Significance
Land plants continuously contact beneficial, commensal, and pathogenic microbes in soil via their roots. There is limited knowledge as to how the totality of root-associated microbes (i.e., the microbiome) is shaped by various factors or its pattern of acquisition in the root. Using rice as a model, we show that there exist three different root niches hosting different microbial communities of eubacteria and methanogenic archaea. These microbial communities are affected by geographical location, soil source, host genotype, and cultivation practice. Dynamics of the colonization pattern for the root-associated microbiome across the three root niches provide evidence for rapid acquisition of root-associated microbiomes from soil, and support a multistep model wherein each root niche plays a selective role in microbiome assembly.
Keywords: microbiomes, rice, soil microbial communities, methane cycling, microbiome assembly
Abstract
Plants depend upon beneficial interactions between roots and microbes for nutrient availability, growth promotion, and disease suppression. High-throughput sequencing approaches have provided recent insights into root microbiomes, but our current understanding is still limited relative to animal microbiomes. Here we present a detailed characterization of the root-associated microbiomes of the crop plant rice by deep sequencing, using plants grown under controlled conditions as well as field cultivation at multiple sites. The spatial resolution of the study distinguished three root-associated compartments, the endosphere (root interior), rhizoplane (root surface), and rhizosphere (soil close to the root surface), each of which was found to harbor a distinct microbiome. Under controlled greenhouse conditions, microbiome composition varied with soil source and genotype. In field conditions, geographical location and cultivation practice, namely organic vs. conventional, were factors contributing to microbiome variation. Rice cultivation is a major source of global methane emissions, and methanogenic archaea could be detected in all spatial compartments of field-grown rice. The depth and scale of this study were used to build coabundance networks that revealed potential microbial consortia, some of which were involved in methane cycling. Dynamic changes observed during microbiome acquisition, as well as steady-state compositions of spatial compartments, support a multistep model for root microbiome assembly from soil wherein the rhizoplane plays a selective gating role. Similarities in the distribution of phyla in the root microbiomes of rice and other plants suggest that conclusions derived from this study might be generally applicable to land plants.
Land plants grow in soil, placing them in direct proximity to a high abundance of microbial diversity (1). Plants and microbes have both adapted to use their close association for their mutual benefit. Critical nutrients are converted to more usable forms by microbes before assimilation by plants (2–4). In turn, bacteria in the rhizosphere receive carbon metabolites from the plant through root exudates (5). Beneficial soil microbes also contribute to pathogen resistance, water retention, and synthesis of growth-promoting hormones (6–8).
Recent studies have used high-throughput sequencing to provide new insights into the bacterial composition and organization of different plant microbiomes, including Arabidopsis, Populus, and maize (9–14). Detailed characterization of the core root microbiome of Arabidopsis (9–11) showed that the dominant phyla inside the root (the endosphere) are much less diverse than the phyla in the soil around the root (the rhizosphere), and a potential core root microbiome could be identified. In Arabidopsis, the endophytic microbiome exhibits some genotype-dependent variation within the species and an increased variation when other related species are examined (9–11). A recent study in maize examined microbiome variation across many different inbred lines at different sites and found a large variation arising from geographical location between three different states in the United States and a relatively smaller dependence on the genotype (12). Although the microbiomes examined in the maize study consisted of combined rhizospheric and endospheric microbes (12), a study in poplar found that the variation between locations in two different states affected both rhizospheric and endospheric microbes (14).
These studies have opened the way toward a new understanding of the composition and structure of plant microbiomes and the factors that affect them. However, this understanding is still at the initial stages, and several key questions are as yet unanswered. One such question regards the mechanism of microbiome acquisition and assembly in plants. Unlike animals, where the gut microbiome is assembled internally and is transmissible through birth (15, 16), the root microbiome is predominantly assembled from the external microbes in the soil. Based on the composition of the endospheric and rhizopheric microbiomes, it has been proposed that plants might assemble their microbiomes in two steps, with the first step involving a general recruitment to the vicinity of the root and a second step for entry inside the root that involves species-specific genetic factors (7). Although this is a plausible hypothesis, direct support for this model through detailed dynamic studies has not yet been provided. Additionally, the role of the root surface or rhizoplane, which forms the critical interface between plants and soil, remains poorly understood, and the microbial composition of the rhizoplane in relation to those of the rhizosphere and endosphere is unknown.
To address some of these questions, we have undertaken an exhaustive characterization of the root-associated microbiome of rice. Rice is a major crop plant and a staple food for half of the world’s population. Metagenomic and proteomic approaches have been used to identify different microbial genes present in the rice microbiome (17, 18), but an extensive characterization of microbiome composition and variation has not been performed. Rice cultivation also contributes to global methane, accounting for an estimated 10–20% of anthropogenic emissions, due to the growth of methanogenic archaea in the vicinity of rice roots (19). Here we have used deep sequencing of microbial 16S rRNA genes to detect over 250,000 operational taxonomic units (OTUs), with a structural resolution of three distinct compartments (rhizosphere, rhizoplane, and endosphere) and extending over multiple factors contributing to variation, both under controlled greenhouse conditions as well as different field environments. The large datasets from the different conditions sampled in this study were used for identification of putative microbial consortia involved in processes such as methane cycling. Through dynamic studies of the microbiome composition, we provide insights into the process of root microbiome assembly.
Results
Root-Associated Microbiomes Form Three Spatially Separable Compartments Exhibiting Distinct and Overlapping Microbial Communities.
Sterilized rice seeds were germinated and grown under controlled greenhouse conditions in soil collected from three rice fields across the Central Valley of California (SI Appendix, Fig. S1). We analyzed the bacterial and archaeal microbiomes from three separate rhizocompartments: the rhizosphere, rhizoplane, and endosphere (Fig. 1A). Because the root microbiome has been shown to correlate with the developmental stage of the plant (10), the root-associated microbial communities were sampled at 42 d (6 wk), when rice plants from all genotypes were well-established in the soil but still in their vegetative phase of growth. For our study, the rhizosphere compartment was composed of ∼1 mm of soil tightly adhering to the root surface that is not easily shaken from the root (SI Appendix, Fig. S2). The rhizoplane compartment microbiome was derived from the suite of microbes on the root surface that cannot be removed by washing in buffer but is removed by sonication (SI Appendix, Materials and Methods). The endosphere compartment microbiome, composed of the microbes inhabiting the interior of the root, was isolated from the same roots left after sonication. Unplanted soil pots were used as a control to differentiate plant effects from general edaphic factors.
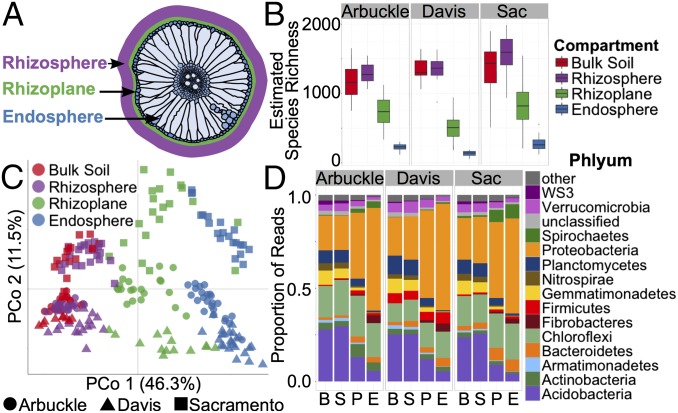
Fig. 1.
Root-associated microbial communities are separable by rhizocompartment and soil type. (A) A representation of a rice root cross-section depicting the locations of the microbial communities sampled. (B) Within-sample diversity (α-diversity) measurements between rhizospheric compartments indicate a decreasing gradient in microbial diversity from the rhizosphere to the endosphere independent of soil type. Estimated species richness was calculated as eShannon_entropy. The horizontal bars within boxes represent median. The tops and bottoms of boxes represent 75th and 25th quartiles, respectively. The upper and lower whiskers extend 1.5× the interquartile range from the upper edge and lower edge of the box, respectively. All outliers are plotted as individual points. (C) PCoA using the WUF metric indicates that the largest separation between microbial communities is spatial proximity to the root (PCo 1) and the second largest source of variation is soil type (PCo 2). (D) Histograms of phyla abundances in each compartment and soil. B, bulk soil; E, endosphere; P, rhizoplane; S, rhizosphere; Sac, Sacramento.
The V4-V5 region of the 16S rRNA gene was amplified using PCR and sequenced using the Illumina MiSeq platform. A total of 10,554,651 high-quality sequences was obtained with a median read count per sample of 51,970 (range: 2,958–203,371; Dataset S2). The high-quality reads were clustered using >97% sequence identity into 101,112 microbial OTUs. Low-abundance OTUs (<5 total counts) were discarded, resulting in 27,147 OTUs. The resulting OTU counts in each library were normalized using the trimmed mean of M values method. This method was chosen due to its sensitivity for detecting differentially abundant taxa compared with traditional microbiome normalization techniques such as rarefaction and relative abundance (20). Measures of within-sample diversity (α-diversity) revealed a diversity gradient from the endosphere to the rhizosphere (Fig. 1B and Dataset S4). Endosphere communities had the lowest α-diversity and the rhizosphere had the highest α-diversity. The mean α-diversity was higher in the rhizosphere than bulk soil; however, the difference in α-diversity between these two compartments cannot be considered as statistically significant (Wilcoxon test; Dataset S4).
Unconstrained principal coordinate analyses (PCoAs) of weighted and unweighted UniFrac distances were performed to investigate patterns of separation between microbial communities (SI Appendix, Materials and Methods). The UniFrac distance is based on taxonomic relatedness, where the weighted UniFrac (WUF) metric takes abundance of taxa into consideration whereas the unweighted UniFrac (UUF) does not and is thus more sensitive to rare taxa. In both the WUF and UUF PCoAs, the rhizocompartments separate across the first principal coordinate, indicating that the largest source of variation in root-associated microbial communities is proximity to the root (Fig. 1C, WUF and SI Appendix, Fig. S4, UUF). Moreover, the pattern of separation is consistent with a gradient of microbial populations from the exterior of the root, across the rhizoplane, and into the interior of the root. Permutational multivariate analysis of variance (PERMANOVA) corroborates that rhizospheric compartmentalization comprises the largest source of variation within the microbiome data when using a WUF distance metric (46.62%, P < 0.001; Dataset S5A). PERMANOVA using a UUF distance, however, describes rhizospheric compartmentalization as having the second largest source of variation behind soil type (18.07%, P < 0.001; Dataset S5H). In addition to PERMANOVA, we also performed partial canonical analysis of principal coordinates (CAP) on both the WUF and UUF metrics to quantify the variance attributable to each experimental variable (SI Appendix, Materials and Methods). This technique differs from unconstrained PCoA in that technical factors can be controlled for in the analysis and the analysis can be constrained to any factor of interest to better understand the quantitative impact of the factor on the microbial composition. Using this technique to control for soil type, cultivar, and technical factors (biological replicate, sequencing batch, and planting container), we found that in agreement with the PERMANOVA results, microbial communities vary significantly between rhizocompartments (34.2% of variance, P = 0.005, WUF, SI Appendix, Fig. S5A and 22.6% of variance, P = 0.005, UUF, SI Appendix, Fig. S5C).
There are notable differences in the proportions of various phyla across the compartments that are consistent across every tested soil (Fig. 1D). The endosphere has a significantly greater proportion of Proteobacteria and Spirochaetes than the rhizosphere or bulk soil, whereas Acidobacteria, Planctomycetes, and Gemmatimonadetes are mostly depleted in the endosphere compared with the bulk soil or rhizosphere (Wilcoxon test; Dataset S6). The reduction in relative abundance of these phyla across the compartments is consistent with the observation that microbial diversity decreases from the rhizosphere to the endosphere.
Association of Significantly Enriched OTUs with Different Rhizocompartments.
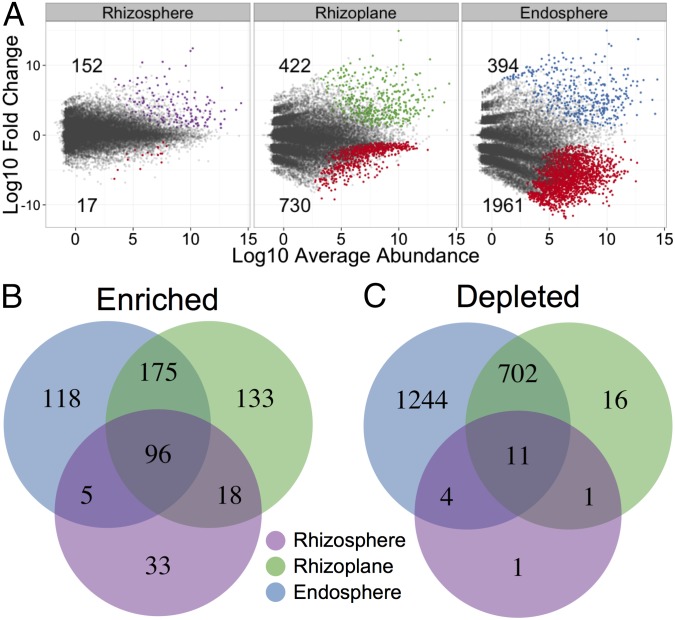
To identify OTUs that are correlated with community separation between the compartments, we conducted differential OTU abundance analysis by fitting a generalized linear model with a negative binomial distribution to normalized values for each of the 27,147 OTUs in the greenhouse experiment and testing for differential abundance using a likelihood ratio test (Dataset S7). Using OTU counts from unplanted soil as a control and an adjusted P value cutoff of 0.01, there were 578 OTUs that were significantly enriched in at least one compartment. The rhizosphere was the most similar to bulk soil, as indicated by the “tail” in the MA plot (Fig. 2A); however, an enrichment effect in the rhizosphere is implied by the high ratio of statistically significant enriched OTUs compared with depleted OTUs (152 vs. 17). In comparison, the rhizoplane enriches for many OTUs while simultaneously depleting a larger proportion of OTUs (422 vs. 730). The endosphere is the most exclusive compartment, enriching for 394 OTUs while depleting 1,961 OTUs (Fig. 2A).
Fig. 2.
Rhizocompartments are enriched and depleted for certain OTUs. (A) Enrichment and depletion of the 27,147 OTUs included in the greenhouse experiment for each rhizospheric compartment compared with bulk soil controls as determined by differential abundance analysis. Each point represents an individual OTU, and the position along the y axis represents the abundance fold change compared with bulk soil. (B) Numbers of differentially enriched OTUs between each compartment compared with bulk soil. (C) Numbers of differentially depleted OTUs between each compartment.
There were noteworthy overlaps in differentially abundant OTUs between the compartments (Fig. 2 B and C). The OTUs enriched in the rhizosphere are very successful at colonizing the root, as 119 out of the 152 OTUs enriched in the rhizosphere are also enriched in either the rhizoplane or endosphere communities or both (Fig. 2B). There was a set of 96 OTUs mainly consisting of Bacteroidetes, Firmicutes, Chloroflexi, and Betaproteobacteria that were differentially enriched in all rhizocompartments compared with bulk soil (SI Appendix, Fig. S6A). The Betaproteobacterial OTUs that are enriched in every rhizocompartment correspond mainly to Rhodocyclaceae and Comamonadaceae (SI Appendix, Fig. S6B). OTUs belonging to the genus Pleomorphomonas are also enriched in all of the rhizocompartments (Dataset S7). Some members within the Pleomorphomonas genus are capable of nitrogen fixation (21–23). The rhizoplane and endosphere were the most similar rhizocompartments, sharing 271 enriched OTUs. Most of the OTUs enriched between the rhizoplane and endosphere compartments belonged to Alpha-, Beta-, and Deltaproteobacterial classes, Chloroflexi, and Bacteroidetes. Not surprisingly, a subset of the OTUs enriched in the endosphere and rhizoplane belong to Fibrobacteres and Spirochaetes, phyla that are associated with cellulose degradation (24, 25).
It was possible to quantify exclusionary effects of each compartment by analyzing OTU abundance relative to bulk soil (Fig. 2A). The rhizosphere had a small effect on excluding microbes, as only 17 OTUs were significantly depleted compared with bulk soil. These OTUs were mainly in the Proteobacteria and Acidobacteria phyla (SI Appendix, Fig. S7). Many more OTUs were reduced in relative abundance in the rhizoplane (730 OTUs, mainly Acidobacteria and Planctomycetes; SI Appendix, Fig. S7), and even more OTUs (1,961 OTUs, mainly Acidobacteria, Planctomycetes, Chloroflexi, and Verrucomicrobia; SI Appendix, Fig. S7) were reduced in the endosphere. There are considerable overlaps in the OTUs that are excluded from each compartment (Fig. 2C). Nearly all of the OTUs depleted from the rhizosphere are also depleted in the rhizoplane and endosphere communities. The rhizoplane shares 713 of the 1,961 OTUs that are significantly depleted from the endosphere. These results indicate that plant-controlled changes in the rhizospheric soil are the first level of exclusion of microbial colonization of the root and that selectivity at the rhizoplane might act effectively as a gate for controlling entry of the microbes into the root endosphere. Similar patterns were present when each soil type was analyzed separately (SI Appendix, Fig. S8 B–G).
Microbial Communities Colonizing Rhizocompartments Vary by Soil Type.
To investigate how soil variation might affect the root microbiome, plants were grown in soil collected from rice fields at three locations from across the California Sacramento Valley: Davis, Sacramento, and Arbuckle (SI Appendix, Fig. S1). Both the Arbuckle and Sacramento fields primarily grow rice and were drained 3 wk before soil collection. The Davis site had previously grown rice for several years; however, it was not cultivated the year before soil collection. By growing the plants in controlled greenhouse conditions, we aimed to control for climatic variations between the sites and identify only changes attributable to the different soils.
α-Diversity measurements comparing microbial communities between each soil type revealed a small but significant difference in diversity (1.54% variance explained, P = 2.54E-5, ANOVA; Dataset S3). The two cultivated fields (Arbuckle and Sacramento) had significantly higher diversities in the rhizoplane and endosphere microbial communities than the uncultivated Davis field (Fig. 1B and Dataset S4). PCoA shows that rhizosphere samples from plants grown in the distinct soils separate along the second axis and that the separation pattern manifests in every compartment (Fig. 1C, WUF and SI Appendix, Fig. S4, UUF). The Arbuckle and Davis rhizosphere microbiomes were most similar to each other despite being the most geographically separated. PERMANOVA using the WUF distance supports the PCoA results that the soil effect describes the second largest source of variation in the tested factors of the experiment (Dataset S5A). PERMANOVA using the UUF distance measure indicates that the soil effect has the largest source of variation in the factors tested (20.90%, P < 0.001; Dataset S5H). CAP analysis constrained to soil type and controlling for rhizocompartment, cultivar, and technical factors agreed with the PERMANOVA result in that soil type explained the second largest source of variation in the microbial communities behind compartment when using the WUF metric (20.2%, P = 0.005; SI Appendix, Fig. S5B) and the largest source of variation when using the UUF metric (26.7%, P = 0.005; SI Appendix, Fig. S5D). This discrepancy is likely due to differences between the WUF and UUF distance metric: Soil type might have more of an effect on frequency of rare taxa than abundant taxa, and thus the UUF metric has a larger effect size for soil type. Compartments of plants grown in distinct soils have commonalities in differentially abundant OTUs (Dataset S9), sharing 92 endosphere-enriched OTUs, 71 rhizoplane-enriched OTUs, and 10 rhizosphere OTUs (SI Appendix, Fig. S8 J, I, and H, respectively, and SI Appendix, Fig. S9). In agreement with the PCoA analysis, Davis and Arbuckle shared a significant overlap in OTUs enriched in the endosphere and rhizoplane (P = 2.22 × 10−16 and 7.86 × 10−7, respectively, hypergeometric test; SI Appendix, Fig. S8 I and J) but not the rhizosphere (P = 0.52, hypergeometric test; SI Appendix, Fig. S8H). The Sacramento soil did not share significant overlaps in compartment-enriched OTUs with the other sites.
The enrichment/depletion effects within each rhizosphere compartment vary by soil. Rhizosphere compartments of plants in Davis and Arbuckle soils exhibited higher enrichment/depletion ratios (72/3 and 53/17, respectively) than plants in Sacramento soil (78/116) (SI Appendix, Fig. S8A). The level of enrichment is similar between each soil in the rhizosphere; however, the depletion level is higher in Sacramento soil than in Arbuckle or Davis. Chemical analysis of the soils showed that the nutrient compositions of the soils did not show any exceptional trends (Dataset S7). The Davis and Arbuckle fields were similar in pH and nitrate, magnesium, and phosphorus content, whereas the Arbuckle and Sacramento fields were similar in potassium, calcium, and iron content. Taken together, these results indicate that each soil contains a different pool of microbes and that the plant is not restricted to specific OTUs but instead draws from available OTUs in the pool to organize its microbiome. Nevertheless, the distribution of phyla across the different compartments was similar for all three soil types (Fig. 1D), suggesting that the overall recruitment of OTUs is governed by a set of factors that result in a consistent representation of phyla independent of soil type.
Microbial Communities in the Rhizocompartments Are Influenced by Rice Genotype.
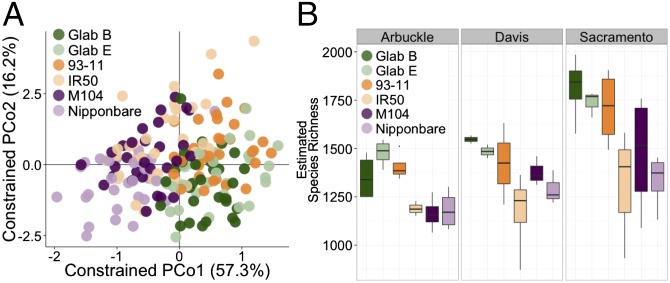
To investigate the relationship between rice genotype and the root microbiome, domesticated rice varieties cultivated in widely separated growing regions were tested. Six cultivated rice varieties spanning two species within the Oryza genus were grown for 42 d in the greenhouse before sampling. Asian rice (Oryza sativa) cultivars M104, Nipponbare (both temperate japonica varieties), IR50, and 93-11 (both indica varieties) were grown alongside two cultivars of African cultivated rice Oryza glaberrima, TOg7102 (Glab B) and TOg7267 (Glab E). PERMANOVA indicated that rice genotype accounted for a significant amount of variation between microbial communities when using WUF (2.41% of the variance, P < 0.001; Dataset S5A) and UUF (1.54% of the variance, P < 0.066; Dataset S5H); however, visual representations for clustering patterns of the genotypes were not evident on the first two axes of unconstrained PCoA ordinations (SI Appendix, Fig. S10). We then used CAP analysis to quantify the effect of rice genotype on the microbial communities. By focusing on rice cultivar and controlling for compartment, soil type, and technical factors, we found that genotypic differences in rice have a significant effect on root-associated microbial communities (5.1%, P = 0.005, WUF, Fig. 3A and 3.1%, P = 0.005, UUF, SI Appendix, Fig. S11A). Ordination of the resulting CAP analysis revealed clustering patterns of the cultivars that are only partially consistent with genetic lineage for both the WUF and UUF metrics. The two japonica cultivars clustered together and the two O. glaberrima cultivars clustered together; however, the indica cultivars were split, with 93-11 clustering with the O. glaberrima cultivars and IR50 clustering with the japonica cultivars.
Fig. 3.
Host plant genotype significantly affects microbial communities in the rhizospheric compartments. (A) Ordination of CAP analysis using the WUF metric constrained to rice genotype. (B) Within-sample diversity measurements of rhizosphere samples of each cultivar grown in each soil. Estimated species richness was calculated as eShannon_entropy. The horizontal bars within boxes represent median. The tops and bottoms of boxes represent 75th and 25th quartiles, respectively. The upper and lower whiskers extend 1.5× the interquartile range from the upper edge and lower edge of the box, respectively. All outliers are plotted as individual points.
To analyze how the genotypic effect manifests in individual rhizocompartments, we separated the whole dataset to focus on each compartment individually and conducted CAP analysis controlling for soil type and technical factors. The rhizosphere had the greatest genotypic effect on the microbiome (30.3%, P = 0.005, WUF, SI Appendix, Fig. S11B and 10.5%, P = 0.005, UUF, SI Appendix, Fig. S11E). The clustering patterns of the cultivars in the rhizosphere were similar to the clustering patterns exhibited when conducting CAP analysis on the whole data using all rhizocompartments. Again, the japonica and O. glaberrima cultivars clustered separately, whereas the indica cultivars were split between the japonica and O. glaberrima clusters. This clustering pattern is maintained in the rhizoplane communities (SI Appendix, Fig. S11 C and F); however, it breaks down in the endosphere compartment communities, which coincidently are the least affected by rice genotype (12.8%, P = 0.005, WUF, SI Appendix, Fig. S11D and 8.5%, P = 0.028, UUF, SI Appendix, Fig. S11G). α-Diversity measurements within the rhizosphere show a notable difference between the cultivars (P = 3.12E-06, ANOVA), with the O. glaberrima cultivars exhibiting high diversity relative to the japonica cultivars, especially in Arbuckle soil (Fig. 3B and Dataset S11). Again, the two japonica cultivars were more similar to the indica cultivar IR50, and the two O. glaberrima cultivars were more similar to the indica cultivar 93-11. These patterns in α-diversity were not evident when examining other compartments (SI Appendix, Fig. S12). To explain which OTUs accounted for the genotypic effects in each rhizocompartment, we performed differential OTU abundance analyses between the cultivars (Dataset S12). In total, we found 125 OTUs that were affected by the plant genotype in at least one rhizocompartment. The rhizosphere had the most OTUs that were significantly impacted by genotype (SI Appendix, Fig. S13). This is consistent with the results from PERMANOVA and the CAP analyses.
Geographical Effects on the Microbiomes of Field-Grown Plants.
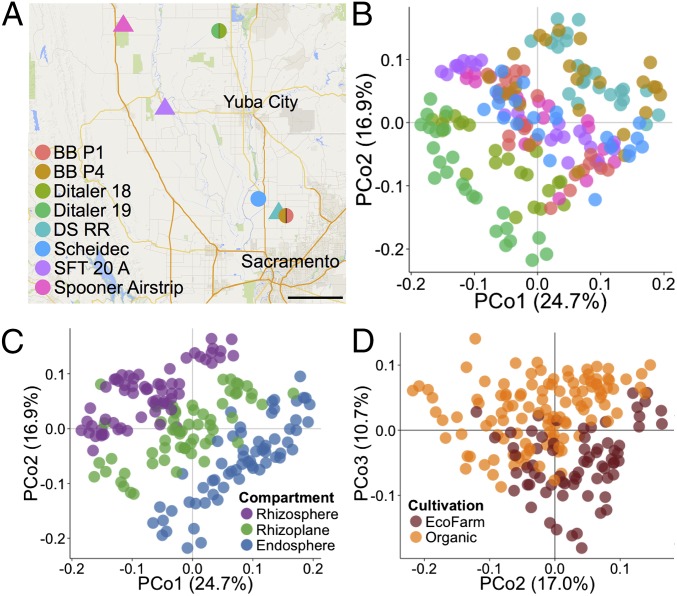
We sought to determine whether the results from greenhouse plants were generalizable to cultivated rice and to investigate other factors that might affect the microbiome under field conditions. We therefore characterized the root-associated microbiomes of field rice plants distributed across eight geographically separate sites across California’s Sacramento Valley (Fig. 4A). These eight sites were operated under two cultivation practices: organic cultivation and a more conventional cultivation practice termed “ecofarming” (see below). Because genotype explained the least variance in the greenhouse data, we limited the analysis to one cultivar, S102, a California temperate japonica variety that is widely cultivated by commercial growers and is closely related to M104 (26). Field samples were collected from vegetatively growing rice plants in flooded fields and the previously defined rhizocompartments were analyzed as before. Unfortunately, collection of bulk soil controls for the field experiment was not possible, because planting densities in California commercial rice fields are too high to find representative soil that is unlikely to be affected by nearby plants. Amplification and sequencing of the field microbiome samples yielded 13,349,538 high-quality sequences (median: 54,069 reads per sample; range: 12,535–148,233 reads per sample; Dataset S13). The sequences were clustered into OTUs using the same criteria as the greenhouse experiment, yielding 222,691 microbial OTUs and 47,983 OTUs with counts >5 across the field dataset.
Fig. 4.
Root-associated microbiomes from field-grown plants are separable by cultivation site, rhizospheric compartment, and cultivation practice. (A) Map depicting the locations of the field experiment collection sites across California’s Central Valley. Circles represent organic-cultivated sites whereas triangles represent ecofarm-cultivated sites. (Scale bar, 10 mi.) (B) PCoA using the WUF method colored to depict the various sample collection sites. (C) Same PCoA in B colored by rhizospheric compartment. (D) Same PCoA in B and C depicting second and third axes and colored by cultivation practice.
We found that the microbial diversity of field rice plants is significantly influenced by the field site. α-Diversity measurements of the field rhizospheres indicated that the cultivation site significantly impacts microbial diversity (SI Appendix, Fig. S14A, P = 2.00E-16, ANOVA and Dataset S14). Unconstrained PCoA using both the WUF and UUF metrics showed that microbial communities separated by field site across the first axis (Fig. 4B, WUF and SI Appendix, Fig. S14B, UUF). PERMANOVA agreed with the unconstrained PCoA in that field site explained the largest proportion of variance between the microbial communities for field plants (30.4% of variance, P < 0.001, WUF, Dataset S5O and 26.6% of variance, P < 0.001, UUF, Dataset S5P). CAP analysis constrained to field site and controlled for rhizocompartment, cultivation practice, and technical factors (sequencing batch and biological replicate) agreed with the PERMANOVA results in that the field site explains the largest proportion of variance between the root-associated microbial communities in field plants (27.3%, P = 0.005, WUF, SI Appendix, Fig. S15A and 28.9%, P = 0.005, UUF, SI Appendix, Fig. S15E), suggesting that geographical factors may shape root-associated microbial communities.
Rhizospheric Compartmentalization Is Retained in Field Plants.
Similar to the greenhouse plants, the rhizospheric microbiomes of field plants are distinguishable by compartment. α-Diversity of the field plants again showed that the rhizosphere had the highest microbial diversity, whereas the endosphere had the least diversity for all fields tested (SI Appendix, Fig. S14A and Dataset S15). PCoA of the microbial communities from field plants using the WUF and UUF distance metrics showed that the rhizocompartments separate across PCo 2 (Fig. 4C, WUF and SI Appendix, Fig. S14C, UUF). PERMANOVA indicated that the separation in the rhizospheric compartments explained the second largest source of variation of the factors that were tested (20.76%, P < 0.001, WUF, Dataset S5O and 7.30%, P < 0.001, UUF, Dataset S5P). CAP analysis of the field plants’ microbiomes constrained to the rhizocompartment factor and controlled for field site, cultivation practice, and technical factors agreed with PERMANOVA that a significant proportion of the variance between microbial communities is explained by rhizocompartment (20.9%, P = 0.005, WUF, SI Appendix, Fig. S15C and 10.9%, P = 0.005, UUF, SI Appendix, Fig. S15G).
Taxonomic distributions of phyla for the field plants were overall similar to the greenhouse plants: Proteobacteria, Chloroflexi, and Acidobacteria make up the majority of the rice microbiota. The taxonomic gradients from the rhizosphere to the endosphere are maintained in the field plants for Acidobacteria, Proteobacteria, Spirochaetes, Gemmatimonadetes, Armatimonadetes, and Planctomycetes. However, unlike for greenhouse plants, the distribution of Actinobacteria generally showed an increasing trend from the rhizosphere to the endosphere of field plants (SI Appendix, Fig. S14E and Dataset S16).
We again performed differential abundance tests between the OTUs in the compartments of field-grown plants (SI Appendix, Fig. S16). We found a set of 32 OTUs that were enriched in the endosphere compartment between every cultivation site, potentially representing a core field rice endospheric microbiome (SI Appendix, Fig. S17). The set of 32 OTUs consisted of Deltaproteobacteria in the genus Anaeromyxobacter and Spirochaetes, Actinobacteria, and Alphaproteobacteria in the family Methylocystaceae. Interestingly, 11 of the 32 core field endosphere OTUs were also found to be enriched in the endosphere compartment of greenhouse plants (SI Appendix, Fig. S18). Three of these OTUs were classifiable at the family level. These OTUs consisted of taxa in the families Kineosporiaceae, Rhodocyclaceae, and Myxococcaceae, all of which are also enriched in the Arabidopsis root endosphere microbiome (10).
Cultivation Practice Results in Discernible Differences in the Microbiomes.
The rice fields that we sampled from were cultivated under two practices, organic farming and a variation of conventional cultivation called ecofarming (27). Ecofarming differs from organic farming in that chemical fertilizers, fungicide use, and herbicide use are all permitted but growth of transgenic rice and use of postharvest fumigants are not permitted. Although cultivation practice itself does significantly affect α-diversity of the rhizospheric compartments overall (P = 0.008, ANOVA; Dataset S14), there is also a significant interaction between the cultivation practice used and the rhizocompartments (P = 3.52E-07, ANOVA; Dataset S14), indicating that the α-diversities of some rhizocompartments are affected differentially by cultivation practice. The α-diversity within the rhizosphere compartment varied significantly by cultivation practice, with the mean α-diversity being higher in ecofarmed rhizospheres than organic rhizospheres (P = 0.001, Wilcoxon test; Dataset S14), whereas not in the endosphere and rhizoplane microbial communities (P = 0.51 and 0.75, respectively, Wilcoxon tests; Dataset S14). Under nonconstrained PCoA, the cultivation practices are separable across principal coordinates 2 and 3 for both the WUF metric (Fig. 4D) and UUF metric (SI Appendix, Fig. S14D). PERMANOVA of the microbial communities was in agreement with the PCoAs in that cultivation practice has a significant impact on the rhizospheric microbial communities of field rice plants (8.47%, P < 0.001, WUF, Dataset S5O and 6.52%, P < 0.001, UUF, Dataset S5P). CAP analysis of the field plants constrained to cultivation practice agreed with the PERMANOVA results that there are significant differences between microbial communities from organic and ecofarmed rice plants (6.9% of the variance, P = 0.005, WUF, SI Appendix, Fig. S15D and 7.0% of the variance, UUF, P = 0.005, SI Appendix, Fig. S15H).
To better understand which taxa account for the separation between organic and ecofarmed cultivated plants, we used a generalized linear model with a negative binomial distribution to identify OTUs that had significantly different abundance between the two cultivation practices in each rhizocompartment (SI Appendix, Fig. S19 and Dataset S18). Notably, organic cultivation farms were enriched for Alphaproteobacteria, Actinobacteria, and Gemmatimonadetes whereas ecofarmed samples were enriched in Deltaproteobacteria, Chloroflexi, and Spirochaetes (SI Appendix, Fig. S20). We examined OTUs from genera that have potential implications in promoting plant growth and appear to depend upon cultivation practice. OTUs belonging to known nitrogen-fixing genera were enriched in organically cultivated samples including the cyanobacterium Anabaena and the Alphaproteobacterial genera Azospirillum and Rhodobacter (SI Appendix, Fig. S21A). Organically cultivated samples were also enriched for Actinobacterial OTUs belonging to the genus Streptomyces (SI Appendix, Fig. S21 B and C). Species within Streptomyces are known for their wide diversity of secondary metabolite production, many of which include antibiotic compounds (28). Microbial communities from ecofarmed rhizocompartments were enriched for OTUs that play a part in methane cycling belonging to the families Syntrophorhabdaceae (SI Appendix, Fig. S21 D and E) and Syntrophaceae. Members from both of these families are known for syntrophic growth with methanogenic archaea (see below), providing these archaea with H2 and formate essential for methane production (29–31).
Microbes Involved in Methane Emissions.
Global rice cultivation accounts for a significant proportion of annual greenhouse gas emissions, primarily methane (32). Methane (CH4) emitted from rice paddies is mostly synthesized by methanogenic archaea. Although the majority of the CH4 produced in rice paddy soil is oxidized by CH4-using eubacteria (33), much of the remaining CH4 diffuses through the soil into the plant roots and is expelled via the aerenchyma tissue of the rice plant itself (19). We found a large difference in abundance of methanogenic archaea between the field plants and the greenhouse plants. Methanogenic archaea were in much higher abundance in the field plants than in the greenhouse plants, indicating that either the greenhouse conditions are not a suitable growth habitat for methanogenic archaea or that the soils used for the greenhouse study had a low initial abundance of methanogens (SI Appendix, Fig. S22 A–D).
Methanogenic archaea require anaerobic conditions to function, and thus we did not expect methanogens to inhabit the endosphere or the rhizoplane. Surprisingly, the field plants exhibited high relative abundances of OTUs assigned to the genus Methanobacterium in the endosphere and rhizoplane that were comparable to or greater than the relative abundance of Methanobacterium in the rhizosphere (SI Appendix, Fig. S22A). This result is unexpected, given that the interior of rice roots is expected to have the highest levels of O2 relative to other spatial compartments when grown in flooded soil. This pattern is not maintained with OTUs in other methanogenic genera including Methanosarcina, Methanocella, and Methanosaeta, however (SI Appendix, Fig. S22 B–D). Relative abundance of these genera in field plants was always highest in the rhizosphere, where the O2 concentration is expected to be relatively lower. To further confirm the presence of methanogenic archaea in the endosphere, we used an alternative approach by PCR amplification of the methyl coenzyme-M reductase (mcrA) gene, a gene essential for CH4 production (34), from endosphere samples. By cloning and sequencing the mcrA amplicons, we again found that sequences derived from Methanobacteriaceae were in higher relative abundance in the endosphere than in the rhizosphere of field plants, whereas Methanosarcina sequences exhibited the opposite distribution (Dataset S19).
The relatively oxygenated environment within the rice roots might be expected to be suitable for CH4 oxidation by methanotrophs. We found that in general the methanotrophic bacteria followed the expected trend of distribution across the rhizocompartments, with the relative abundance lowest in the rhizosphere and highest in the endosphere (SI Appendix, Fig. S22 E and F). The field plants had higher abundances of methanotrophic bacteria compared with greenhouse plants, consistent with the low levels of methanogenic archaea and presumptive low levels of CH4 production under greenhouse growth.
Identification of Coabundance Networks of OTUs.
The sequence-based characterization of plant-associated microbiomes has primarily focused on how individual taxa within the microbiome associate with the host plant; however, the complex interactions that occur between taxa in the context of microbial communities are not revealed through this approach. Identification of microbial consortia is important for understanding their biological impact on the plant, but has been hindered by the inability to culture the vast majority of the microbes. It is likely that an important reason for the inability to grow many microbial species in pure culture can be attributed to consortium dynamics (35). Microbes cooperate in networks, supplying each other with critical nutrients for growth and survival. For example, methanogenic archaea cooperate with syntrophic partners to obtain H2 and formate for CH4 synthesis; hydrogen consumption by the methanogen allows the secondary fermentation process by the syntroph to become energetically favorable (29, 35).
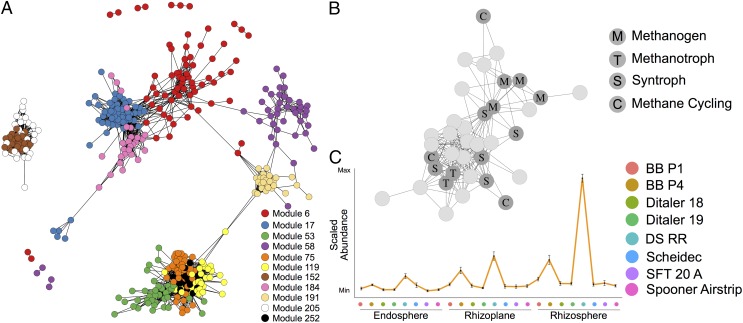
We hypothesized that it might be possible to identify consortia that are involved in CH4 cycling by generating OTU coabundance modules from the list of differentially abundant OTUs from the field experiment. We used an approach similar to gene coexpression network construction to generate the OTU coabundance modules. Briefly, pairwise Pearson correlations between OTUs determined to be differentially abundant between experimental factors included in the field experiment (10,848 OTUs; Datasets S16 and S17) were calculated and used as a distance metric for hierarchical clustering into a dendrogram, which was then dynamically pruned to form 284 coabundance modules (Dataset S20). The resulting modules were queried for clusters containing taxa that belonged to methanogenic archaea including Methanobacterium, Methanosarcina, Methanocella, and Methanosaeta. We identified 15 modules containing methanogenic OTUs with a correlation of 0.6 or greater to other OTUs within the same module (11 shown in Fig. 5A; Dataset S21). Modules containing methanogenic taxa were enriched for OTUs with methanotrophic, syntrophic, and CH4 cycling potential (hypergeometric test, Dataset S22; Fig. 5B and SI Appendix, Fig. S23). Many of the remaining OTUs in the methanogenic modules have limited taxonomic information or functional information available, thus making functional inference difficult. Taken together, these results show that an OTU coabundance network approach is successful in generating associations that can recapitulate empirical data, and therefore might have predictive value for identifying novel microbial associations.
Fig. 5.
OTU coabundance network reveals modules of OTUs associated with methane cycling. (A) Subset of the entire network corresponding to 11 modules with methane cycling potential. Each node represents one OTU and an edge is drawn between OTUs if they share a Pearson correlation of greater than or equal to 0.6. (B) Depiction of module 119 showing the relationship between methanogens, syntrophs, methanotrophs, and other methane cycling taxonomies. Each node represents one OTU and is labeled by the presumed function of that OTU’s taxonomy in methane cycling. An edge is drawn between two OTUs if they have a Pearson correlation of greater than or equal to 0.6. (C) Mean abundance profile for OTUs in module 119 across all rhizocompartments and field sites. The position along the x axis corresponds to a different field site. Error bars represent SE. The x and y axes represent no particular scale.
Acquisition of Microbiomes by the Root.
We have shown that rice seedlings grown in field soil in the greenhouse acquire microbiomes that exhibit characteristics similar to those of field rice plants, in terms of the general distribution of phyla in the rhizocompartments. To understand the dynamics of microbiome acquisition from soil, we performed a time-series experiment. We transplanted sterilely germinated seedlings of the japonica M104 cultivar into soil collected from the rice field in Davis in the greenhouse and collected samples at increasing intervals (0, 1, 2, 3, 5, 8, and 13 d). To monitor general changes in the soil microbial communities, we sampled from pots containing unplanted soil in the same container at each time point.
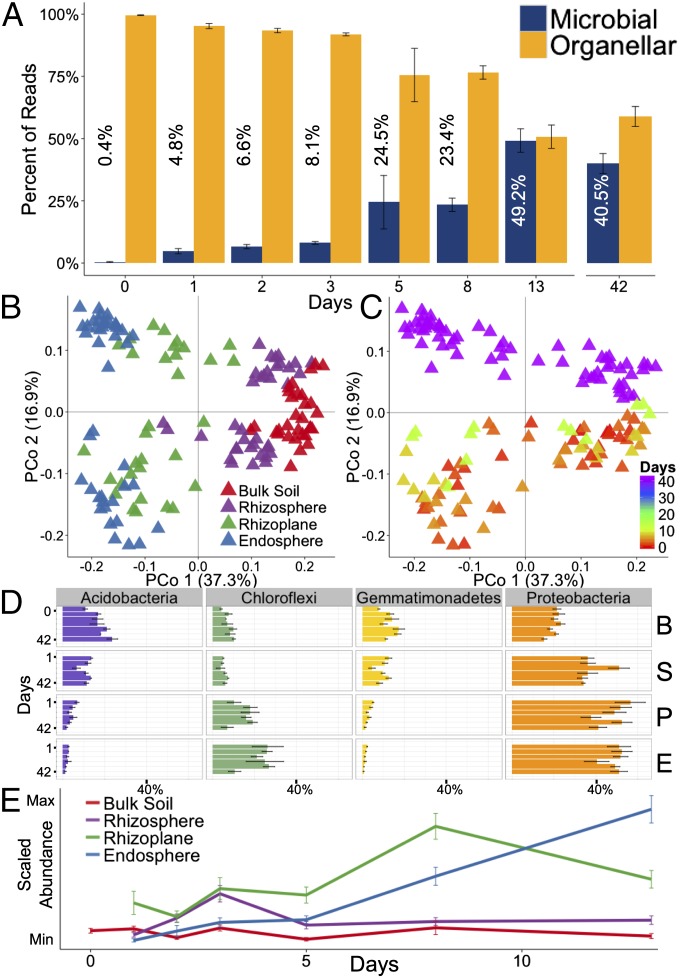
After collection, 16S rRNA gene sequencing of the samples from different compartments was performed as before. We used the proportion of microbial reads to organellar (plastidial and mitochondrial) reads to analyze microbial abundance in the endosphere over time (Fig. 6A). Using this technique, we confirmed the sterility of seedling roots before transplantation. We found that microbial penetrance into the endosphere occurred at or before 24 h after transplantation and that the proportion of microbial reads to organellar reads increased over the first 2 wk after transplantation (Fig. 6A). To further support the evidence for microbiome acquisition within the first 24 h, we sampled root endospheric microbiomes from sterilely germinated seedlings before transplanting into Davis field soil as well as immediately after transplantation and 24 h after transplantation (SI Appendix, Fig. S24). The root endospheres of sterilely germinated seedlings, as well as seedlings transplanted into Davis field soil for 1 min, both had a very low percentage of microbial reads compared with organellar reads (0.22% and 0.71%), with the differences not statistically significant (P = 0.1, Wilcoxon test). As before, endospheric microbial abundance increased significantly, by >10-fold after 24 h in field soil (3.95%, P = 0.05, Wilcoxon test). We conclude that brief soil contact does not strongly increase the proportion of microbial reads, and therefore the increase in microbial reads at 24 h is indicative of endophyte acquisition within 1 d after transplantation.
Fig. 6.
Time-series analysis of root-associated microbial communities reveals distinct microbiome colonization patterns. (A) Ratios of microbial to organellar (plastidial and mitochondrial) 16S rRNA gene reads in the endosphere after transplantation into Davis soil. The 42-d time point corresponds to the earlier greenhouse experiment data (Fig. 1) subsetted to M104 in Davis soil. Mean percentages of the ratios are depicted with each bar. (B) PCoA of the time-series experiment and the greenhouse experiment subsetted to plants growing in Davis soil and colored by rhizospheric compartment. (C) The same PCoA as in B colored by collection day after transplantation into soil. (D) Average relative abundance for select phyla over the course of microbiome acquisition. (E) Average abundance profile of 53 out of the 92 core endosphere-enriched OTUs in each rhizospheric compartment. Error bars represent SE.
α-Diversity significantly varied by rhizocompartment (P < 2E-16; Dataset S23) and there was a significant interaction between rhizocompartment and collection time (P = 0.042; Dataset S23); however, when each rhizocompartment was analyzed individually, the bulk soil was the only compartment that showed a significant amount of variation in α-diversity over time (SI Appendix, Fig. S25 and Dataset S23). The above results suggest that a diverse microbiota can begin to colonize the rhizoplane and endosphere as early as 24 h after transplanting into soil. We next evaluated how β-diversities shift over time in each rhizocompartment. We compared the time-series microbial communities with the previous greenhouse experiment microbial communities of M104 in Davis soil (Fig. 6 B and C). β-Diversity measurements of the time-series data indicated that microbiome samples from each compartment are separable by time. Furthermore, the rhizoplane and endosphere microbiomes from the later time point in the time-series data (13 d) approach the endosphere and rhizoplane microbiome compositions for plants that have been grown in the greenhouse for 42 d.
There are slight shifts in the distribution of phyla over time; however, there are significant distinctions between the compartments starting as early as 24 h after transplantation into soil (Fig. 6D, SI Appendix, Figs. S24B and S26, and Dataset S24). Because each phylum consists of diverse OTUs that could exhibit very different behaviors during acquisition, we next examined the dynamics and colonization patterns of specific OTUs within the time-course experiment. The core set of 92 endosphere-enriched OTUs obtained from the previous greenhouse experiment (SI Appendix, Fig. S9C) was analyzed for relative abundances at different time points (Fig. 6E). Of the 92 core endosphere-enriched microbes present in the greenhouse experiment, 53 OTUs were detectable in the endosphere in the time-course experiment. The average abundance profile over time revealed a colonization pattern for the core endospheric microbiome. Relative abundance of the core endosphere-enriched microbiome peaks early (3 d) in the rhizosphere and then decreases back to a steady, low level for the remainder of the time points. Similarly, the rhizoplane profile shows an increase after 3 d with a peak at 8 d with a decline at 13 d. The endosphere generally follows the rhizoplane profile, except that relative abundance is still increasing at 13 d. These results suggest that the core endospheric microbes are first attracted to the rhizosphere and then locate to the rhizoplane, where they attach before migration to the root interior. To summarize, microbiome acquisition from soil appears to occur relatively rapidly, initiating within 24 h and approaching steady state within 14 d. The dynamics of accumulation suggest a multistep process, in which the rhizosphere and rhizoplane are likely to play key roles in determining the compositions of the interior and exterior components of the root-associated microbiome (Discussion).
Discussion
Factors Affecting the Composition of Root-Associated Microbiomes.
The data presented here provide a characterization of the microbiome of rice, involving the combination of finer structural resolution and deeper sequencing than previous plant microbiome studies and using both controlled greenhouse and field studies covering a geographical range of cultivation. Specifically, we have been able to characterize in-depth the compositions of three distinct rhizocompartments—the rhizosphere, rhizoplane, and endosphere—and gain insights into the effects of external factors on each of these compartments. We note that a detailed characterization of plant rhizoplane microbiota in relation to the rhizosphere and the endosphere has not been previously attempted. To achieve this, we successfully adapted protocols for removal of rhizoplane microbes from the endosphere of Arabidopsis roots (9, 10). Because the fractional abundance of organellar reads in the rhizosphere, rhizoplane, and endosphere exhibits a clear increasing gradient (SI Appendix, Fig. S27), we hypothesize that we are isolating the rhizoplane fraction via disruption of the rhizodermis, consistent with direct EM observations on Arabidopsis roots following sonication (9, 10). The fine structure approach we have used combined with depth of sequencing allowed us to analyze over 250,000 OTUs, an order of magnitude greater than in any single plant species to date. Under controlled greenhouse conditions, the rhizocompartments described the largest source of variation in the microbial communities sampled (Dataset S5A). The pattern of separation between the microbial communities in each compartment is consistent with a spatial gradient from the bulk soil across the rhizosphere and rhizoplane into the endosphere (Fig. 1C). Similarly, microbial diversity patterns within samples hold the same pattern where there is a gradient in α-diversity from the rhizosphere to the endosphere (Fig. 1B). Enrichment and depletion of certain microbes across the rhizocompartments indicates that microbial colonization of rice roots is not a passive process and that plants have the ability to select for certain microbial consortia or that some microbes are better at filling the root colonizing niche. Similar to studies in Arabidopsis, we found that the relative abundance of Proteobacteria is increased in the endosphere compared with soil, and that the relative abundances of Acidobacteria and Gemmatimonadetes decrease from the soil to the endosphere (9–11), suggesting that the distribution of different bacterial phyla inside the roots might be similar for all land plants (Fig. 1D and Dataset S6). Under controlled greenhouse conditions, soil type described the second largest source of variation within the microbial communities of each sample. However, the soil source did not affect the pattern of separation between the rhizospheric compartments, suggesting that the rhizocompartments exert a recruitment effect on microbial consortia independent of the microbiome source.
By using differential OTU abundance analysis in the compartments, we observed that the rhizosphere serves an enrichment role for a subset of microbial OTUs relative to bulk soil (Fig. 2). Further, the majority of the OTUs enriched in the rhizosphere are simultaneously enriched in the rhizoplane and/or endosphere of rice roots (Fig. 2B and SI Appendix, Fig. S16B), consistent with a recruitment model in which factors produced by the root attract taxa that can colonize the endosphere. We found that the rhizoplane, although enriched for OTUs that are also enriched in the endosphere, is also uniquely enriched for a subset of OTUs, suggesting that the rhizoplane serves as a specialized niche for some taxa. Conversely, the vast majority of microbes depleted in the rhizoplane are also depleted in the endosphere (Fig. 2C and SI Appendix, Fig. S16C), suggesting that the selectivity for colonization of the interior occurs at the rhizoplane and that the rhizoplane may serve an important gating role for limiting microbial penetrance into the endosphere. It is important to note that the community structure we observe in each compartment is likely not simply caused by the plant alone. Microbial community structural differences between the compartments may be attributable also to microbial interactions involving both competition and cooperation.
In the case of field plants, we observed that the largest source of microbiome separation was due to cultivation site, rather than the spatial compartments (Dataset S5 O and P). These results are in contrast to the controlled greenhouse experiment where the soil effect was the second largest source of variation, suggesting the geography may be more important for determining the composition of the root microbiome than soil structure alone (Dataset S5A). These results differ from the results in the maize microbiome study, where microbial communities showed clear separation by state but not very much by geographic location within the same state (12). However, we note that in our study the locations within California were separated by distances of up to ∼125 km, vs. a maximum separation of ∼40 km in the intrastate locations of the maize study. Other factors that might account for the different results in our study include the number of field sites examined (eight, vs. three intrastate fields examined in the maize study), increased sequencing depth, different resolution because spatial compartments in maize roots were not separately analyzed, or possibly intrinsic differences between cultivated rice and maize.
Our design of the field experiment allowed us to test for cultivation practice effects on the rice root-associated microbiome, specifically between organic cultivation and ecofarming, a more conventional cultivation practice. We found that cultivation practice described a significant amount of variation between the microbiomes, and that this effect was exhibited across every rhizospheric compartment (Fig. 4D and SI Appendix, Fig. S14D). These results are in contrast to the maize microbiome study, where an organically cultivated field had no significant separation from a conventionally cultivated field in the same state (12). Again, the difference in results between the two studies might be due to differences in sequencing depth and structural resolution or to intrinsic differences between the crops.
Microbial Consortia Associated with Methane Cycling in Rice Fields.
Rice cultivation is a large contributor to agricultural CH4 emissions due to the anaerobic conditions resulting from flooded fields. Currently, the model for CH4 emissions under rice cultivation is that CH4 is formed in the soil away from the roots and diffuses through the soil to the rice root, where it is expelled via the aerenchyma in the roots (19, 32, 36). We used our dataset to examine the abundances of taxa related to CH4 cycling. As predicted, we observed that the relative abundance of some methanogens (Methanocella, Methanosarcina, and Methanosaeta) is higher in the rhizosphere than in the endosphere (SI Appendix, Fig. S22). However, the relative abundance of the methanogen Methanobacterium was found to be typically equal or higher in the endosphere than in the rhizosphere. This is an unexpected finding, because the root interior is relatively O2-rich and therefore unfavorable for anaerobic archaea. How these archaea might survive within the root and whether they are contributing to CH4 production at this interior location are questions that should be investigated. These results indicate that our understanding of rice-associated CH4 production is still incomplete, and emphasize the need for more detailed studies to elucidate the different players and their roles in the process, perhaps using metagenomic and metatranscriptomic sequencing to better understand the metabolic potential of endosphere microbial communities.
An approach toward unraveling methanogen interactions with rice roots that we have explored is the construction of OTU coabundance networks. We hypothesized that microbes producing or using CH4 might form consortia that would be revealed through this analysis. The sequencing depth and variation provided by the field experiment allowed us to test this approach by generating clusters of OTUs and identification of clusters containing methanogenic archaea. Inspection of individual OTUs within such clusters revealed additional taxonomies known to be involved in CH4 cycling, such as methanotrophic, syntrophic, and CH4-cycling eubacteria (Fig. 5 and SI Appendix, Fig. S23). The clusters also contained OTUs with no known role in CH4 cycling. Possibly these OTUs represent novel players that have been revealed through this approach, although larger-scale studies with additional field sites will be needed to validate the significance of their associations. Even with this limited scale, the results of this analysis are very encouraging. This approach might be generalizable for culture-independent identification of other microbial consortia involved in different types of geochemical cycles that interact with plant roots. However, this approach might be more applicable to those microbial lineages that are tightly linked to certain traits, such as CH4 cycling, and might not be successful when a trait is much more widespread independent of lineage, such as nitrogen fixation.
A Multistep Model for Microbiome Acquisition.
Studies on mammals have established that the fetus at birth already has a microbiome derived from the mother, and that diet and other environmental factors further shape the gut microbiome (37–39). For plants, the process of microbiome acquisition is not well-understood (7). Microbiome transmission through the embryo seems unlikely, because surface sterilization of the seeds resulted in axenic plants (Fig. 6A and SI Appendix, Fig. S23). Our success in obtaining in-depth sequence coverage for all three different spatial compartments in the root provided us with an opportunity to address this question by carrying out a time-staged profiling of the microbial compositions of the compartments during the early stages of microbiome acquisition. We find that microbiome acquisition from soil is rapid, namely that rice plants begin to assemble an endospheric microbiome within a day after transplantation from sterile media to soil, and the relative level approaches steady state within 2 wk. The 13-d endosphere and rhizoplane microbiomes were most similar to the older, 42-d, microbiomes from the previous greenhouse experiment that we consider to represent steady state, as the latter microbiomes are from well-established vegetatively growing plants (Fig. 6 B and C). Because separate batches of soil were used between the greenhouse and the acquisition experiment, it is important to note that the microbiomes may not be directly comparable across different experiments, due to seasonal variation between soil batches (9–11). The patterns of relative abundance in phyla between the compartments (e.g., higher proportions of Acidobacteria in the rhizosphere than endosphere and lower proportions of Proteobacteria and Chloroflexi in the rhizosphere than endosphere) are evident within 24 h after the rice plants were transplanted into soil (Fig. 6C and SI Appendix, Figs. S24B and S26). Based on studies of the steady-state microbiomes of the rhizosphere and endosphere, it has been proposed that plants might assemble their microbiomes in two steps, with the first step involving a general recruitment to the vicinity of the root and a second step for entry inside the root that involves species-specific genetic factors (7). The dynamics of microbiome acquisition in our study provide experimental support for this model, in that the general concept of recruitment and assembly as separate steps is consistent with our data. We observed an initial enrichment in the rhizosphere, consistent with the attraction of a diverse set of microbes to the vicinity of the plant, followed by slower rates of accumulation of OTUs in the endosphere (Fig. 6E). The selectivity of the latter process is also implied by the extensive depletion of rhizospheric OTUs in the endosphere. However, when rhizoplane microbiome data are taken into consideration, microbiome acquisition in the root appears to be a more complex multistep process, in which the rhizoplane plays a key role. After initial recruitment to the rhizosphere, only a subset of these microbes initially recruited to the rhizosphere are bound to the root surface at the rhizoplane, suggesting selectivity for direct physical association with the root. This selection may occur by the plant, or may occur through the ability to form biofilms, as certain microbes are known to form biofilms along root surfaces (40, 41). A further selective step must operate to account for the additional depletion of rhizoplane OTUs from the endosphere at steady state, implying that binding is not sufficient for entry. Binding at the rhizoplane might, however, act as a necessary prerequisite for endospheric OTUs, as the time course shows that accumulation of OTUs at the rhizoplane precedes that in the endosphere. We suggest that the rhizoplane serves a critical gating role; of the microbes that are attracted to the rhizosphere, only a subset can bind the rhizoplane, and a fraction of these are permitted to enter and proliferate in the endosphere. Each of these steps likely involves molecular signals from the plant, presumptively components of root exudates and possibly cell-wall components or membrane proteins. The signals could consist of general plant metabolites as well as species- and genotype-specific molecules. In the model proposed by Bulgarelli et al. (7), genotype signals are proposed to regulate entry into the endosphere. However, in our study, the rhizosphere composition showed the largest variation in the comparison of different cultivars, suggesting that the genotypic factors appear to also act at the level of general microbial recruitment. In summary, the dynamic shifts in microbiomes across each compartment indicate that the actions of three or more selective steps at distinct spatial locations from the root interior, and responding to multiple signals from the plant, coordinate the assembly of the root microbiome.
Materials and Methods
Microbial communities from the rhizosphere, rhizoplane, and root endosphere of various rice cultivars grown in the greenhouse and field were profiled by amplifying fragments of the 16S rRNA gene and sequenced using the Illumina MiSeq platform. The sequence analysis was carried out using the QIIME pipeline (42) and all statistical analyses were performed using R v3.1.0 (43). Further details of materials and methodology are explained in SI Appendix, Materials and Methods. The raw sequencing reads for this project were submitted to the National Center for Biotechnology Information Short Read Archive under accession no. SRP044745. The custom scripts used for analyzing the data presented here can be found at github.com/ricemicrobiome/edwards-et-al.-2014.
Supplementary Material
Acknowledgments
We thank Lance Benson and Jessica Lundberg of Lundberg Family Farms; George Tibbetts and Michael Lear for generously providing samples and access to their field facilities; Derek Lundberg and Jeffery Dangl (The University of North Carolina) for sharing their protocol for root sonication prior to publication; Kelsey Galimba, John Jaeger, Cassandra Ramos, and Paul Tisher for technical assistance at the early stages of the project; Gurdev Khush and Kate Scow for valuable advice; and members of the J.A.E. and V.S. laboratories for helpful discussions. J.A.E. and V.S. acknowledge the support of National Science Foundation Awards DBI-0923806 and IOS-1444974; J.E. acknowledges partial support from the Henry A. Jastro Graduate Research Award; and C.S.-M. acknowledges support from the University of California Institute for Mexico (UCMEXUS)/Consejo Nacional de Ciencia y Tecnología (CONACYT) and Secretaría de Educación Pública (Mexico).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the National Center for Biotechnology Information Short Read Archive (accession no. SRP044745).
See Commentary on page 2299.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414592112/-/DCSupplemental.
References
- 1.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308(5721):554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, et al. A soil bacterium regulates plant acquisition of iron via deficiency-inducible mechanisms. Plant J. 2009;58(4):568–577. doi: 10.1111/j.1365-313X.2009.03803.x. [DOI] [PubMed] [Google Scholar]
- 3.Long SR. Rhizobium-legume nodulation: Life together in the underground. Cell. 1989;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- 4.Bolan N. A critical review on the role of mycorrhizal fungi in the uptake of phosphorus by plants. Plant Soil. 1991;134(2):189–207. [Google Scholar]
- 5.Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol. 2006;57:233–266. doi: 10.1146/annurev.arplant.57.032905.105159. [DOI] [PubMed] [Google Scholar]
- 6.Berendsen RL, Pieterse CM, Bakker PA. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012;17(8):478–486. doi: 10.1016/j.tplants.2012.04.001. [DOI] [PubMed] [Google Scholar]
- 7.Bulgarelli D, Schlaeppi K, Spaepen S, Ver Loren van Themaat E, Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- 8.Mendes R, et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science. 2011;332(6033):1097–1100. doi: 10.1126/science.1203980. [DOI] [PubMed] [Google Scholar]
- 9.Bulgarelli D, et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature. 2012;488(7409):91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 10.Lundberg DS, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488(7409):86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schlaeppi K, Dombrowski N, Oter RG, Ver Loren van Themaat E, Schulze-Lefert P. Quantitative divergence of the bacterial root microbiota in Arabidopsis thaliana relatives. Proc Natl Acad Sci USA. 2014;111(2):585–592. doi: 10.1073/pnas.1321597111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiffer JA, et al. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA. 2013;110(16):6548–6553. doi: 10.1073/pnas.1302837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottel NR, et al. Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl Environ Microbiol. 2011;77(17):5934–5944. doi: 10.1128/AEM.05255-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shakya M, et al. A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE. 2013;8(10):e76382. doi: 10.1371/journal.pone.0076382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palmer C, Bik EM, DiGiulio DB, Relman DA, Brown PO. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5(7):e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci USA. 2010;107(26):11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sessitsch A, et al. Functional characteristics of an endophyte community colonizing rice roots as revealed by metagenomic analysis. Mol Plant Microbe Interact. 2012;25(1):28–36. doi: 10.1094/MPMI-08-11-0204. [DOI] [PubMed] [Google Scholar]
- 18.Knief C, et al. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012;6(7):1378–1390. doi: 10.1038/ismej.2011.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neue H-U. Methane emission from rice fields. Bioscience. 1993;43(7):466–474. [Google Scholar]
- 20.McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLOS Comput Biol. 2014;10(4):e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Im W-T, Kim S-H, Kim MK, Ten LN, Lee S-T. Pleomorphomonas koreensis sp. nov., a nitrogen-fixing species in the order Rhizobiales. Int J Syst Evol Microbiol. 2006;56(Pt 7):1663–1666. doi: 10.1099/ijs.0.63499-0. [DOI] [PubMed] [Google Scholar]
- 22.Madhaiyan M, et al. Pleomorphomonas diazotrophica sp. nov., an endophytic N-fixing bacterium isolated from root tissue of Jatropha curcas L. Int J Syst Evol Microbiol. 2013;63(Pt 7):2477–2483. doi: 10.1099/ijs.0.044461-0. [DOI] [PubMed] [Google Scholar]
- 23.Xie C-H, Yokota A. Pleomorphomonas oryzae gen. nov., sp. nov., a nitrogen-fixing bacterium isolated from paddy soil of Oryza sativa. Int J Syst Evol Microbiol. 2005;55(Pt 3):1233–1237. doi: 10.1099/ijs.0.63406-0. [DOI] [PubMed] [Google Scholar]
- 24.Ransom-Jones E, Jones DL, McCarthy AJ, McDonald JE. The Fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb Ecol. 2012;63(2):267–281. doi: 10.1007/s00248-011-9998-1. [DOI] [PubMed] [Google Scholar]
- 25.Leschine SB. Cellulose degradation in anaerobic environments. Annu Rev Microbiol. 1995;49:399–426. doi: 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- 26.Kim S-I, Tai TH. Identification of SNPs in closely related temperate japonica rice cultivars using restriction enzyme-phased sequencing. PLoS ONE. 2013;8(3):e60176. doi: 10.1371/journal.pone.0060176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lundberg Family Farms 2014 Organic vs. eco-farmed. Available at www.lundberg.com/commitment/ecofarmed.aspx. Accessed July 14, 2014.
- 28.Watve MG, Tickoo R, Jog MM, Bhole BD. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol. 2001;176(5):386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 29.Stams AJ, Plugge CM. Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat Rev Microbiol. 2009;7(8):568–577. doi: 10.1038/nrmicro2166. [DOI] [PubMed] [Google Scholar]
- 30.Gray ND, et al. The quantitative significance of Syntrophaceae and syntrophic partnerships in methanogenic degradation of crude oil alkanes. Environ Microbiol. 2011;13(11):2957–2975. doi: 10.1111/j.1462-2920.2011.02570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schink B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol Mol Biol Rev. 1997;61(2):262–280. doi: 10.1128/mmbr.61.2.262-280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Conrad R. The global methane cycle: Recent advances in understanding the microbial processes involved. Environ Microbiol Rep. 2009;1(5):285–292. doi: 10.1111/j.1758-2229.2009.00038.x. [DOI] [PubMed] [Google Scholar]
- 33.Holzapfel-Pschorn A, Conrad R, Seiler W. Production, oxidation and emission of methane in rice paddies. FEMS Microbiol Lett. 1985;31(6):343–351. [Google Scholar]
- 34.Thauer RK. Biochemistry of methanogenesis: A tribute to Marjory Stephenson. Microbiology. 1998;144(Pt 9):2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 35.McInerney MJ, Sieber JR, Gunsalus RP. Syntrophy in anaerobic global carbon cycles. Curr Opin Biotechnol. 2009;20(6):623–632. doi: 10.1016/j.copbio.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee HJ, Kim SY, Kim PJ, Madsen EL, Jeon CO. Methane emission and dynamics of methanotrophic and methanogenic communities in a flooded rice field ecosystem. FEMS Microbiol Ecol. 2014;88(1):195–212. doi: 10.1111/1574-6941.12282. [DOI] [PubMed] [Google Scholar]
- 37.Gueimonde M, et al. Effect of maternal consumption of Lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J Pediatr Gastroenterol Nutr. 2006;42(2):166–170. doi: 10.1097/01.mpg.0000189346.25172.fd. [DOI] [PubMed] [Google Scholar]
- 38.Vaishampayan PA, et al. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koenig JE, et al. Succession of microbial consortia in the developing infant gut microbiome. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4578–4585. doi: 10.1073/pnas.1000081107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134(1):307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walker TS, et al. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004;134(1):320–331. doi: 10.1104/pp.103.027888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. R Core Team (2012) R: A Language and Environment for Statistical Computing (R Found Stat Comput, Vienna)
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.