Brain biopsy from a child with unknown cause of encephalopathy was deep-sequenced. Astrovirus VA1/HMO-C was identified, highly divergent from human astroviruses and 95% identical to astrovirus previously associated with encephalitis. Findings suggest astrovirus VA1/HMO-C is an under-recognized cause of viral encephalitis.
Keywords: RNASeq, astrovirus, deep sequencing, encephalopathy, pathogen discovery
Abstract
Background. An 18-month-old boy developed encephalopathy, for which extensive investigation failed to identify an etiology, 6 weeks after stem cell transplant. To exclude a potential infectious cause, we performed high-throughput RNA sequencing on brain biopsy.
Methods. RNA-Seq was performed on an Illumina Miseq, generating 20 million paired-end reads. Nonhost data were checked for similarity to known organisms using BLASTx. The full viral genome was sequenced by primer walking.
Results. We identified an astrovirus, HAstV-VA1/HMO-C-UK1(a), which was highly divergent from human astrovirus (HAstV 1–8) genotypes, but closely related to VA1/HMO-C astroviruses, including one recovered from a case of fatal encephalitis in an immunosuppressed child. The virus was detected in stool and serum, with highest levels in brain and cerebrospinal fluid (CSF). Immunohistochemistry of the brain biopsy showed positive neuronal staining. A survey of 680 stool and 349 CSF samples identified a related virus in the stool of another immunosuppressed child.
Conclusions. The discovery of HAstV-VA1/HMO-C-UK1(a) as the cause of encephalitis in this case provides further evidence that VA1/HMO-C viruses, unlike HAstV 1–8, are neuropathic, particularly in immunocompromised patients, and should be considered in the differential diagnosis of encephalopathy. With a turnaround from sample receipt to result of <1 week, we confirm that RNA-Seq presents a valuable diagnostic tool in unexplained encephalitis.
(See the Editorial Commentary by Calistri and Palù on pages 889–91, and Brief Report by Naccache et al on pages 919–23.)
Encephalitis is characterized by inflammation in the brain parenchyma associated with clinical evidence of brain dysfunction [1, 2]. The majority of encephalitis cases are thought to be viral in origin; however, in up to 63% of cases a causative agent is not found [3]. Cases with unknown etiology pose difficulties in patient management, particularly when a patient is immunosuppressed, because the treatment for infectious and noninfectious cases differs greatly, with the latter sometimes requiring additional immunosuppression, which can be detrimental if a pathogen is present.
To investigate a case of encephalopathy in an immunosuppressed 18-month-old boy for which no cause could be found by traditional techniques, we applied RNA sequencing technologies to a brain biopsy. Deep sequencing of the transcriptome has previously been used to identify viral flora and disease-associated pathogens in human tissues [4–8]. A key feature of this methodology is that it does not make prior assumptions about the type of pathogen, but has the potential to detect RNA from all species.
Here we describe the use of this strategy in a routine diagnostic setting to identify astrovirus (AstV) of the VA1 and human-mink-ovine–like (HMO)-C subgroup as the likely causative agent in this case. The result, which was obtained within 6 days of brain biopsy, informed subsequent patient management. To investigate further the need for routine diagnostic testing for VA1/HMO-C viruses, we screened 680 samples of stool and 349 of cerebrospinal fluid (CSF) from undiagnosed cases of diarrhea and encephalitis, respectively. Our data underline the clinical significance of VA1/HMO-C AstV as a clinically significant pathogen in immunocompromised patients.
METHODS
Patient and Samples
Patient A, an 18-month-old boy with cartilage hair hypoplasia and associated immunodeficiency due to RMRP mutations, underwent a peripheral blood stem cell transplant on 28 March 2013 from a 10/10 human leukocyte antigen–matched unrelated donor following conditioning with fludarabine (150 mg/m2 over 5 days), melphalan (140 mg/m2 day –2), and alemtuzumab (1 mg/kg over 5 days) for in vivo T-cell depletion. Ciclosporin A and mycophenolate mofetil were given as graft-vs-host disease (GVHD) prophylaxis.
Transplant was uncomplicated with neutrophil engraftment achieved by day 13. Persistent stool viruses (sapovirus, adenovirus) were detected by polymerase chain reaction (PCR). Adenoviremia occurred from day 18 onward with peak load of 201 697 copies/mL on day 21. This required treatment with cidofovir (5 mg/kg 1 dose) and donor-derived adenovirus-specific cytotoxic T-lymphocytes (1 × 104/kg total CD3+ cells) on day 28. Adenovirus load declined to 820 copies/mL 12 days after infusion (Supplementary Figure 1). Two weeks later, the child became acutely unwell with irritability, dystonia, and reduced consciousness. Encephalopathy secondary to drug toxicity, infectious complications, or inflammation associated with immune reconstitution were considered in the differential diagnosis. Ciclosporin was stopped on day 43 and replaced by methylprednisolone therapy (1 mg/kg).
Magnetic resonance imaging of the brain showed progressive changes with loss of volume (Supplementary Figure 2), and electroencephalography revealed increased amplitudes of ongoing slow activity over posterior regions with no focal features or epileptiform abnormalities. PCR of CSF 2 days, 2 weeks, and 1 month after onset of symptoms did not detect adenovirus, herpes simplex virus type 1 or 2, varicella zoster virus, cytomegalovirus, Epstein-Barr virus, BK virus, norovirus, sapovirus, astrovirus (HAstV 1–8), rotavirus, enterovirus, measles, parechovirus, JC virus, human herpesvirus type 6 or 7, or Toxoplasma. Brain biopsy performed 8 weeks after neurological deterioration was also negative for these pathogens as well as 18S (fungal) and 16S (bacterial) ribosomal RNA gene PCR assays. Histological examination of the brain (described below) showed an unusual pattern of pathology, but the differential diagnosis included encephalitis of infectious etiology. Withdrawal of immunosuppression was associated with the development of skin GVHD (grade 2, stage 3) requiring methylprednisolone (2 mg/kg/day). As a result, kinetics of T-cell reconstitution were reduced with a CD3 count of 0.35 × 109/L at 6 months, and in the context of ongoing neurological impairment with recurrent respiratory and gastrointestinal complications, the patient died 9 months after transplant.
Deep Sequencing
Total RNA was purified from a fresh brain biopsy and polyA RNA separated for sequencing library preparation. Rapid RNA-Seq was performed on an Illumina Miseq. Nonhost data, assembled into contigs, and remaining nonassembled reads that showed only partial homology to protein database sequences using BLASTx were analyzed using a Bayesian mixture model [9] to obtain a concise summary of the species present in the mixture. This approach overcomes the limitation of existing methods for analysis of short read data, which are not designed to detect pathogens at low levels. In this context, we use a Monte Carlo Markov chain (MCMC) strategy to identify the subset of species that best explains the data. Multiple MCMC chains are run in parallel using a population MCMC strategy to speed up convergence. Each potential species is then assigned a posterior probability that quantifies the support for the presence of this infectious agent.
Pan-Astrovirus PCR and Sequencing
To confirm the presence of AstV in the brain biopsy, a pan-astrovirus heminested PCR assay was carried out as described previously [10].
PCR products were capillary sequenced using BigDye 3.1 (Life Technologies) and an ABI 3130. Sequencing data was visualized and analyzed with SeqMan Pro version 11.1 (DNA Star). The consensus sequence was checked for similarity to known AstV sequences using BLASTn [11].
Real-time Reverse Transcription PCR
Contig sequences generated from deep sequencing were used to design a real-time reverse transcription PCR assay specific to the AstV identified in this study, named AstV-contig. Primers are shown in Supplementary Table 1.
An AstV-contig PCR assay was used to retrospectively test all stored samples for patient A during his hospital admission. These samples were from stool (n = 17), nasopharyngeal aspirate (NPA) (n = 12), urine (n = 1), sera (n = 2), and CSF (n = 3).
To investigate the possible source of infection, all stored specimens from patients on the same ward as patient A, which were collected from 2 weeks before to 2 weeks after patient A's first positive AstV-contig PCR result, were tested by the AstV-contig PCR assay. This included 18 samples (14 stool and 4 urine) from 9 patients. For patient B, who had a single positive stool sample, an additional 14 stools, 23 NPAs, and 3 sera were tested by the AstV-contig PCR.
Genome Sequencing
Twelve tiled primer sets were designed to cover the full AstV genome. They were designed using the full genome sequences for AstV-VA1 and -SG and partial sequences for HMO-C (genbank accession numbers FJ973620, GQ891990, GQ415662, GQ441191, GQ441192), as these showed 95%–98% identity with the pan-astrovirus PCR product from the brain biopsy. Primer sequences are shown in Supplementary Table 1. The terminal 3′ end of the genome was obtained by rapid amplification of complementary DNA (cDNA) ends (RACE, Invitrogen).
The consensus sequence was checked for similarity to known AstV sequences using BLAST and used for phylogenetic analysis.
Phylogenetic Analysis
Clustal W was used to align a 432 nucleotide region of the RNA-dependent RNA polymerase (RdRp) and 2460 nucleotide region of the capsid gene with 39 other AstV sequences. Sequences included representatives of the classical human astroviruses HAstV1–8, bat, mink, ovine, and, to our knowledge, all of the recently described novel HAstVs MLB-1–3, HMO-A–C, VA-1–4, and -SG. Turkey AstV was used as an outgroup. The aligned regions correspond to nucleotides 3432–3864 and 3628–4042 for the RdRp alignment and 4382–6733 and 4210–6486 for the capsid of AstV-SG and HAstV-1, respectively.
The RdRp phylogenetic tree was reconstructed using the neighbor-joining method [12] with 1000 bootstrap replicates [13]. The evolutionary distances were computed using the maximum composite likelihood method [14] and are in units of the number of base substitutions per site. All positions containing gaps and missing data were eliminated. Evolutionary analyses were conducted in MEGA5 [15].
Immunohistochemistry
Sections of the brain biopsy were stained using AstV-specific antibody as used previously [16], kindly provided by Professor Ian Lipkin. The immunohistochemistry was performed using a titer of 1:1000 on a Leica Bond Max automated system stained with antigen retrieval solution 2, for 10 minutes.
Epidemiology
To determine the local prevalence of AstV VA1/HMO-C, we tested 686 stool and 349 CSF samples from immunocompetent and immunocompromised patients. Stool samples were collected December 2013–February 2014 and CSF samples from September 2013 to March 2014. Samples were screened using the AstV-contig PCR assay and a second, more broad-range assay, VA1-ORF1. Primer and probe sequences are in Supplementary Table 1.
RESULTS
Deep Sequencing
The RNA-Seq data consisted of 20 588 062 paired-end reads, of which 92% passed quality control (nonrepetitive sequence and average Phred scaled quality score >15) (Table 1). Reads that could not be assigned to the human genome (77 915 reads, 0.4% of total reads) were potentially of foreign origin. Our mixture model identified 2 viruses with confidence, 1 of which was the phiX bacteriophage (79% of nonhost reads) that was used for quality control of the Illumina high-throughput sequencing. Five assembled contigs (44 reads) and 2 nonassembled reads (therefore 46 reads overall) were assigned to the AstV VA1 sequence (genbank accession number FJ973620) with 97% sequence identity and a probability score of 1. After adding the AstV VA1 and the bacteriophage phiX species in our mixture model, we found no evidence for any additional viruses in the data.
Table 1.
Deep-Sequencing Read Data
| Data | Read Count | Total Reads | Nonhuman Reads |
|---|---|---|---|
| Raw data | 20 588 062 | 100% | |
| Quality control passed data | 18 877 676 | 91.69% | |
| Nonhuman data | 77 915 | 0.38% | |
| PhiX control | 61 758 | 0.3% | 79% |
| Astrovirus | 46 | 0.0002% | 0.06% |
| Environmental bacteriaa | 470 | 0.002% | 0.6% |
| Unidentified | 15 377 | 0.07% | 20% |
| Humanb | 264 | 0.001% | 0.3% |
aSphingobium yanoikuyae, Afipia broomeae, Roseomonas cervicalis, Massilia timonae, Achromobacter piechaudii, Acinetobacter species, Serratia odorifera, Citrobacter youngae, Escherichia coli, Propionibacterium acnes, Sphingobacterium spiritivorum, Streptococcus mitis.
bReads that were not filtered out at the nucleotide level (BLASTn) but identified at the amino acid level (BLASTx).
In addition to viral sequences, 0.6% (<500 reads) of the nonhost reads were assigned to environmental bacterial species. A fraction (0.3%) of the remaining reads was assigned to human sequences initially absent from our sequence database. The remainder (20%) of the nonhuman data did not match any database sequence reliably, which could result from sequencing errors or species without homology to known sequences (Table 1).
The AstV assigned contigs range from 167 to 471 bp in length. On the amino acid level, the percentage identity they share with AstV-VA1 viral proteins is 96%–100%. The contigs were mapped with BLASTn against the genome of AstV VA1, showing that the reads map to the 5′ and 3′ ends of the viral genome (Supplementary Figure 3A). The assembled contigs map equally well to the 3′ and 5′ ends of AstV-SG and the 3′ end of HMO-C (genbank accession numbers GQ891990 and GQ415662). We have named the AstV identified in this study HAstV-VA1/HMO-C-UK1(a).
Full Genome Analysis and Phylogeny
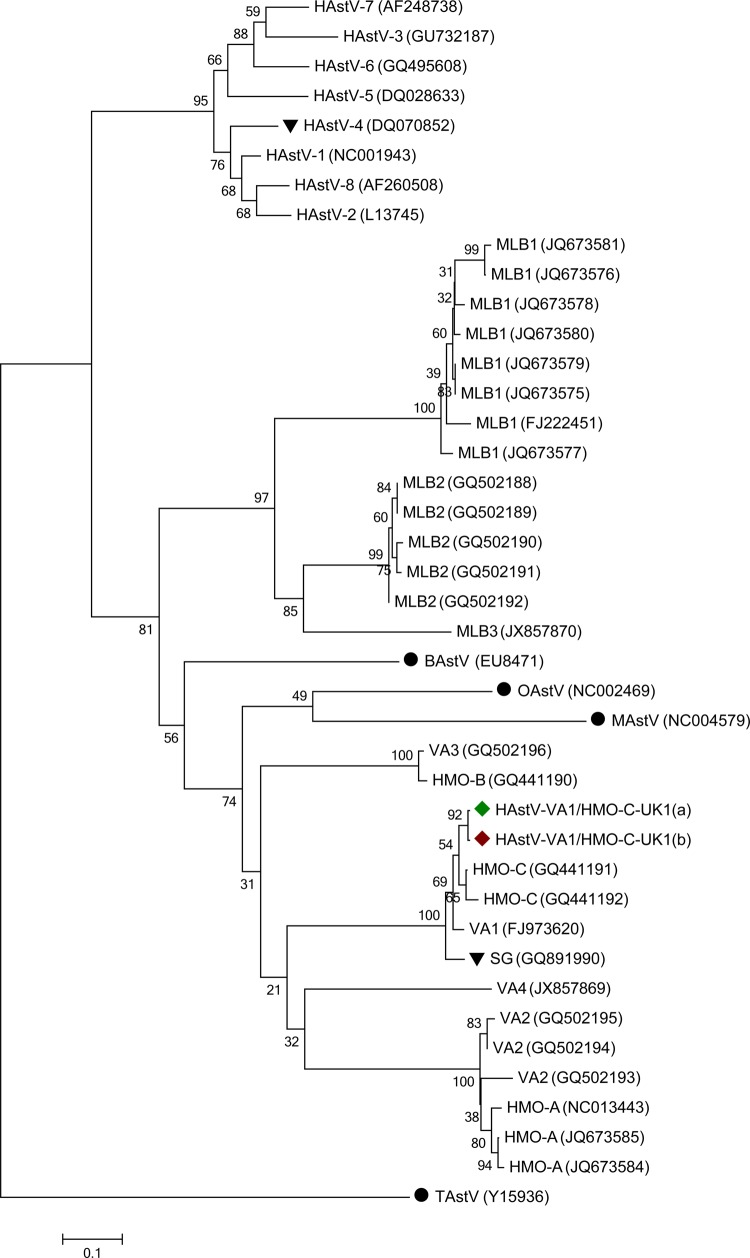
Phylogenetic analysis of the RdRp gene places HAstV-VA1/HMO-C-UK1(a) in a cluster with HMO-C, -VA1, and -SG (Figure 1) with 98%, 97%, and 95% nucleotide sequence identity, respectively (Supplementary Figure 4A). This group of viruses is more closely related to mink and ovine AstV sequences than HAstV 1–8. The phylogenetic relationship between HAstV-VA1/HMO-C-UK1(a) and other AstVs is unchanged when analyzing RdRp, capsid, or full genome (Figure 1 and Supplementary Figure 4; phylogenies for capsid and full genome not shown), suggesting that recombination with other known strains has not occurred.
Figure 1.
Phylogenetic relationship of human astrovirus (HAstV)–VA1/HMO-C-UK1(a) ( ) and -UK1(b) (
) and -UK1(b) ( ) to other astroviruses (AstVs) based on RNA-dependent RNA polymerase nucleotide sequences.
) to other astroviruses (AstVs) based on RNA-dependent RNA polymerase nucleotide sequences.  indicates other AstV species that have been reported in patients with neurological disease.
indicates other AstV species that have been reported in patients with neurological disease.  indicates sequences of animal origin, not human. Scale bar represents the number of base substitutions per site. Abbreviations: BAstV, bat astrovirus; MAstV, mink astrovirus; OAstV, ovine astrovirus; TAstV, turkey astrovirus.
indicates sequences of animal origin, not human. Scale bar represents the number of base substitutions per site. Abbreviations: BAstV, bat astrovirus; MAstV, mink astrovirus; OAstV, ovine astrovirus; TAstV, turkey astrovirus.
The 6563 nucleotide (excluding the 5′ untranslated region) HAstV-VA1/HMO-C-UK1(a) genome (GenBank accession number KJ920196) has 95% sequence identity with AstV-SG (Supplementary Figure 4B), which has previously been described in a fatal case of encephalitis in an immunosuppressed patient [16]. Conversely, the percentage sequence identity of HAstV-VA1/HMO-C-UK1(a) and classical human astrovirus (HAstV-1), normally associated with diarrhea, is <47%.
Confirmation by PCR
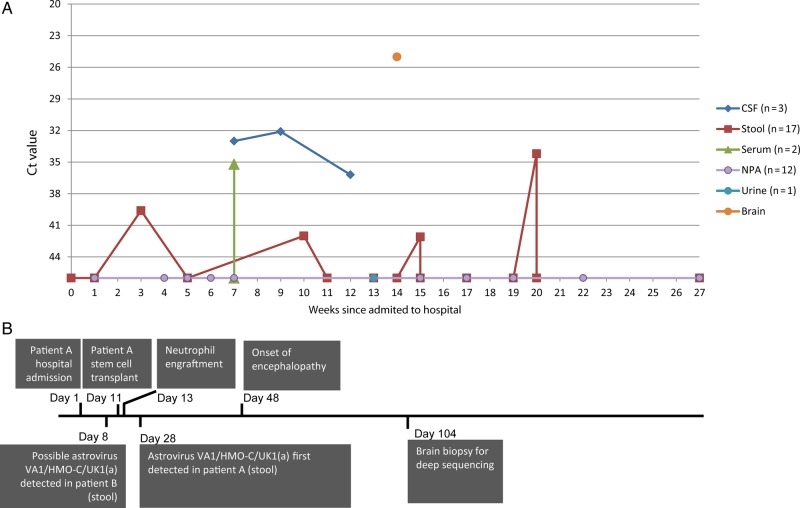
cDNA from the brain biopsy was positive by pan-astrovirus and both real-time PCR assays, confirming the presence of AstV in the brain tissue. A 560-bp amplicon generated by the former showed 95%, 97%, and 98% identity to AstVs SG, VA1, and HMO-C, respectively. The results of the real-time astrovirus PCR on 35 stored samples from patient A show the highest viral titer (cycle threshold [Ct] 25) in the brain biopsy, consistently positive CSF, and intermittently positive stool and serum (Figure 2 and Supplementary Table 2).
Figure 2.
A, AstV-contig polymerase chain reaction results for all retrospectively tested samples from patient A. Data points at the base of the y-axis indicate astrovirus (HAstV)–VA1/HMO-C-UK1(a) RNA not detected. Cycle threshold (Ct) values represent the cycle number at which amplification was detected and thus have an inverse relationship with viral titer; a smaller Ct value indicates a higher titer. In clinical practice, Ct >38 may be considered equivocal but, in the context of other positive results, is considered a true positive. A difference of 3 Ct values is approximately equivalent to a 10-fold difference in viral load. B, Clinical and diagnostic timeline. Abbreviations: AstV, astrovirus; CSF, cerebrospinal fluid; HAstV, human astrovirus; NPA, nasopharyngeal aspirate.
Histopathology
Routine histopathology investigations of the brain biopsy showed frequent neuronal apoptosis demonstrated by neuronal karyorrhexis (Figure 3A). There was microglial activation confirmed on CD68 immunohistochemistry (Figure 3B). There was diffuse activation of microglial but also small clusters, some of which were associated with neurons, and occasional areas suggestive of neuronophagia were seen. However, lymphocytes were inconspicuous on microscopy and immunohistochemistry (Figure 3C), and there were no acute inflammatory cells. Extensive astrocytosis (Figure 3D) was confirmed on glial fibrillary acidic protein immunohistochemistry.
Figure 3.
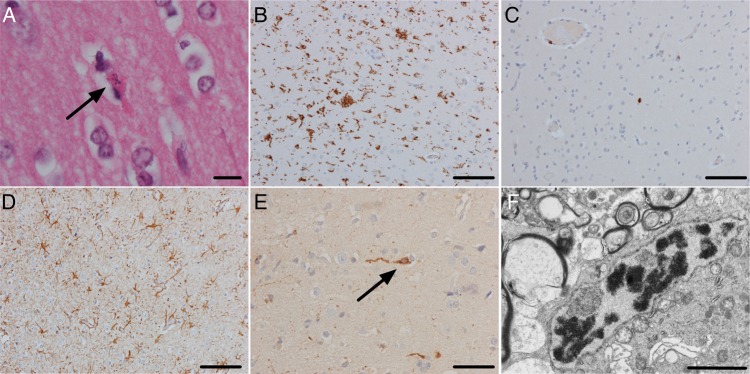
The brain biopsy showed extensive neuronal apoptosis in the cerebral cortex as demonstrated by pyknotic and karyorrhectic neurons (A, arrow) (hematoxylin and eosin [H&E], scale bar, 10 µm). There was a brisk microglial reaction demonstrated by CD68 immunohistochemistry with some nodules of microglia (B; scale bar, 100 µm). In contrast, there was no significant lymphocytic reaction (C, CD3 immunohistochemistry; scale bar, 100 µm). There was an extensive astrocytosis (D, glial fibrillary acidic protein [GFAP]; scale bar, 100 µm). Immunohistochemistry with an antibody against astrovirus showed extensive staining of cell bodies and processes in the neuropil (E; scale bar, 50 µm). Some of the positive cells had the morphology of pyramidal neurons (arrow). Electron microscopy showed a rare focus of crystalline material but no viral particles (F; scale bar, 2 µm).
Electron microscopy showed frequent microglial activation and microglial surrounding neurons. Many neurons appeared shrunken and electron-dense. Viral material was not seen, but a rare focus of crystalline material was found in neurons (Figure 3F).
Immunohistochemistry
Immunohistochemistry for AstV was positive, staining cell bodies and processes within the neuropil (Figure 3E). Several of the positive cells had the morphology of pyramidal neurons.
qPCR Screening Within the Ward
Results for screening of patients from the same ward identified 1 (1/14) positive stool sample (Ct 38; Supplementary Table 3) from a single patient (patient B), collected 1 week after patient A was admitted and 2 weeks before patient A's first positive sample. There was insufficient residual sample for repeat testing by quantitative PCR or for sequencing; therefore, the positive result could not be confirmed. No other stored samples from patient B were positive.
qPCR Screening Within Local and Other Hospitals
Wider screening identified 1 (1/686) positive stool and no (0/349) positive CSFs (Supplementary Table 3). The single positive stool came from an immunosuppressed child, patient C, with diarrhea. The full genome, HAstV-VA1/HMO-C-UK1(b) (genbank accession number KJ920197), was sequenced by primer walking and included in the phylogenetic analysis (Figure 1 and Supplementary Figure 4). HAstV-VA1/HMO-C-UK1(b) had 99.4% identity to HAstV-VA1/HMO-C-UK1(a).
DISCUSSION
Encephalopathy occurring in immunodeficient subjects or recipients of stem cell transplants can present a diagnostic dilemma; distinguishing infection from other potential causes such as drug toxicity or GVHD can be difficult. In the case outlined here of encephalitis occurring in an immunosuppressed 18-month-old boy, having excluded common pathogens using conventional methods, we resorted to deep sequencing of brain tissue from which we were able to identify astrovirus, HAstV-VA1/HMO-C-UK1(a). Based on phylogeny, we propose that this virus belongs to the VA1 and HMO-C group of astroviruses. VA1/HMO-C astroviruses are part of a divergent group that is related to Astroviridae found in association with neurological illnesses in mink [17] and cattle [18]. Our finding represents the second report of an AstV in the VA1/HMO-C group causing fatal encephalitis, which confirms a pathogenic role for this virus group likely to have been previously underrecognized in immunocompromised patients. The first case described, AstV-SG, is phylogenetically close to HAstV-VA1/HMO-C-UK1(a) and was also found by deep sequencing of brain tissue from a 15-year-old boy with X-linked agammaglobulinemia who developed unexplained encephalitis [16]. Two more distantly related viruses in this phylum, HMO-A and -B, were detected in stool from children with nonpolio flaccid paralysis; however, healthy controls were not included in the study, so the clinical significance of HMO-A and -B is unclear. Other related HMO-C viruses have been associated with diarrhea in children living in Nepal [10] and the United States [19], suggesting a global distribution of this virus.
The neuropathology on brain biopsy was distinctive with frequent neuronal apoptosis associated with a microglial reaction but lacking a significant lymphocytic inflammatory response. Whereas the pattern of microglial reaction raised the possibility of viral encephalitis, the pattern of neuronal cell death would be unusual in most typical viral encephalitides.
The pattern of both gastrointestinal and neurological infections caused by VA1/HMO-C viruses contrasts with that of conventional human Astroviridae, HAstV 1–8, which are primarily associated with gastroenteritis. For HAstV-VA1/HMO-C-UK1(a), RNA in the brain was confirmed by 3 different PCR assays and we were able to generate full-genome sequences using overlapping PCR. The positive immunohistochemistry for the capsid protein provided further confirmation of replication competent virus in the brain. The highest titers of viral RNA were found in the brain tissue, and RNA was consistently detected in the CSF. Significantly lower levels of viral RNA were found in stool and serum (plasma and whole blood were not available), the latter confirming viremic spread. By contrast, 1 case of HAstV-4 has been identified in brain tissue from a patient with severe combined immunodeficiency following a hematopoietic stem cell transplant [20]; however, this was associated with much higher levels of viral RNA in the stool, significantly lower levels in the brain, and no immunohistochemical evidence of viral proteins in brain, although intestinal staining was positive.
A cross-sectional community study of diarrhea identified HMO-C viruses in 2% of cases of diarrhea and 1% of controls, compared with 13% of cases due to HAstV [10]. This low prevalence fits with data from our study of 680 stool and 349 CSF samples from all ages that identified only 2 patients (patients B and C), both immunocompromised, with evidence of the virus in the stool. In patient B, the infection may have been transient or even caused by contamination, as the viral load was at the limit of detection of the assay and none of 42 (16 stool, 3 serum, 23 NPA) additional samples from the child were positive. Whole-genome sequencing of the AstV from the second patient, patient C, which we have termed HAstV-VA1/HMO-C-UK1(b), revealed it to be 99.4% similar to patient A's virus, which is phylogenetically closer than has been observed for other co-circulating VA1/HMO-C viruses . The result suggests that transmission may have occurred between patients A and C, although without additional sequences from the United Kingdom this cannot be confirmed. Because infection in patients A and C occurred many months apart and they were never together on the same ward, transmission, if it occurred, is likely to have been indirect. Similar to HAstV 1–8, antibodies against HMO-C viruses have been shown to increase with age, reaching a prevalence of 65% in adults in the United States [21]. Taken together, these data suggest that in healthy individuals, VA1/HMO-C AstV infection is common but generally asymptomatic. By contrast, our data support VA1/HMO-C as a neuropathic agent in immunocompromised patients and one that should be looked for in cases of encephalitis.
In summary, we have used state-of-the-art deep sequencing in a routine diagnostic setting together with PCR and immunohistochemistry to identify AstV VA1/HMO-C as the cause of encephalitis of unknown origin occurring in an immunocompromised child and a possible case of nosocomial spread. We have used deep-sequencing methodologies in a diagnostic setting to identify the causative agent in unknown encephalitis within 6 days of receiving the specimen. The result excluded drug toxicity and immune-mediated inflammation as causes for the clinical picture, and informed a decision to reduce immunosuppression. This case illustrates the utility of deep sequencing in a clinical setting to identify unexpected or novel pathogens.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online (http://cid.oxfordjournals.org). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. Written consent for publication was obtained from the patient's family, with thanks to the Great Ormond Street (GOS) Hospital / Institute of Child Health Biomedical Research Council and GOS Hospital Special Trustees.
Disclaimer. The views expressed in this publication are those of the authors and not necessarily those of the National Health Service (NHS), the National Institute for Health Research (NIHR) or the Department of Health.
Financial support. This work was supported by NIHR (grant number NIHR-HCS-D12-03-15 to J. R. B.), the NIHR GOS Hospital Biomedical Research Centre and the NIHR Biomedical Research Centre at University College London Hospitals NHS Foundation Trust and University College London.
Potential conflicts of interest. All authors: No potential conflicts of interest.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Thompson C, Kneen R, Riordan A, Kelly D, Pollard AJ. Encephalitis in children. Arch Dis Child. 2012;97:150–61. doi: 10.1136/archdischild-2011-300100. [DOI] [PubMed] [Google Scholar]
- 2.Koskiniemi M, Korppi M, Mustonen K, et al. Epidemiology of encephalitis in children. A prospective multicentre study. Eur J Pediatr. 1997;156:541–5. doi: 10.1007/s004310050658. [DOI] [PubMed] [Google Scholar]
- 3.Glaser CA, Honarmand S, Anderson LJ, et al. Beyond viruses: clinical profiles and etiologies associated with encephalitis. Clin Infect Dis. 2006;43:1565–77. doi: 10.1086/509330. [DOI] [PubMed] [Google Scholar]
- 4.Foulongne V, Sauvage V, Hebert C, et al. Human skin microbiota: high diversity of DNA viruses identified on the human skin by high throughput sequencing. PLoS One. 2012;7:e38499. doi: 10.1371/journal.pone.0038499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johansson H, Bzhalava D, Ekstrom J, Hultin E, Dillner J, Forslund O. Metagenomic sequencing of “HPV-negative” condylomas detects novel putative HPV types. Virology. 2013;440:1–7. doi: 10.1016/j.virol.2013.01.023. [DOI] [PubMed] [Google Scholar]
- 6.Handley SA, Thackray LB, Zhao G, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–66. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mokili JL, Dutilh BE, Lim YW, et al. Identification of a novel human papillomavirus by metagenomic analysis of samples from patients with febrile respiratory illness. PLoS One. 2013;8:e58404. doi: 10.1371/journal.pone.0058404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Willner D, Furlan M, Haynes M, et al. Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One. 2009;4:e7370. doi: 10.1371/journal.pone.0007370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morfopoulou S, Plagnol V. Bayesian mixture analysis for metagenomic community profiling. bioRxiv. 2014 doi: 10.1093/bioinformatics/btv317. doi:http:/dx.doi.org/10.1101/007476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kapoor A, Li L, Victoria J, et al. Multiple novel astrovirus species in human stool. J Gen Virol. 2009;90(pt 12):2965–72. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 12.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–5. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Quan PL, Wagner TA, Briese T, et al. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis. 2010;16:918–25. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol. 2010;48:4392–6. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li L, Diab S, McGraw S, et al. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis. 2013;19:1385–92. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finkbeiner SR, Li Y, Ruone S, et al. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol. 2009;83:10836–9. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wunderli W, Meerbach A, Gungor T, et al. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One. 2011;6:e27483. doi: 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burbelo PD, Ching KH, Esper F, et al. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS One. 2011;6:e22576. doi: 10.1371/journal.pone.0022576. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.