the structure of ribosomal RNA in healthy living cells and discovered that stably assembled 30S subunits exist predominantly in the inactive conformation, with structural differences localized in the functionally important decoding region. Disrupting the ability to interconvert between active and inactive conformations compromised translation in cells. In-cell RNA structure probing supports a model in which “inactive” 30S subunits comprise an abundant in-cell state that regulates ribosome function.
Keywords: SHAPE, conformational change, in vivo, ribosome, 16S rRNA
Abstract
It was shown decades ago that purified 30S ribosome subunits readily interconvert between “active” and “inactive” conformations in a switch that involves changes in the functionally important neck and decoding regions. However, the physiological significance of this conformational change had remained unknown. In exponentially growing Escherichia coli cells, RNA SHAPE probing revealed that 16S rRNA largely adopts the inactive conformation in stably assembled, mature 30S subunits and the active conformation in translating (70S) ribosomes. Inactive 30S subunits bind mRNA as efficiently as active subunits but initiate translation more slowly. Mutations that inhibited interconversion between states compromised translation in vivo. Binding by the small antibiotic paromomycin induced the inactive-to-active conversion, consistent with a low-energy barrier between the two states. Despite the small energetic barrier between states, but consistent with slow translation initiation and a functional role in vivo, interconversion involved large-scale changes in structure in the neck region that likely propagate across the 30S body via helix 44. These findings suggest the inactive state is a biologically relevant alternate conformation that regulates ribosome function as a conformational switch.
Forty-five years ago, Zamir, Elson, and their colleagues reported that purified 30S subunits of the ribosome undergo a readily reversible conformational change between “active” and “inactive” states and proposed that this conformational rearrangement might mimic a natural process (1). Noller and coworkers used chemical probing to show that this conformational change occurs in the neck and decoding center regions of the 16S ribosomal RNA (rRNA) and has “the appearance of a reciprocal interconversion between two differently structured states” (2). Recent structural analyses indicate that the protein-free 16S rRNA adopts alternative base-paired conformations in the neck region that are conserved among diverse eubacterial and archeal organisms (3). The ability to sample multiple conformations in this region is also conserved in eukaryotes (4). The original studies on the inactive and active states noted that probing ribosomes in cells might allow the biological roles of these states to be established (1, 2). Here we make use of recent innovations in in-cell RNA SHAPE (selective 2′-hydroxyl acylation analyzed by primer extension) probing (5) to interrogate the structure of 16S rRNA in free 30S subunits, in actively translating ribosomes, and in mutant ribosomes in exponentially growing Escherichia coli.
Results
In Vivo SHAPE Probing of Ribosomal States.
We used in vivo SHAPE (5, 6) to probe the RNA structure in exponentially growing E. coli cells and then halted translation by rapidly pouring the cells over ice (7). Experiments were performed with the SHAPE reagent 1M7, which readily enters cells and either reacts with RNA or undergoes inactivation by hydrolysis over ∼2 min. Probing is thus rapid, no explicit quench step is required, and the experiment is performed under mild conditions compatible with recovery of intact cellular ribonucleoprotein complexes. Ribosomal components were separated by velocity sedimentation through a sucrose gradient. We observed well-defined peaks corresponding to polysomes, indicating that in-cell probing and subsequent ribosome component fractionation did not disrupt the interaction between ribosomes and mRNA (Fig. 1A).
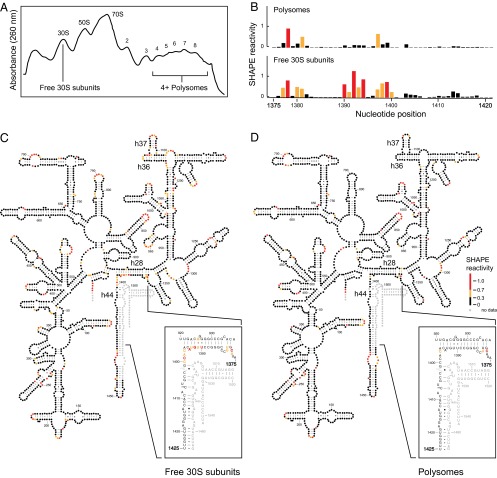
Fig. 1.
In vivo SHAPE analysis of ribosome complexes. (A) Ribosome complexes from E. coli cells, probed with 1M7, partitioned on sucrose density gradients. Peaks corresponding to 30S and 50S ribosomal subunits, 70S ribosomes, and polysomes containing four to eight ribosomes are indicated; the top of the gradient is on the left. (B) SHAPE reactivity profiles for 16S rRNA from polysomes (Top) and free 30S subunits (Bottom). (C and D) SHAPE reactivities for 16S rRNA isolated from (C) free 30S subunits and (D) polysomes superimposed on the conventional secondary structure. Nucleotides are shown as circles, colored by SHAPE reactivity (see scale). As shown in the Insets, reactivities differ in the neck region, and reactivities for the 16S rRNA in free 30S subunits are inconsistent with the conventional structure.
Fractions were obtained corresponding to free 30S subunits, 70S ribosomes, and polysomes (at least four ribosomes per mRNA). For each sample, the rRNA was isolated and primer extension used to quantify the 1M7 reactivities of 16S rRNA nucleotides (Fig. 1B). SHAPE reactivities provide a model-free measure of local nucleotide flexibility (8). The vast majority of nucleotides (∼94%) had similar SHAPE reactivities (within 0.3 SHAPE units) in 30S subunits, 70S ribosomes, and polysomes (Fig. 1 C and D and Fig. S1). Reactivity patterns were compared with the expected accessibility of nucleotides in the conventional RNA secondary structure (9). The reactivity patterns for 70S ribosomes and polysomes were nearly identical and were fully consistent with the conventional secondary structure (Fig. 1D). In the free 30S subunits, 43 out of the 45 helices in the 16S rRNA had SHAPE reactivities consistent with the conventional RNA secondary structure (Fig. 1C).
An RNA Conformational Change Differentiates Free 30S Subunits and Translating Ribosomes.
Critically, two regions in the 16S rRNA isolated from free 30S subunits had reactivity profiles inconsistent with the conventional secondary structure model. One spans half of helix 28 (h28) and the second involves h36 (Fig. 1 C). These helices form part of the neck in the 30S subunit, adjacent to the decoding site. In the conventional structure, h28 is formed in part by base pairing between nucleotides 923–927 and 1390–1393, and the in vivo SHAPE data from polysomes are consistent with this secondary structure (Fig. 1 B, Top, and 1D). In contrast, in the free 30S subunit, nucleotides 1390–1394 are modified by SHAPE (Fig. 1 B, Bottom), indicating that they are not constrained by stable base-pairing interactions. Nucleotides in one strand of h36 (positions 1081–1083) were also strongly reactive, suggesting that in free 30S subunits these nucleotides are not involved in base pairing. These data indicate that the conformation of the neck region differs in translating and free 30S subunits.
A few other regions had higher SHAPE reactivity in free 30S subunits than in polysomes (Fig. S1). These include nucleotides in loops centered on positions 790, 965, 1092, and 1109. The latter two loops interact with one another as part of the h35/h36/h37 region that packs against the neck helix h28 (Fig. S2). Increased reactivity of the 790 loop in the free 30S subunit may be due to initiation factor 3 (IF3) binding or to the absence of the P-site tRNA or the 50S subunit (10, 11). Higher SHAPE reactivities in the 965 loop likely reflect a vacant P site in the free subunit (11).
The free 30S fraction likely contained mature subunits that have participated in previous rounds of translation as well as newly assembled 30S subunits. There was a short lag between the time when cells were modified by 1M7 and when translation was stopped and ribosomes isolated. We therefore examined, and ruled out, contributions from (i) immature 30S subunits that were modified before complete assembly (5) and (ii) 70S ribosomes that were modified but dissociated during purification to sediment in the 30S peak. To examine whether the alternate 16S rRNA structure resulted from an immature 30S species, cells were incubated with rifampicin to halt transcription; under these conditions, 30S species assemble fully (5). SHAPE reactivities for the 16S rRNA from the free 30S peak from cells treated with and without rifampicin were nearly identical (Fig. 2A; Pearson’s linear r = 0.92). To test whether dissociation of 70S ribosomes contributed to the free 30S fraction, we treated cells with the antibiotic chloramphenicol, which binds to the 50S subunit, prevents peptidyl transfer and subsequent translation, and stabilizes 70S ribosomes (12). Again, SHAPE reactivities for 16S rRNA in free 30S subunits plus and minus the drug were nearly identical (Fig. S3; Pearson’s linear r = 0.90). Finally, SHAPE reactivities for the 16S rRNA probed in cells treated with both rifampicin and chloramphenicol did not differ significantly from untreated cells. These data indicate that the SHAPE reactivity profile we observe for 16S rRNA in the 30S peak in exponentially growing cells reflects mature 30S subunits.
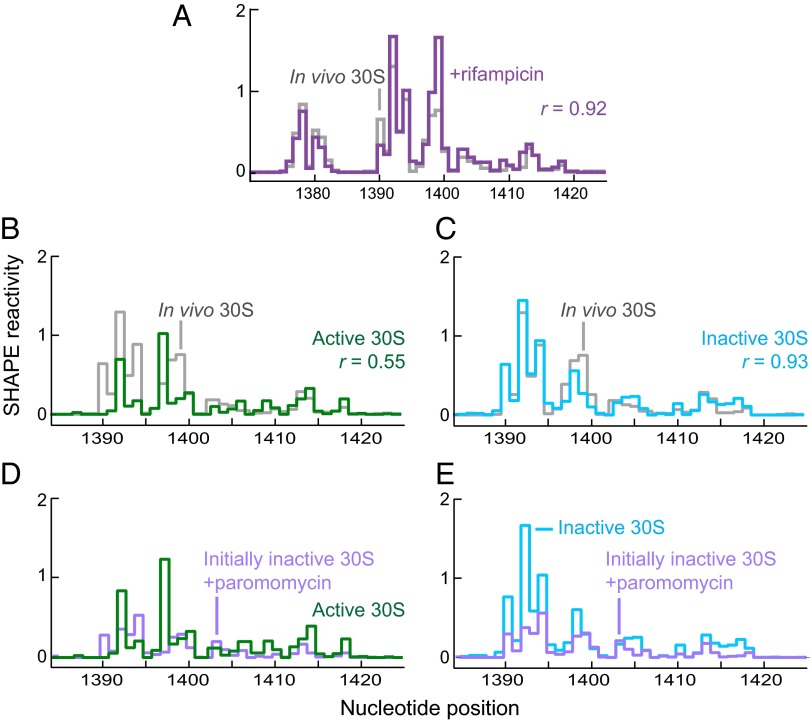
Fig. 2.
16S rRNA structure in 30S subunits in vivo resembles the in vitro inactive state, and paromomycin induces the switch from inactive to active conformation. (A) Histograms comparing in-cell SHAPE reactivities for 16S rRNA from free 30S subunits isolated from cells during log-phase growth (gray) with 16S rRNA reactivities from free 30S subunits isolated from cells treated with rifampicin (purple). (B and C) Histograms comparing in-cell SHAPE reactivities for 16S rRNA isolated from 30S subunits (gray) with reactivities for 16S rRNA from isolated 30S subunits treated in vitro under conditions that yield (B) active (green) and (C) inactive (cyan) 30S subunits. (D and E) Histograms comparing SHAPE reactivities for 16S rRNA from inactive 30S subunits treated in vitro with paromomycin (purple) with those from subunits obtained in vitro under conditions that yield (D) active (green) and (E) inactive (cyan) 30S subunits.
In Vivo 16S rRNA Structure in Free 30S Subunits Corresponds to the Inactive State.
Purification of ribosomal subunits for biochemical and structural studies generally includes a step in which ribosomes are dialyzed against buffer containing a low Mg2+ concentration (1–2 mM). This condition dissociates the subunits and induces a major structural change in the 30S subunit (termed “inactivation”); this conformational change interferes with tRNA binding to the P site. Incubation of these inactive subunits at 42 °C in the presence of high Mg2+ concentrations (10–20 mM) promotes reversal of the structural change (termed “activation”) and recreates the state that efficiently binds tRNA (13). We probed purified 30S subunits under these two conditions in vitro. The SHAPE profile for the in-cell state of 16S rRNA in free 30S subunits was very different from the active state but was highly similar to the inactive state (Fig. 2 B and C).
The antibiotic paromomycin binds to helix h44 at the internal loop formed by A1408, A1492, and A1493 (Fig. 3A, boxed nucleotides) (14) and stimulates ribosome subunit association at low-magnesium ion concentrations (15). We hypothesized that binding by paromomycin might shift the equilibrium of the 16S rRNA from the inactive to the active state. We probed the structure of the 16S rRNA in inactivated 30S subunits in vitro before and after incubation with paromomycin and compared the SHAPE reactivity profiles to that of activated subunits. Addition of paromomycin caused the inactive state to adopt a conformation similar to the active state but had essentially no effect when added to 30S subunits in the active state (Fig. 2 D and E and Fig. S2). The largest differences in the inactive versus active state occurred at nucleotides 1391–1398, a region outside the site where paromomycin binds. Thus, the small free energy increment provided by paromomycin binding, which stabilizes h44, is sufficient to shift the equilibrium from an inactive to an active-like state.
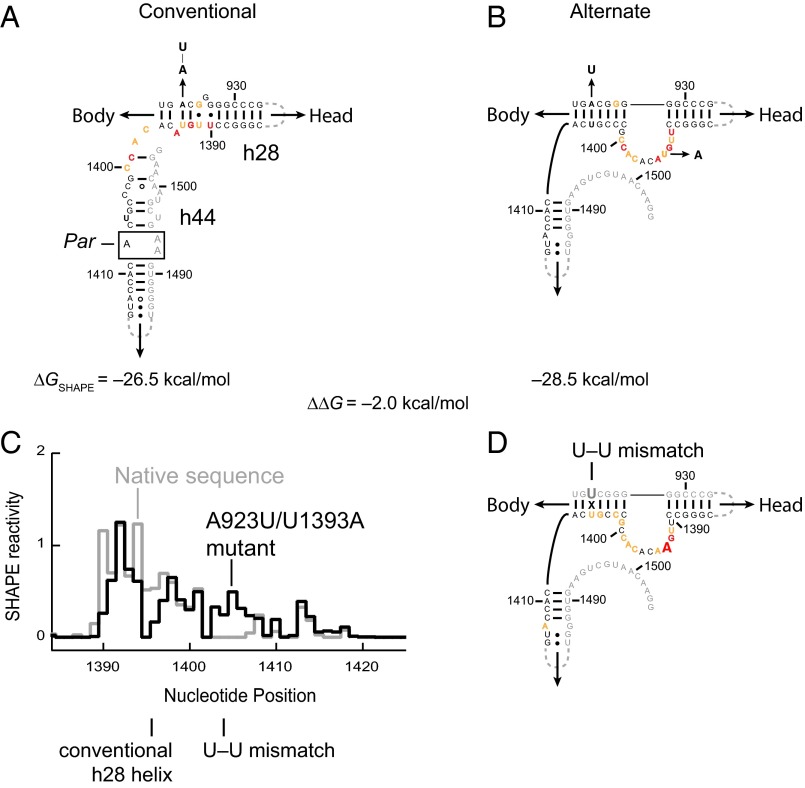
Fig. 3.
Alternate secondary structure for 16S rRNA in free 30S subunits in vivo. (A) Conventional base-pairing model and (B) SHAPE-supported alternate model (3) of 16S rRNA. SHAPE reactivity profile for 16S rRNA from free 30S subunits isolated after in vivo SHAPE probing are superimposed on each structure model. (C) Histograms comparing 16S rRNA SHAPE reactivities for the native sequence (gray) and A923U/U1393A mutant (black) from free 30S subunits isolated after in vivo modification. Mutant-specific structural landmarks are shown. (D) SHAPE reactivities for the mutant 16S rRNA superimposed on the alternate base-pairing model. The two mutated nucleotides are shown in a larger font. The experiments shown in A and B versus C and D were performed on wild-type and ∆7 prrn E. coli cells, respectively; a full set of comparisons showing SHAPE reactivities superimposed on both conventional and alternate secondary structure models are shown in Figs. S2 and S5. Nucleotides are colored by SHAPE reactivity using the red, yellow, and black scale shown in Fig. 1.
The Inactive State Contains an Alternative Helix.
The h28–h44 region is conspicuously lacking sequence covariation (16). Nucleotides in these helices are highly conserved, and sequence variations that occur do not specifically support (or contradict) formation of these helices. Sequence alignment of the 16S rRNAs from E. coli, Clostridium difficile, and Haloferax volcanii, facilitated by structural information based on SHAPE data (3), supports formation of a specific structure at the h28–h44 junction that differs from that of the conventional structure (Fig. 3B). This conformation involves a register shift. Nucleotides 1402–1408, which form an irregular helix at the beginning of h44 in the conventional structure, pair with positions 921–927 (within h28 in the conventional structure), and nucleotides 1390–1401 form a loop. In-cell SHAPE reactivities for 16S rRNA in free 30S subunits correspond closely to the alternate base-pairing model: Nucleotides proposed to pair in the alternate h28 were unreactive, and nucleotides 1390–1401 were reactive (Fig. 3 A and B). SHAPE reactivities for the active and inactive states prepared in vitro also agreed with the conventional and the proposed alternate base-pairing models, respectively (Fig. S4).
The alternate h28 conformation predominates for the 16S rRNA when ribosomal proteins are removed (3, 17), suggesting that it is more thermodynamically stable than the conventional conformation. We estimated the relative stabilities of the two rRNA conformations using both nearest neighbor interactions and the pseudo-free energy change term based on SHAPE reactivities (17, 18). The difference in stabilities between conformations in the ribosome neck region was estimated to be 2.0 kcal/mol (Fig. 3 A and B). Thus, the alternate conformation is more stable than the conventional one, but only by a small increment, consistent with a low-energy barrier and with the observed ability of paromomycin binding to induce the transition from the inactive to an active state.
The base-pair substitution A923U/U1393A does not alter the estimated stability of the conventional h28 but should disrupt the alternate helix (replacing an A–U pair with a U–U mismatch; Fig. 3 A and B). This mutation was introduced into h28 and tested using a specialized ribosome system that allows the effect of 16S rRNA mutations to be quantified without affecting cell growth (19). Translation efficiency was reduced fivefold, consistent with a role for the conformational switch in translation initiation (full results of these experiments are summarized in Table S1). We also analyzed the in-cell structures of the wild-type and mutant 16S rRNA from free 30S subunits isolated from cells containing a single rRNA operon (∆7 prrn). Superposition of the SHAPE reactivities on secondary structure models for each state shows that nucleotides in the alternate helix (positions 1403–1407) became reactive precisely at the site of the introduced U–U mismatch (Fig. 3 C and D and Fig. S5). Critically, by making a mutation in the 921–927 strand of the alternate h28, we observe a specific increase in SHAPE reactivity precisely in the 1402–1408 strand, providing very strong support for the existence of this interaction in vivo. Conversely, nucleotides 1395–1396 became less reactive in the mutant, consistent with stabilization of the conventional h28 through destabilization of the alternate conformation (Fig. 3C and Fig. S5). Collectively, these data support formation of the proposed alternate h28 helix conformation in free 30S subunits in cells.
30S Subunits Purified from Cells Bind mRNA Efficiently but Initiate Translation Slowly.
We examined mRNA binding by conventional active and inactive 30S subunits and by free 30S subunits purified directly from cells using a filter binding assay; we used poly(U) and m292, a model mRNA with a complex sequence, in the assay. All three classes of 30S subunits bound poly(U) similarly and rapidly, at ≥5 min–1 (Fig. 4A, filled symbols), consistent with early studies (13). Binding to the m292 mRNA was notably slower (∼0.06 min–1) for all three classes of 30S subunits, and the extent of binding was twofold higher for purified intracellular 30S subunits than for the conventionally purified active or inactive 30S subunits (Fig. 4B, open symbols). The free intracellular 30S (in the alternate conformation) thus binds mRNA efficiently and distinguishes between poly(U) and a mixed-sequence mRNA in roughly the same way as do conventionally purified active and inactive state subunits.
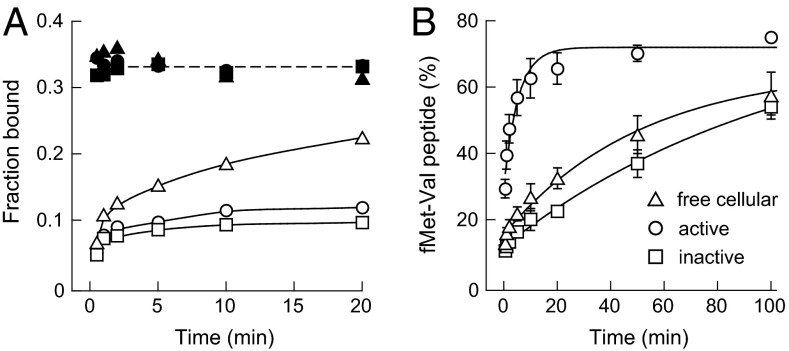
Fig. 4.
mRNA binding and translation initiation activities of 30S subunits. (A) mRNA binding, assayed by nitrocellulose filter binding. poly(U) and m292 mRNAs are shown in filled and open symbols, respectively. (B) Translation initiation measured by dipeptide formation. Data were fit to a single-exponential function; for active, inactive, and free cellular 30S subunits, kapp was 0.38 ± 0.10, 0.0093 ± 0.0012, and 0.020 ± 0.005 min–1, respectively. Error bars show the SEM from three or more independent experiments.
We next examined the ability of conventional inactive and active subunits and free 30S subunits purified from cells to initiate translation in vitro. 30S subunits were incubated with the components required to form the 70S initiation complex and synthesize an fMet–Val dipeptide, and the amount of dipeptide formed was quantified as a function of time. Activated 30S subunits synthesized peptide at a rate of 0.38 min–1 (Fig. 4B, circles). Conventionally purified inactive 30S subunits gave an overall initiation rate of 0.0093 min–1 (Fig. 4B, squares). When free 30S subunits were purified directly from cells and exchanged into reaction buffer, translation initiation rates were 0.02 min–1, twofold faster than the rate of conventionally purified inactive 30S subunits (Fig. 4B, triangles). We then tested whether activation or inactivation procedures converted the free 30S subunits, isolated directly from cells, to the active or inactive state, respectively. Neither treatment appreciably changed the translation initiation activity of free 30S subunits (Table S2). This lack of activation suggests that some physical barrier slows free cellular 30S particles during translation initiation.
Discussion
Our data indicate that 16S rRNA in free 30S ribosomal subunits in rapidly growing E. coli adopts an alternate structure that closely resembles the inactive conformation of this subunit identified decades ago. The intrinsic barrier between this inactive state and the active one is low, consistent with the ability of paromomycin binding to shift the equilibrium toward the active state and with formation of nearly isoenergetic base pairs in the conventional and alternate structures (Figs. 2 D and E and 3). Addition of rifampicin before in-cell probing did not alter the SHAPE reactivity pattern, indicating that mature 30S subunits in cells spend significant time in the inactive conformation. The ability of the small ribosome subunit to form the alternate rRNA structure appears to be conserved in the 16S rRNAs from bacteria and archaea (3) and in the 18S rRNA in eucarya (4). Our in vivo probing experiments were performed over short periods of time in rapidly growing E. coli cells; hence, these free 30S particles represent an abundant, natural population of subunits in the cell. What functional role, then, does the inactive or alternate conformation play?
Conformational variability in h28 and h44 has been implicated in multiple stages in 30S subunit translational functions including maturation, initiation, and turnover. Structural changes in the h44 region accompany 30S subunit maturation (20), and the alternate conformation is the functional substrate for the Ksg methyltransferase (21) and YjeQ (22) biogenesis factors. Multiple factors that regulate both quality control (23) and translation initiation (24) bind at sites that overlap with the h28 and h44 helices, suggesting that structural changes involving the conventional and alternate h28–h44 helix conformations could be modulated by or govern accessibility to ribosome-binding assembly and translation factors. The alternate conformation also binds mRNA (Fig. 4A), suggesting potential roles in translation initiation. Finally, the current model for ribosome turnover emphasizes passive control in which free subunits accumulate as cell growth slows, and the alternate conformation could affect 30S subunit turnover, as 16S rRNA degradation involves initial endonucleolytic cleavage between h27 and h28 (25).
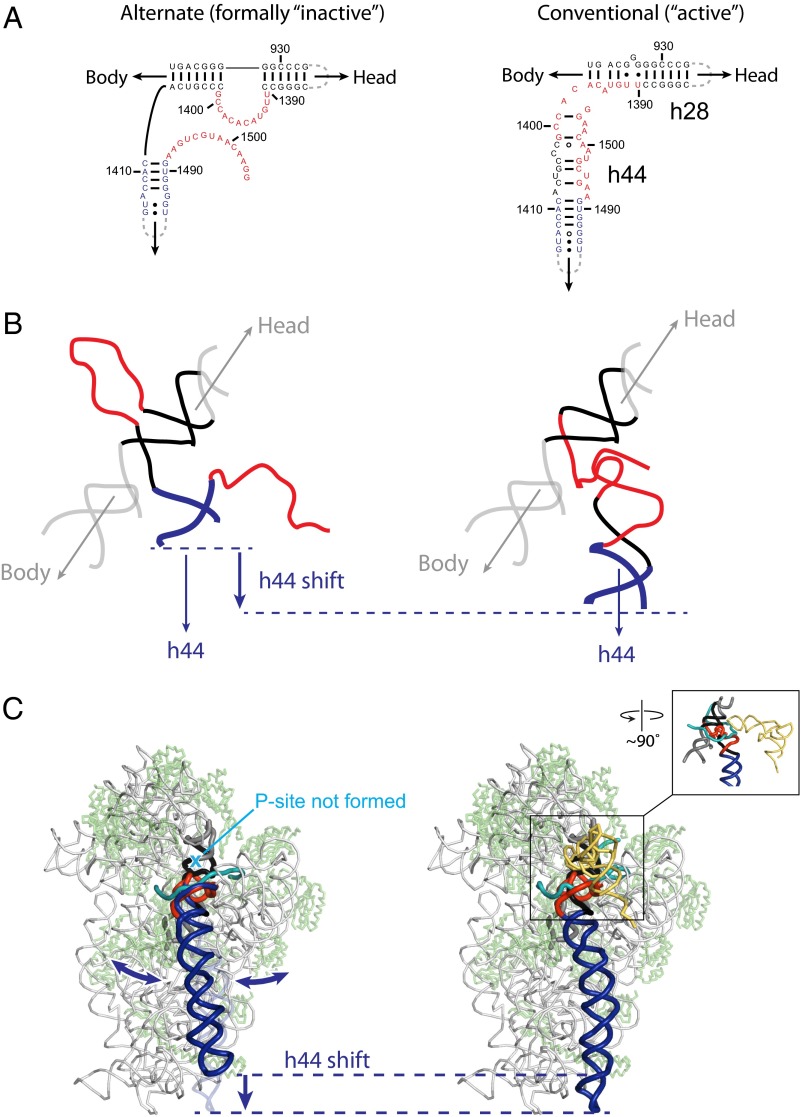
The conformational switch between conventional and alternate conformations provides a structural framework for interpreting this extensive body of information, emphasizing important roles for the h28–h44 region in many elements of ribosome function. Formation of the alternate h28 has the effect of shortening h44 or changing the linkage between this helix and the rest of the 30S subunit (Fig. 5 A and B). This conformational change likely dislodges h44 from the body of the ribosome (Fig. 5C), consistent with the absence of density for h44 in cryo-EM structures of the inactive 30S subunit (21). This simple conformational switch would yield a subunit that does not bind tRNA in the P site and that would be unable to form the extensive interface with the 50S subunit. The functions of the inactive subunit could then be regulated by diverse interactions with h28 and h44 or adjacent to their interaction sites within the 30S subunit.
Fig. 5.
Structural and mechanistic consequences for formation of the alternate secondary structure in the 16S rRNA neck helices. (A) Secondary structures for alternate and conventional models of the 16S rRNA neck helices. (B) 3D models for the alternate (Left) and conventional (Right) structures. The alternate and conventional models were based on discrete molecular dynamics and crystallographic structures (26), respectively. (C) Illustration of the position for h44 in the context of the 30S subunit for the alternate and conventional conformations of the 16S rRNA. In the diagram of the alternate state, double-headed arrows emphasize likely conformational dynamics of h44. Inset illustrates area highlighted in B plus tRNA (yellow) and mRNA (cyan).
The shift between inactive and active states of the 30S subunit (1) was one of the first conformational rearrangements discovered in a cellular ribonucleoprotein complex. It has now become routine to “activate” purified 30S subunits before their use in most biochemical and crystallographic experiments. Thus, researchers make the assumption that ribosome function begins with the 30S subunit in a conformation competent to bind both tRNA and mRNA and initiate translation. Our findings reveal that the alternate conformation predominates for free 30S subunits in bacterial cells and suggest that the alternate conformation serves as a switch to activate multiple ribosome functions.
Methods
E. coli cells (DH5α or Δ7 prrn) were grown to midlogarithmic phase (OD600 ∼0.6) at 37 °C, subjected to SHAPE probing with 1M7 [dissolved in anhydrous DMSO; final concentrations of the reagent and organic cosolvent were 5 mM and 3% (vol/vol), respectively], and allowed to react for 2–5 min at 37 °C. No-reagent controls were performed in parallel. Ribosome subunits were subsequently purified by sucrose sedimentation gradient fractionation (7), and sites of chemical modification in the 16S rRNA were resolved by capillary electrophoresis (5). For SHAPE probing performed with purified subunits, inactivation and reactivation of 30S subunits was performed as described (2). Detailed descriptions of in-cell ribosome probing, probing following antibiotic treatment, mutant ribosome construction and in vivo analysis, thermodynamic calculations (Fig. S6), and structure modeling are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank C. Squires and S. Quan for E. coli strain SQZ10. This work was supported by National Science Foundation Grants MCB-1121024 (to K.M.W.) and MCB-1243997 (to K.F.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1411514112/-/DCSupplemental.
References
- 1.Zamir A, Miskin R, Elson D. Interconversions between inactive and active forms of ribosomal subunits. FEBS Lett. 1969;3(1):85–88. doi: 10.1016/0014-5793(69)80103-1. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Van Stolk BJ, Douthwaite S, Noller HF. Interconversion of active and inactive 30 S ribosomal subunits is accompanied by a conformational change in the decoding region of 16 S rRNA. J Mol Biol. 1986;191(3):483–493. doi: 10.1016/0022-2836(86)90143-9. [DOI] [PubMed] [Google Scholar]
- 3.Lavender CA, et al. Model-free RNA sequence and structure alignment informed by SHAPE probing reveals a conserved alternate secondary structure for 16S rRNA. PLoS Comp Biol. 2015 doi: 10.1371/journal.pcbi.1004126. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swiatkowska A, et al. Kinetic analysis of pre-ribosome structure in vivo. RNA. 2012;18(12):2187–2200. doi: 10.1261/rna.034751.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McGinnis JL, Weeks KM. Ribosome RNA assembly intermediates visualized in living cells. Biochemistry. 2014;53(19):3237–3247. doi: 10.1021/bi500198b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tyrrell J, McGinnis JL, Weeks KM, Pielak GJ. The cellular environment stabilizes adenine riboswitch RNA structure. Biochemistry. 2013;52(48):8777–8785. doi: 10.1021/bi401207q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bommer U, et al. Ribosomes and polysomes. In: Graham J, Rickwoods D, editors. Subcellular Fractionation: A Practical Approach. IRL Press at Oxford Univ Press; Oxford: 1996. pp. 271–301. [Google Scholar]
- 8.Weeks KM, Mauger DM. Exploring RNA structural codes with SHAPE chemistry. Acc Chem Res. 2011;44(12):1280–1291. doi: 10.1021/ar200051h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cannone JJ, et al. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dallas A, Noller HF. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol Cell. 2001;8(4):855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 11.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313(5795):1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 12.Flessel CP, Ralph P, Rich A. Polyribosomes of growing bacteria. Science. 1967;158(3801):658–660. doi: 10.1126/science.158.3801.658. [DOI] [PubMed] [Google Scholar]
- 13.Zamir A, Miskin R, Vogel Z, Elson D. The inactivation and reactivation of Escherichia coli ribosomes. Methods Enzymol. 1974;30:406–426. doi: 10.1016/0076-6879(74)30042-0. [DOI] [PubMed] [Google Scholar]
- 14.Carter AP, et al. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature. 2000;407(6802):340–348. doi: 10.1038/35030019. [DOI] [PubMed] [Google Scholar]
- 15.Hirokawa G, Kaji H, Kaji A. Inhibition of antiassociation activity of translation initiation factor 3 by paromomycin. Antimicrob Agents Chemother. 2007;51(1):175–180. doi: 10.1128/AAC.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shang L, Xu W, Ozer S, Gutell RR. Structural constraints identified with covariation analysis in ribosomal RNA. PLoS ONE. 2012;7(6):e39383. doi: 10.1371/journal.pone.0039383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106(1):97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hajdin CE, et al. Accurate SHAPE-directed RNA secondary structure modeling, including pseudoknots. Proc Natl Acad Sci USA. 2013;110(14):5498–5503. doi: 10.1073/pnas.1219988110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abdi NM, Fredrick K. Contribution of 16S rRNA nucleotides forming the 30S subunit A and P sites to translation in Escherichia coli. RNA. 2005;11(11):1624–1632. doi: 10.1261/rna.2118105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jomaa A, et al. Understanding ribosome assembly: The structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA. 2011;17(4):697–709. doi: 10.1261/rna.2509811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boehringer D, O’Farrell HC, Rife JP, Ban N. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J Biol Chem. 2012;287(13):10453–10459. doi: 10.1074/jbc.M111.318121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jomaa A, et al. Cryo-electron microscopy structure of the 30S subunit in complex with the YjeQ biogenesis factor. RNA. 2011;17(11):2026–2038. doi: 10.1261/rna.2922311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karbstein K. Quality control mechanisms during ribosome maturation. Trends Cell Biol. 2013;23(5):242–250. doi: 10.1016/j.tcb.2013.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Myasnikov AG, Simonetti A, Marzi S, Klaholz BP. Structure-function insights into prokaryotic and eukaryotic translation initiation. Curr Opin Struct Biol. 2009;19(3):300–309. doi: 10.1016/j.sbi.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 25.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: Comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17(2):338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332(6032):981–984. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.