Significance
FLOWERING LOCUS T (FT) acts as a mobile floral activator that is synthesized in leaf and transported to shoot apex. A Rice FT-like protein, Heading date 3a (Hd3a), requires interaction with 14-3-3 proteins and transcription factor FD to induce flowering. We confirm that Hd3a and its interactors, as well as their transcriptional target, coexist in the shoot apex at the appropriate time during floral transition. RNA-sequencing analysis of shoot apices from wild-type and RNA-interference plants for FT-like genes showed that 58% of classified transposable elements are transcribed, and >200 are down-regulated in response to FT-like. Our results indicate a link between reproductive development and transposon behavior in the shoot apical meristem, supporting and extending recent evidence for such a link during gametophyte development.
Keywords: floral transition, FT-like proteins, shoot apical meristem, transposable elements, rice
Abstract
Floral induction is a crucial developmental step in higher plants. Florigen, a mobile floral activator that is synthesized in the leaf and transported to the shoot apex, was recently identified as a protein encoded by FLOWERING LOCUS T (FT) and its orthologs; the rice florigen is Heading date 3a (Hd3a) protein. The 14-3-3 proteins mediate the interaction of Hd3a with the transcription factor OsFD1 to form a ternary structure called the florigen activation complex on the promoter of OsMADS15, a rice APETALA1 ortholog. However, crucial information, including the spatiotemporal overlap among FT-like proteins and the components of florigen activation complex and downstream genes, remains unclear. Here, we confirm that Hd3a coexists, in the same regions of the rice shoot apex, with the other components of the florigen activation complex and its transcriptional targets. Unexpectedly, however, RNA-sequencing analysis of shoot apex from wild-type and RNA-interference plants depleted of florigen activity revealed that 4,379 transposable elements (TEs; 58% of all classifiable rice TEs) were expressed collectively in the vegetative and reproductive shoot apex. Furthermore, in the reproductive shoot apex, 214 TEs were silenced by florigen. Our results suggest a link between floral induction and regulation of TEs.
Floral induction has been studied extensively over the years, and the use of molecular genetics in the model species Arabidopsis thaliana has been particularly successful in identifying genes and genetic pathways that are involved in flower development (1–3). Several key flowering genes sense environmental conditions and activate the proper expression of genes that are essential for floral induction. Once floral induction occurs, further key genes positively and negatively regulate each other in the shoot apex and activate flower identity genes, eventually leading to the formation of flowers (1–3).
One of the critical molecules for floral induction is florigen, a mobile floral activator that is produced in leaves and transported to the shoot apex. Many studies now indicate that proteins encoded by Arabidopsis FLOWERING LOCUS T (FT) and its orthologs in other species are florigens (4–8). Transcription of florigen genes is tightly regulated by environmental conditions, and florigen protein is transported from leaves to the shoot apex where it induces the expression of downstream genes to ensure proper flower development. Despite its important role in floral induction, many questions about florigen remain unresolved. They include its molecular function, mechanism of transport, localization in the shoot apex, and the identities of its downstream genes.
Rice is a short-day (SD) plant and has two florigens, Heading date 3a (Hd3a) and RICE FLOWERING LOCUS T1 (RFT1), which predominate, respectively, under SD and long-day (LD) conditions (7, 9). When GFP fusions of Hd3a and RFT1 are expressed under their own promoters, GFP fluorescence is clearly detected in the shoot apex (7, 9). We have shown (10) that the interaction of Hd3a with OsFD1, a basic leucine-zipper domain-containing transcription factor orthologous to Arabidopsis FD (11), is mediated by 14-3-3 proteins (GF14s) and that Hd3a–14-3-3–OsFD1 forms a ternary structure called the florigen activation complex (FAC) on C-box elements in the promoter of OsMADS15, a rice AP1 ortholog (12).
Individual meristems of rice can be manually dissected with relative ease under a microscope (Fig. S1). We exploited this ability to address two questions about florigen (i) How are Hd3a and its direct target OsMADS15 distributed in the inflorescence meristem during flower development? (ii) Which genes are regulated by Hd3a in the shoot apex during the early stage of floral induction?
In this study, we made a detailed analysis of Hd3a and OsMADS15 protein localization in the shoot apical meristem (SAM) during flower formation. Then, while addressing the second question, we made unexpected findings that link transposable element (TE) activation and silencing with floral induction in the rice shoot apex. Our results highlight TEs as potential regulators of floral induction and flower development, and we discuss our findings in relation to other known types of TE-related regulation in plants.
Results
Hd3a Protein Localization in the SAM During Flower Formation and the Role of Hd3a in Inflorescence Development.
To study Hd3a localization in the SAM during floral induction and inflorescence development, we used stable transgenic rice plants expressing Hd3a–GFP under the control of the endogenous Hd3a promoter (pHd3a:Hd3a–GFP). In a previous analysis of pHd3a:Hd3a–GFP plants, we could detect GFP fluorescence in the shoot apex 1–3 d after the transition from the vegetative to the reproductive stage, but were unable to monitor either earlier stages of the floral transition or later stages, when inflorescence branch primordia form. We obtained T2 or T3 progeny of pHd3a:Hd3a–GFP plants for this study and observed their shoot apices at 20–27 d after germination (DAG) for vegetative meristems and at 32–38 DAG for reproductive meristems. The developmental stage of each shoot apex was identified morphologically and classified as vegetative (V) through four stages of the early reproductive phase (R1–R4) and later floral development (Fig. 1).
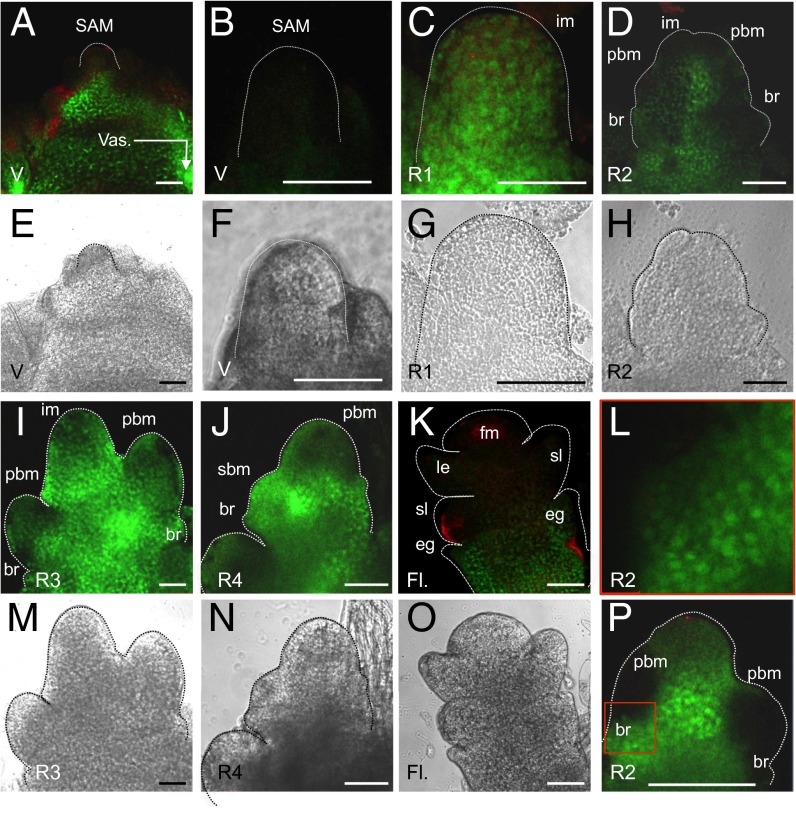
Fig. 1.
Hd3a protein localization in the rice shoot apex during the transition from vegetative to reproductive stage. GFP fluorescence (A–D, I–L, and P) and bright-field images (E–H and M–O) of SAMs in pHd3a:Hd3a-GFP transgenic plants are shown. Development of the mature vegetative meristem (at 20–27 DAG) and different stages (R1–R4) of the inflorescence meristem (observed at 30–38 DAG and assigned to each stage by morphological characteristics) and FM (observed at >40–45 DAG) were analyzed. (A, B, E, and F) V, vegetative-stage plants at 27 DAG. At this time the plants display a meristem before the transition to reproductive development. Vas, stem vasculature. (C and G) R1, initial stage of floral transition with first bract formation. im, inflorescence meristem. (D, H, L, and P; L is an enlarged image of the area indicated by the red square in P) R2, early stage of primary branch meristem initiation. br, bract; pbm, primary branch meristem. (I and M) R3, late stage of primary panicle branch initiation. (J and N) R4, final stage of primary branch formation and initiation of secondary branch meristem (sbm). (K and O) Fl., floral organ development after R4. e.g., empty glume; fm, floret meristem; le, lemma; sl, sterile lemma. The red staining in the images indicates autofluorescence. (Scale bars, 50 µm.)
In the V stage, the SAM was devoid of GFP (Fig. 1 A, B, E, and F), but GFP signal was detected in the upper part of the stem vasculature beneath the meristem at 25–27 DAG before flowering (7) (Fig. 1 A and E). This finding indicates that, before Hd3a–GFP protein arrives at the SAM, it moves through the regions just beneath the SAM (13). At the transition from V to R1 (reproductive meristem) stages, when the SAM undergoes accelerated cell division for longitudinal expansion (14), GFP fluorescence was then clearly visible throughout the meristem (Fig. 1 C and G). At the next stage (R2), several bract primordia and primary branch meristems began to develop in the meristem (Fig. 1 D, H, and P). GFP signal was observed in the entire shoot apex, but was slightly weaker at the top of the inflorescence meristem and in newly developing primary branch meristems (Fig. 1 D, H, and P). At the R3 stage, GFP was still observed throughout the shoot apex, from which multiple primary branch meristems emerged (Fig. 1 I and M). At R4, Hd3a–GFP protein continued to accumulate in the primary branch meristems and the lower part of the shoot apex, whereas the signal became weak within the meristem (Fig. 1 J and N). Furthermore, the appearance of GFP in secondary branch meristems (Fig. 1 J and N) suggested that, in addition to its role in floral induction, Hd3a participates in inflorescence development. The relatively weak accumulation of Hd3a in the inflorescence meristem may help to keep the meristem in an undifferentiated state and establish the inflorescence architecture. After the R4 stage, branch meristem transformed into floral meristem (FM; spikelet meristem and floret meristem in rice), and floral organ differentiation began (Fig. 1 K and O). GFP fluorescence was detected in the bract-like organs, called empty glumes, at the lower part of the flowers, but its absence in the upper organs indicated a restricted localization of Hd3a in the floral organs (Fig. 1 K and O). GFP fluorescence appeared as diffuse and speckle-like structures in the meristem cells (Fig. 1 L and P), suggesting that Hd3a is present in both cytoplasm and nucleus. These results indicate that Hd3a protein persists in the inflorescence meristems during the floral transition.
Hd3a mRNA is absent from the vegetative and R1–R2 inflorescence meristems (7), whereas Hd3a protein was detected in these meristems (Fig. 1 C and D) (7), indicating the transport of Hd3a from leaves to the meristems. To examine whether this transportation continues at the later stages of the inflorescence development, we compared the level of Hd3a mRNA in apical tissue with that in leaves at 40 DAG, corresponding to the time when the meristems reach the R4 and FM stages. Hd3a mRNA accumulation was negligible in the meristems (R3–R4 and FM; Fig. 2A), whereas it was still abundant in the leaves (Fig. 2A), indicating that Hd3a continues to be transported from the leaf to the shoot apex during establishment of the inflorescence meristem, as well as of the primary and secondary branch meristems.
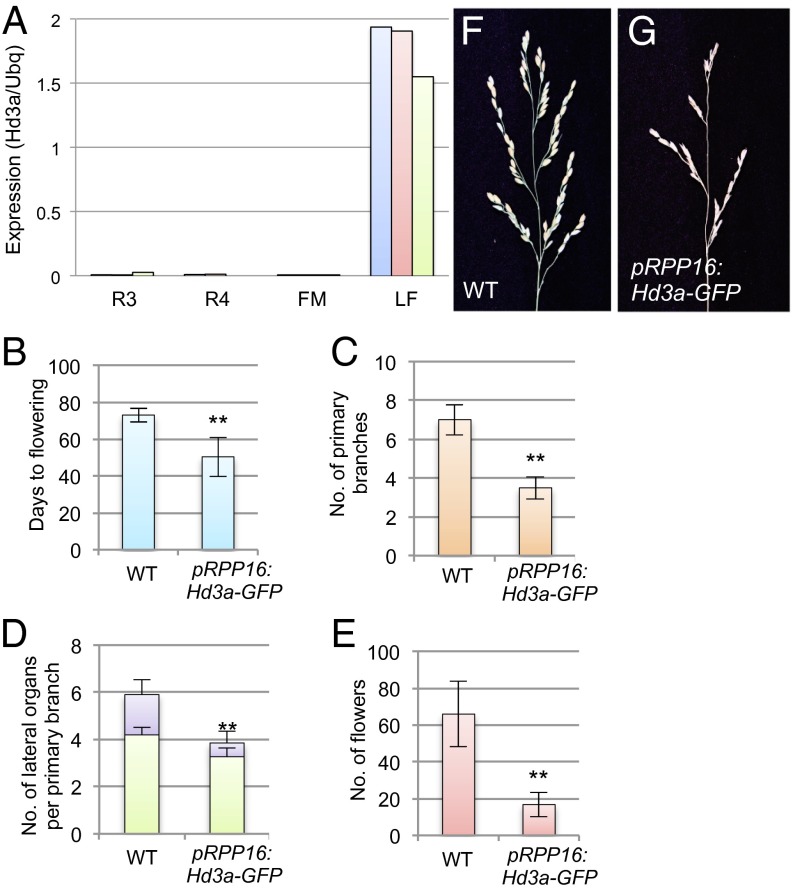
Fig. 2.
Inflorescence phenotype of Hd3a-overexpressing plants. (A) Expression of Hd3a in inflorescence meristems at stages R3 and R4, in FM, and in leaves at 40 DAG (LF), measured by qRT-PCR. Each of the blue, red, and green bars for each stage indicates a single sample for a replicate. (B–G) Inflorescence phenotypes of pRPP16:Hd3a–GFP. (B–E) Days to flowering (B), number of primary branches (C), number of lateral organs per primary branch (D), and number of flowers (E). In D, green and purple bars indicate number of flowers and number of secondary branches, respectively. (F and G) Panicle morphology of WT (F) and pRPP16:Hd3a–GFP transgenic plants (G). **P < 0.01 (significant difference by t test for B–E).
In Arabidopsis, FT activity promotes precocious conversion of inflorescence meristem into FM, changing the indeterminate inflorescence to the determinate inflorescence (15–17). In rice, Heading date 1 (Hd1) and Early heading date 1 (Ehd1), which are upstream regulators of Hd3a, control inflorescence architecture (18). Because Hd3a–GFP persists in developing inflorescence meristems (Fig. 1), we tested whether Hd3a also regulates inflorescence architecture in rice. We examined the inflorescence phenotypes of transgenic plants expressing Hd3a–GFP under the phloem-specific RPP16 promoter (pRPP16:Hd3a–GFP) (19), which enables us to increase Hd3a expression in the tissue where the Hd3a promoter is active and should minimize the effect of Hd3a expression outside the phloem (7). As described, pRPP16:Hd3a–GFP plants flowered earlier than wild type (WT; Fig. 2B) (7, 10). Hd3a–GFP expression reduced the numbers of primary branches (Fig. 2C) and of secondary branches that developed on the primary branches (Fig. 2D), resulting in a reduced number of flowers (Fig. 2 E–G). Suppression of Hd3a by RNA interference (RNAi) did not affect the number of branches, suggesting that it plays a redundant role with its paralog RFT1 in panicle architecture (Fig. S2). The number of flowers was less severely reduced by Hd3a RNAi, probably because of the longer cultivation under SD conditions and the weak induction of RFT1 at the later stage (>50 DAG) of plant growth under Hd3a suppression (20). We were unable to examine inflorescences in Hd3a RFT1 double-RNAi plants, because simultaneous suppression of Hd3a and RFT1 completely blocks floral transition, and the inflorescence never emerges (20). These results suggest that, in addition to its role in the vegetative-to-reproductive transition in the SAM, Hd3a promotes the inflorescence-to-flower transition in the reproductive meristem to control inflorescence architecture.
Localization of OsMADS15 in the SAM During Flower Formation.
Because OsMADS15 is a direct target of the FAC (10), a comparison of Hd3a localization and OsMADS15 protein expression in the SAM should yield interesting information on the relationship between these two proteins during floral induction. To study spatiotemporal patterns of OsMADS15 expression, we generated OsMADS15–mOrange transgenic rice plants by gene targeting (21, 22). A gene encoding mOrange fluorescent protein was inserted downstream of OsMADS15 to generate an in-frame OsMADS15–mOrange fusion gene at the endogenous OsMADS15 locus (Fig. 3Q; for more detail, see Fig. S3 A and B). The levels of both endogenous OsMADS15 and OsMADS15–mOrange mRNAs were low in leaves and high in inflorescence meristem, indicating that OsMADS15–mOrange expression is regulated similarly to the endogenous gene (Fig. S3C).
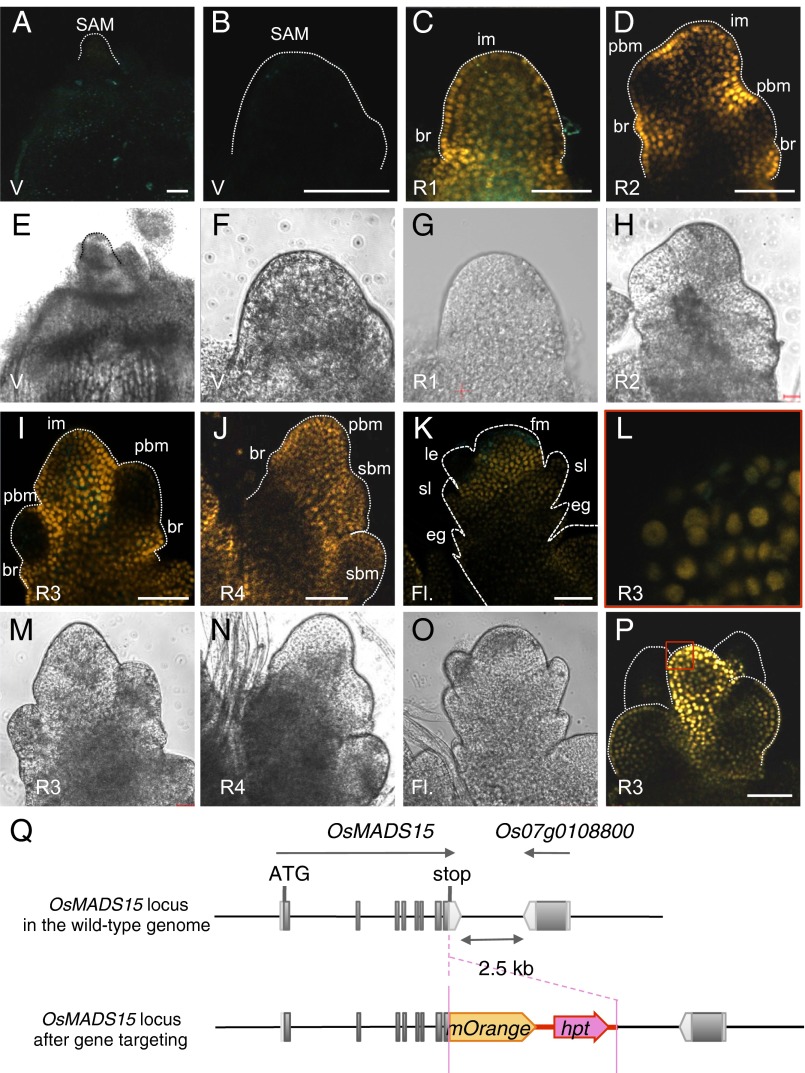
Fig. 3.
OsMADS15 protein localization in the rice shoot apex during the transition from vegetative to reproductive stage. Confocal images of OsMADS15–mOrange gene targeting plants at the indicated stages are shown. Stages and abbreviations are as described in the Fig. 1 legend. (A, B, E, and F) V, vegetative-stage plants at 27 DAG. (C and G) R1. (D and H) R2. (I, L, M, and P) R3. (J and N) R4. (K and O) Fl. (L) Enlarged view of the R3 stage meristem (red square in P) to illustrate the accumulation of mOrange in the nuclei. (Scale bars, 50 µm.) (Q) Schematic diagrams of OsMADS15–mOrange gene targeting with representative features. Dark gray boxes, exons; light gray boxes, UTRs. The direction of transcription is indicated by arrows. Orange pentagon, mOrange coding sequence; red arrow, expression cassette of hygromycin phosphotransferase (hpt) for selection of targeting allele.
At the V stage, no mOrange fluorescence was detected in the SAM (Fig. 3 A, B, E, and F) or the stem vasculature beneath the meristem, although abundant Hd3a–GFP signal was visible in the latter regions, suggesting that Hd3a does not function to activate OsMADS15 transcription there (Fig. 3 A and E, compared with Fig. 1 A and E). At the transition from the V to the R1 stage, mOrange fluorescence was clearly detected throughout the elongating meristem (Fig. 3 C and G). At the R2 stage, the mOrange signal was observed in the inflorescence meristem but was weak in the newly developing primary branch meristems (Fig. 2 D and H), similar to the Hd3a–GFP signal (Fig. 1 D and H). At the R3 stage, the mOrange signal persisted throughout the shoot apex, whereas primary branch meristems displayed very low levels of mOrange protein expression (Fig. 3 I and M). At R4, OsMADS15–mOrange protein accumulated in the entire primary branch meristems and newly formed secondary branch meristems (Fig. 3 J and N), consistent with the pattern of Hd3a–GFP accumulation and its role in inflorescence development (Figs. 1 J and N and 2). After the R4 stage, when the inflorescence meristem turned into FM, mOrange fluorescence was detected in the FM and, to a lesser extent, in the bract-like organ (sterile lemma). However, the lower organs, where Hd3a–GFP protein accumulated (Fig. 1K), were devoid of mOrange fluorescence, suggesting that in the FMs OsMADS15 expression was not responsive to Hd3a activity (Fig. 3 K and O). mOrange fluorescence was clearly observed in the nucleus of the meristem cells (Fig. 3 L and P). Together, these results indicate that the localization of Hd3a–GFP and OsMADS15–mOrange proteins overlaps extensively in the SAM during floral induction.
Expression of OsFD1, GF14b, and OsMADS15 Genes in the Rice Shoot Apex During Floral Induction.
To study the expression of OsFD1, GF14b (which encodes one of the eight rice 14-3-3 proteins), and OsMADS15 at the shoot apex, in situ hybridizations were performed at the V, R1, and R3 stages (Fig. 4 A–L). Expression of OsFD1 was detected at the V stage and in the entire shoot apex at the R1 and R3 stages (Fig. 4 A–C, compared with Fig. 4D as the negative control). Similarly, GF14b expression was detected at all three stages (Fig. 4 E–G, compared with Fig. 4H as the negative control). In contrast, OsMADS15 was not expressed at the V stage but was induced in the entire shoot apex at the R1 and R3 stages (Fig. 4 I–K, compared with Fig. 4L as the negative control).
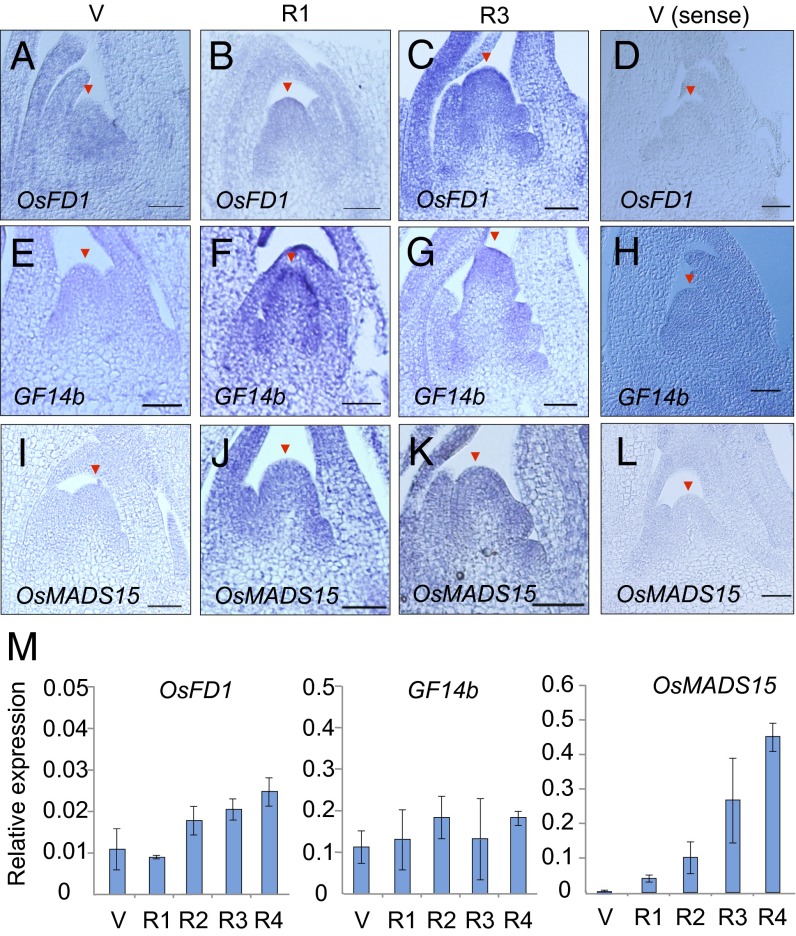
Fig. 4.
Expression of OsFD1, GF14b, and OsMADS15 genes in the rice shoot apex during floral induction. (A–L) In situ localization of OsFD1 (A–D), GF14b (E–H), and OsMADS15 (I–L) mRNAs in the shoot apex of WT plants at stage V (A, E, and I), R1 (B, F, and J), and R3 (C, G, and K), probed by antisense probes. (D, H, and L) Hybridizations with sense probes at stage V are shown as negative controls. Arrowheads indicate the top of the SAM (V stage) or inflorescence meristem (R stages). (Scale bars, 50 µm.) (M) Expression levels of OsFD1 (Left), GF14b (Center), and OsMADS15 (Right) mRNAs relative to that of ubiquitin mRNA, measured by qPCR. Error bars indicate SD for three biological replicates.
RNA was extracted from individual meristems at the five developmental stages V–R4 (Fig. 1 A–N), and the expression levels of selected genes were analyzed by quantitative PCR (qPCR). OsFD1 and GF14b were expressed at the V stage, and their expression persisted through the R4 stage of inflorescence meristem development (Fig. 4M). These results indicate that when Hd3a protein reaches the SAM, rice FD and 14-3-3 proteins are already present there. OsMADS15 expression was induced at the R1 stage, and its expression level increased from R1 through R4 stages (Fig. 4M).
The SAM of Hd3a–RFT1 Double-RNAi Plants Fails to Progress to the Reproductive Stage.
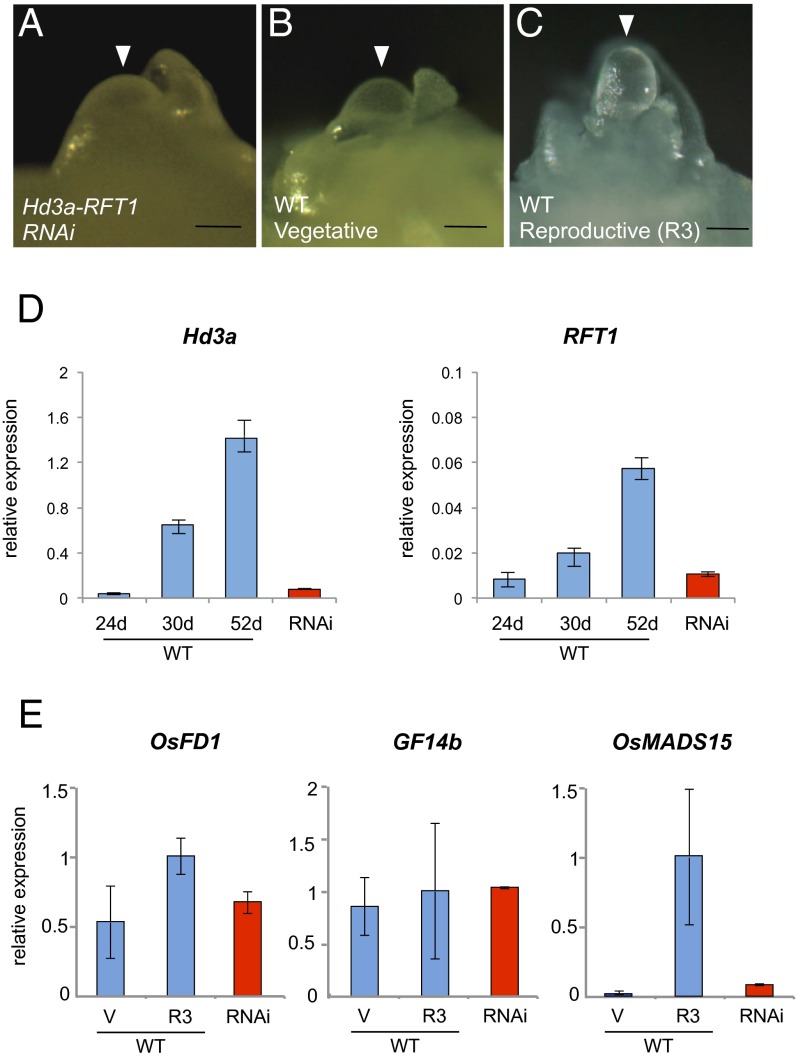
Arabidopsis plants lacking both FT and its homolog Twin Sister of FT (TSF) are able to flower, although flowering is delayed (23–25). FT/TSF-independent floral induction in Arabidopsis is permitted by genetic pathways that bypass a requirement for florigen (25–27). In contrast, rice plants in which two FT orthologs, Hd3a and RFT1, are knocked down by RNAi do not flower within almost a year after regeneration under natural conditions, suggesting that rice plants absolutely require florigen for flowering and that Hd3a and RFT1 are the only functional florigens in rice (20). Because rice plants can grow for several years if proper temperature and nutrition are supplied, Hd3a–RFT1 double-RNAi plants have been grown for >4 y in the greenhouse. They never flowered during this period, confirming our previous results (20). We next examined, in 4-y-old Hd3a–RFT1 RNAi plants, the morphology of the SAM and the expression there of genes involved in floral induction. SAM morphology in these plants (Fig. 5A) was essentially the same as that at the V stage of WT plants (Fig. 5B), indicating that in the absence of Hd3a and RFT1, rice plants were developmentally arrested and the SAM was not converted to the inflorescence meristem. The morphology of the R3-stage SAM was well elongated, indicating its floral conversion (Fig. 5C). The expression levels of Hd3a and RFT1 in leaves of the 4-y-old RNAi plants were lower than those in WT plants grown under SD conditions for >30 d, when expression levels of these two florigen genes increased (Fig. 5D). Expression of OsFD1, GF14b, and OsMADS15 in the SAM of double-RNAi plants was analyzed and compared to that in the inflorescence and primary branch meristems of WT plants at the V and R3 stages. Expression levels of OsFD1 and GF14b in the SAM of double-RNAi plants were similar to those in both the V and R3 stages of WT plants (Fig. 5E). Expression of OsMADS15 was very low in double-RNAi plants, similar to that at the V stage of WT plants (Fig. 5E) and in contrast to the high level seen at the R3 stage of WT plants. Together, these results indicate that suppressing Hd3a and RFT1 expression in the leaves prevents floral induction, changes in SAM morphology, and induction of the rice AP1 ortholog in the SAM.
Fig. 5.
Analysis of Hd3a–RFT1 double-RNAi plants. (A–C) Shoot apex of a double-RNAi plant (A) and of WT plants at stages V (B) and R3 (C). (Scale bars, 50 µm.) (D) Expression of Hd3a and RFT1 in leaves of WT (blue bars) sampled at 24 DAG (d), 30d, and 52d and in double-RNAi plants (red bars) sampled at 60 d after transplantation. Expression is relative to that of ubiquitin mRNA. (E) Expression levels in the SAM of mRNAs for OsFD1 (Left), GF14b (Center), and OsMADS15 (Right) in double-RNAi plants (red bars) compared with those in WT plants (blue bars). Relative expression was calculated by normalizing the level in WT plants at the R3 stage to 1. Error bars indicate SD for three biological replicates.
Analysis of Genes Induced by Florigen Hd3a and RFT1 in the Shoot Apex.
Although transcriptome analysis during floral induction has been performed in Arabidopsis, tomato, and soybean (28–31), and microarray analysis to compare vegetative and reproductive meristems in rice (32), no comparison has been made by using RNA sequencing (RNA-seq) between WT and florigen-depleted meristems in rice. Therefore, to identify genes either up- or down-regulated by florigen Hd3a and RFT1, we isolated shoot apices of Hd3a–RFT1 double-RNAi plants under a dissecting microscope. For WT plants, shoot apices at the early reproductive stage (R1 and R2 stages combined) were isolated. RNAs were then extracted and compared by using RNA-seq analysis. Three independent shoot apex samples were prepared, and RNAs were isolated from each sample; the analysis was therefore performed in triplicate. After linear amplification of cDNA (33), the resulting six libraries were sequenced by using Illumina GAIIx (Table S1). In total, 159 Hd3a/RFT1–up-regulated genes (i.e., genes whose expression was reduced in Hd3a/RFT1–RNAi plants) and 743 down-regulated genes, whose expression was changed in the opposite manner, were identified [false discovery rate (FDR) < 0.05; fold change > 2; Dataset S1]. A comparison of assigned Gene Ontology (GO) terms between up- and down-regulated genes revealed no substantial overall difference (34) (Fig. S4).
Genes Up-Regulated During the Early Stage of Floral Induction.
A total of 159 genes showed more than a twofold increase in expression in the shoot apex at the early stage of floral induction by Hd3a and RFT1 (Fig. 6A and Dataset S1). The list included the OsMADS15 gene, suggesting that the RNA-seq data were valid. The MADS-box genes OsMADS14, OsMADS18, and PAP2/OsMADS34 have been identified by microarray analysis in previous reports (32). Of these, OsMADS34 is particularly important for inflorescence development. Promoter activity of OsMADS34 overlaps with the localization of Hd3a–GFP (Fig. 1) in the reproductive meristem at stage R1, and mutation of OsMADS34 partially suppresses the phenotypes of prolC:Hd3a–GFP (32).
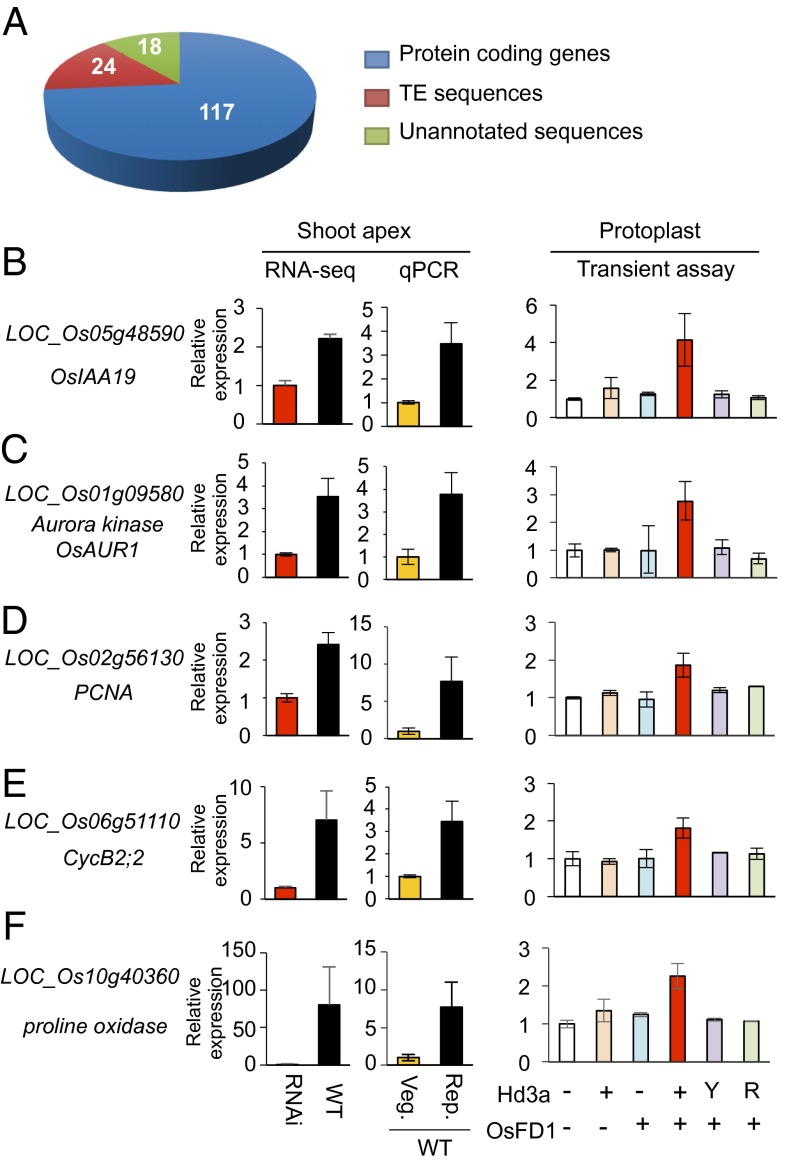
Fig. 6.
Genes up-regulated during the early stage of floral induction. (A) Number and kinds of genes up-regulated during the early stage of floral induction. (B–F) Validation of selected protein-coding genes that were up-regulated by florigen. The ID and annotation of the validated genes are indicated to the left of graphs showing relative expression of the five indicated genes at the shoot apex, examined by RNA-seq and validated by qPCR. For RNA-seq, vegetative meristem of RNAi plants and reproductive meristem of WT plants are compared, and the expression in RNAi was set as 1 for normalization. For qPCR validation, meristems at vegetative (Veg.) and reproductive (Rep.) stages from WT plants were examined, and the expression in Veg. was set as 1 for normalization. Error bars indicate SD for three biological replicates. The graphs in Right display qPCR results from transient assays using rice protoplasts. Protoplasts were transfected with (+) or without (−) expression vectors for Hd3a and its mutants (Y, Y87H; R, the double mutation of R64K and R132K) and OsFD1. The expression levels of the indicated genes were examined. Expression levels in protoplasts lacking both Hd3a and OsFD1 expression vectors were set as 1 for normalization of the results from other combinations of transfected vectors. Error bars indicate SD for three biological replicates.
Of the these MADS-box genes, OsMADS14 and OsMADS34 appeared to be up-regulated in WT compared with Hd3a/RFT1 RNAi, but they were excluded by our statistical analysis because of variable expression values among biological replicates.
One group of genes we identified was those involved in cell division. Because cell division in the meristem accelerates immediately after the arrival of Hd3a in the shoot apex (Fig. 1 F and G) (14), these genes may be involved in this process. We selected an auxin-signaling gene, OsIAA19, and three cell-cycle–related genes for confirmation by qPCR because auxin and cell division seem to play roles in organ development during the floral transition (35, 36).
Interestingly, OsIAA19 was up-regulated (Fig. 6B), suggesting that auxin may be involved in the early stages of floral induction. The three cell-cycle–related genes, Aurora kinase (Fig. 6C), PCNA (Fig. 6D), and CycB2;2, were also up-regulated (Fig. 6E). Further confirmation of the RNA-seq results was obtained by qPCR analysis of a gene encoding proline oxidase, selected randomly from the list of up-regulated genes (Fig. 6F). To examine whether these selected up-regulated genes were similarly regulated by the FAC, we conducted transient transcription assays in rice protoplasts (Fig. 6 B–F), in which cotransformation of the protoplasts with expression vectors for Hd3a and OsFD1 results in the formation of the FAC by Hd3a, OsFD1, and endogenous 14-3-3 proteins, and in the activation of downstream gene expression (10). The results revealed that up-regulation of the selected genes required both Hd3a and OsFD1 and was abolished in an Hd3a mutant having two mutations, R64K and R132K, at the 14-3-3 interaction surface (Fig. 6 B–F). A second control was the Hd3a Y87H mutant, which is orthologous to an FT mutant whose function is converted to that of a floral repressor (37) and which abolishes Hd3a activity in OsMADS15 activation (10). Up-regulation of all genes analyzed was eliminated by this mutation (Fig. 6 B–F). Collectively, these results show that the six selected genes require functional Hd3a and the FAC for their up-regulation.
The Majority of Down-Regulated Genes Are TE Sequences.
We found 205 protein-coding genes whose expression was lower in WT than in RNAi plants, meaning that endogenous Hd3a/RFT1 down-regulates them (Fig. 7A). The list included genes involved in a variety of cellular processes (Dataset S1). Here, we focus on an unexpected finding regarding TEs.
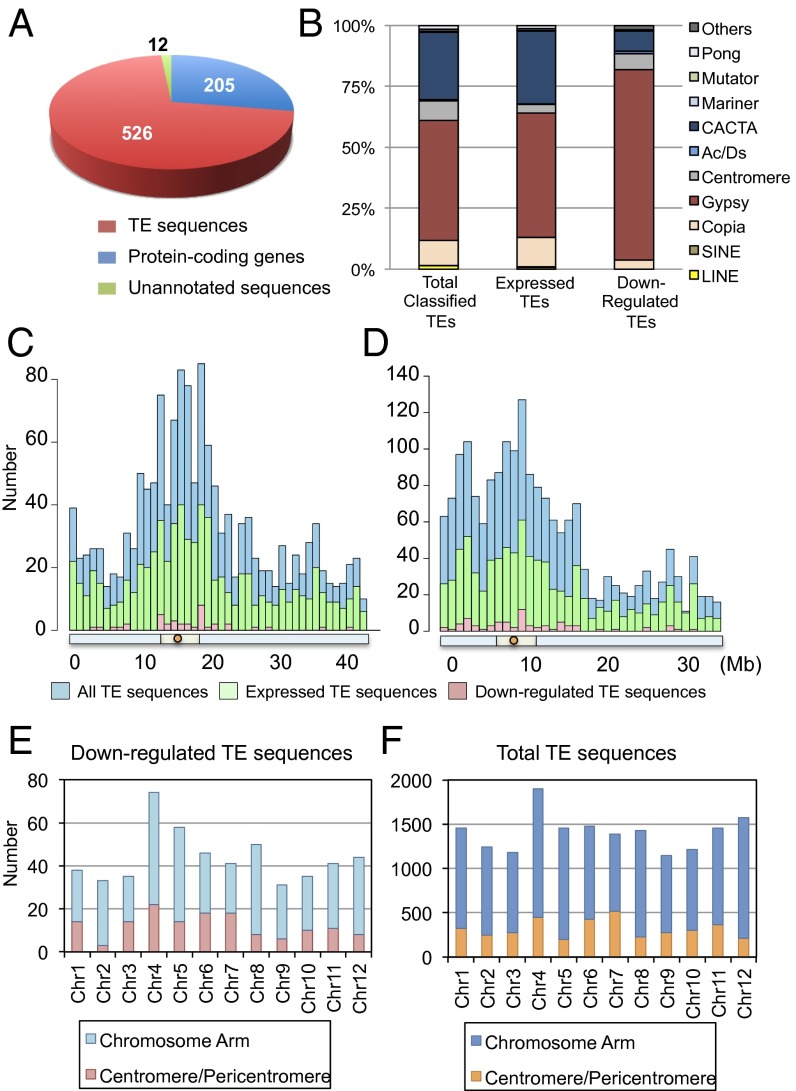
Fig. 7.
Characterization of down-regulated TE sequences. (A) Number and kinds of genes down-regulated during the early stage of floral induction. (B) Composition of total rice TEs, TEs expressed in the shoot apex, and TEs down-regulated by Hd3a florigen. Down-regulated TEs were classified into known groups of transposons and retrotransposons, and unclassified TEs were not included. (C and D) Distribution of all TE sequences, those expressed in the vegetative shoot apex, and those down-regulated, along rice chromosomes 1 (C) and 4 (D). Red circles, light green bars, and light blue bars below the histograms indicate centromeres, pericentromeric regions, and chromosome arms, respectively. (E and F) Distribution of down-regulated (E) and total (F) TE sequences in different regions of rice chromosomes.
Surprisingly, 526 (70.8%) of the 743 down-regulated transcripts were from TE sequences (Fig. 7A), indicating that transcription of multiple transposons is suppressed by Hd3a in the shoot apex during floral induction. Down-regulated TE sequences were classified into known groups of transposons and retrotransposons (Fig. 7B). The rice genome annotation we used (MSU7) includes 16,941 TE sequences, comprising 7,549 classifiable and 9,392 unclassifiable TEs. We refer below to classifiable TEs as “TEs,” to unclassifiable sequences as “unclassified TEs,” and to the total of both categories as “TE sequences.” Among the 526 down-regulated TE sequences, 214 were TEs. Because the remaining unclassified TEs could not be grouped into known TE types, they were excluded from Fig. 7B; however, they are included in Fig. 7 C–F to illustrate the entire picture of TE sequences.
We then defined “expressed TE sequences” as those whose expression was detected in all three replicates of both WT and RNAi. Among the total of 16,941 TE sequences in the rice genome, 8,045 (∼50%) were expressed according to our RNA-seq analysis. When we consider the classified TEs, among the total of 7,549, there were 4,379 (58%) that were expressed. As shown in Fig. 7B, by far the most actively transcribed group of TEs in the shoot apex were gypsy-like retrotransposons, CACTA transposons, and copia-like retrotransposons. The relative abundance of each group of TEs in the total number of rice TEs and in the number that were expressed was similar. Interestingly, however, there was a difference between the relative abundance of those down-regulated in the reproductive shoot apex and those expressed in the vegetative shoot apex. Approximately 75% of the down-regulated TEs were gypsy-like retrotransposons, and this group was, proportionately, more silenced than CACTA transposons and copia-like retrotransposons in the reproductive shoot apex (Fig. 7B). The distribution of silenced TE sequences (classified and unclassified) along the chromosome was similar to the relative abundance of total TE sequences and those expressed in the shoot apex along the chromosome (Fig. 7C for chromosome 1, the longest chromosome, and Fig. 7D for chromosome 4, as an example of a chromosome with a large heterochromatin region). Silenced TE sequences were distributed in all 12 chromosomes (Fig. 7E). They occurred in both the centromere/pericentromere and the chromosome arms, and their proportion relative to all transposons was similar in each chromosome (Fig. 7 E and F).
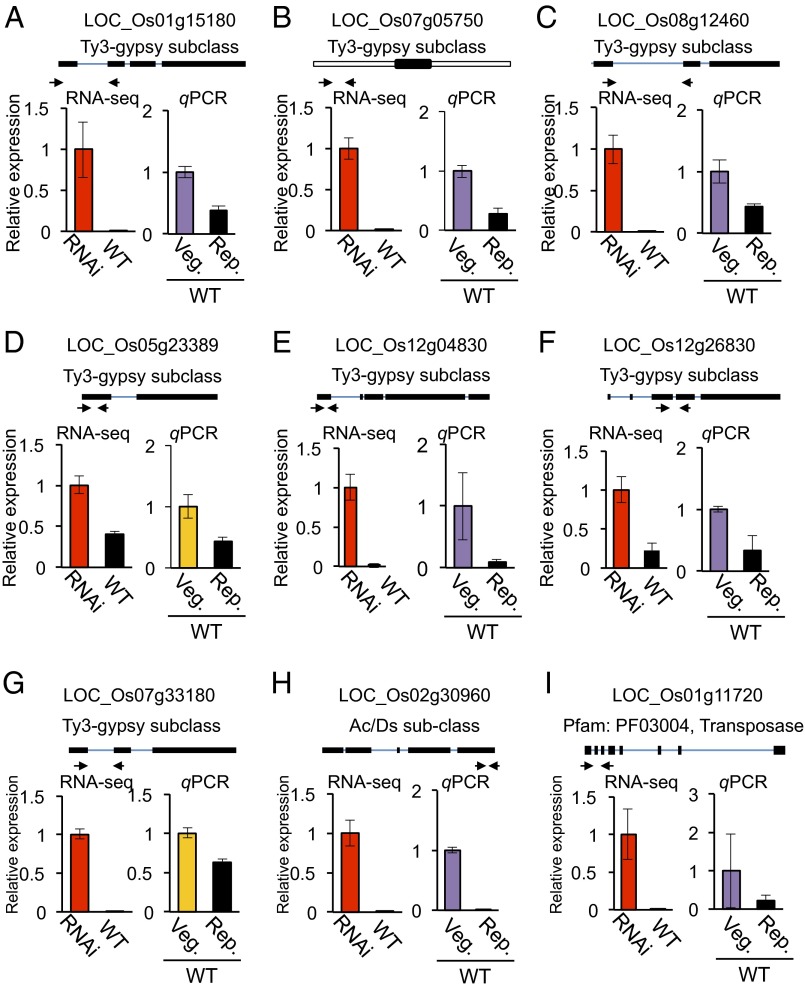
Down-regulation of nine representative TE sequences was confirmed by qPCR (for classified TEs, Fig. 8 A–H, and for unclassified, Fig. 8I). All nine were down-regulated at the reproductive shoot apex compared with the vegetative shoot apex, confirming the results of RNA-seq analysis and thus its reliability.
Fig. 8.
(A–I) Validation of down-regulated TE sequences. Selected TE sequences that were down-regulated during floral induction by Hd3a florigen are shown. Upper indicates the locations of qPCR primers (arrows). Thick and thin lines indicate exons and introns, and the open boxes in B indicate UTRs. Lower shows RNA-seq (Left) and qPCR (Right) data for WT-vegetative (Veg.) and WT-reproductive (Rep.) samples.
Discussion
Localization of Hd3a–GFP and OsMADS15–mOrange Overlaps During Floral Induction and Inflorescence Development.
The spatiotemporal pattern of florigen accumulation in the meristem and developing primordia has not been investigated. In this study, we focused on three critical aspects of Hd3a–GFP localization in the rice inflorescence meristem: (i) the temporal relationship between Hd3a accumulation and floral transition in the meristem; (ii) Hd3a–GFP accumulation in the developing primordia during inflorescence development; and (iii) the overlap of Hd3a–GFP localization with the expression of OsMADS15 and the other components of the FAC in the meristem.
In the vegetative stage, Hd3a–GFP signal was not visible in the SAM, but accumulated in the stem vasculature just beneath the meristem (Fig. 1 A and B). Concomitant with the initiation of reproductive growth, Hd3a–GFP was clearly detected in the entire SAM (Fig. 1C). OsMADS15–mOrange was also expressed in the SAM at a very early stage of the reproductive transition (Fig. 3C), consistent with the essential role of Hd3a as the trigger of the floral transition in rice (20).
Florigen function during floral induction has been well studied, but its role in inflorescence development has not been analyzed. Localization of Hd3a–GFP in the developing inflorescence may provide an important clue for a link between Hd3a function and inflorescence development. The Hd3a–GFP signal persisted in the meristem and emerging primordia in later stages of the reproductive meristem (Fig. 1 I–N). The weaker Hd3a–GFP signal in the inflorescence meristem (Fig. 1D) could explain the indeterminate nature of the rice inflorescence meristem, because overproduction of FT orthologs often results in the conversion of indeterminate to determinate inflorescence (15, 16, 38, 39). The Hd3a–GFP signal was rather weak in the newly emerging branch primordia (Fig. 1I), but became stronger at the later stage of branch development (Fig. 1J). This finding may reflect termination of the branch meristem and its conversion into the FM. Consistent with this observation, Hd3a–GFP expression from the phloem-specific RPP16 promoter reduced inflorescence branching through precocious conversion of the branch meristem to the FM (Fig. 2). This result is the opposite phenotype of the pap2 mutant, which is defective in the OsMADS34 in rice (40). Because OsMADS34 is one of the earliest inducible genes at the floral transition, and genetically interacts with Hd3a (32), regulation of OsMADS34 by Hd3a should be important for future study.
Hd3a forms the FAC with 14-3-3 proteins and OsFD1 to activate OsMADS15 and promote flowering (10). However, whether all of the FAC components and downstream OsMADS15 expression coexist in the SAM has not been examined. The data presented here on Hd3a–GFP localization (Fig. 1), OsMADS15–mOrange localization (Fig. 3), and in situ hybridization and qRT-PCR for GF14b and OsFD1 transcripts (Fig. 4) indicate that all these components are expressed in the same regions of the SAM. Before the arrival of Hd3a–GFP at the meristem, OsMADS15 mRNA and OsMADS15–mOrange were not detected in the meristem (Fig. 3 A and B), whereas GF14b and OsFD1 were expressed there (Fig. 4 A–H and M). When Hd3a–GFP reached the SAM, OsMADS15 expression began in the SAM. These data support the idea that Hd3a triggers the floral transition via formation of the FAC in the SAM to activate OsMADS15.
Transposons Are Highly Expressed in the Shoot Apex of both Vegetative and Early Reproductive Stages.
Surprisingly, 58% (4,379/7,549) of classifiable rice TEs was expressed in the vegetative shoot apex (Fig. 7B). In the early reproductive shoot apex, although transposon silencing (∼5%; 214 of 4,379 classifiable TEs silenced) occurs, considerable numbers of TEs were expressed (Fig. 7B). Based on pyrosequencing analysis of microdissected SAM in maize, it was shown that in the vegetative SAM, ∼14% of the expressed transcripts were transposon-related genes (41). This phenomenon is known as developmental relaxation of TE silencing (DRTS), and several examples are known in plants (42). DRTS in pollen vegetative nuclei and female gametophytes has recently been reported, and epigenetic regulators are involved in the phenomenon in these organs (42–45). In endosperm, regulation of imprinted genes is closely associated with DRTS (46, 47), and in Arabidopsis male gametocytes, a high level of DRTS has been reported to occur (48). In maize, the transition from juvenile to adult phase during vegetative growth is accompanied by DRTS in the MuDR element (49). This DRTS is regulated by epigenetic changes during the juvenile-to-adult transition, implying the importance of developmental transition as a trigger for DRTS.
Among known plant DRTS events, the DRTS we found in rice shoot apices seems to be the most abundant in number. In the rice vegetative shoot apex, 4,379 of 7,549 known TEs (58%) were expressed, the majority of which were retrotransposons. In the reproductive shoot apex, although ∼5% of them were silenced, most were transcriptionally active. These results suggest that, contrary to the idea that TEs are generally silenced because of their detrimental effects, a large number are expressed in specific organs at specific developmental times. Although the number of expressed TEs is large, it is likely that most of them do not transpose because of the presence of posttranscriptional regulation (50). However, it is possible that a fraction of them may be transposed to new locations. Insertion of newly active retrotransposons is one mechanism for increasing the genome size of a species; it is known, for example, that Oryza australiensis has doubled its genome size mainly by increasing the number of retrotransposons (51).
Why does massive DRTS occur in the SAM at both vegetative and reproductive stages? Because small RNAs may be generated from transcribed TEs by the RNAi machinery and potentially regulate TE activation or protein-coding genes by multiple epigenetic mechanisms (52, 53), TE-derived small RNAs may have an impact on the regulation of gene expression in the SAM, which is an important group of cells for generating both flowers and male and female gametes. A possible explanation for the massive DRTS in the SAM is that plants transcribe the majority of TEs in the reproductive shoot apex, producing the full repertoire of small RNAs. This explanation implies that the TE-related sequences comprise a substantial proportion of all small RNAs (53–55). These small RNAs then reorganize the genome through the RNA-dependent DNA methylation pathway (52) to ensure that those TEs that have deleterious effects will not be transcribed in later stages of gamete production.
Activation of TEs can be induced by many different epigenetic mechanisms (50, 56). These include disturbance of the epigenetic program (57, 58), environmental signals (58, 59), and the presence of a transposon activator encoded by a transposon (60). Whether the massive DRTS that occurs in the rice SAM is caused by a similar or a completely different molecular mechanism will be an interesting question to address.
Transposon Silencing During the Floral Transition in the Shoot Apex.
Because our original aim in this study was to identify which genes are differentially regulated during the transition from vegetative to reproductive phase in the shoot apex, we were particularly interested in genes encoding functions that potentially regulate floral induction. Up-regulated genes, as expected, included those related to transcription, the cell cycle and cell division, and auxin signaling. To our surprise, however, the majority of the down-regulated genes were TE-related sequences, and the majority of these were retrotransposons. They aligned to locations throughout the 12 chromosomes, and their distribution does not seem to have any special features (Fig. 7 C–F). However, Gypsy transposons were overrepresented in the silenced TEs (Fig. 7B). Therefore, there may be specificity in transposon silencing in the SAM at the early reproductive stage. Such silencing of a large number of transposons in a specific organ at a specific developmental stage appears to be a unique phenomenon. Furthermore, in the present case, a flower-inducing signal, florigen, acts as the trigger for this phenomenon. We do not yet have any clear explanation for this behavior. However, we can offer three ideas about the meaning of transposon silencing in the shoot apex. First, if retrotransposons generate small RNAs that regulate TE activity and the expression of protein-coding genes, down-regulation of some of these retrotransposons could derepress the expression of genes that are silenced in the vegetative shoot apex during the transition to the reproductive phase. In this way, transposon silencing would activate genes that are important for flower development. This hypothesis fits the “controlling elements” theory for transposons put forward by Barbara McClintock, who proposed that transposons control development (61). Such trans-regulation of gene activity by TEs has been reported in the Arabidopsis ddm1 mutant (53). ddm1 mutant plants expresses a Gypsy-type retrotransposon, Athila, whose mRNA is processed into small 21- or 22-nucleotide (nt) RNAs. These small RNAs are loaded onto ARGONAUTE1 (AGO1) protein, and the resultant AGO1–small RNA complex cleaves a subset of transcripts with partial sequence complementarity to the Athila-derived small RNAs (53). Second, transposon silencing may suppress the transposition of specific TE-related sequences in the early reproductive shoot apex to minimize any harmful effects of TE transposition. Third, transposon silencing in the reproductive shoot apex may shut down the production of regulatory small RNAs, which, otherwise, could later move to meiotic organs or gametes and disturb gene regulation in these important organs.
Although the biological meaning of the dramatic DRTS, which occurs in both the vegetative shoot apex and the reproductive shoot apex, as well as the transposon silencing found in the reproductive shoot apex of rice, is not known, the roles of TEs in reproductive development may be much broader than they are currently thought to be. To better understand these interesting phenomena, we need to determine which TEs are involved, what kinds of small RNAs are produced and silenced, and which protein-coding genes are affected by TE-derived small RNAs.
Materials and Methods
Plant Materials and Growth Conditions.
The Japonica rice cultivar Norin 8 was used as WT. pHd3a:Hd3a–GFP plants were as described (7). Plants were grown in climate chambers at 70% humidity under SD conditions with daily cycles of 10 h of light at 27 °C and 14 h of dark at 25 °C. Light was provided by fluorescent white light tubes (400–700 nm, 100 μmol m−2⋅s−1). For RNA-seq and real-time qRT-PCR, 8–10 shoot apices were collected and pooled as a single biological replicate, and three biological replicates were prepared for experiments.
OsMADS15–mOrange Gene Targeting.
Gene targeting of the OsMADS15 locus was conducted as described, with some modifications (21). Briefly, the binary vector for knock-in targeting was constructed by cloning the PCR-amplified OsMADS15 genomic region into a vector carrying the mOrange coding sequence (Fig. S2A). After Agrobacterium-mediated transformation, calli were selected for hygromycin resistance. Calli with incomplete targeting were first eliminated by the expression of the diphtheria toxin A gene on the vector (21), and correctly targeted genotypes were identified by PCR using genomic DNA from calli as a template. Two lines of calli were successfully obtained as gene targeting lines (Fig. S2B). The primers used are listed in Table S2.
RNA Extraction and qPCR.
Total RNA from shoot apices was extracted by using TRIzol reagent (Invitrogen) and treated with DNase I (Invitrogen). cDNA was synthesized from this RNA by using SuperScript II reverse transcriptase (Invitrogen). cDNA was used for quantitative analysis of gene expression, performed with SYBR Green PCR Master Mix (Applied Biosystems) and gene-specific primers (Table S2). Data were collected by using the ABI PRISM 7000 sequence detection system in accordance with the manufacturer's instruction manual.
RNA Sequencing.
Double Hd3a–RFT1 RNAi plants were as described (20). Total RNAs were extracted from SAMs of WT and Hd3a–RFT1 RNAi plants by using TRIzol reagent at 35 d after sowing for WT and 35 d after transplanting for RNAi plants. The extracted RNA was quantified by using a 2100 Bioanalyzer (Agilent) and converted to cDNA by using the Ovation RNA-Seq Kit (NuGEN) (33). RNA-seq libraries were prepared according to the Illumina library preparation kit protocol and sequenced on an Illumina GAIIx to generate single-end reads of 36 nt. This procedure was repeated in triplicate.
Data Analysis.
Rice genome sequences and annotated gene models were downloaded from the MSU Rice Genome Annotation Project Database and Resource (Version 7.0; rice.plantbiology.msu.edu). The single-end reads were aligned to the rice genome by using TopHat (Version 2.0.4; ref. 62) with default parameters because repetitive sequences including TEs can align to many genome locations. If a read matches several positions in the reference genome with the same score, multiple alignments up to 20 positions are allowed. When there are >20 alignments, 20 randomly chosen alignments are output. Avadis NGS (Agilent), a tool for determining differential expression in sequencing data, was used to quantify the changes occurring in mRNA expression levels between WT and Hd3a–RFT1 RNAi plants. The read count for a gene is calculated with correction for multiple alignments. After normalization by the trimmed mean of M values method (63), differentially expressed genes were extracted by moderated t test between WT and RNAi samples (64) with multiple comparison correction. A FDR (65) of <0.05 was chosen as the cutoff for determining whether differential gene expression was significant. For GO analysis, GO terms for each gene were defined according to the MSU Rice Genome Annotation Project Database and Resource (Version 7.0). Differences in GO enrichment between up- and down-regulated genes were analyzed by WEGO (34). To analyze TE-related sequences, expressed TEs were defined as those that were aligned by at least one read in all of the triplicates. TEs were classified according to the MSU Rice Genome Annotation Project Database and Resource (Version 7.0).
Confocal Laser-Scanning Microscopy.
Transgenic rice plant tissues were visualized by using Zeiss LSM 700 and 710 confocal laser-scanning microscopes. PCR-selected T2- and T3-generation transgenic plants were grown in a growth chamber. Shoot apices were manually dissected and suspended in a drop of water on a covered glass slide. For observation of GFP or mOrange signals, fluorescence was excited with a 488- or 514-nm argon laser, and emission images were collected in the 490- to 550-nm or 540- to 600-nm range, respectively. Fluorescence signals were separated from background noise by using an emission fingerprinting linear unmixing function.
RNA in Situ Hybridization.
RNA in situ hybridization was performed by using described methods (9). Plasmids carrying full-length cDNAs were linearized and used as templates for making digoxygenin-labeled antisense probes.
Supplementary Material
Acknowledgments
We thank I. Smith for critical reading of the manuscript; J. Naritomi, Y. Tamaki, Y. Konomi, Y. Ohno, M. Kanda, S. Toyoda, T. Nakajima, S. Moritoh, M. Ogawa, Y. Ikeda, T. Miyamoto, and H. Saitoh for technical assistance. S.T. was supported by a Japan Society for the Promotion of Science fellowship. This work was supported in part by MEXT Grants-in-Aid for Specially Promoted Research Project 24000017 (to K.S.); the Plant Global Education Project of Nara Institute of Science and Technology (T.S. and T.K.); Grants-in-Aid for Young Scientists A Project 24688001; the Program for Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industry; and Grant-in-Aid for Scientific Research on Innovative Areas Project 26113713 (to H.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences have been deposited in the DDBJ Sequence Read Archive, trace.ddbj.nig.ac.jp/dra/index_e.html (accession no. DRA002310).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1417623112/-/DCSupplemental.
References
- 1.Amasino R. Seasonal and developmental timing of flowering. Plant J. 2010;61(6):1001–1013. doi: 10.1111/j.1365-313X.2010.04148.x. [DOI] [PubMed] [Google Scholar]
- 2.Huijser P, Schmid M. The control of developmental phase transitions in plants. Development. 2011;138(19):4117–4129. doi: 10.1242/dev.063511. [DOI] [PubMed] [Google Scholar]
- 3.Andrés F, Coupland G. The genetic basis of flowering responses to seasonal cues. Nat Rev Genet. 2012;13(9):627–639. doi: 10.1038/nrg3291. [DOI] [PubMed] [Google Scholar]
- 4.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316(5827):1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 5.Lin MK, et al. FLOWERING LOCUS T protein may act as the long-distance florigenic signal in the cucurbits. Plant Cell. 2007;19(5):1488–1506. doi: 10.1105/tpc.107.051920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr Biol. 2007;17(12):1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 7.Tamaki S, Matsuo S, Wong HL, Yokoi S, Shimamoto K. Hd3a protein is a mobile flowering signal in rice. Science. 2007;316(5827):1033–1036. doi: 10.1126/science.1141753. [DOI] [PubMed] [Google Scholar]
- 8.Jaeger KE, Pullen N, Lamzin S, Morris RJ, Wigge PA. Interlocking feedback loops govern the dynamic behavior of the floral transition in Arabidopsis. Plant Cell. 2013;25(3):820–833. doi: 10.1105/tpc.113.109355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Komiya R, Yokoi S, Shimamoto K. A gene network for long-day flowering activates RFT1 encoding a mobile flowering signal in rice. Development. 2009;136(20):3443–3450. doi: 10.1242/dev.040170. [DOI] [PubMed] [Google Scholar]
- 10.Taoka K, et al. 14-3-3 proteins act as intracellular receptors for rice Hd3a florigen. Nature. 2011;476(7360):332–335. doi: 10.1038/nature10272. [DOI] [PubMed] [Google Scholar]
- 11.Tsuji H, Nakamura H, Taoka K, Shimamoto K. Functional diversification of FD transcription factors in rice, components of florigen activation complexes. Plant Cell Physiol. 2013;54(3):385–397. doi: 10.1093/pcp/pct005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taoka K, Ohki I, Tsuji H, Kojima C, Shimamoto K. Structure and function of florigen and the receptor complex. Trends Plant Sci. 2013;18(5):287–294. doi: 10.1016/j.tplants.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Yoo SC, et al. Phloem long-distance delivery of FLOWERING LOCUS T (FT) to the apex. Plant J. 2013;75(3):456–468. doi: 10.1111/tpj.12213. [DOI] [PubMed] [Google Scholar]
- 14.Ikeda K, Sunohara H, Nagato Y. Developmental course of inflorescence and spikelet in rice. Breed Sci. 2004;54(2):147–156. [Google Scholar]
- 15.Kardailsky I, et al. Activation tagging of the floral inducer FT. Science. 1999;286(5446):1962–1965. doi: 10.1126/science.286.5446.1962. [DOI] [PubMed] [Google Scholar]
- 16.Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286(5446):1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- 17.Niwa M, et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell. 2013;25(4):1228–1242. doi: 10.1105/tpc.112.109090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endo-Higashi N, Izawa T. Flowering time genes Heading date 1 and Early heading date 1 together control panicle development in rice. Plant Cell Physiol. 2011;52(6):1083–1094. doi: 10.1093/pcp/pcr059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Asano T, et al. Rpp16 and Rpp17, from a common origin, have different protein characteristics but both genes are predominantly expressed in rice phloem tissues. Plant Cell Physiol. 2002;43(6):668–674. doi: 10.1093/pcp/pcf083. [DOI] [PubMed] [Google Scholar]
- 20.Komiya R, Ikegami A, Tamaki S, Yokoi S, Shimamoto K. Hd3a and RFT1 are essential for flowering in rice. Development. 2008;135(4):767–774. doi: 10.1242/dev.008631. [DOI] [PubMed] [Google Scholar]
- 21.Terada R, Urawa H, Inagaki Y, Tsugane K, Iida S. Efficient gene targeting by homologous recombination in rice. Nat Biotechnol. 2002;20(10):1030–1034. doi: 10.1038/nbt737. [DOI] [PubMed] [Google Scholar]
- 22.Dang TT, Shimatani Z, Kawano Y, Terada R, Shimamoto K. Gene editing a constitutively active OsRac1 by homologous recombination-based gene targeting induces immune responses in rice. Plant Cell Physiol. 2013;54(12):2058–2070. doi: 10.1093/pcp/pct147. [DOI] [PubMed] [Google Scholar]
- 23.Yamaguchi A, Kobayashi Y, Goto K, Abe M, Araki T. TWIN SISTER OF FT (TSF) acts as a floral pathway integrator redundantly with FT. Plant Cell Physiol. 2005;46(8):1175–1189. doi: 10.1093/pcp/pci151. [DOI] [PubMed] [Google Scholar]
- 24.Jang S, Torti S, Coupland G. Genetic and spatial interactions between FT, TSF and SVP during the early stages of floral induction in Arabidopsis. Plant J. 2009;60(4):614–625. doi: 10.1111/j.1365-313X.2009.03986.x. [DOI] [PubMed] [Google Scholar]
- 25.Wang JW, Czech B, Weigel D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell. 2009;138(4):738–749. doi: 10.1016/j.cell.2009.06.014. [DOI] [PubMed] [Google Scholar]
- 26.Wahl V, et al. Regulation of flowering by trehalose-6-phosphate signaling in Arabidopsis thaliana. Science. 2013;339(6120):704–707. doi: 10.1126/science.1230406. [DOI] [PubMed] [Google Scholar]
- 27.Yamaguchi N, et al. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science. 2014;344(6184):638–641. doi: 10.1126/science.1250498. [DOI] [PubMed] [Google Scholar]
- 28.Schmid M, et al. Dissection of floral induction pathways using global expression analysis. Development. 2003;130(24):6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- 29.Jung CH, Wong CE, Singh MB, Bhalla PL. Comparative genomic analysis of soybean flowering genes. PLoS ONE. 2012;7(6):e38250. doi: 10.1371/journal.pone.0038250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park SJ, Jiang K, Schatz MC, Lippman ZB. Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci USA. 2012;109(2):639–644. doi: 10.1073/pnas.1114963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torti S, et al. Analysis of the Arabidopsis shoot meristem transcriptome during floral transition identifies distinct regulatory patterns and a leucine-rich repeat protein that promotes flowering. Plant Cell. 2012;24(2):444–462. doi: 10.1105/tpc.111.092791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kobayashi K, et al. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. Plant Cell. 2012;24(5):1848–1859. doi: 10.1105/tpc.112.097105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hsieh TF, et al. Regulation of imprinted gene expression in Arabidopsis endosperm. Proc Natl Acad Sci USA. 2011;108(5):1755–1762. doi: 10.1073/pnas.1019273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ye J, et al. WEGO: A web tool for plotting GO annotations. Nucleic Acids Res. 2006;34(web server issue):W293–W297. doi: 10.1093/nar/gkl031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pautler M, Tanaka W, Hirano HY, Jackson D. Grass meristems I: Shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 2013;54(3):302–312. doi: 10.1093/pcp/pct025. [DOI] [PubMed] [Google Scholar]
- 36.Tanaka W, Pautler M, Jackson D, Hirano HY. Grass meristems II: Inflorescence architecture, flower development and meristem fate. Plant Cell Physiol. 2013;54(3):313–324. doi: 10.1093/pcp/pct016. [DOI] [PubMed] [Google Scholar]
- 37.Hanzawa Y, Money T, Bradley D. A single amino acid converts a repressor to an activator of flowering. Proc Natl Acad Sci USA. 2005;102(21):7748–7753. doi: 10.1073/pnas.0500932102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lifschitz E, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103(16):6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krieger U, Lippman ZB, Zamir D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat Genet. 2010;42(5):459–463. doi: 10.1038/ng.550. [DOI] [PubMed] [Google Scholar]
- 40.Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant Cell Physiol. 2010;51(1):47–57. doi: 10.1093/pcp/pcp166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohtsu K, et al. Global gene expression analysis of the shoot apical meristem of maize (Zea mays L.) Plant J. 2007;52(3):391–404. doi: 10.1111/j.1365-313X.2007.03244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martínez G, Slotkin RK. Developmental relaxation of transposable element silencing in plants: Functional or byproduct? Curr Opin Plant Biol. 2012;15(5):496–502. doi: 10.1016/j.pbi.2012.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Slotkin RK, et al. Epigenetic reprogramming and small RNA silencing of transposable elements in pollen. Cell. 2009;136(3):461–472. doi: 10.1016/j.cell.2008.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoft VK, et al. Function of the DEMETER DNA glycosylase in the Arabidopsis thaliana male gametophyte. Proc Natl Acad Sci USA. 2011;108(19):8042–8047. doi: 10.1073/pnas.1105117108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ibarra CA, et al. Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science. 2012;337(6100):1360–1364. doi: 10.1126/science.1224839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gehring M, Bubb KL, Henikoff S. Extensive demethylation of repetitive elements during seed development underlies gene imprinting. Science. 2009;324(5933):1447–1451. doi: 10.1126/science.1171609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hsieh TF, et al. Genome-wide demethylation of Arabidopsis endosperm. Science. 2009;324(5933):1451–1454. doi: 10.1126/science.1172417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C, et al. Meiosis-specific gene discovery in plants: RNA-Seq applied to isolated Arabidopsis male meiocytes. BMC Plant Biol. 2010;10:280. doi: 10.1186/1471-2229-10-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li H, Freeling M, Lisch D. Epigenetic reprogramming during vegetative phase change in maize. Proc Natl Acad Sci USA. 2010;107(51):22184–22189. doi: 10.1073/pnas.1016884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirouze M, Paszkowski J. Epigenetic contribution to stress adaptation in plants. Curr Opin Plant Biol. 2011;14(3):267–274. doi: 10.1016/j.pbi.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 51.Piegu B, et al. Doubling genome size without polyploidization: Dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res. 2006;16(10):1262–1269. doi: 10.1101/gr.5290206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Law JA, Jacobsen SE. Molecular biology. Dynamic DNA methylation. Science. 2009;323(5921):1568–1569. doi: 10.1126/science.1172782. [DOI] [PubMed] [Google Scholar]
- 53.McCue AD, Nuthikattu S, Slotkin RK. Genome-wide identification of genes regulated in trans by transposable element small interfering RNAs. RNA Biol. 2013;10(8):1379–1395. doi: 10.4161/rna.25555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tran RK, et al. Chromatin and siRNA pathways cooperate to maintain DNA methylation of small transposable elements in Arabidopsis. Genome Biol. 2005;6(11):R90. doi: 10.1186/gb-2005-6-11-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Havecker ER, et al. The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010;22(2):321–334. doi: 10.1105/tpc.109.072199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lisch D, Bennetzen JL. Transposable element origins of epigenetic gene regulation. Curr Opin Plant Biol. 2011;14(2):156–161. doi: 10.1016/j.pbi.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 57.Mirouze M, et al. Selective epigenetic control of retrotransposition in Arabidopsis. Nature. 2009;461(7262):427–430. doi: 10.1038/nature08328. [DOI] [PubMed] [Google Scholar]
- 58.Ito H, et al. An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature. 2011;472(7341):115–119. doi: 10.1038/nature09861. [DOI] [PubMed] [Google Scholar]
- 59.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461(7267):1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 60.Fu Y, et al. Mobilization of a plant transposon by expression of the transposon-encoded anti-silencing factor. EMBO J. 2013;32(17):2407–2417. doi: 10.1038/emboj.2013.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fincham JR, Sastry GR. Controlling elements in maize. Annu Rev Genet. 1974;8:15–50. doi: 10.1146/annurev.ge.08.120174.000311. [DOI] [PubMed] [Google Scholar]
- 62.Kim D, Salzberg SL. TopHat-Fusion: An algorithm for discovery of novel fusion transcripts. Genome Biol. 2011;12(8):R72. doi: 10.1186/gb-2011-12-8-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11(3):R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 65.Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J Roy Stat Soc B Met. 1995;57(1):289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.