Abstract
Ageing and lifespan of organisms are determined by complicated interactions between their genetics and the environment, but the cellular mechanisms remain controversial. There have been a number of studies suggesting that cellular energy metabolism and free radical dynamics affect lifespan, implicating mitochondrial function. Recently, Shen et al.1 provided apparent mechanistic insight by reporting that mitochondrial oscillations of ‘free radical production’, called ‘mitoflashes’, in the pharynx of 3-day old Caenorhabditis elegans correlated inversely with lifespan. The interpretation of ‘mitoflashes’ as ‘bursts of superoxide’ radicals assumes that circularly permuted yellow fluorescent protein (cpYFP) is a reliable indicator of mitochondrial superoxide2. This interpretation has been criticised because experiments and theoretical considerations both show that changes in cpYFP fluorescence are due to alterations in pH, not superoxide3-7. We now provide direct evidence that purified cpYFP is completely unresponsive to superoxide. Therefore ‘mitoflashes’ do not reflect superoxide generation and are not evidence for a link between mitochondrial free radical dynamics and lifespan.
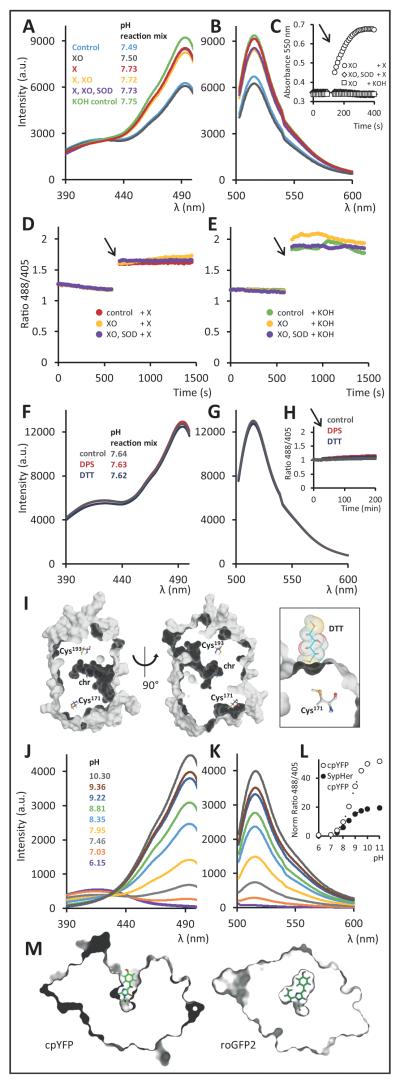
We carried out experiments with purified recombinant cpYFP sensor protein to test whether it responds to superoxide (Fig. 1A-E). Exposure of cpYFP to a superoxide-generating system (xanthine (X) and xanthine oxidase (XO)) slightly changed the excitation and emission spectra. However, the same change occurred when cpYFP was incubated with the individual assay constituents in the absence of superoxide production, or when superoxide dismutase (SOD) was added to degrade superoxide (Fig. 1A,B). The cytochrome c reduction assay confirmed that superoxide is produced by the X/XO system, and is abolished by SOD (Fig. 1C). Xanthine is dissolved in KOH, causing a small increase in pH upon addition. Indeed, there was an excellent correlation between spectral changes and resulting assay pH upon addition of xanthine (i.e. KOH) (Fig. 1A,B). In time course assays in which superoxide generation was started upon introduction of xanthine (Fig. 1D) addition of KOH as the solvent control for xanthine (Fig. 1E) gave the same increase in fluorescence ratio. Extended reductive or oxidative treatment with thiol redox agents (dithiothreitol (DTT, a reducing agent) and 2,2′-dipyridyl disulfide (DPS, an oxidizing agent)) did not alter the spectral behaviour (Fig. 1F,G,H), consistent with structural information suggesting that both Cys residues are buried inside the mature protein and are unlikely to be accessible for thiol redox chemistry (Fig. 1I). Likewise, reductive pre-treatment with DTT under inert atmosphere, followed by DTT removal did not impact on the outcome of the superoxide assays. Further variation of experimental variables, including pre-incubation conditions, pH buffer systems and a 100-fold range of sensor concentrations, did not lead to any rapid, reversible change in cpYFP sensor signal required for superoxide-related ‘mitoflashes’, as long as the pH and halide ion concentrations were kept constant.
Figure. Spectroscopic and structural analysis of the ‘mitoflash’ probe cpYFP.
(A) cpYFP fluorescence excitation spectra (emission at 515 nm) and (B) emission spectra (excitation at 488 nm) upon addition of X (xanthine 2 mM), XO (xanthine oxidase 100 mU/mL), SOD (bovine Cu/Zn superoxide dismutase 600 U/mL) and KOH (solvent control for xanthine, note pH increase), and the in situ pH of the assayed 200 μL reaction mix. (C) Cytochrome c reduction detected by absorption at 550 nm to measure superoxide generation in response to the X/XO system in the presence and absence of SOD and in response to KOH as solvent control for X. Cyt c 100 μM; arrows indicates X or KOH addition. (D) The response of cpYFP excitation ratio (488/405 nm) to superoxide generation. The arrow indicates introduction of X to constitute the X/XO system. Controls contained either SOD in addition or no XO. (E) The same assays were performed with KOH introduction as solvent control for X. (F) cpYFP fluorescence excitation spectra and (G) emission spectra upon 24 h incubation with DTT (dithiothreitol; 10 mM) and DPS (2,2′-dipyridyl disulfide; 1 mM) and the in situ pH of the assayed 200 μL reaction mix. (H) cpYFP fluorescence ratio upon DTT and DPS addition over 3 h; arrow indicates DTT or DPS addition. (I) Cross-sections through a surface model of cpYFP (left and middle). Chromophore (chr) and cysteines are represented as ball-and-stick models. The Cys171 thiol is relatively close to the protein surface, but unlikely to be accessible to solutes as indicated by the docking of a DTT molecule to the protein surface (right). (J) cpYFP fluorescence excitation spectra (emission at 515 nm) and (K) emission spectra (excitation at 488 nm) in response to pH as determined in situ after the measurements. (L) pH dependence of cpYFP excitation ratio (488/405 nm; normalized to pH 7.0) as compared to the cpYFP part of the pH sensor SypHer. (M) Sectional views through volume models of cpYFP and roGFP2, with the clipping plane parallel to and just above the chromophore phenoxy ring. The chromophore is represented as a ball-and-stick model. Data in panels A-H and J-L are background corrected and experiments were repeated at least 5 times with consistent results.
‘Mitoflashes’ can be fully explained by the extraordinary pH sensitivity of cpYFP, which has a pKa of ≈8.7 (upon excitation at 488 nm, the wavelength at which flashes are observed) and shows a >50-fold change in fluorescence ratio between pH 7 and 10, similar to the structurally related pH-sensor SypHer (Fig. 1J,K,L)8. In the mitochondrial matrix a resting pH (≈7.9) close to sensor pKa and a limited pH buffering capacity mean that even minor perturbations will elicit a pronounced sensor response (Fig. 1A,B,D,E). The cpYFP pH sensitivity is due to the structural perturbation caused by the circular permutation. A large cleft in the β-barrel exposes the pH-active phenoxy group of the chromophore (Fig. 1M, left panel), which is concealed in non-permuted GFP-based biosensors (Fig 1M, right panel).
Based on this evidence using purified cpYFP and earlier studies in cells and isolated mitochondria5,6,9, the ‘mitoflash’ phenomenon cannot be attributed to bursts of mitochondrial superoxide. In accordance with the pH responsiveness of the probe, recent work with different sensors suggests that ‘mitoflash’ events indicate brief periods of alkalinisation in individual mitochondria, possibly as a result of acceleration in proton pumping, triggered by mitochondrial fusion initiation and/or a change in ion homeostasis6,9,10.
The debate about the nature of ‘mitoflashes’ has focussed on in situ evidence which has left space for interpretation on both sides. Critics have pointed out the implausibility of ‘superoxide flashes’ based on mitochondrial energetics3,5,9, the absence of a plausible chemical mechanism for the reversible interaction between cpYFP and superoxide4,7, and the fact that the pH sensor SypHer also detects ‘mitoflashes’6,10. These arguments have been countered by data suggesting a correlation of ‘mitoflashes’ with the response of chemical ROS probes11-13, the notion that the pH probe SypHer may also respond to superoxide17, and the suggestion that a ‘mitoflash’ represents a mixture of superoxide burst and pH transient11,13. Ultimate resolution of the debate has been hampered by the use of different biological systems and the complexity of mitochondrial physiology, where matrix pH and free radical release are connected via the electron transport chain and linked to several other parameters such as availability of respiratory substrates, membrane potential, redox and ion homeostasis, and mitochondrial morphology2,5-7,10,14-16. Here we resolve the controversy by a thorough analysis of the fundamental properties of the ‘mitoflash’ sensor cpYFP. Previous work already excluded the suggestion that the pH probe SypHer responds to superoxide6. We now provide definitive evidence that cpYFP itself does not respond to superoxide and that flashes recorded by cpYFP do not represent superoxide bursts. Of course, sudden changes in mitochondrial physiology may still include altered free radical levels. While the ‘mitoflash’ phenomenon may reflect an important feature of mitochondrial function that deserves further mechanistic analysis the interpretation of the events by Shen et al. lacks a biophysical foundation and ‘mitoflashes’ cannot serve as evidence for free radical involvement in determining lifespan.
Methods
cpYFP was purified from Escherichia coli Origami (DE3) and Rosetta 2 (DE3) 24 h after induction at 20 °C and assayed at 10, 25 and 1000 μg protein/mL using a Jasco spectrofluorimeter FP8300 and a BMG Labtech Clariostar plate reader. Detector gain was adjusted for individual experiments. Buffers contained 100 mM NaCl, 1 mM Na2EDTA and 100 mM Tris-HCl, pH 7.5 (for thiol redox and superoxide assays; degassed and under argon for thiol redox treatments) or 100 mM Tris-TES (for pH assays). All reagents were dissolved in assay buffer, except for xanthine (100x stock in 1 M KOH, base required for solubility) and xanthine oxidase (118x (NH4)2SO4 suspension as delivered by Sigma). Protein structures (PDB entries 3O78 and 1JC1) were rendered using PyMOL.
Contributor Information
Markus Schwarzländer, Institute of Crop Science and Resource Conservation (INRES), University of Bonn, Friedrich-Ebert-Allee 144, 53113 Bonn, Germany, markus.schwarzlander@uni-bonn.de.
Stephan Wagner, Institute of Crop Science and Resource Conservation (INRES), University of Bonn, Friedrich-Ebert-Allee 144, 53113 Bonn, Germany.
Yulia G. Ermakova, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, 117997, Russia
Vsevolod V. Belousov, Shemyakin-Ovchinnikov Institute of Bioorganic Chemistry, Russian Academy of Sciences, Moscow, 117997, Russia
Rafael Radi, Center for Free Radical and Biomedical Research, Departamento de Bioquímica, Facultad de Medicina, Universidad de la República, Avda. General Flores 2125, 11800 Montevideo, Uruguay.
Joseph S. Beckman, Linus Pauling Institute, Environmental Health Sciences Center, Department of Biochemistry and Biophysics, Oregon State University, Corvallis, Oregon 97331, USA
Garry R. Buettner, The University of Iowa, Department of Radiation Oncology and Interdisciplinary Graduate Program in Human Toxicology, and ESR Facility, College of Medicine, Med Labs B180K, Iowa City, IA 52242-1181, USA
Nicolas Demaurex, Department of Cell Physiology and Metabolism, University of Geneva, 1, rue Michel-Servet, Geneva 4 CH-1211, Switzerland.
Michael R. Duchen, Department of Cell and Developmental Biology and Consortium for Mitochondrial Research, University College London, Gower Street, London WC1E 6BT, UK
Henry J. Forman, Life and Environmental Sciences Unit, University of California, Merced, 5200 N. Lake Road, Merced, CA 95344, USA; Andrus Gerontology Center of the Davis School of Gerontology, University of Southern California, 3715 McClintock Avenue, Los Angeles, CA 90089-0191, USA
Mark D. Fricker, Department of Plant Sciences, University of Oxford, South Parks Road, Oxford OX1 3RB, UK
David Gems, Institute of Healthy Ageing, and Department of Genetics, Evolution and Environment, University College London, London WC1E 6BT, UK.
Andrew P. Halestrap, School of Biochemistry and The Bristol Heart Institute, University of Bristol, University Walk, Bristol BS8 1TD, UK
Barry Halliwell, Department of Biochemistry, National University of Singapore, Singapore 117597, Singapore.
Ursula Jakob, Molecular, Cellular and Developmental Biology Department, University of Michigan, Ann Arbor, MI 48109-1048, USA.
Iain G. Johnston, Department of Mathematics, South Kensington Campus, Imperial College London, London SW7 2AZ, UK
Nick S. Jones, Department of Mathematics, South Kensington Campus, Imperial College London, London SW7 2AZ, UK
David C. Logan, Université d’Angers & INRA & Agrocampus Ouest, UMR 1345 Institut de Recherche en Horticulture et Semences, Angers, F-49045, France
Bruce Morgan, Division of Redox Regulation, German Cancer Research Center (DKFZ), DKFZ-ZMBH Alliance, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany.
Florian L. Müller, Department of Cancer Biology, University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA
David G. Nicholls, Buck Institute for Research on Aging, 8001 Redwood Blvd, Novato California 94945, USA
S. James Remington, Department of Physics, Institute of Molecular Biology, University of Oregon, Eugene, Oregon 97403-1229, USA.
Paul T. Schumacker, Department of Pediatrics, Division of Neonatology, Northwestern University, Feinberg School of Medicine, Chicago, Illinois, IL60611, USA
Christine C. Winterbourn, Centre for Free Radical Research, Department of Pathology, University of Otago, Christchurch, PO Box 4345, Christchurch, New Zealand
Lee J. Sweetlove, Department of Plant Sciences, University of Oxford, South Parks Road, Oxford OX1 3RB, UK
Andreas J. Meyer, Institute of Crop Science and Resource Conservation, University of Bonn, Friedrich-Ebert-Allee 144, 53113 Bonn, Germany
Tobias P. Dick, Division of Redox Regulation, German Cancer Research Center (DKFZ), DKFZ-ZMBH Alliance, Im Neuenheimer Feld 280, 69120 Heidelberg, Germany
Michael P. Murphy, MRC Mitochondrial Biology Unit, Hills Road, Cambridge CB2 0XY, UK
References
- 1.Shen EZ, et al. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature. 2014;508:128–132. doi: 10.1038/nature13012. doi:10.1038/nature13012. [DOI] [PubMed] [Google Scholar]
- 2.Wang W, et al. Superoxide flashes in single mitochondria. Cell. 2008;134:279–290. doi: 10.1016/j.cell.2008.06.017. doi:10.1016/j.cell.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muller FL. A critical evaluation of cpYFP as a probe for superoxide. Free Radical Biology & Medicine. 2009;47:1779–1780. doi: 10.1016/j.freeradbiomed.2009.09.019. doi:10.1016/j.freeradbiomed.2009.09.019. [DOI] [PubMed] [Google Scholar]
- 4.Ezerina D, Morgan B, Dick TP. Imaging dynamic redox processes with genetically encoded probes. Journal of Molecular and Cellular Cardiology. 2014;73:43–49. doi: 10.1016/j.yjmcc.2013.12.023. doi:10.1016/j.yjmcc.2013.12.023. [DOI] [PubMed] [Google Scholar]
- 5.Schwarzländer M, Logan DC, Fricker MD, Sweetlove LJ. The circularly permuted yellow fluorescent protein cpYFP that has been used as a superoxide probe is highly responsive to pH but not superoxide in mitochondria: implications for the existence of superoxide ‘flashes’. Biochemical Journal. 2011;437:381–387. doi: 10.1042/BJ20110883. doi:10.1042/BJ20110883. [DOI] [PubMed] [Google Scholar]
- 6.Santo-Domingo J, Giacomello M, Poburko D, Scorrano L, Demaurex N. OPA1 promotes pH flashes that spread between contiguous mitochondria without matrix protein exchange. The EMBO Journal. 2013;32:1927–1940. doi: 10.1038/emboj.2013.124. doi:10.1038/emboj.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwarzländer M, et al. Mitochondrial ‘flashes’: a radical concept repHined. Trends in Cell Biology. 2012;22:503–508. doi: 10.1016/j.tcb.2012.07.007. doi:10.1016/j.tcb.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Poburko D, Santo-Domingo J, Demaurex N. Dynamic regulation of the mitochondrial proton gradient during cytosolic calcium elevations. The Journal of Biological Chemistry. 2011;286:11672–11684. doi: 10.1074/jbc.M110.159962. doi:10.1074/jbc.M110.159962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzländer M, et al. Pulsing of membrane potential in individual mitochondria: a stress-induced mechanism to regulate respiratory bioenergetics in Arabidopsis. The Plant Cell. 2012;24:1188–1201. doi: 10.1105/tpc.112.096438. doi:10.1105/tpc.112.096438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Azarias G, Chatton JY. Selective ion changes during spontaneous mitochondrial transients in intact astrocytes. PLoS One. 2011;6:e28505. doi: 10.1371/journal.pone.0028505. doi:10.1371/journal.pone.0028505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei-LaPierre L, et al. Respective contribution of mitochondrial superoxide and pH to mitochondria-targeted circularly permuted yellow fluorescent protein (mt-cpYFP) flash activity. J Biol Chem. 2013;288:10567–10577. doi: 10.1074/jbc.M113.455709. doi:10.1074/jbc.M113.455709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pouvreau S. Superoxide flashes in mouse skeletal muscle are produced by discrete arrays of active mitochondria operating coherently. PLoS One. 2010;5:e13035. doi: 10.1371/journal.pone.0013035. doi:10.1371/journal.pone.0013035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, et al. Superoxide constitutes a major signal of mitochondrial superoxide flash. Life Sci. 2013;93:178–186. doi: 10.1016/j.lfs.2013.06.012. doi: 10.1016/j.lfs.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 14.Ma Q, et al. Superoxide flashes: early mitochondrial signals for oxidative stress-induced apoptosis. J Biol Chem. 2011;286:27573–27581. doi: 10.1074/jbc.M111.241794. doi:10.1074/jbc.M111.241794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breckwoldt MO, et al. Multiparametric optical analysis of mitochondrial redox signals during neuronal physiology and pathology in vivo. Nat Med. 2014;20:555–560. doi: 10.1038/nm.3520. doi: 10.1038/nm.3520. [DOI] [PubMed] [Google Scholar]
- 16.Hou T, Wang X, Ma Q, Cheng H. Mitochondrial flashes: new insights into mitochondrial ROS signaling and beyond. J Physiol (Lond.) 2014;592:3703–3713. doi: 10.1113/jphysiol.2014.275735. doi:10.1113/jphysiol.2014.275735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quatresous E, Legrand C, Pouvreau S. Mitochondria-targeted cpYFP: pH or superoxide sensor? J Gen Physiol. 2012;140:567–570. doi: 10.1085/jgp.201210863. doi:10.1085/jgp.201210863. [DOI] [PMC free article] [PubMed] [Google Scholar]