Abstract
Background
Circulating microRNA (miRNA) are promising biomarkers for diagnosing and prognosticating numerous diseases. Reports have demonstrated controversial or even contradictory conclusions in studies on circulating microRNA. This study aimed to evaluate the potential bias of using different reference genes for analyzing circulating microRNAs in the same malignant digestive diseases.
Material/Methods
We measured plasma concentrations of U6-snRNA, let-7a, miRNA-21, miRNA-106a, miRNA-155, miRNA-219, miRNA-221, and miRNA-16 in patients with hepatocellular carcinoma (HCC), gastric carcinoma (GC), hepatic cirrhosis, hepatitis B, and healthy volunteers using quantitative real-time polymerase chain reaction (qPCR). The GeNorm, Normfinder, BestKeeper, and Comparative ΔCq algorithms integrated in RefFinder were used to screen the most suitable reference genes from the candidates. The 4 commonly used statistical evaluation software packages provided different results regarding the stability of the candidate reference genes.
Results
RefFinder revealed miRNA-106a and miRNA-21 as the most stably expressed reference genes, with comprehensive stability values of 1.189 and 1.861, respectively. U6-snRNA was the most unstable nucleic acid in our data. When 5 normalization strategies were compared using U6-snRNA, serum volume, miRNA-106a, miRNA-21, or the mean value of miRNA-106a and miRNA-21, obvious expression bias was detected in almost all target microRNAs. Intriguingly, all these normalization strategies indicated that circulating miRNA-155 is greatly upregulated in patients with HCC and GC, but downregulated in benign hepatic disease.
Conclusions
Single reference genes used without justification in plasma microRNAs produce significant analysis bias or even erroneous results. Circulating miRNA-155 may be a promising non-invasive biomarker for discriminating malignant digestive tumors from the corresponding benign diseases.
MeSH Keywords: Carcinoma, Hepatocellular; Genes, vif; MicroRNAs; Real-Time Polymerase Chain Reaction
Background
MicroRNAs (miRNA) are the most promising disease biomarkers in plasma. They are 18–22 nucleotide-long, single-stranded RNA molecules that regulate gene expression and influence the physiologic and pathologic processes of cells. MicroRNA deregulation plays an important role in the disproportionate proliferation and suppressed apoptosis of malignant cells. Abnormal microRNA expression has been demonstrated in numerous diseases, including tumors [1]. Serum microRNAs are highly stable under harsh conditions, including heating, pH alteration, extended storage, and freeze-thaw cycles, without any significant differences compared with untreated serum samples [2–4]. Therefore, plasma/serum microRNAs are the next generation of serum markers for disease diagnosis, prognosis, and therapy, especially for various cancers [5–7]. However, studies on circulating microRNA have demonstrated controversial or even contradictory conclusions, even for the same disease. For example, let-7a is reportedly decreased in GC patients [8,9], but it is increased in hepatocellular carcinoma (HCC) [10], and could be used as reference genes in cervical tissues [11]. Researchers have not established a consensus on whether miRNA-21 distinguishes HCC and other malignant tumors from benign liver diseases and healthy individuals [12–17].
The expression levels of a target gene in samples is usually determined by quantitative real-time PCR (qPCR) and normalized to a reference gene, which allows adjusting the variations in measurements of different samples. The ideal reference gene remains at consistent levels in all detected samples, irrespective of the disease states, thereby allowing samples to be compared even with different quantities or qualities. Using improper reference genes for normalization can result in evaluation bias during data analysis. To our knowledge, there is no current consensus on the use of reference genes in qPCR to analyze circulating microRNA. Aside from ethnic and individual differences and the inconsistent disease process and/or activity, the lack of a unified reference gene and uniform rules for selecting reference genes may explain the lack of consensus [15]. In this study, we aimed to evaluate the potential bias of using different reference genes for analyzing circulating microRNA in patients with HCC and GC and their corresponding healthy controls. Consequently, we detected and analyzed microRNA, such as miRNA-106a, miRNA-16, miRNA-221, miRNA-219, let-7a, miRNA-155, miRNA-21, and the commonly used reference gene U6-snRNA.
Material and Methods
Sample collection
Randomly selected plasma samples were routinely collected from patients at the Changhai Hospital between 1 January 2012 and 31 July 2012. Venous blood was collected in EDTA anticoagulation vacuum tubes before any medical treatment. Samples were then centrifuged (10 min, 2500 g), separated, and then stored at −80°C within 2 h until analysis. This study cohort included 120 individuals, including 30 patients with non-metastatic HCC (18 males, 12 females; median age 60, range 39–76 years), 30 gastric cancer patients without metastasis (GC, 16 males, 14 females; median age, 58 range 39–78 years), 20 patients with hepatic cirrhosis (11 males, 9 females; median age 60, range 38–72 years), 20 patients with hepatitis B (11 males, 9 females; median age 59, range 36–69 years), and 20 healthy volunteers (11 males, 9 females; median age 58, range 38–75 years). Samples of patients with hepatic cancers and gastric cancers were all collected before surgeries. All these cancers were diagnosed pathologically and without metastasis. No positive serum or nucleic acid indicators of other types of hepatitis viruses were detected. Patients with primary hepatic cancers were all of HCC and had histories of hepatitis B virus with serum test results of small 3 positives (positive HBsAg, HBeAb, and HBcAb) or big 3 positives (positive HBsAg, HBeAg, and HBcAb), regardless of positive or negative serum hepatitis B virus DNA. Age- and sex- matched gastric cancer patients served as a cancer control group, who were without histories of hepatitis B, other types of hepatitis, or hepatic cirrhosis. Pathological types of gastric cancers were all adenocarcinomas or undifferentiated carcinomas. Patients with hepatic cirrhosis were all diagnosed clinically and had histories of hepatitis B rather than other types of hepatitis. They were all age- and sex- matched with the hepatic cancer group. Patients with hepatitis B virus were diagnosed via quantitative tests of hepatitis B virus DNA and were all age- and sex-matched with the hepatic cancer group. Written informed consent was obtained from each subject, and the study was approved by the local ethics committee.
RNA isolation, reverse transcription, and quantitative real-time PCR
An Applied Biosystems stemloop RT/PCR kit (AB) and TaqMan microRNA Assays were used to detect and quantify selected mature microRNA from fresh-frozen samples using sequence-specific primers from the TaqMan Assay Plates. All PCR experiments were carried out on an ABI Prism 7900HT (Applied Biosystems, USA) in 384-well plates. The specificity of the PCR products was confirmed through melting curve analysis. Each run also included water blanks and genomic DNA as negative controls.
Data analysis
The Cq values from triplicate wells of each sample were used to calculate intra-assay precision, and PCR amplification efficiency was determined as previously described [15]. The stabilities of candidate reference microRNAs were evaluated using RefFinder, a user-friendly web-based comprehensive tool for evaluating and screening reference genes from extensive experimental datasets. RefFinder integrates the currently available major computational programs (GeNorm [18], Normfinder [19], BestKeeper [20], and comparative delta-Cq method algorithms [21]) to separately and comprehensively compare and rank the tested candidate reference genes (http://www.leonxie.com/referencegene.php?type=reference) [15]. The expression of the target microRNAs relative to those of selected normalized was calculated using the ΔΔCq method, as previously described [22]. When normalized to serum volume, the average Cq values for each of the microRNAs in all 120 samples were used as references in the ΔΔCq. Statistical analyses were carried out using the Kruskal-Wallis test on GraphPad Prism 5.01.
Results
Characteristics of candidate reference genes quantified by qPCR
Based on literature screening, we used the small nuclear RNA U6-snRNA and 7 microRNAs (Table 1). Let-7a, U6-snRNA, miRNA-21 miRNA-106a, miRNA-221, and miRNA-16 were used as reference genes to normalize the circulating microRNA data from published articles [14,15]. We evaluated miRNA-219 and miRNA-155 simultaneously, together with the candidate reference genes supposed to be analyzed as target genes when different normalization strategies were compared in this study. The expression levels of these genes were determined using TaqMan qPCR assays, as described in the Methods section. All of the qPCR assays showed high amplification efficiency and low intra-assay variations (Table 1). However, the lower copy number when using plasma resulted in lower but acceptable intra-assay precision for miRNA-221 and miRNA-219 (Table 1).
Table 1.
The assay performance of selected microRNAs.
| Gene name | Accession | Sequence | Length (nt) | PCR efficiency (%) | Intra-assay Precision | |
|---|---|---|---|---|---|---|
| Mean CV (%) | Min-Max (%) | |||||
| let-7a | MIMAT0000062 | UGAGGUAGUAGGUUGUAUAGUU | 22 | 98 | 0.38 | 0.01–1.41 |
| miR-106a | MIMAT0000103 | AAAAGUGCUUACAGUGCAGGUAG | 23 | 96 | 0.41 | 0.08–1.95 |
| miR-155 | MIMAT0000646 | UUAAUGCUAAUCGUGAUAGGGGU | 23 | 99 | 0.38 | 0.04–1.09 |
| miR-16 | MIMAT0000069 | UAGCAGCACGUAAAUAUUGGCG | 22 | 97 | 0.51 | 0.04–2.05 |
| miR-21 | MIMAT0000076 | UAGCUUAUCAGACUGAUGUUGA | 22 | 97 | 0.36 | 0.04–1.69 |
| miR-219 | MIMAT0000276 | UGAUUGUCCAAACGCAAUUCU | 21 | 95 | 1.61 | 0.09–4.49 |
| miR-221 | MIMAT0004568 | ACCUGGCAUACAAUGUAGAUUU | 22 | 96 | 1.71 | 0.11–6.23 |
| U6-snRNA | NR_002752.2 | Omitted due to length | 107 | Not detected | 0.44 | 0.01–0.99 |
CV – coefficient of variation.
Evaluation of expression gene stability in plasma
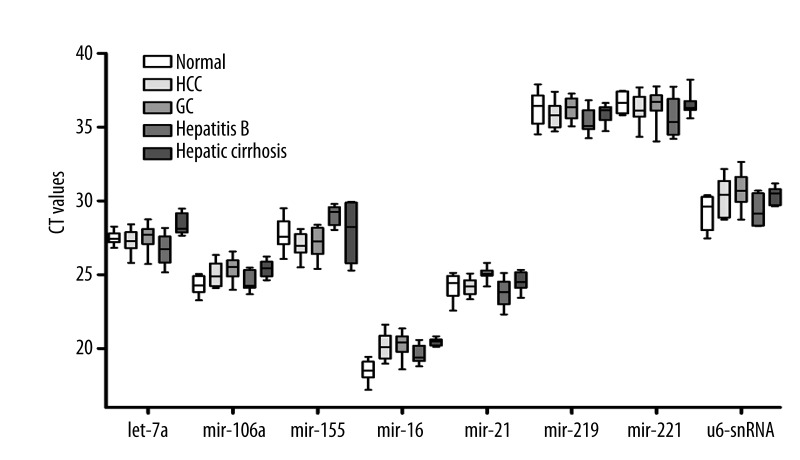
We also investigated whether a single gene could serve as a reference for the different genes among all the patients and control volunteers. All 8 genes were measured separately in the 120 samples. The 8 microRNAs exhibited wide expression ranges, with Cq values between 17.19 and 37.89, as shown in Figure 1. The expression levels of miRNA-219 and miRNA-221 were relatively low, with mean Cq values of 35.93 and 36.30, respectively. According to the Cq values, the candidate reference microRNAs did not significantly differ among HCC, hepatitis B, hepatic cirrhosis, GC patients, and healthy controls (P>0.05).
Figure 1.
Expression of candidate reference genes in plasma samples from patients and healthy volunteer. QPCR analyses were performed on the 120 samples collected. The expression levels of the candidate reference genes are given as absolute Cq values. Boxes represent the lower and upper quartiles with the medians as horizontal lines; whiskers depict the min and max Cq values. No statistical differences were found among the different sources of plasma in all the quantified target microRNAs and U6-snRNA.
The expression stability of microRNAs and U6-snRNA was then tested using 4 well-established, complementary statistical methods for selecting reference genes (GeNorm, Normfinder, BestKeeper, and the comparative ΔCq method) integrated using the RefFinder software. RefFinder exports the individual stability values of each gene designated by the 4 algorithms, and calculates the comprehensive gene stability of specific candidates as the final stability value. Lower stability values indicate higher expression stability. When the selected genes were evaluated separately in each group, all these algorithms demonstrated highly consistent gene stabilities (Table 2). miRNA-21, let-7a, and miRNA-106a were the most stable microRNAs among patients with malignant or benign diseases and among the healthy controls, although different stability values were obtained because of different algorithms. The more stable microRNAs in patients with different disease were also highly stable in the healthy controls. Interestingly, the commonly used U6-snRNA was consistently the least stable gene in all groups. The similarity of the stability profiles among patients and healthy controls provided evidence for further analysis.
Table 2.
Selected genes in the plasma of patients and healthy controls ranked by stability.
| Rank* | HCC | Hepatitis B | Hepatic cirrhosis | Normal | ||||
|---|---|---|---|---|---|---|---|---|
| Genes | Stability | Genes | Stability | Genes | Stability | Genes | Stability | |
| 1 | miR-21 | 1.32 | miR-21 | 1.73 | miR-21 | 1.57 | miR-21 | 1.32 |
| 2 | let-7a | 2.51 | let-7a | 1.86 | let-7a | 1.73 | let-7a | 1.68 |
| 3 | miR-106a | 2.83 | miR-106a | 1.86 | miR-106a | 1.86 | miR-106a | 2.63 |
| 4 | miR-16 | 3.50 | miR-16 | 4.23 | miR-16 | 4.73 | miR-16 | 3.66 |
| 5 | miR-219 | 3.66 | miR-219 | 5.12 | miR-219 | 4.86 | miR-219 | 4.73 |
| 6 | miR-155 | 5.24 | miR-155 | 5.23 | miR-155 | 5.63 | miR-155 | 6.48 |
| 7 | miR-221 | 6.74 | miR-221 | 6.24 | miR-221 | 6.48 | miR-221 | 6.96 |
| 8 | u6-snRNA | 8.00 | u6-snRNA | 8.00 | u6-snRNA | 8.00 | u6-snRNA | 7.44 |
Denotes microRNAs ranked according to comprehensive gene stability value.
We re-analyzed the data from the 120 samples using RefFinder to identify the reference microRNAs suitable for both patients and healthy volunteers. ΔCT, BestKeeper, Normfinder, and the comprehensive stability values indicated that miRNA-106a and miRNA-21 were the most stable microRNAs (Table 3). GeNorm, however, indicated that miRNA-16 was slightly more stable than miRNA-21. Let-7a, more stable when separately analyzed in the patients and the healthy controls, was less stable than miRNA-106a and miRNA-16. miRNA-221 and miRNA-155 were the least stable microRNAs. U6-snRNA was confirmed as unstable for normalization. Therefore, miRNA-106a and miRNA-21 are relatively suitable for normalizing the current dataset.
Table 3.
Stability of candidate reference microRNAs in patients with liver diseases and healthy volunteers.
| Rank | Genes | Comprehensive | Delta CT | BestKeeper | Normfinder | GeNorm |
|---|---|---|---|---|---|---|
| 1 | hsa-mir-21 | 1.00 | 1.01 | 0.668 | 0.427 | 0.672 |
| 2 | hsa-mir-106a | 2.21 | 1.00 | 0.606 | 0.447 | 0.728 |
| 3 | hsa-mir-219 | 2.63 | 1.08 | 0.735 | 0.588 | 0.672 |
| 4 | hsa-let-7a | 3.46 | 1.17 | 0.801 | 0.619 | 0.779 |
| 5 | hsa-mir-221+ | 5.48 | 1.08 | 0.701 | 0.848 | 0.835 |
| 6 | hsa-mir-16 | 5.48 | 1.20 | 0.785 | 0.771 | 0.891 |
| 7 | hsa-u6-snRNA | 7.00 | 1.65 | 1.061 | 1.448 | 1.082 |
| 8 | hsa-mir-155 | 8.00 | 1.69 | 1.114 | 1.507 | 1.235 |
MicroRNAs are ranked according to comprehensive gene stability.
Different reference genes affect the variation profiles of the target microRNA in plasma
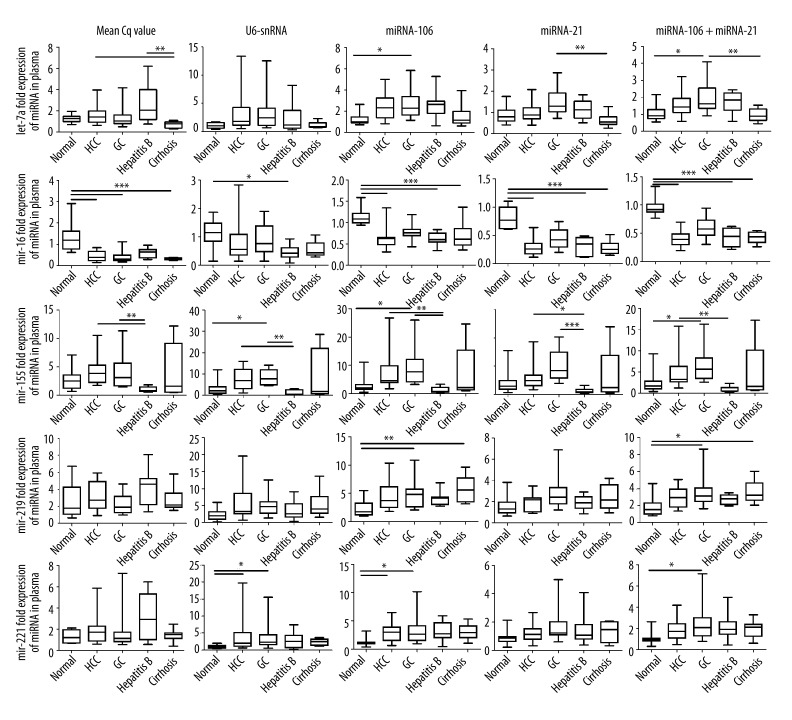
We also examined the effect of different normalizers on the expression profile of target microRNA. We compared 5 normalization strategies to evaluate the potential bias in the quantitative analyses of circulating microRNA. We selected U6-snRNA, miRNA-106a, miRNA-21, and the geometric mean of miRNA-106a and miRNA-21 as reference genes. Normalizing with the serum volume, the once commonly used normalization strategy was also evaluated in this study. The remaining microRNAs, let-7a, miRNA-16, miRNA-219, miRNA-221, and miRNA-155, were selected as target microRNAs and quantified via the ΔΔCq method using 5 different standardized strategies. Slight differences in let-7a expression were observed between the patients and the normal controls (Figure 2). However, selecting miRNA-106a for normalization revealed the higher let-7a expression in the GC patients compared with the healthy controls. All 5 normalization methods indicated that miRNA-16 is less expressed in the patients than in the normal volunteers. Normalization using U6-snRNA, however, barely detected the statistical differences discovered using the other 4 methods. Although they had different degrees of variation, all 5 methods indicated that miRNA-155 is sharply upregulated in the plasma of patients with malignant tumors, especially GC patients, but downregulated in benign liver diseases. The different normalization strategies also provided different statistical expression profiles for miRNA-221 and miRNA-219 between patients and healthy volunteers. The obvious bias cannot be eliminated, even when the geometric mean of the 2 most stable reference microRNAs (miRNA-106a and miRNA-21) was used as reference. These results demonstrate unexpected bias and erroneous disease research using commonly accepted reference genes, such as U6-snRNA. In addition, the results show that erroneous interpretations of the target gene variation cannot be completely avoided, even when using the reference genes recommended by professional screening software for individual or comprehensive candidate reference genes.
Figure 2.
Reference gene selection intensively affects the results of relative qRT-PCR. Relative quantities of plasma microRNAs in patients and healthy volunteers were determined by different standardized methods. Expression levels of plasma let-7a, miRNA-16, miRNA-219, miRNA-221, and miRNA-155 were normalized by sample volume, U6-snRNA, miRNA-21, miRNA-106a, or the geometric mean of miRNA-21 and miRNA-106a, respectively. Boxplots show interquartile ranges and medians; whiskers indicate the min to max ranges. Normal: healthy controls; HCC: hepatic carcinoma; GC: gastric cancer. Asterisks indicate significant differences between groups (*, P<0.05; **, P<0.01; ***, P<0.001).
Discussion
Accurate microRNA determination is an essential step in understanding their functional significance in physiologic and pathologic processes, as well as in using them as biomarkers for diagnosing and prognosticating various diseases and for evaluating therapeutic effects. Studies on circulating microRNA as a new generation of non-invasive biomarkers have increased rapidly after a tumor-associated microRNA was first found in the serum of patients in 2008 [23]. Relative qRT-PCR is a fast and reliable method for detecting subtle but meaningful variations in microRNA levels in different populations, but it requires stably expressed endogenous genes as internal references [24]. “Classical” reference genes, such as 5S and U6-snRNA, are always used in relative quantification [25]. However, the suitability of “classical” reference genes needs to be verified, with the recently increasing number of controversial views [14,15,24,25]. An increasing number of studies have reported different results using the same target genes in the same diseases, with the use of different individual reference genes as a potential cause [14,16,17].
In this study, we first reviewed the published literature and selected microRNAs used as reference genes in different laboratories. In addition to U6-snRNA, let-7a has been used as a reference gene in breast cancer and colorectal cancer studies, miRNA-106a has been used in kidney cancer patients, and miRNA-16 and miRNA-221 have recently been identified as suitable reference genes for analyzing serum/plasma microRNA in GC and B cell lymphoma patients [15,23]. We also used target microRNAs that might be distinctly expressed in the plasma of HCC patients, patients with benign liver disease, and healthy persons. Patients with GC were also enrolled as tumor controls to evaluate the stability of the reference genes in different cancers and the specificity of the target microRNAs. We evaluated whether these suggested reference genes are stable in a single experiment system and determined the potential bias of selecting these suggested reference genes for detecting these target microRNAs. The stability of the selected genes was evaluated using RefFinder, which integrates 4 commonly used complementary statistical approaches (GeNorm, Normfinder, BestKeeper, and comparative delta-Cq method algorithms).
Although miRNA-106a is unsuitable as a reference gene for prostate disease [14] and miRNA-21 was a marker for necroinflammatory activity with or without HCC [13], RefFinder revealed miRNA-106a and miRNA-21 as the 2 most stable reference genes for plasma microRNA analysis among patients with benign or malignant hepatic diseases, GC patients, and healthy controls. The effects of different reference genes on target genes are then shown using the remaining microRNAs. Previous studies have proposed the use of global expression means (using the same volume of liquid sample) [15], small RNAs (such as U6-snRNA and 5S-RNA) [25], or microRNAs to normalize microRNA expression. In this study, we compared the 3 commonly used strategies in the same detection system. Different normalization strategies demonstrated obvious expression bias in almost all the target microRNAs, even when the 2 most stable reference microRNAs were used. For example, let-7a was reportedly downregulated in the plasma of GC patients [9], but increased in HCC patients [10]. In our data, however, we detected a slight difference in the expression of let-7a among HCC, GC, benign hepatic disease, and the normal controls. When the most stable miRNA-106a was selected as the normalizer, even higher let-7a expression was identified in GC patients. These results emphasize the unstable effect of different reference genes on the relative quantification of circulating microRNAs.
To the best of our knowledge, no widely recognized rules for normalization strategy have been established. Thus, we need to determine whether these “reference genes” are constantly expressed in the blood of healthy individuals and in patients with different diseases. We also need to verify whether these commonly used and reported “reference genes” undergo the same biological procedure with target genes and minimize the discrimination of operation efficiencies during extraction, reverse transcription, and amplification. Our data, as well as those of other studies, indicate that these “reference genes” are not always constantly expressed and do not undergo the same procedure. For example, miRNA-16 was used as a reference gene in GC patients and controls, but was differently expressed in the serum of patients with PCA and benign prostate hyperplasia [26]. In the current study, however, all 5 normalization methods demonstrated decreased miRNA-16 expression among patients with malignant disease and those with benign disease. The different lengths of small RNAs compared with microRNAs also resulted in divergent efficiencies in microRNA extraction, reverse transcription, and amplification [27]. Moreover, normalization can be difficult between microRNAs themselves if microRNAs originate from different extracellular transport systems (e.g., ago-bound versus exosomal), in which the stability of the microRNAs are different [23,28]. The divergent expression of commonly used reference genes indicates that reference genes cannot be simply transposed from one study to another without validating the specifics in each experimental system. Recently, several software packages, such as GeNorm, Normfinder, BestKeeper, and the comparative ΔCq method, constructed using different algorithms, were adapted to evaluate and determine the most suitable reference genes [15,18–21]. However, the reference gene selected by the software did not perform as well as expected because different laboratories always screen out different suitable reference genes or reference gene combinations, and the reference genes selected in one laboratory are not always used correctly in other studies [15,24,25,29]. As shown in our data (Table 3), different evaluation software packages recommend different reference genes, even in the same experimental system. Therefore, when properly selecting and using the appropriate reference gene evaluation software, uniform guidelines to obtain reliable and comparable relative quantification of circulating microRNA are also needed.
miRNA-155 is highly expressed in gastric and hepatic cancer tissues [30,31], but downregulated in gastric cancer cells [32]. Intriguingly, all 5 normalization strategies in this study indicated that plasma miRNA-155 is greatly upregulated in patients with malignant liver and gastric tumors, but downregulated in those with benign disease compared with those in healthy volunteers. The significant variation in miRNA-155 expression between malignant and benign disease indicates a promising clinical application in diagnosing and treating different tumors. However, further studies are needed to confirm these results in larger clinical samples.
Conclusions
In summary, although relatively small samples were included in this study, our research confirmed bias and erroneous results in the relative quantification of circulating microRNA using qPCR when unsuitable reference genes were used. The “classical” reference genes and those used in other studies cannot be used directly in new studies without validation. For the first time, we compared and found that the commonly used statistical evaluation software for reference genes may not always give consistent recommendations, and thus need applicable guidelines. Finally, a consensus should be reached on selecting reference genes before circulating microRNAs are further quantified using relative qPCR for any purpose, to avoid the continued emergence of divergent and contradictory conclusions in studies on microRNA. As an alternative, using justified reference genes (e.g., the geometric mean of multiple carefully selected reference genes as described in our study and others [18]) might more accurately describe qPCR expression profiling of plasma miRNA. The absolute quantification, however, of circulating microRNAs with synthetic microRNAs [9] using qPCR may be the final solution. We are the first to report that circulating miRNA-155 is greatly upregulated in the plasma of patients with liver and gastric malignant tumors, but downregulated in benign hepatic disease, which may be a promising non-invasive biomarker for malignant tumors.
Footnotes
Source of support: This research was supported by grants from the Wujieping Medical Foundation for laboratory diagnosis of liver disease (LDWMF-SY-2011A003) and the Science and Technology Commission of Shanghai Municipality (114119b2600)
References
- 1.Lages E, Ipas H, Guttin A, et al. MicroRNAs: molecular features and role in cancer. Front Biosci (Landmark Ed) 2012;17:2508–40. doi: 10.2741/4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 3.Jung M, Schaefer A, Steiner I, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin Chem. 2010;56:998–1006. doi: 10.1373/clinchem.2009.141580. [DOI] [PubMed] [Google Scholar]
- 4.McDonald JS, Milosevic D, Reddi HV, et al. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin Chem. 2011;57:833–40. doi: 10.1373/clinchem.2010.157198. [DOI] [PubMed] [Google Scholar]
- 5.Starkey Lewis PJ, Merz M, et al. Serum microRNA biomarkers for drug-induced liver injury. Clin Pharmacol Ther. 2012;92:291–93. doi: 10.1038/clpt.2012.101. [DOI] [PubMed] [Google Scholar]
- 6.Thum T, Mayr M. Review focus on the role of microRNA in cardiovascular biology and disease. Cardiovasc Res. 2012;93:543–44. doi: 10.1093/cvr/cvs085. [DOI] [PubMed] [Google Scholar]
- 7.Healy NA, Heneghan HM, Miller N, et al. Systemic mirnas as potential biomarkers for malignancy. Int J Cancer. 2012;131:2215–22. doi: 10.1002/ijc.27642. [DOI] [PubMed] [Google Scholar]
- 8.Yang Q, Jie Z, Cao H, et al. Low-level expression of let-7a in gastric cancer and its involvement in tumorigenesis by targeting RAB40C. Carcinogenesis. 2011;32:713–22. doi: 10.1093/carcin/bgr035. [DOI] [PubMed] [Google Scholar]
- 9.Tsujiura M, Ichikawa D, Komatsu S, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102:1174–79. doi: 10.1038/sj.bjc.6605608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sukata T, Sumida K, Kushida M, et al. Circulating microRNAs, possible indicators of progress of rat hepatocarcinogenesis from early stages. Toxicol Lett. 2011;200:46–52. doi: 10.1016/j.toxlet.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 11.Shen Y, Li Y, Ye F, et al. Identification of miR-23a as a novel microRNA normalizer for relative quantification in human uterine cervical tissues. Exp Mol Med. 2011;43:358–66. doi: 10.3858/emm.2011.43.6.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomimaru Y, Eguchi H, Nagano H, et al. Circulating microRNA-21 as a novel biomarker for hepatocellular carcinoma. J Hepatol. 2012;56:167–75. doi: 10.1016/j.jhep.2011.04.026. [DOI] [PubMed] [Google Scholar]
- 13.Bihrer V, Waidmann O, Friedrich-Rust M, et al. Serum microRNA-21 as marker for necroinflammation in hepatitis C patients with and without hepatocellular carcinoma. PLoS One. 2011;6:e26971. doi: 10.1371/journal.pone.0026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanders I, Holdenrieder S, Walgenbach-Brunagel G, et al. Evaluation of reference genes for the analysis of serum miRNA in patients with prostate cancer, bladder cancer and renal cell carcinoma. Int J Urol. 2012;19:1017–25. doi: 10.1111/j.1442-2042.2012.03082.x. [DOI] [PubMed] [Google Scholar]
- 15.Song J, Bai Z, Han W, et al. Identification of suitable reference genes for qPCR analysis of serum microRNA in gastric cancer patients. Dig Dis Sci. 2012;57:897–904. doi: 10.1007/s10620-011-1981-7. [DOI] [PubMed] [Google Scholar]
- 16.Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem. 2012;58:610–18. doi: 10.1373/clinchem.2011.172767. [DOI] [PubMed] [Google Scholar]
- 17.Shen J, Hruby GW, McKiernan JM, et al. Dysregulation of circulating microRNAs and prediction of aggressive prostate cancer. Prostate. 2012;72:1469–77. doi: 10.1002/pros.22499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:Research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64:5245–50. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper – Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26:509–15. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 21.Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang G, Qin Q, Zhang P, et al. Reverse signaling using an inducible costimulator to enhance immunogenic function of dendritic cells. Cell Mol Life Sci. 2009;66:3067–80. doi: 10.1007/s00018-009-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–75. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 24.Kwon MJ, Oh E, Lee S, et al. Identification of novel reference genes using multiplatform expression data and their validation for quantitative gene expression analysis. PLoS One. 2009;4:e6162. doi: 10.1371/journal.pone.0006162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peltier HJ, Latham GJ. Normalization of microRNA expression levels in quantitative RT-PCR assays: identification of suitable reference RNA targets in normal and cancerous human solid tissues. RNA. 2008;14:844–52. doi: 10.1261/rna.939908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mahn R, Heukamp LC, Rogenhofer S, et al. Circulating microRNAs (miRNA) in serum of patients with prostate cancer. Urology. 2011;77:1265.e9–16. doi: 10.1016/j.urology.2011.01.020. [DOI] [PubMed] [Google Scholar]
- 27.Ratert N, Meyer HA, Jung M, et al. Reference miRNAs for miRNAome analysis of urothelial carcinomas. PLoS One. 2012;7:e39309. doi: 10.1371/journal.pone.0039309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turchinovich A, Weiz L, Langheinz A, Burwinkel B. Characterization of extracellular circulating microRNA. Nucleic Acids Res. 2011;39:7223–33. doi: 10.1093/nar/gkr254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim QE, Zhou L, Ho YK, et al. snoU6 and 5S RNAs are not reliable miRNA reference genes in neuronal differentiation. Neuroscience. 2011;199:32–43. doi: 10.1016/j.neuroscience.2011.10.024. [DOI] [PubMed] [Google Scholar]
- 30.Huang YH, Lin KH, Chen HC, et al. Identification of postoperative prognostic microRNA predictors in hepatocellular carcinoma. PLoS One. 2012;7:e37188. doi: 10.1371/journal.pone.0037188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim BH, Hong SW, Kim A, et al. Prognostic implications for high expression of oncogenic microRNAs in advanced gastric carcinoma. J Surg Oncol. 2013;107:505–10. doi: 10.1002/jso.23271. [DOI] [PubMed] [Google Scholar]
- 32.Li CL, Nie H, Wang M, et al. microRNA-155 is downregulated in gastric cancer cells and involved in cell metastasis. Oncol Rep. 2012;27:1960–66. doi: 10.3892/or.2012.1719. [DOI] [PubMed] [Google Scholar]