Significance
We have recently identified the sequence of the ether lipid-cleaving enzyme alkylglycerol monooxygenase. Like nitric oxide synthases and aromatic amino acid hydroxylases, alkylglycerol monooxygenase needs tetrahydrobiopterin as cofactor. Whereas the former enzymes have well-established roles in the cell, the physiology of alkylglycerol monooxygenase is still not clear. Here we show its regulation in murine macrophage differentiation and its dependence on the cofactor tetrahydrobiopterin in live murine macrophage-like RAW264.7 cells. Upon modulation of the activity of alkylglycerol monooxygenase and the key enzyme in tetrahydrobiopterin biosynthesis, we observe extensive changes in various lipid classes ranging from ether lipids to far more complex lipids. These findings point to an important role of tetrahydrobiopterin in cellular lipid homeostasis.
Keywords: alkylglycerols, lipidomics, macrophages, RAW264.7, tetrahydrobiopterin
Abstract
Tetrahydrobiopterin is a cofactor synthesized from GTP with well-known roles in enzymatic nitric oxide synthesis and aromatic amino acid hydroxylation. It is used to treat mild forms of phenylketonuria. Less is known about the role of tetrahydrobiopterin in lipid metabolism, although it is essential for irreversible ether lipid cleavage by alkylglycerol monooxygenase. Here we found intracellular alkylglycerol monooxygenase activity to be an important regulator of alkylglycerol metabolism in intact murine RAW264.7 macrophage-like cells. Alkylglycerol monooxygenase was expressed and active also in primary mouse bone marrow-derived monocytes and “alternatively activated” M2 macrophages obtained by interleukin 4 treatment, but almost missing in M1 macrophages obtained by IFN-γ and lipopolysaccharide treatment. The cellular lipidome of RAW264.7 was markedly changed in a parallel way by modulation of alkylglycerol monooxygenase expression and of tetrahydrobiopterin biosynthesis affecting not only various ether lipid species upstream of alkylglycerol monooxygenase but also other more complex lipids including glycosylated ceramides and cardiolipins, which have no direct connection to ether lipid pathways. Alkylglycerol monooxygenase activity manipulation modulated the IFN-γ/lipopolysaccharide–induced expression of inducible nitric oxide synthase, interleukin-1β, and interleukin 1 receptor antagonist but not transforming growth factor β1, suggesting that alkylglycerol monooxygenase activity affects IFN-γ/lipopolysaccharide signaling. Our results demonstrate a central role of tetrahydrobiopterin and alkylglycerol monooxygenase in ether lipid metabolism of murine macrophages and reveal that alteration of alkylglycerol monooxygenase activity has a profound impact on the lipidome also beyond the class of ether lipids.
Alkylglycerols are a subclass of the vast family of ether lipids that are essential membrane constituents (e.g., in neurons), are important for spermatogenesis, and protect the eye from cataract (1). Furthermore, they are also involved in many signaling pathways (2). The best-understood ether lipid is platelet-activating factor (PAF; 1-O-alkyl-2-acetyl-sn-glycero-3-phosphocholine), a pleiotropic proinflammatory mediator (3). The first step in PAF degradation removes the acetyl group to produce lyso-platelet–activating factor (lyso-PAF) (4), which can be recycled for PAF production via the remodeling pathway.
The only known enzyme able to cleave the ether bond in saturated alkylglycerols with a free hydroxyl group at sn-2 (lyso-alkylglycerols) is alkylglycerol monooxygenase (AGMO), a highly hydrophobic mixed-function oxidase that degrades alkylglycerols into an aldehyde and a glycerol derivative (5). AGMO was first described in 1964 as a tetrahydrobiopterin-dependent enzyme (6). It is an exceptionally labile integral membrane protein, which could not be purified thus far. In 2010, we assigned a predicted ORF to AGMO by a pattern-based bioinformatics approach to select candidates and experimental validation using heterologous expression of these ORFs (5). In homogenates, AGMO was shown to crucially depend on tetrahydrobiopterin as cofactor (7, 8). This cofactor resembles the vitamins riboflavin and folic acid but can be synthesized in the animal body in a three-step enzymatic cascade that is initiated by GTP cyclohydrolase 1 (GCH1). In addition to its role in AGMO, tetrahydrobiopterin serves well-known cofactor roles in nitric oxide synthesis and aromatic amino acid hydroxylation. It is also used to treat mild forms of phenylketonuria (for a review, see ref. 9).
AGMO is highly expressed and active in various murine and rat tissues, but very few cell lines express AGMO activity (5, 10). A particularly robust and high activity could be detected in the murine macrophage-like cell line RAW264.7 (5). In this cell line and in mouse peritoneal macrophages, AGMO mRNA levels and activity were recently described to be down-regulated by the inflammatory mediator bacterial lipopolysaccharide (LPS) coinciding with increased lyso-PAF and PAF levels (11). The impact of AGMO activity alteration on cellular lipids has so far only been addressed by overexpression of the enzyme in HEK293 cells (11).
In this study, we investigated AGMO activity and expression in primary murine bone marrow-derived monocytes during their differentiation into two extremes of macrophage differentiation, namely M1 and M2 macrophages, by pro- and antiinflammatory cytokines, respectively. M1 or “classically activated” macrophages secrete high levels of interleukin (IL) 12, whereas M2 or “alternatively activated” macrophages comprise a range of antiinflammatory and immunomodulatory profiles and are characterized by high IL-10 levels (12). Furthermore, we manipulated both AGMO and GCH1 expression in RAW264.7 cells by lentiviral overexpression or small hairpin RNA interference technology to investigate (i) whether AGMO represents the main source of alkylglycerol degrading activity in these cells, (ii) whether the intracellular concentration of its cofactor tetrahydrobiopterin determines alkylglycerol degradation by intact cells, and (iii) in which way these manipulations affect the cellular lipidome.
Results
AGMO Expression and Activity in Primary Mouse Bone Marrow Monocytes/Macrophages.
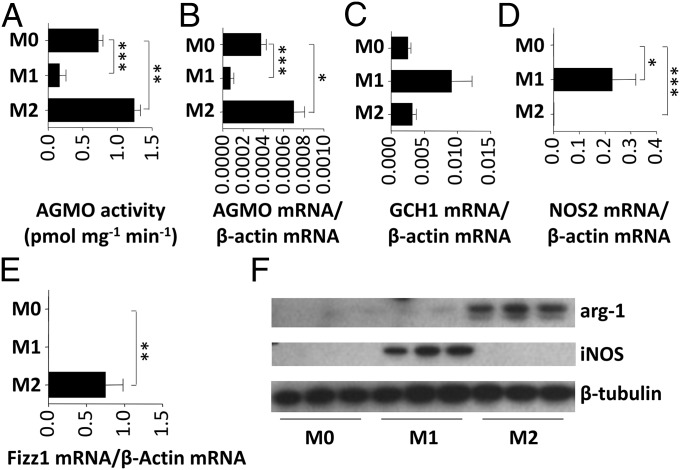
Murine bone marrow-derived monocytes were differentiated for 48 h toward unpolarized (M0; no cytokines added), classically activated (M1; stimulated with LPS and IFN-γ), and alternatively activated (M2; stimulated with IL-4) macrophages. In M0 macrophages, AGMO enzymatic activity (Fig. 1A) and mRNA (Fig. 1B) could readily be detected. Induction of the M1 phenotype led to a strong down-regulation of activity (4.4-fold, P < 0.001) and mRNA expression (5.0-fold, P < 0.001) (Fig. 1 A and B, respectively). In contrast, macrophages stimulated toward an M2 phenotype had significantly higher AGMO activity (1.7-fold, P < 0.01) and expression (1.8-fold, P < 0.05) after 48 h (Fig. 1 A and B). GCH1 showed a trend toward up-regulation in M1 macrophages (Fig. 1C). Another tetrahydrobiopterin-dependent enzyme, inducible nitric oxide synthase (iNOS, NOS2), was not detectable in M0 (below 8.8 × 10−5 mRNA per β-actin mRNA), but was strongly up-regulated in M1 macrophages (0.23 ± 0.09 mRNA per β-actin mRNA, P < 0.05) and detectable in M2 macrophages (1.8 × 10−3 ± 1.0 × 10−4 mRNA per β-actin mRNA, P < 0.001) (Fig. 1D). iNOS Western blots showed up-regulation in M1 only (Fig. 1F). Fizz1 [resistin-like alpha (retnla)] mRNA and arginase-1 protein, both markers for M2 macrophages (13), were not detectable in M0 and M1 but highly expressed in M2 (Fig. 1 E and F).
Fig. 1.
Bone marrow-derived monocytes/macrophages from six to nine mice were differentiated individually from M0 to M1 with IFN-γ/LPS and to M2 with IL-4 for 48 h. (A) AGMO activity was analyzed by a fluorescence-based HPLC microassay (10). (B–E) AGMO (B), GCH1 (C), inducible nitric oxide synthase (D), and Fizz1 (retnla) expression (E) was determined by qPCR using TaqMan technology. Expression data were normalized to β-actin. (F) iNOS and arginase-1 (arg-1) protein expression was analyzed by Western blotting with β-tubulin as loading control. Data are presented as means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Data were obtained in three independent experiments.
Modulation of AGMO Activity in the Murine Macrophage Cell Line RAW264.7.
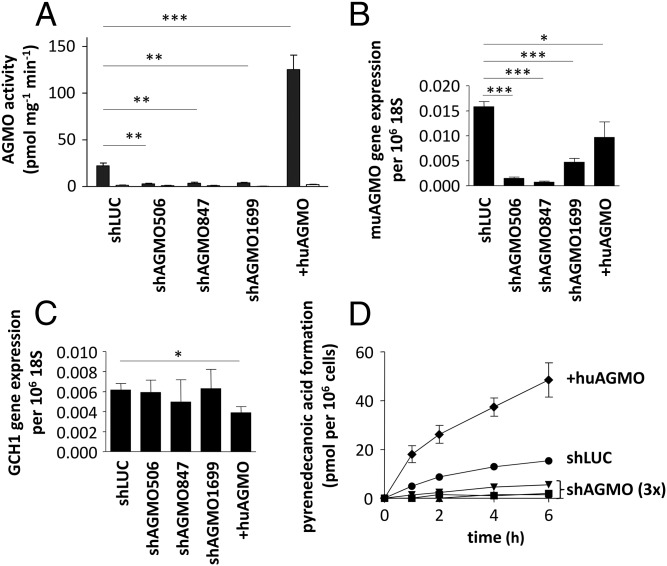
To test whether AGMO is the major cellular alkylglycerol cleaving activity, we transduced the murine macrophage cell line RAW264.7 with three constructs (shAGMO506, shAGMO847, and shAGMO1699). This resulted in significantly diminished (∼10-fold) AGMO activity (P < 0.01; Fig. 2A) and expression (P < 0.001; Fig. 2B) compared with control RAW264.7 cells infected with a short hairpin RNA (shRNA) against the unrelated protein firefly luciferase (shLUC; Fig. 2A). We also created a cell line overexpressing FLAG-tagged human AGMO, which had significantly elevated (6.1-fold, P < 0.001) enzymatic activity (+huAGMO; Fig. 2A) and, as expected, expressed human AGMO mRNA (0.017 mRNA per 106 18S mRNA). GCH1 expression (Fig. 2C) was unchanged in all three knockdown lines and only slightly reduced compared with shLUC (P = 0.0388) in the overexpression line +huAGMO. With these lines, we investigated the ability of intact RAW264.7 cells to cleave alkylglycerols using a live-cell assay (14) (Fig. 2D). Addition of pyrenedecylglycerol—a fluorescently labeled alkylglycerol that closely mimics the natural hexadecylglycerol substrate (10)—to the culture media of RAW264.7 control cells (shLUC) resulted in the formation of pyrenedecanoic acid in the culture supernatants. Cells overexpressing human AGMO (+huAGMO) had a 3.2-fold increased formation of pyrenedecanoic acid. In contrast, all three RNAi cell lines showed a strongly reduced formation of the fluorescent acid (11.4-, 21.3-, and 3.2-fold, respectively). When comparing time-resolved pyrenedecanoic acid formation between shLUC and the overexpression cell line as well as the knockdown cell lines, differences in formation were highly significant for all of them (P < 0.001). Changes in pyrenedecanoic acid formation were not due to an altered cell viability as measured by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide test (shLUC: 102.2 ± 3.1%; shAGMO506: 101.2 ± 3.0%; shAGMO847: 96.3 ± 5.6%; shAGMO1699: 107.0 ± 7.2%; +huAGMO: 98.1 ± 6.3%). As an additional control, we determined the stability of pyrenedecanoic acid in the supernatant in parallel series. Comparable amounts of pyrenedecanoic acid were still present in the supernatants at 6 h after addition (shLUC: 74.0 ± 2.1%; shAGMO506: 63.2 ± 2.1%; shAGMO847: 80.0 ± 2.2%; shAGMO1699: 65.3 ± 1.1%; +huAGMO: 67.6 ± 2.3%). Thus, the observed differences in the generation of pyrenedecanoic acid are most likely due to different expression levels of AGMO.
Fig. 2.
Biochemical effects of AGMO manipulation in RAW264.7 murine macrophages. (A) AGMO activity was quantified in homogenates of shAGMO506, shAGMO847, shAGMO1699, +huAGMO, and shLUC as control (n = 7–13) using a fluorescence-based HPLC microassay (10). (B and C) Expression of murine (mu)AGMO (B) and murine GCH1 (C) was analyzed in 6–12 parallels by qPCR using TaqMan technology and related to 18S. (D) Alkylglycerol cleavage of living shLUC (circles), shAGMO506 (squares), shAGMO847 (triangles), shAGMO1699 (inverted triangles), and +huAGMO (rotated squares) at 0, 1, 2, 4, and 6 h. Data are presented as means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05, **P < 0.01, ***P < 0.001. Data were obtained in at least three independent experiments.
Manipulation of GCH1 Activity in RAW264.7 by Lentiviral Knockdown.
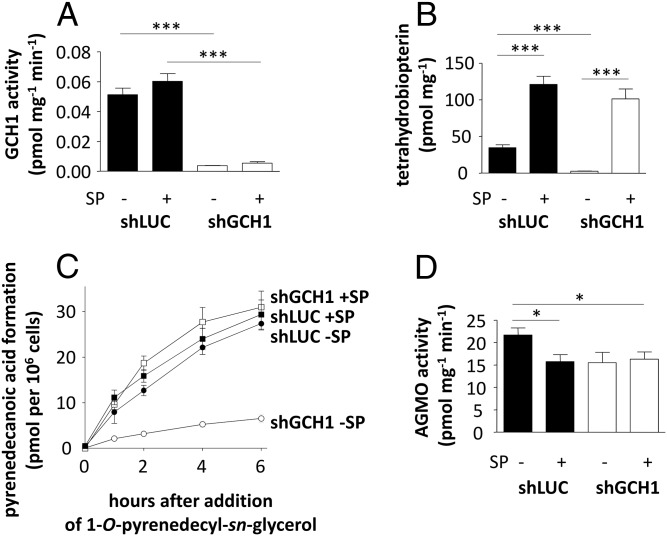
We next investigated the impact of GCH1 expression and tetrahydrobiopterin levels on AGMO activity in RAW264.7 in our live-cell assay. Lentiviral GCH1 knockdown resulted in 14.0-fold reduced GCH1 activity (P < 0.001; Fig. 3A), significantly diminished intracellular tetrahydrobiopterin levels (14.2-fold, P < 0.001; Fig. 3B), and a significant reduction in pyrenedecanoic acid formation (Fig. 3C) compared with shLUC. Addition of sepiapterin (SP), a GCH1-independent tetrahydrobiopterin precursor, readily restored the diminished tetrahydrobiopterin levels (P < 0.001; Fig. 3B) as well as pyrenedecanoic acid formation (Fig. 3C) without impact on GCH1 activity (Fig. 3A) measured in protein fractions freed from low-molecular weight compounds. Pyrenedecanoic acid formation per time unit (Fig. 3C) was highly significantly different between shLUC and shGCH1 (P < 0.001). AGMO activity in cell homogenates measured with saturating amounts of tetrahydrobiopterin was essentially unchanged (Fig. 3D), indicating that the AGMO protein can be expressed at normal levels in tetrahydrobiopterin-depleted cells but does not function any more due to lack of its cofactor. This is in contrast to phenylalanine hydroxylase protein, which needs the presence of cofactor to be stabilized (15).
Fig. 3.
Biochemical effects of GCH1 manipulation in RAW264.7 murine macrophages. (A and B) GCH1 activity (A) and tetrahydrobiopterin content (B) were analyzed in control line shLUC (filled bars) and shGCH1 (open bars) in the presence and absence of sepiapterin. (C) Alkylglycerol cleaved by living shLUC (filled symbols) and shGCH1 (open symbols) cells in the absence (circles) and presence (squares) of tetrahydrobiopterin precursor SP at 0, 1, 2, 4, and 6 h. (D) AGMO activity was quantified in homogenates of shGCH1 and shLUC as control in presence and absence of SP (n = 6) using a fluorescence-based HPLC microassay (10). Data are presented as means ± SEM. Statistical analysis was performed using Student’s t test. *P < 0.05, ***P < 0.001. Data were obtained in three independent experiments.
Impact of AGMO Activity Manipulation on IFN-γ/LPS–Induced Nitric Oxide Synthase and Selected Cytokines.
To investigate whether RAW264.7 cells show functional changes upon AGMO activity manipulation, we investigated the induction of nitric oxide synthesis by IFN-γ/LPS (Fig. S1). Whereas AGMO overexpression enhanced nitrite formation (3.3-fold, P < 0.001), iNOS protein (2.6-fold, P < 0.001), and NOS2 mRNA expression (6.0-fold, P < 0.001), AGMO knockdown led to the opposite trend (Fig. S1 A–C), which was significant for nitrite (1.6-fold reduction, P < 0.01; Fig. S1D). GCH1 knockdown showed effects comparable to AGMO knockdown, and could be partially reversed by sepiapterin treatment. Expression of two other selected IFN-γ/LPS–regulated genes, IL-1β and IL-1 receptor antagonist (IL-1RA), were also changed similarly to NOS2, whereas expression of transforming growth factor beta 1 (TGF-β1) remained unchanged by both IFN-γ/LPS treatment and AGMO manipulation (Fig. S1 E–G).
Targeted Analysis of 1-O-Hexadecyl-sn-Glycerol.
A targeted LC-MS/MS method was used to measure the effect of AGMO and GCH1 modulation on the levels of a physiological AGMO substrate, 1-O-hexadecyl-sn-glycerol (Fig. S2). The metabolite was just detectable in lipid extracts from the shLUC control cell line and AGMO-overexpressing cells but was increased in AGMO knockdown cells. Depletion of tetrahydrobiopterin in the shGCH1 line had a comparable effect (P < 0.05), resulting in strong accumulation of 1-O-hexadecyl-sn-glycerol. This effect could be reversed by supplementation with SP (Fig. S2).
Targeted Analysis of PAF and Lyso-PAF.
To test whether PAF and lyso-PAF levels were influenced by AGMO knockdown (shAGMO506) or overexpression (+huAGMO) as well as GCH1 knockdown in the absence and presence of SP (shGCH1 −SP, shGCH1 +SP), we performed a targeted analysis of these two molecules with the use of deuterated standards and extracts from comparable amounts of cellular material (Fig. S3 and Table S1). Surprisingly, no differences among the groups were detected.
Untargeted Lipidomic Analysis.
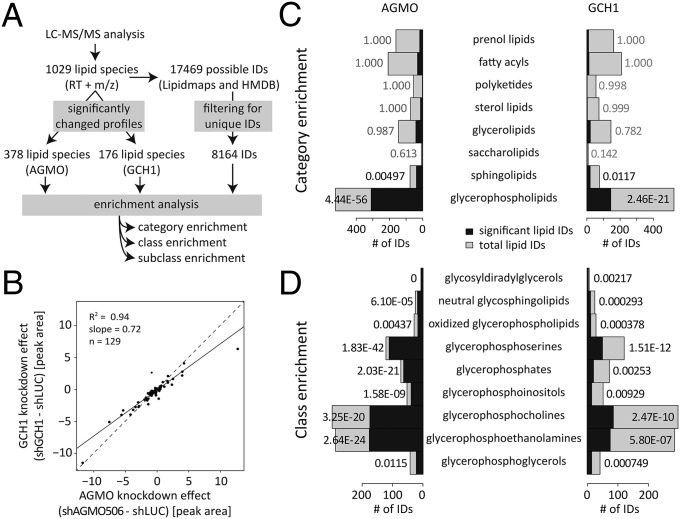
We detected 1,029 lipid species by exact mass and retention time, out of which 378 lipids had a significantly altered profile in the AGMO knockdown and 176 lipids had a significantly altered profile in the GCH1 knockdown (Fig. 4A). Principal-component analysis (PCA) showed a clear clustering (Fig. S4). One hundred and twenty-nine of the significantly altered levels of the 176 compounds associated with GCH1 knockdown were also significantly changed by AGMO knockdown, with a good positive correlation between the two independent approaches to modulate AGMO activity in the living cells (Fig. 4B).
Fig. 4.
Effects of AGMO and GCH1 knockdown on the RAW264.7 lipidome. (A) Scheme summarizing data processing for lipid identification and enrichment analysis. RT, retention time. (B) Comparison of the effects of AGMO and GCH1 knockdown. (C) Statistical enrichment analysis for LIPID MAPS categories of AGMO (shAGMO506, shLUC, and +huAGMO; Left) and GCH1 (shGCH1, shLUC, and shGCH1 +SP; Right). All eight LIPID MAPS lipid categories are shown. The total number of identified lipid species from each category is represented by the size of the bar, which is further classified between significantly changed profiles (P < 0.05, black color) and nonsignificantly changed profiles (gray color). (D) Statistical enrichment analysis for LIPID MAPS classes. Only significantly enriched classes are shown (P value of enrichment <0.01; number of identified lipid species >2). See Table S2 for details.
The most pronounced fold changes were seen in two groups of peaks. One group of signals with an m/z of around 350 was only detected in the AGMO and GCH1 knockdown cells (20- to 30-fold above the detection limit) but was missing in extracts from cells with functional AGMO (shLUC, +huAGMO, and shGCH1 +SP). These signals could be assigned to four species of alkylglycerols (with alkyl side chains of 16–22 carbon atoms) that are good substrates for AGMO. Surprisingly, there was a second group of lipid species completely absent in the AGMO or GCH1 knockdown cells and only present in cells with functional AGMO. These were reduced 10- to 50-fold to background levels by the AGMO knockdown and comprised two types of lipids. One was in an m/z range around 750, which could be assigned to glycosylated ceramides (five species). The other was in an m/z range of about 1,450, which could be assigned to cardiolipins (two species).
Lipid Enrichment Analysis.
Whereas an exact identification was possible for the free alkylglycerols, glycosylated ceramides, and cardiolipins, for many other peaks identification remained ambiguous despite the use of high-resolution mass spectrometry (Fig. 4A). We therefore applied an enrichment analysis to the data to examine whether a LIPID Metabolites And Pathways Strategy (LIPID MAPS) category, class, or subclass was found statistically enriched among the possible identifications of the significantly altered peaks (Fig. 4A). Among the eight categories of lipids, only glycerophospholipids and sphingolipids were significantly affected (Fig. 4C). The eight categories can be further subdivided into a total of 49 classes, 9 of which were significantly enriched (Fig. 4D). Eight of these significantly enriched classes originated from the glycerophospholipids category, and one class, the neutral glycosphingolipids, from the sphingolipid category, showed significant enrichment in both the AGMO and GCH1 experiments. In total, 151 subclasses emerge from the 49 classes listed in the LIPID MAPS nomenclature system. Out of these, 38 subclasses were found to be significantly enriched in either the AGMO or GCH1 experiment or both. A large number of these subclasses, namely 27 out of 38, belonged to the significantly enriched classes of the glycerophospholipids category. Two enriched subclasses belonged to two different classes (the neutral glycosphingolipids and phosphosphingolipids) of the sphingolipid category. The remaining nine subclasses belonged to other categories (Table S2). Because different molecular species of a given subclass displayed different experimental variability, not all molecular species of a subclass emerged as significantly changed (Table S2).
The category of glycerolipids comprising the class of monoradylglycerols that contains the subclass of monoalkylglycerols (i.e., the four species found to be most significantly changed; see above) did not appear to be significantly enriched, because alkylglycerols with side-chain carbon atoms 16–22 were not yet listed in the version of LIPID MAPS used.
Cluster Enrichment.
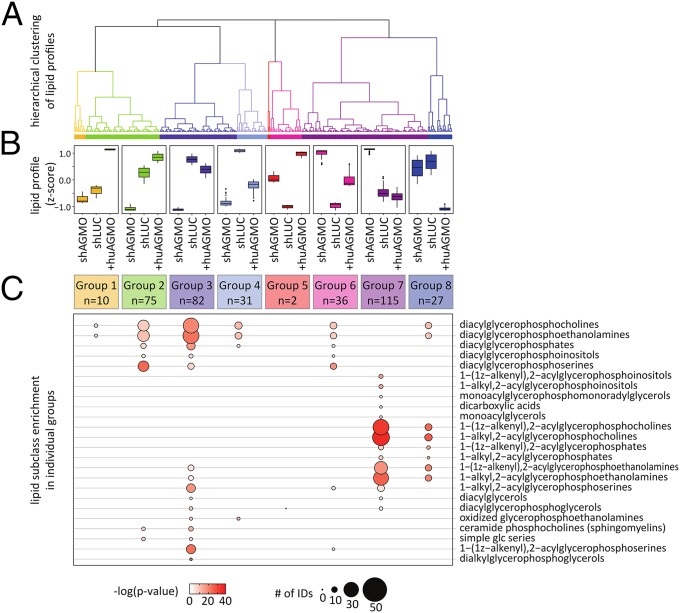
Hierarchical clustering analysis divided lipid profile changes in the cell lines with manipulated AGMO activity (shAGMO and +huAGMO; Fig. 5A) into eight different groups with distinct profile changes upon AGMO knockdown and overexpression (Fig. 5B). As an example, lipids combined in group 7 (which is the largest cluster, with 115 IDs) accumulated strongly in the AGMO knockdown (shAGMO; median of fold change: 2.9; 25th percentile: 1.9; 75th percentile: 6.6) but remained largely unchanged when AGMO was overexpressed (+huAGMO; median of fold change: 0.80; 25th percentile: 0.50; 75th percentile: 1.1). In contrast, lipids in groups 2 (n = 75; median of fold change: 0.42; 25th percentile: 0.089; 75th percentile: 0.64) and 3 (n = 82; median of fold change: 0.36; 25th percentile: 0.19; 75th percentile: 0.44) were strongly reduced when AGMO was knocked down (shAGMO). In +huAGMO cells, the same pattern of accumulation (group 1; n = 10; median of fold change: 1.5; 25th percentile: 1.4; 75th percentile: 2.0) and depletion (group 8; n = 27; median of fold change: 0.41; 25th percentile: 0.24; 75th percentile: 0.53) in lipid levels could be found.
Fig. 5.
Cluster analysis and subclass enrichment of lipid species affected by AGMO knockdown. (A) Unsupervised hierarchical clustering of measured lipids (LC-MS peaks) according to their coregulation in cells with manipulated AGMO activity (shAGMO506, shAGMO, shLUC, and +huAGMO). The clustered profiles were divided into eight different groups with distinctive behavior. (B) Lipid concentration changes in the respective groups. Each box is calculated from all lipids belonging to the respective group, each with peak area data from five biological replicates. Z scores were determined based on the mean and SD of all data for the respective lipid. (C) Lipid subclass enrichment for all lipid IDs corresponding to LC-MS peaks in groups 1–8. Only significantly enriched subclasses for AGMO modulation are shown (P value of enrichment <0.05; number of identified lipid species >3). Circle size represents the number of lipid species matching the respective subclass. Enrichment P values are indicated by the intensity of red filling of the circles. See Table S3 for details.
We performed enrichment analysis in all eight clusters to link lipid categories, classes, and subclasses with certain lipid profile changes (Fig. 5C; for details, see Table S3). Each group had a distinct pattern of enriched subclasses, implying that AGMO manipulation causes specific profile changes in different lipid subclasses. The subclass enrichment scores for group 7 suggest that AGMO knockdown led to an intracellular accumulation of a diverse range of 1-alkyl- and 1-alkenyl-phospholipids including phosphatidylcholines, phosphatidylethanolamines, phosphatidylinositols, and phosphatidic acids. Simultaneously, knockdown of AGMO resulted in reduction of the respective diacyl-phospholipid species (see groups 2 and 3; Fig. 5C).
Discussion
Here we show by shRNA-mediated knockdown of AGMO that activity manipulation affects a surprisingly high number of lipid species in the lipidome of RAW264.7 also beyond the class of ether lipids. Knockdown of tetrahydrobiopterin synthesis in intact cells greatly affects their ability to metabolize added labeled alkylglycerol and leads to changes in the lipidome comparable to direct AGMO knockdown (Fig. 4B). These data thus establish for the first time, to our knowledge, that intracellular tetrahydrobiopterin is an absolute requirement for alkylglycerol degradation. We find that both AGMO expression and activity are up-regulated by differentiation of primary murine bone marrow-derived macrophages to the M2 type, and confirm its down-regulation by inflammatory stimuli [M1 type (11)].
Because lyso-PAF had been suggested to be a substrate of AGMO (16), it was speculated that this enzyme might limit PAF synthesis in cells (8, 11, 17). However, the data presented in Fig. S3 clearly show that down-regulation of AGMO alone is not sufficient to account for the effect of LPS on PAF concentrations in RAW264.7 cells (11). Apparently, intact RAW264.7 cells compensate the altered flux of 1-O-alkyl-lysophosphatidylcholines through AGMO by synthesizing or degrading alkyl-acyl-phosphatidylcholines without changing the concentrations of 1-O-alkyl-lysophosphatidylcholines, presumably by a feedback regulatory mechanism. An ∼50% decline in the content of alkyl-acyl-phosphatidylcholine lipids as well as PAF had previously been observed in HEK293 cells upon introduction of recombinant AGMO by transfection (11). In these transiently transfected cells, however, the differences of AGMO activity were at least changed by two orders of magnitude, because HEK293 cells do not express AGMO without transfection. In contrast, in RAW264.7 cells, lentivirus-mediated stable overexpression increased AGMO activity only by about fivefold, because AGMO activity is already high in untreated cells.
One of the most striking effects of AGMO and GCH1 knockdown on the lipidome of RAW264.7 cells was the accumulation of free alkylglycerols, which are very low in untreated cells (Fig. S2). This demonstrates that free alkylglycerols are the main endogenous AGMO substrates. Pathways leading to free alkylglycerol in cells are poorly characterized and may require a lysophospholipase D and a phosphatase (18) or a lysophospholipase C similar or identical to ENPP6, which has been shown to yield alkenylglycerols (19).
Most of the changes in the lipidome resulting from AGMO or GCH1 knockdown can be interpreted as a result of free alkylglycerol accumulation, because they resemble the effect of feeding alkylglycerol to liver cells (20). Fig. S5 shows a simplified scheme of ether metabolism. Upon inhibition of AGMO, alkylglycerols accumulate and feed to ether lipid biosynthesis by known routes (reviewed in ref. 21) to alkyl-acyl- and alkenyl-acyl-phospholipids, at the expense of diacyl-phospholipids (Fig. 5C). We even see the striking lack of glycosylated ceramides, which had also been observed in alkylglycerol-fed liver cells. Another substance class eliminated from the lipidome by AGMO knockdown was cardiolipins, which had not been noted in the liver lipidome after alkylglycerol feeding (20). The mechanism of all these striking changes with no obvious connection to ether lipid pathways is currently unknown. An allosteric effect of alkylglycerols on enzymes of these distant pathways might be one plausible explanation. Cluster analysis groups 4–6 (Fig. 5C), where knockdown and overexpression of AGMO affect lipid levels of a subclass in a similar manner, cannot be readily interpreted by a simple hypothesis. It remains to be seen whether this results from experimental noise or a complex regulatory mechanism.
In RAW267.4 cells, we observed an enhancement of IFN-γ/LPS–mediated nitric oxide formation by AGMO overexpression and the reverse trend by AGMO or GCH1 knockdown. These results give a mechanistic explanation for the previously unexplained finding of an inhibition of the expression (and not only the activity) of NOS2 in murine macrophages by pharmacological inhibition of tetrahydrobiopterin biosynthesis (22). Genetic inactivation of GCH1 in mouse leukocytes, however, did not lead to a comparable effect (23), maybe due to compensatory mechanisms active in mouse development. The parallel behavior of NOS2 and two other selected IFN-γ/LPS–regulated genes in the RAW264.7 cells observed in this work, but not of a gene that is not regulated by this stimulus (Fig. S1), suggests that AGMO/tetrahydrobiopterin might affect the IFN-γ/LPS signal in a proinflammatory way. Because AGMO itself is down-regulated by the proinflammatory stimulus IFN-γ/LPS, this may be part of a complex regulatory network. Alterations of the plasma profile observed in tumor-bearing mice upon pharmacological inhibition of tetrahydrobiopterin biosynthesis (24) are compatible with such a proinflammatory mechanism of AGMO/tetrahydrobiopterin. On the other hand, submicromolar amounts of alkylglycerols, the substrates of AGMO, have been reported to act on lymphocytes by increasing IFN-γ and decreasing IL-4 formation (25), thus acting in a proinflammatory way, and their degradation by AGMO would be antiinflammatory. The findings on AGMO action and regulation presented in this work do not allow a simple assignment to pro- or antiinflammatory, a situation reminiscent of the partly conflicting observations made on the role of nitric oxide in the immune response (26). The complex interplay of the two pterin-dependent enzymes iNOS (high in M1) and AGMO (high in M2) observed here in RAW264.7 remains to be investigated in primary macrophages.
Genome-wide data approaches have collected diverse pieces of information on biological effects of AGMO. In congenital heart disease in humans, AGMO (TMEM195) is one of 277 candidate genes (27). In IGF/insulin-like signaling in Caenorhabditis elegans, AGMO (BE10.2) is a top hit out of 41 mediating daf2 gene action (28). Whether the prominent human SNP rs2191349 associated with high fasting glucose (29), which lies between the AGMO and the DGKB gene, mediates its effect via AGMO remains to be seen. We show here that AGMO activity alteration in cells, either by direct AGMO knockdown or overexpression or by manipulating the biosynthesis of its cofactor tetrahydrobiopterin, impacts on a wide range of lipids, some of which are known to influence signaling events. It is attractive to speculate that this might cause the diverse biological actions of AGMO found by genome-wide techniques.
Materials and Methods
Materials and methods are described briefly here. For a more detailed description, see SI Materials and Methods.
Cell-Culture Techniques.
All animal studies were conducted with ethical approval from the Local Ethical Review Committee and in accordance with UK Home Office regulations [Guidance on the Operation of the Animals (Scientific Procedures) Act, 1986]. Bone marrow-derived macrophages were prepared from C57bl/6J femur and tibia bone marrow and differentiated for 48 h with 10 ng/mL IFN-γ + 100 ng/mL LPS or with 20 ng/mL IL-4. RAW264.7 lines were cultured under standard conditions.
Biochemical Assays.
AGMO activity in homogenates was analyzed using a fluorescent alkylglycerol by HPLC (30). AGMO activity in live cells was measured by feeding a fluorescent alkylglycerol, collecting supernatants, and analyzing the fluorescent acid product released from the cells (14). GCH1 activity was analyzed according to ref. 31 by monitoring the amount of neopterin derivatives formed from GTP using HPLC. Cellular tetrahydrobiopterin was quantified using iodine oxidation of supernatants in acid and base (32) using HPLC with fluorescence detection. Gene expression was analyzed by quantitative PCR (qPCR), transcribing total RNA into cDNA, and analysis by qPCR using TaqMan technology. Western blots were performed by Bis-Tris or SDS gels, transferred to PVDF membranes, and stained with the respective antibodies.
Modulation of AGMO and GCH1 Activity in RAW264.7 Macrophages by Lentiviral Constructs.
Three individual shRNAs against AGMO (shAGMO506, shAGMO847, shAGMO1699) and one shRNA against GCH1 (shGCH1) were used to generate RAW264.7 knockdown cell lines according to ref. 33. A control line expressing shRNA against luciferase (shLUC) was generated similarly. An AGMO-overexpressing line (+huAGMO) was created using the ORF of human AGMO.
Lipid Extraction from Cells and LC-MS/MS Analysis.
Lipids were extracted with chloroform/methanol 2:1 (vol/vol), dried, and taken up in isopropanol/acetonitrile/water 2:1:1 (vol/vol/vol). For targeted LC-MS/MS analysis, lipids were separated on an Agilent 1290 LC system and analyzed with a triple-quadrupole Agilent 6460 mass spectrometer (for parameters see Table S4). For PAF and lyso-PAF, deuterated standards were added before extraction. For untargeted LC-MS, lipidomics lipids were separated by reversed-phase ultraperformance liquid chromatography (UPLC) and analyzed by high-resolution mass spectrometry in positive and negative electrospray ionization (ESI) mode using an ionKey/MS system with an ACQUITY UPLC M-Class, the ionKey source, and an iKey CSH C18 column (Waters Corporation) coupled to a SYNAPT G2-Si (Waters).
Data Processing and Analysis.
For the untargeted lipidomic approach, data processing was conducted using Progenesis QI (Nonlinear Dynamics). A combination of analysis of the variance (ANOVA) and multivariate statistics identified lipids most responsible for differences between our sample groups. Compounds were identified using the Human Metabolome Database, version 3.5 (HMDB; www.hmdb.ca) (34) and LIPID MAPS (www.lipidmaps.org) as well as fragmentation patterns, retention times, and ion mobility-derived collision cross-sections versus commercially available reference standards.
Lipid Enrichment Analysis in RAW264.7 with AGMO and GCH1 Modulation.
Compounds identified from HMDB version 3.5 and LIPID MAPS were categorized to LIPID MAPS categories/classes/subclasses. For significance criteria, a P value cutoff of 0.01 and a number of compounds for every category/class/subclass more than two were used.
Statistical Analysis.
Unless indicated otherwise, data are presented as means ± SEM. Data were compared by Student’s t test and one-way ANOVA with Bonferroni’s multiple comparison, as mentioned in the figure legends, using GraphPad Prism 5.01. P values <0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
The authors thank Petra Loitzl, Rita Holzknecht, and Nina Madl for expert technical help. This study was supported by the Austrian Science Funds (J3264 to K.W., J3341 to M.A.K., P22406 to E.R.W.), the Autonomous Province of Bolzano/Bozen–South Tyrol (Division for the Promotion of Education, Universities and Research) (K.W.), Tiroler Wissenschaftsfonds UNI-0404/958 (to M.A.K.), and the Austrian Academy of Science (DOC fellowship to M.A.K.). K.M.C. is supported by the British Heart Foundation (RE/08/004/23915 and RG/07/003/23133) and by the National Institute for Health Research Oxford Biomedical Research Centre.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. C.N.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1414887112/-/DCSupplemental.
References
- 1.Gorgas K, Teigler A, Komljenovic D, Just WW. The ether lipid-deficient mouse: Tracking down plasmalogen functions. Biochim Biophys Acta. 2006;1763(12):1511–1526. doi: 10.1016/j.bbamcr.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 2.Warne TR, Buchanan FG, Robinson M. Growth-dependent accumulation of monoalkylglycerol in Madin-Darby canine kidney cells. Evidence for a role in the regulation of protein kinase C. J Biol Chem. 1995;270(19):11147–11154. doi: 10.1074/jbc.270.19.11147. [DOI] [PubMed] [Google Scholar]
- 3.Zimmerman GA, McIntyre TM, Prescott SM, Stafforini DM. The platelet-activating factor signaling system and its regulators in syndromes of inflammation and thrombosis. Crit Care Med. 2002;30(Suppl 5):S294–S301. doi: 10.1097/00003246-200205001-00020. [DOI] [PubMed] [Google Scholar]
- 4.Prescott SM, Zimmerman GA, Stafforini DM, McIntyre TM. Platelet-activating factor and related lipid mediators. Annu Rev Biochem. 2000;69:419–445. doi: 10.1146/annurev.biochem.69.1.419. [DOI] [PubMed] [Google Scholar]
- 5.Watschinger K, et al. Identification of the gene encoding alkylglycerol monooxygenase defines a third class of tetrahydrobiopterin-dependent enzymes. Proc Natl Acad Sci USA. 2010;107(31):13672–13677. doi: 10.1073/pnas.1002404107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tietz A, Lindberg M, Kennedy EP. A new pteridine-requiring enzyme system for the oxidation of glyceryl ethers. J Biol Chem. 1964;239(12):4081–4090. [PubMed] [Google Scholar]
- 7.Taguchi H, Armarego WL. Glyceryl-ether monooxygenase [EC 1.14.16.5]. A microsomal enzyme of ether lipid metabolism. Med Res Rev. 1998;18(1):43–89. doi: 10.1002/(sici)1098-1128(199801)18:1<43::aid-med3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 8.Watschinger K, Werner ER. Alkylglycerol monooxygenase. IUBMB Life. 2013;65(4):366–372. doi: 10.1002/iub.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Werner ER, Blau N, Thöny B. Tetrahydrobiopterin: Biochemistry and pathophysiology. Biochem J. 2011;438(3):397–414. doi: 10.1042/BJ20110293. [DOI] [PubMed] [Google Scholar]
- 10.Werner ER, Hermetter A, Prast H, Golderer G, Werner-Felmayer G. Widespread occurrence of glyceryl ether monooxygenase activity in rat tissues detected by a novel assay. J Lipid Res. 2007;48(6):1422–1427. doi: 10.1194/jlr.D600042-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokuoka SM, Kita Y, Shindou H, Shimizu T. Alkylglycerol monooxygenase as a potential modulator for PAF synthesis in macrophages. Biochem Biophys Res Commun. 2013;436(2):306–312. doi: 10.1016/j.bbrc.2013.05.099. [DOI] [PubMed] [Google Scholar]
- 12.Benoit M, Desnues B, Mege JL. Macrophage polarization in bacterial infections. J Immunol. 2008;181(6):3733–3739. doi: 10.4049/jimmunol.181.6.3733. [DOI] [PubMed] [Google Scholar]
- 13.Misson P, van den Brûle S, Barbarin V, Lison D, Huaux F. Markers of macrophage differentiation in experimental silicosis. J Leukoc Biol. 2004;76(5):926–932. doi: 10.1189/jlb.0104019. [DOI] [PubMed] [Google Scholar]
- 14.Keller MA, et al. Studying fatty aldehyde metabolism in living cells with pyrene-labeled compounds. J Lipid Res. 2012;53(7):1410–1416. doi: 10.1194/jlr.D025650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thöny B, Ding Z, Martínez A. Tetrahydrobiopterin protects phenylalanine hydroxylase activity in vivo: Implications for tetrahydrobiopterin-responsive hyperphenylalaninemia. FEBS Lett. 2004;577(3):507–511. doi: 10.1016/j.febslet.2004.10.056. [DOI] [PubMed] [Google Scholar]
- 16.Lee TC, Blank ML, Fitzgerald V, Snyder F. Substrate specificity in the biocleavage of the O-alkyl bond: 1-Alkyl-2-acetyl-sn-glycero-3-phosphocholine (a hypotensive and platelet-activating lipid) and its metabolites. Arch Biochem Biophys. 1981;208(2):353–357. doi: 10.1016/0003-9861(81)90519-1. [DOI] [PubMed] [Google Scholar]
- 17.Snyder F. Platelet-activating factor: The biosynthetic and catabolic enzymes. Biochem J. 1995;305(Pt 3):689–705. doi: 10.1042/bj3050689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawasaki T, Snyder F. The metabolism of lyso-platelet-activating factor (1-O-alkyl-2-lyso-sn-glycero-3-phosphocholine) by a calcium-dependent lysophospholipase D in rabbit kidney medulla. Biochim Biophys Acta. 1987;920(1):85–93. doi: 10.1016/0005-2760(87)90314-6. [DOI] [PubMed] [Google Scholar]
- 19.Sakagami H, et al. Biochemical and molecular characterization of a novel choline-specific glycerophosphodiester phosphodiesterase belonging to the nucleotide pyrophosphatase/phosphodiesterase family. J Biol Chem. 2005;280(24):23084–23093. doi: 10.1074/jbc.M413438200. [DOI] [PubMed] [Google Scholar]
- 20.Bergan J, et al. The ether lipid precursor hexadecylglycerol causes major changes in the lipidome of HEp-2 cells. PLoS ONE. 2013;8(9):e75904. doi: 10.1371/journal.pone.0075904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watschinger K, Werner ER. Orphan enzymes in ether lipid metabolism. Biochimie. 2013;95(1):59–65. doi: 10.1016/j.biochi.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bogdan C, et al. 2,4-Diamino-6-hydroxypyrimidine, an inhibitor of tetrahydrobiopterin synthesis, downregulates the expression of iNOS protein and mRNA in primary murine macrophages. FEBS Lett. 1995;363(1-2):69–74. doi: 10.1016/0014-5793(95)00284-g. [DOI] [PubMed] [Google Scholar]
- 23.McNeill E, et al. Regulation of iNOS function and cellular redox state by macrophage Gch1 reveals specific requirements for tetrahydrobiopterin in NRF2 activation. Free Radic Biol Med. 2014;79C:206–216. doi: 10.1016/j.freeradbiomed.2014.10.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pickert G, et al. Inhibition of GTP cyclohydrolase attenuates tumor growth by reducing angiogenesis and M2-like polarization of tumor associated macrophages. Int J Cancer. 2013;132(3):591–604. doi: 10.1002/ijc.27706. [DOI] [PubMed] [Google Scholar]
- 25.Qian L, et al. Alkylglycerols modulate the proliferation and differentiation of non-specific agonist and specific antigen-stimulated splenic lymphocytes. PLoS ONE. 2014;9(4):e96207. doi: 10.1371/journal.pone.0096207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2(10):907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 27.Zaidi S, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498(7453):220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Samuelson AV, Carr CE, Ruvkun G. Gene activities that mediate increased life span of C. elegans insulin-like signaling mutants. Genes Dev. 2007;21(22):2976–2994. doi: 10.1101/gad.1588907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dupuis J, et al. DIAGRAM Consortium; GIANT Consortium; Global BPgen Consortium; Anders Hamsten on behalf of Procardis Consortium; MAGIC investigators New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Watschinger K, et al. Catalytic residues and a predicted structure of tetrahydrobiopterin-dependent alkylglycerol mono-oxygenase. Biochem J. 2012;443(1):279–286. doi: 10.1042/BJ20111509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werner ER, Wachter H, Werner-Felmayer G. Determination of tetrahydrobiopterin biosynthetic activities by high-performance liquid chromatography with fluorescence detection. Methods Enzymol. 1997;281:53–61. doi: 10.1016/s0076-6879(97)81008-7. [DOI] [PubMed] [Google Scholar]
- 32.Fukushima T, Nixon JC. Chromatographic analysis of pteridines. Methods Enzymol. 1980;66:429–436. doi: 10.1016/0076-6879(80)66485-4. [DOI] [PubMed] [Google Scholar]
- 33.Sigl R, Ploner C, Shivalingaiah G, Kofler R, Geley S. Development of a multipurpose GATEWAY-based lentiviral tetracycline-regulated conditional RNAi system (GLTR) PLoS ONE. 2014;9(5):e97764. doi: 10.1371/journal.pone.0097764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wishart DS, et al. HMDB 3.0—The Human Metabolome Database in 2013. Nucleic Acids Res. 2013;41(Database issue):D801–D807. doi: 10.1093/nar/gks1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.