Significance
Heteroplasmy is the existence of different mtDNA sequences within an individual due to somatic or inherited mutations, and it has been implicated in many mtDNA-related diseases, other diseases, cancer, and aging. However, little is known about how heteroplasmy varies across different tissues from the same individual; here, we analyzed heteroplasmy variation across the entire mtDNA genome in 12 tissues obtained at autopsy from each of 152 individuals. Our results suggest that in addition to neutral processes and negative selection, positive selection has an important influence on heteroplasmy: As individuals get older, specific alleles are selected for at specific nucleotide positions in specific tissues. The functional consequences of these positively selected somatic mutations may play a role in human health and disease.
Keywords: mtDNA, heteroplasmy, selection, human, tissue variation
Abstract
Heteroplasmy in human mtDNA may play a role in cancer, other diseases, and aging, but patterns of heteroplasmy variation across different tissues have not been thoroughly investigated. Here, we analyzed complete mtDNA genome sequences at ∼3,500× average coverage from each of 12 tissues obtained at autopsy from each of 152 individuals. We identified 4,577 heteroplasmies (with an alternative allele frequency of at least 0.5%) at 393 positions across the mtDNA genome. Surprisingly, different nucleotide positions (nps) exhibit high frequencies of heteroplasmy in different tissues, and, moreover, heteroplasmy is strongly dependent on the specific consensus allele at an np. All of these tissue-related and allele-related heteroplasmies show a significant age-related accumulation, suggesting positive selection for specific alleles at specific positions in specific tissues. We also find a highly significant excess of liver-specific heteroplasmies involving nonsynonymous changes, most of which are predicted to have an impact on protein function. This apparent positive selection for reduced mitochondrial function in the liver may reflect selection to decrease damaging byproducts of liver mitochondrial metabolism (i.e., “survival of the slowest”). Overall, our results provide compelling evidence for positive selection acting on some somatic mtDNA mutations.
Although mtDNA heteroplasmy (intraindividual variability in mtDNA sequences) was initially thought to be rare in humans, studies using next-generation sequencing platforms have documented extensive heteroplasmy at low levels (<2%) (1–4). Heteroplasmy is thought to represent an intermediate stage in the fixation of mtDNA mutations within an individual, and heteroplasmic mtDNA mutations have been implicated in various diseases, cancer, and aging (5–8). Heteroplasmies occur preferentially at positions with a high mutation rate (2, 9), suggesting that mutation and drift are the primary forces influencing heteroplasmy. In addition, elevated levels of nonsynonymous (NS) heteroplasmies within individuals relative to NS polymorphisms among individuals (2) suggest that there is negative selection against heteroplasmies that involve deleterious amino acid changes. In other words, deleterious amino acid changes can be observed as heteroplasmies as long as the allele frequency stays below a certain threshold and “normal” mitochondrial function is maintained; exceeding this threshold results in impaired mitochondrial function, as is commonly observed for disease-associated mtDNA mutations (10).
However, fundamental aspects about the nature of heteroplasmy remain unknown. For example, although certain nucleotide positions (nps) in the mtDNA genome are prone to heteroplasmy (2, 4, 11), it is not clear to what extent these positions simply have a high mutation rate, and thus are more prone to heteroplasmy across all tissues, vs. a role for tissue-specific processes in heteroplasmy (i.e., certain tissues may be more prone to heteroplasmy due to their metabolic requirements, rate of cellular turnover, etc.). Tissue-specific patterns of heteroplasmy are commonly observed with mtDNA mutations associated with disease and are thought to reflect the differing bioenergetic requirements of different tissues (10). Moreover, mice constructed to be heteroplasmic for different haplotypes show tissue-specific patterns of segregation over time (12, 13), suggesting positive selection related to mtDNA function in different tissues. However, these results may be specific to disease-associated mutations and to the artificial mouse constructs. A recent study of heteroplasmy in 10 tissues from two individuals found some tissue-specific patterns that suggested positive selection (11), but, currently, there is little evidence to support a significant role for positive selection as a force promoting heteroplasmy (5, 14).
Previous studies of variation in heteroplasmy across different tissues have been limited in terms of number of individuals and/or tissues studied (1, 2, 11, 15–19). Here, we present the results of the most comprehensive study to date (to our knowledge) of patterns of heteroplasmy in different tissues in humans. Our results suggest a surprisingly significant role for positive selection as a force influencing some somatic heteroplasmic mutations.
Results and Discussion
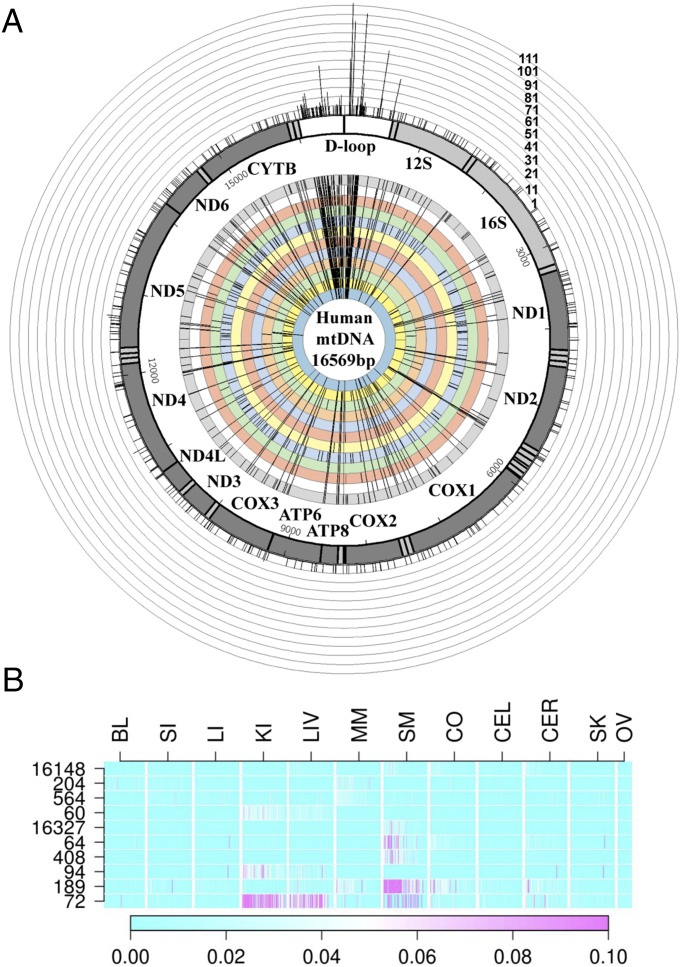
We obtained samples from each of 12 tissues obtained at autopsy from each of 152 individuals, ranging in age at death from 3 d after birth to 96 y (mean age = 56 y; SI Appendix, Fig. S1). Details concerning the major causes of death are provided in SI Appendix, Table S1. The complete mtDNA genome was sequenced from each sample (where sample refers to a single tissue from a single individual) to an average coverage of ∼3,458× (range: 46–36,444×; SI Appendix, Fig. S2).We used stringent criteria (Materials and Methods) to identify 4,577 heteroplasmies with an alternative allele frequency of at least 0.5%, distributed across 393 positions in the mtDNA genome (Fig. 1A and Dataset S1). When counting across all of the tissues from each individual, there are 1,198 heteroplasmies, of which 35% are observed in a single tissue from an individual, 22% are observed in two tissues from an individual, and 43% are observed in three or more tissues from an individual. Although we do not have information as to which heteroplasmies are inherited and which represent somatic mutations, these numbers provide a rough guide, because heteroplasmies in a single tissue are more likely to be somatic mutations, heteroplasmies in three or more tissues are more likely to be inherited (or to have occurred early in development), and heteroplasmies in two tissues are likely to be a mixture of inherited and somatic mutations (because of the high mutation rate for mtDNA, we expect some cases where somatic mutations have occurred at the same position independently in different tissues). Thus, around 43–65% of the heteroplasmies detected in this study are likely to be inherited.
Fig. 1.
Nonrandom distribution of heteroplasmies across positions and tissues. (A) Distribution and occurrence of heteroplasmy at 393 positions across the human mtDNA genome. The length of each bar in the outer ring indicates the number of heteroplasmies observed across all individuals and tissues, whereas the inner rings indicate heteroplasmies for each tissue, ordered as follows from the innermost to outermost ring: ovary, skin, cerebrum, cerebellum, cortex, skeletal muscle, myocardial muscle, liver, kidney, large intestine, small intestine, and blood. (B) Heat plot of the frequency and tissue distribution of heteroplasmy at HFH sites (the 10 sites with the highest number of observed heteroplasmies). Rows are HFH sites; columns are tissues; and each cell contains a line for each individual (ordered from left to right as oldest to youngest in age), with the line color indicating the frequency of heteroplasmy according to the scale. BL, blood; CEL, cerebellum; CER, cerebrum; CO, cortex; KI, kidney; LI, large intestine; LIV, liver; MM, myocardial muscle; OV, ovary; SI, small intestine; SK, skin; SM, skeletal muscle.
Tissue-Specific and Allele-Specific Characteristics of Heteroplasmic Mutations.
Heteroplasmies are not distributed at random across the mtDNA genome but occur preferentially at certain positions (Fig. 1A), mostly in the control region (CR), in agreement with previous studies (1, 2). We focus attention on the 10 positions where heteroplasmy is observed most frequently, because there are sufficient numbers of heteroplasmies for these positions to investigate tissue-related and age-related variation; these high-frequency heteroplasmies (HFHs) account for about 50% of the total number of observed heteroplasmies. Unexpectedly, these HFH positions are strongly tissue-related (Fig. 1B). For example, np 72 shows high levels of heteroplasmy in the liver and kidney, moderate levels in skeletal muscle, and low levels in all other tissues, whereas np 189 shows high levels of heteroplasmy in skeletal muscle but hardly any heteroplasmy in any other tissue. Thus, we do not find a general pattern of particular heteroplasmies at high frequency regardless of the type of tissue (implying a mutation-driven process), and we do not find an elevated frequency of all heteroplasmies in a particular tissue (implying a tissue-driven process). Instead, HFHs are localized to a particular position in a particular tissue, suggesting that heteroplasmy at this position is advantageous (or permitted) in this tissue but not in others.
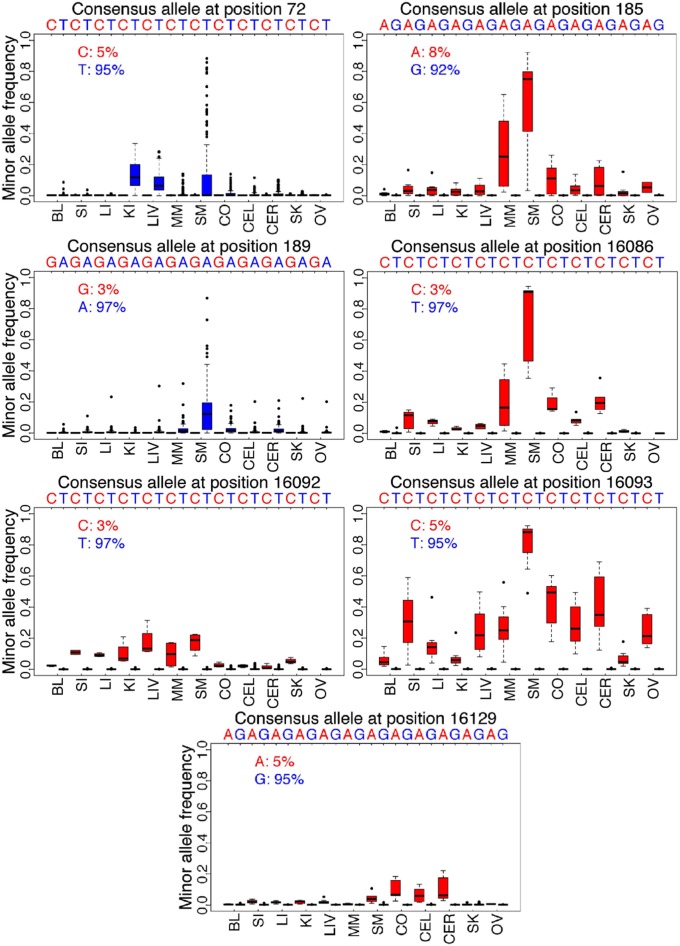
Another unexpected finding is that HFHs are strongly allele-dependent. We identified seven positions for which there is a significant difference in the level of heteroplasmy depending on the consensus allele at that position (Fig. 2). For example, heteroplasmy at np 189 is not only much more frequent in skeletal muscle than in other tissues, but heteroplasmy occurs significantly more frequently in all tissues when the consensus allele (based on the average allele frequency across all tissues) at this position is A than when it is G. Thus, heteroplasmies at np 189 are biased in favor of AT→GC changes and against GC→AT changes. In addition to the seven positions in Fig. 2, there are likely to be other positions with allele-related heteroplasmy, but the overall levels of heteroplasmy are too low to detect a significant effect. Note that at some nps in some tissues the level of heteroplasmy can reach more than 50%, which means that there is a different consensus allele at that np in that tissue than there is at that np in the other tissues. For example, at np 16,093 virtually all individuals with C as the consensus allele in other tissues have T as the consensus allele in skeletal muscle (Fig. 2). Also note that for several positions, heteroplasmy is preferentially observed when the consensus allele is the less frequent allele in the population (e.g., positions 185, 16,086, 16,092, 16,093, 16,129; Fig. 2). Heteroplasmy is thus not only determined by the specific np and the specific tissue but also by the specific consensus allele. Although some of these tissue-specific and allele-specific patterns have been observed in previous studies (SI Appendix, Table S2), the larger scale of the present study enables more thorough investigation of the nature and significance of these patterns. As discussed further below, our results imply positive selection for heteroplasmies involving specific alleles in specific tissues.
Fig. 2.
Allele-related heteroplasmy. Distribution of the alternative allele frequency at seven heteroplasmic sites that exhibit significant differences in levels of heteroplasmy, depending on the consensus allele (defined by the consensus sequence across all tissues for that individual). For each position and tissue, the frequency of heteroplasmy is shown for each of the two observed consensus alleles at that position; the frequency of each allele among all of the individuals is shown in the upper left corner of each plot. Blue indicates the allele that is in higher frequency among the consensus sequences, and red indicates the allele in lower frequency.
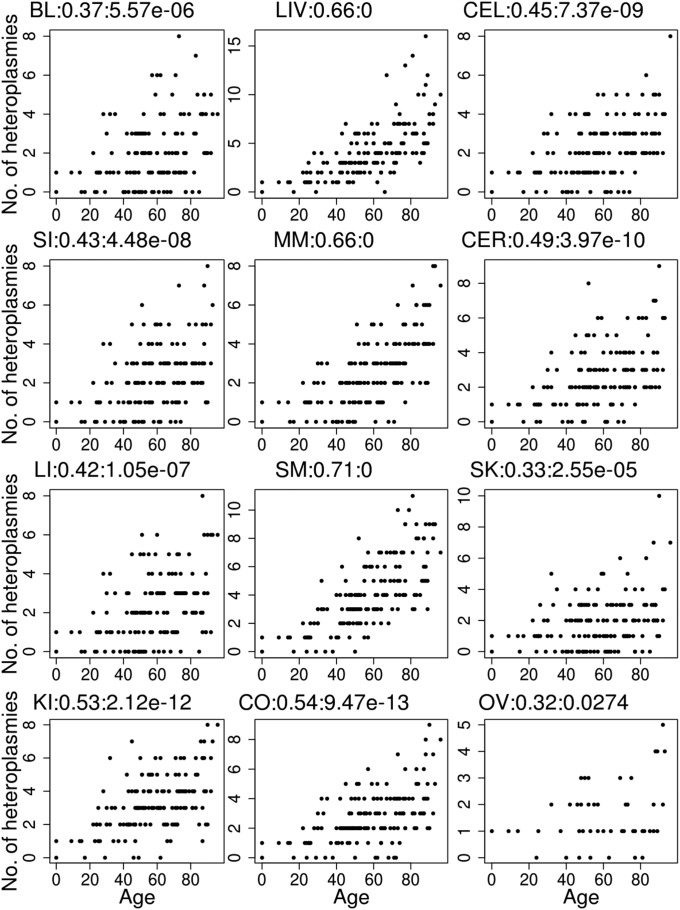
Heteroplasmies are not only tissue-related and allele-related but also age-related. There is a significant correlation between age and the occurrence of heteroplasmy in each individual tissue (Fig. 3), in agreement with previous studies (10, 20, 21). Moreover, many individual HFHs are also significantly correlated with age (SI Appendix, Fig. S3), either across all tissues (e.g., np 64, np 189) or only in some tissues (e.g., np 94, np 408). Overall, the strongest correlation observed is at np 564 in myocardial muscle (r = 0.85, P < 0.0001; SI Appendix, Fig. S3H), where age explains 70% of the heteroplasmy level variation among different individuals. Moreover, some of these age-related mutations are not independent of one another; we found nine pairs of sites in which heteroplasmies co-occurred within the same tissue more often than expected by chance (SI Appendix, Table S3). Previous studies did not find evidence for nonindependence of heteroplasmic sites (11), but these studies involved much smaller sample sizes, and thus lacked sufficient power to detect such associations. The potential functional consequences of interactions between different heteroplasmies in the same tissue need further investigation.
Fig. 3.
Correlation between the number of heteroplasmic sites and age for each tissue. The label for each plot indicates the tissue, correlation coefficient, and P value for the null hypothesis that the correlation coefficient is equal to 0.
The mutational spectrum of heteroplasmies differs from the mutational spectrum of polymorphism data (i.e., differences in the consensus sequences from the same individuals; P < 0.0001, Fisher exact test; SI Appendix, Table S4). The major difference is that transversions are observed more often in heteroplasmies than among consensus sequences, especially for low-level and tissue-specific heteroplasmies (TSHs) (SI Appendix, Table S4). We previously did not find a difference between the mutational spectrum of heteroplasmies vs. polymorphism data (2), which probably is because this previous study involved fewer samples and could only detect heteroplasmies with a relatively high alternative allele frequency. The elevated frequency of transversions among low-frequency heteroplasmies (SI Appendix, Table S4) is probably a further indication of negative selection against heteroplasmies that are likely to have functional consequences; that is, at least some of these transversions presumably have a detrimental effect on mtDNA function and so can be tolerated as low-frequency heteroplasmies but cannot reach “fixation” within an individual, and hence do not appear as polymorphisms among individuals.
Several lines of evidence, including verification of a subset of the results by an independent method, suggest that these unexpected findings regarding TSH are real and not experimental artifacts (SI Appendix, Supplementary Note 1). Mutational biases that might reflect biochemical processes, such as oxidation or deamination, cannot explain the results, because for the seven positions that exhibit significant allele-related heteroplasmy (Fig. 2), four involve preferences for TA→CG changes over CG→TA changes, whereas three involve preferences in the opposite direction, for CG→TA changes over TA→CG changes. The explanation that seems most likely for these tissue-related and allele-related aspects of heteroplasmy is that there is positive selection for the specific alternative allele in the specific tissue, as suggested previously (11). For example, when the consensus allele at np 72 is T, there is presumably selection in favor of a C in the kidney, liver, and (to a lesser extent) skeletal muscle. However, when the consensus allele is already a C, there is no such selection for, and hence no heteroplasmy, involving a T at this position. In some tissues, this selection must be quite strong, because the alternative allele in some individuals becomes the consensus allele in that tissue (Fig. 2).
The functional explanation for this putative tissue-related and allele-related selection remains unknown but may perhaps be related to the different metabolic roles fulfilled by the mitochondria in different tissues (22). Because all of the positions that exhibit this tissue/allele-related behavior are in the CR, it seems likely that regulation of mtDNA stability, replication, and/or transcription is involved, as suggested previously for CR heteroplasmies (11). Indeed, many of the HFH positions in the CR potentially alter the mtDNA secondary structure (SI Appendix, Fig. S4), which suggests that there could be an effect on regulation or control of mtDNA replication and/or transcription.
Excess of NS Heteroplasmies in Liver Tissue.
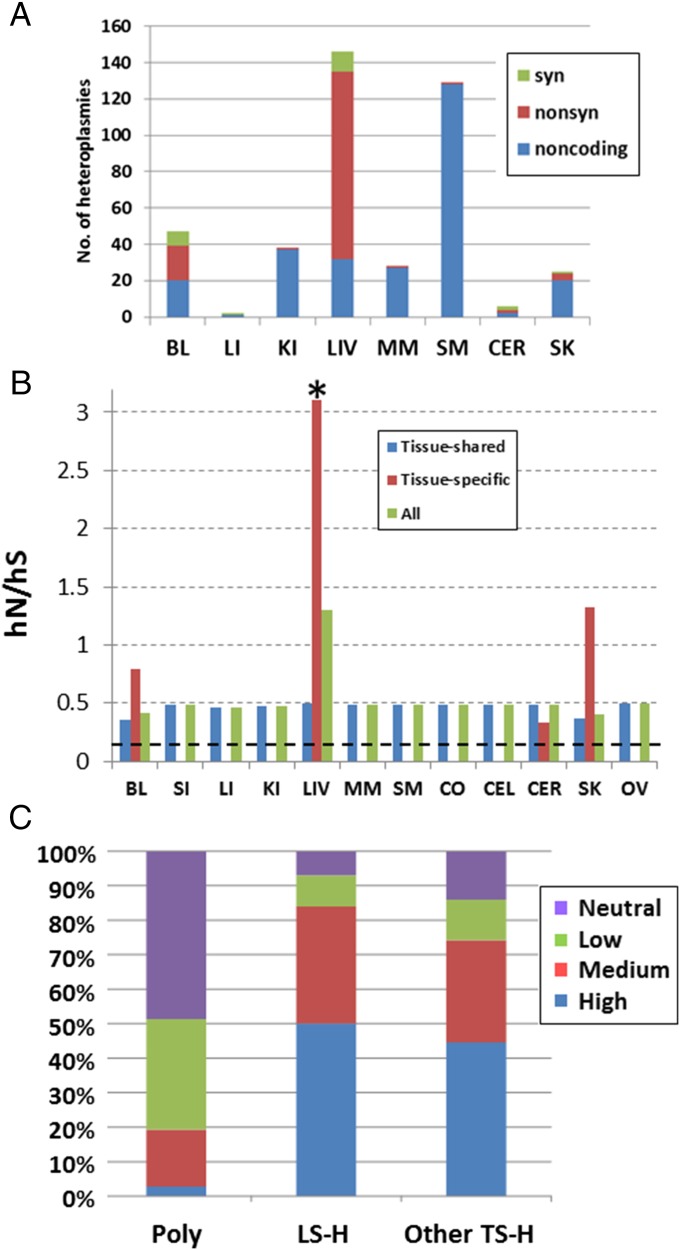
A further striking difference among tissues is observed in the distribution of TSHs in noncoding vs. coding regions, and nonsynonymous (NS) vs. synonymous (S) heteroplasmies. Overall, there are more TSHs in the liver than in any other tissue, and the ratio of TSHs in the CR vs. coding regions varies significantly among tissues (Fig. 4A and SI Appendix, Table S5). Furthermore, there are more TSHs involving NS changes in the liver than in any other tissue (Fig. 4A). To investigate the NS changes further, we calculated the hN/hS ratio, where hN is the number of NS heteroplasmies per NS site and hS is the number of S heteroplasmies per S site (see SI Appendix, Supplementary Note 2 for further discussion of this statistic), for both tissue-shared heteroplasmies and TSHs (Fig. 4B and SI Appendix, Table S5). The hN/hS ratio is elevated with respect to the pN/pS ratio (NS polymorphisms per NS site divided by S polymorphisms per S site, where the polymorphisms are identified from the consensus sequences of the 152 individuals) across all tissues (Fig. 4B). The higher hN/hS ratio is in keeping with previous observations, and probably reflects negative selection against some NS heteroplasmies that prevents them from reaching “fixation” within an individual, and thereby appearing as polymorphisms (2, 4). Surprisingly, the hN/hS ratio for TSHs in the liver is 3.11 (Fig. 4B and SI Appendix, Table S5); hN/hS ratios greater than 1 are indicative of positive selection. To assess the statistical significance of the observed ratio in the liver, we devised a resampling test that takes into account the mutational spectrum of heteroplasmies (details are provided in SI Appendix, Supplementary Note 2), and the empirical P value (based on 100,000 resamplings) is 0.00241. These results suggest that the hN/hS ratio in the liver is significantly greater than 1, and hence there is positive selection for NS heteroplasmies. Calculating the hN/hS ratio per gene (SI Appendix, Table S6) shows that this ratio is highest for ND5, but is substantially elevated for many mtDNA genes, indicating that the putative positive selection for amino acid changes is occurring for many, if not all, of the mtDNA genes in the liver. Moreover, the alternative allele at 82% of all NS heteroplasmies in the liver has never been observed as the consensus allele among human mtDNA sequences (SI Appendix, Table S5), and 84% of the liver-specific amino acid changes are predicted to have a medium or high risk of having an impact on protein function (Fig. 4C). The proportion of liver-specific amino acid changes that are medium/high-risk is slightly more than the proportion of medium/high-risk amino acid changes for other tissues (Fig. 4C), and significantly more than would be expected if NS heteroplasmies were occurring at random with respect to risk of a functional effect (SI Appendix, Supplementary Note 2).
Fig. 4.
Liver exhibits an excess number of NS heteroplasmies. (A) Number of TSHs for each tissue that are noncoding, nonsynonymous (nonsyn), or synonymous (syn). No TSHs were observed for the CO, CEL, SI, or OV. (B) hN/hS ratio for tissue-shared heteroplasmies, TSHs, and all heteroplasmies observed in the mtDNA coding region for each tissue. The dashed horizontal line indicates the pN/pS ratio (i.e., the ratio of NS polymorphisms per NS site to S polymorphisms per S site) observed among the consensus sequences of the individuals in this study. The asterisk indicates that the hN/hS ratio for liver-specific heteroplasmies is significantly greater than 1 (P = 0.00241), based on a resampling test (SI Appendix, Supplementary Note 2), indicating positive selection for NS heteroplasmies in the liver (also SI Appendix, Table S5). (C) Predicted risk (neutral, low, medium, or high) of functional impact for NS polymorphisms observed among the consensus sequences of the individuals in this study, compared with NS TSHs in the liver and in all other tissues. The x-axis labels are as follows: LS-H, liver-specific heteroplasmies; other TS-H, other TSHs; Poly, polymorphisms.
These results therefore suggest that there is positive selection for somatic mutations that decrease mitochondrial function in the liver. Many important metabolic processes occur in the liver that produce DNA-damaging byproducts, and mitochondrial turnover in the liver is quite rapid compared with other tissues (23). We hypothesize that positive selection for presumably detrimental somatic mutations in the liver could therefore reflect selection for a decrease in liver mitochondrial function as a means of avoiding excess damage to liver mitochondria. This putative positive selection for presumably detrimental mutations is consistent with the “survival of the slowest” hypothesis (24), which proposes that there may be positive selection for mutations that reduce mitochondrial damage. However, further studies are needed to investigate the impact of the excess hN/hS ratio and excess number of high/medium-risk somatic mutations on mitochondrial function in the liver.
Conclusions.
We have identified an unexpectedly strong tissue-related and allele-related specificity for mtDNA heteroplasmies. These heteroplasmies are also strongly age-related. In addition, we find a significantly elevated hN/hS ratio in liver tissue. These results indicate that positive selection, in addition to drift and negative selection (2, 4, 14, 25), plays a major role in influencing human mtDNA heteroplasmies. Some of the HFH alleles that we identify are also associated with tumors of specific tissues (SI Appendix, Table S2); whether this association simply reflects the increased frequency of that allele in that tissue during aging, or whether the HFH plays a role in causing the tumor, remains an open question. Evaluating the functional importance and potential impact of this presumed positive selection for somatic mtDNA mutations on human health and disease would now seem to be crucial.
Materials and Methods
Sample Preparation.
Tissue dissections (blood, cerebellum, cerebrum, cortex, kidney, large intestine, liver, myocardial muscle, ovary, skeletal muscle, skin, and small intestine) were performed during autopsy within 48–72 h of death and frozen at −20 °C. The collection of samples and informed consent procedures for this study were approved by the Ethics Commission of the Rheinische Friedrich Wilhelm University Medical Faculty and by the Ethics Commission of the University of Leipzig Medical Faculty. Approximately 25 mg of tissue was mechanically homogenized with a TissueRuptor (Qiagen), and DNA was extracted from the homogenized tissue (or, in the case of blood, from 100 μL) with QIAamp DNA Mini Kits (Qiagen) according to the manufacturer’s protocol.
Sequencing and Assembling.
A double-indexed Illumina sequencing library, with barcodes specific for each sample, was prepared from each extract as described previously (26). Up to 250 libraries were pooled in an equimolar ratio, and mtDNA sequences were enriched via in-solution capture (27). The capture-enriched library pools were then pooled in an equimolar ratio into a single pool (containing 1,732 individual libraries), which was then sequenced on eight lanes on an Illumina HiSeq2000 platform with 95 base pair, paired-end reads. Base-calling was done with IBIS (28). A total of 3.7 billion reads were obtained, which were then filtered as follows: (i) reads were discarded if the index sequence quality score was lower than 10 or if there were more than five bases with a quality score lower than 15, and (ii) forward reads and reverse reads were merged if they completely overlapped (i.e., length of reads greater than length of the molecule). After quality-filtering, reads were first mapped to the human genome (HG19) using the Burrows–Wheeler Aligner (BWA) (29) and those reads that mapped to the nuclear genome were removed. The remaining reads were then mapped to the Revised Cambridge Reference Sequence (rCRS) (30), with the first 550 base pairs copied to the end of the sequence to permit mapping to a circular genome, followed by realignment with the Genomes Analysis Toolkit (GATK) (31). Duplicate reads were then removed by SAMtools (32), along with reads with a mapping quality score lower than 20 and a base quality score lower than 20. The preliminary consensus sequence was called using the majority rule [with N (no call) if the alternative allele frequency was greater than 0.3]. The rCRS was then replaced by the consensus sequence, and the mapping and assembly were redone by means of the same procedure. A total of 48 samples did not have sufficient reads and were discarded from further analysis; the average sequencing coverage for the remaining individuals was 3,458 (range: 46–36,444). The raw sequencing data are publicly available from the European Nucleotide Archive’s Sequence Read Archive through accession no. PRJEB5480.
Heteroplasmy Detection.
In this study, we focus on heteroplasmies involving base substitutions at a single np; heteroplasmies involving indels require different strategies for reliable detection and will be the subject of a subsequent study. A two-stage process was used to identify substitution heteroplasmies. First, we required a minimum alternative allele frequency of 2% on each strand, at least three reads in each direction with the alternative allele, a detecting low-level mutations by utilizing the resequencing error profile of the data (DREEP) quality score (33) of at least 10, no indels in the 3 bp of the flanking sequence in each direction, and a total sequencing coverage that is at least 50× and that is within 20–200% of the genome average. We also excluded the poly-C runs (np 302–316, np 513–526, np 566–573, and np 16,181–16,194). Potential contamination was detected using the following criteria. First, we determined if the alternative alleles at five or more heteroplasmic positions could define an alternative haplogroup; 28 samples were identified by this criterion and removed from further analysis. We then determined if more than 50% of the heteroplasmies identified in a particular sample could be explained by contamination from another sample in the same library, and we also required that more than 70% of the nucleotides from the potential contamination donor at the donor–recipient discrepant positions were detectable. This latter requirement is important, because many heteroplasmies will overlap with polymorphic positions due to the high mutation rate at some positions; for example, sample 1 might have six heteroplasmies, three of which could be derived from sample 2. However, the consensus sequences from sample 1 and sample 2 differ at 20 positions; thus, if the heteroplasmies in sample 1 are derived by contamination from sample 2, we would expect to detect heteroplasmy at more of the positions at which sample 1 and sample 2 differ. An additional three samples were detected by this second criterion, for a total of 31 samples (1.7% of the data, because each sample is a single tissue from a single individual) removed from analysis because of potential contamination. In a previous study (33), this pipeline proved to be highly reliable when applied to the same type of data (false-positive discovery rate is lower than 0.01).
With this procedure, we identified 2,412 primary heteroplasmies. For each primary heteroplasmy in a specific tissue and individual, we then examined the reads covering that np for every other tissue in that individual, and counted this np as a secondary heteroplasmy in that tissue if there was an alternative allele identical to the allele for the primary heteroplasmy with a frequency of at least 0.5% and present in at least three reads in each direction. This procedure identified 2,165 additional heteroplasmies, for a total of 4,577 heteroplasmies at 393 positions (Dataset S1).
Confirmation by Droplet Digital PCR.
We used droplet digital PCR (ddPCR) (34) to confirm independently a subset of the heteroplasmies observed via sequencing. Briefly, the method involves setting up a conventional PCR assay in a 20-μL volume with target DNA, primers, polymerase, nucleotides, buffer, and two allele-specific probes (one for each allele at the position of interest) labeled with different fluorescent tags. The PCR is then partitioned into ∼20,000 individual emulsion droplets, each containing, on average, one template DNA molecule. PCR is then carried out on the individual droplets, and the fluorescence from each droplet is read. Droplets containing either no template molecule or more than one template molecule are discarded, and the output is a count of the number of template molecules carrying either the consensus or the alternative allele. The alternative allele frequency is then determined by dividing the number of template molecules with the alternative allele by the total number of template molecules. We chose eight positions for confirmation (SI Appendix, Tables S7 and S8); these positions included four positions from the CR that exhibited varying degrees of tissue specificity and a wide range of alternative allele frequencies and four positions at which NS heteroplasmies were observed in liver tissue. The ddPCR reactions containing allele-specific probes were prepared and analyzed on a QX200 Droplet Digital PCR System (Bio-Rad) according to protocols supplied by the manufacturer. The annealing temperature was 60 °C in all reactions, and PCR was carried out for 40 cycles.
Secondary Structure.
The secondary structure of mtDNA with different nucleotides was estimated for seven positions: 72, 185, 189, 16,083, 16,092, 16,093, and 16,129. We used Mfold software (35) and np 1–500 as the input sequence for np 72, np 185, and np 189; for the other positions, the input sequence was np 16,024–16,569. In each case, the context sequence was retrieved from one of the samples showing the allele-dependent pattern. If multiple predictions were given for one input, the one showing the smallest change was chosen.
Estimation of hN/hS.
The software KaKs_Calculator (https://code.google.com/p/kaks-calculator/) was used to estimate the number of NS and S sites (based on the human mtDNA genetic code). Heteroplasmies observed at the same position in different individuals were regarded as multiple events, whereas polymorphisms at the same position in different individuals were regarded as a single event.
Prediction of the Impact of NS Mutations.
The functional impact of each NS heteroplasmy/polymorphism was predicted by Mutationassessor (36), release 2. Mutations were classified as high risk, medium risk, low risk, or neutral based on the evolutionary conservation of the affected amino acid in protein homologs.
Identification of New Mutations.
All of the complete mtDNA sequences (9,402 entries) archived on PhyloTree, version 15 (www.phylotree.org/) were downloaded and compared with the rCRS. All nucleotide changes relative to the rCRS, including all haplogroup-defining mutations, were regarded as existing mutations, whereas all other mutations identified in this study but not present in the list of existing mutations were defined as new mutations.
Construction of the Tissue Tree Based on the Sharing of Heteroplasmies.
The distance between tissue A and tissue B, based on the frequency of the alternative allele at shared heteroplasmies, was estimated using the following formula:
where N is the total number of heteroplasmies where either tissue A or tissue B or both present the same alternative allele, fre(A) is the frequency of the alternative allele in tissue A, and fre(B) is the frequency of the alternative allele in tissue B. A neighbor-joining tree was estimated from the distance matrix using programs in the software package PHYLIP (evolution.genetics.washington.edu/phylip.html). TSHs were excluded from this analysis.
Supplementary Material
Acknowledgments
We thank the medical staff at the Institüt für Rechtsmedizin (University of Bonn) for assistance with the sampling; the Sequencing and Bioinformatics Groups in the Department of Evolutionary Genetics (Max Planck Institute for Evolutionary Anthropology) for producing the sequence data; A. Butthoff, E. Macholdt, and V. Lede for technical assistance; M. Meyer for assistance with the ddPCR experiments; and M. Mörl for helpful discussion. This work was funded by the Institüt für Rechtsmedizin in Bonn and by the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the European Nucleotide Archive’s Sequence Read Archive (accession no. PRJEB5480).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1419651112/-/DCSupplemental.
References
- 1.He Y, et al. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464(7288):610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li M, et al. Detecting heteroplasmy from high-throughput sequencing of complete human mitochondrial DNA genomes. Am J Hum Genet. 2010;87(2):237–249. doi: 10.1016/j.ajhg.2010.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Payne BA, et al. Universal heteroplasmy of human mitochondrial DNA. Hum Mol Genet. 2013;22(2):384–390. doi: 10.1093/hmg/dds435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye K, Lu J, Ma F, Keinan A, Gu Z. Extensive pathogenicity of mitochondrial heteroplasmy in healthy human individuals. Proc Natl Acad Sci USA. 2014;111(29):10654–10659. doi: 10.1073/pnas.1403521111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chinnery PF, Hudson G. Mitochondrial genetics. Br Med Bull. 2013;106:135–159. doi: 10.1093/bmb/ldt017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greaves LC, Reeve AK, Taylor RW, Turnbull DM. Mitochondrial DNA and disease. J Pathol. 2012;226(2):274–286. doi: 10.1002/path.3028. [DOI] [PubMed] [Google Scholar]
- 7.Lombès A, Auré K, Bellanné-Chantelot C, Gilleron M, Jardel C. Unsolved issues related to human mitochondrial diseases. Biochimie. 2014;100:171–176. doi: 10.1016/j.biochi.2013.08.012. [DOI] [PubMed] [Google Scholar]
- 8.Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012;12(10):685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoneking M. Hypervariable sites in the mtDNA control region are mutational hotspots. Am J Hum Genet. 2000;67(4):1029–1032. doi: 10.1086/303092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Med. 2013;3(10):a021220. doi: 10.1101/cshperspect.a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Samuels DC, et al. Recurrent tissue-specific mtDNA mutations are common in humans. PLoS Genet. 2013;9(11):e1003929. doi: 10.1371/journal.pgen.1003929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jenuth JP, Peterson AC, Shoubridge EA. Tissue-specific selection for different mtDNA genotypes in heteroplasmic mice. Nat Genet. 1997;16(1):93–95. doi: 10.1038/ng0597-93. [DOI] [PubMed] [Google Scholar]
- 13.Burgstaller JP, et al. MtDNA segregation in heteroplasmic tissues is common in vivo and modulated by haplotype differences and developmental stage. Cell Reports. 2014;7(6):2031–2041. doi: 10.1016/j.celrep.2014.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Durham SE, Samuels DC, Chinnery PF. Is selection required for the accumulation of somatic mitochondrial DNA mutations in post-mitotic cells? Neuromuscul Disord. 2006;16(6):381–386. doi: 10.1016/j.nmd.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 15.Chinnery PF, et al. Nonrandom tissue distribution of mutant mtDNA. Am J Med Genet. 1999;85(5):498–501. [PubMed] [Google Scholar]
- 16.Krjutškov K, et al. Tissue-specific mitochondrial heteroplasmy at position 16,093 within the same individual. Curr Genet. 2014;60(1):11–16. doi: 10.1007/s00294-013-0398-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu VW, Zhang C, Nagley P. Mutations in mitochondrial DNA accumulate differentially in three different human tissues during ageing. Nucleic Acids Res. 1998;26(5):1268–1275. doi: 10.1093/nar/26.5.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanske S, et al. Varying loads of the mitochondrial DNA A3243G mutation in different tissues: Implications for diagnosis. Am J Med Genet A. 2004;130A(2):134–137. doi: 10.1002/ajmg.a.30220. [DOI] [PubMed] [Google Scholar]
- 19.Thèves C, et al. Detection and quantification of the age-related point mutation A189G in the human mitochondrial DNA. J Forensic Sci. 2006;51(4):865–873. doi: 10.1111/j.1556-4029.2006.00163.x. [DOI] [PubMed] [Google Scholar]
- 20.Sondheimer N, et al. Neutral mitochondrial heteroplasmy and the influence of aging. Hum Mol Genet. 2011;20(8):1653–1659. doi: 10.1093/hmg/ddr043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams SL, Mash DC, Züchner S, Moraes CT. Somatic mtDNA mutation spectra in the aging human putamen. PLoS Genet. 2013;9(12):e1003990. doi: 10.1371/journal.pgen.1003990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernández-Vizarra E, Enríquez JA, Pérez-Martos A, Montoya J, Fernández-Silva P. Tissue-specific differences in mitochondrial activity and biogenesis. Mitochondrion. 2011;11(1):207–213. doi: 10.1016/j.mito.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 23.Miwa S, Lawless C, von Zglinicki T. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: Application of a simple dynamic model. Aging Cell. 2008;7(6):920–923. doi: 10.1111/j.1474-9726.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Grey AD. A proposed refinement of the mitochondrial free radical theory of aging. BioEssays. 1997;19(2):161–166. doi: 10.1002/bies.950190211. [DOI] [PubMed] [Google Scholar]
- 25.Stewart JB, Freyer C, Elson JL, Larsson NG. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat Rev Genet. 2008;9(9):657–662. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 26.Kircher M, Sawyer S, Meyer M. Double indexing overcomes inaccuracies in multiplex sequencing on the Illumina platform. Nucleic Acids Res. 2012;40(1):e3. doi: 10.1093/nar/gkr771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maricic T, Whitten M, Pääbo S. Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS ONE. 2010;5(11):e14004. doi: 10.1371/journal.pone.0014004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kircher M, Stenzel U, Kelso J. Improved base calling for the Illumina Genome Analyzer using machine learning strategies. Genome Biol. 2009;10(8):R83. doi: 10.1186/gb-2009-10-8-r83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrews RM, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23(2):147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 31.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. 1000 Genome Project Data Processing Subgroup The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li M, Stoneking M. A new approach for detecting low-level mutations in next-generation sequence data. Genome Biol. 2012;13(5):R34. doi: 10.1186/gb-2012-13-5-r34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hindson BJ, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: Application to cancer genomics. Nucleic Acids Res. 2011;39(17):e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.