Abstract
This study tested the hypothesis that reciprocal communication occurs between macrophages and cultured human endometrial stromal cells and that this communication may contribute to the pathology of endometriosis. An endometrial stromal cell line (T-HESC) was treated with macrophage conditioned medium (CM) ± estradiol+progesterone. Macrophages were treated without or with T-HESC CM. DNA microarray identified 716 differentially expressed genes in T-HESC cells in response to factors secreted by macrophages. Up-regulated genes in T-HESC included IL8/CXCL8, MMP3, phospholamban, CYR61/CCN1, CTGF/CCN2, tenascin C, and NNMT, whereas integrin alpha 6 was down-regulated. In contrast, 15 named genes were differentially expressed in macrophages in response to factors secreted by endometrial stromal cells. The data document reciprocal communication between macrophages and endometrial stromal cells and suggest that interaction with macrophages stimulates the expression of genes in endometrial stromal cells that may support the establishment of endometriosis.
Keywords: endometriosis, T-HESC, macrophage, endometrial stromal cells
Introduction
Although endometriosis is considered a benign disease, the implantation of endometrial tissue in the peritoneal cavity causes significant pain, debility, and infertility1, 2. It has been proposed that the immune system is faulty in women who develop endometriosis3, 4 and endometriosis is clearly an inflammatory disease1, 2. Bone marrow-derived cells are plentiful in endometriotic explants and in the peritoneal fluid of women with the disease5-7, yet the immune system fails to destroy endometrial cells in the peritoneal cavity of those women. The presence of cytokines, growth factors, and chemokines secreted by bone marrow-derived cells alters the peritoneal environment in endometriosis and it has been proposed that these factors contribute to the development of endometriosis7-11.
Yet, despite the abundance of macrophages and monocytes found in peritoneal fluid and endometriotic lesions of women with endometriosis, little is known regarding the role of these cells in the etiology of endometriosis or their interplay with endometrial cells. In order for endometriosis lesions to develop, endometrial cells must survive transit from the uterus to the peritoneal cavity in the absence of a blood supply, adhere to the peritoneum or ovarian surface, invade through the surface of the tissue to which they adhere, develop a new vascular supply, and grow. We hypothesize that communication between macrophages and endometrial cells can stimulate events associated with the development of endometriosis. More specifically, we theorize that interactions with macrophages tip the balance for endometrial cells from death to survival, adhesion, invasion, neovascularization, and growth. We have shown that monocytes and endometrial stromal cells communicate directly with each other12. The current study was designed to test the hypothesis that reciprocal communication occurs between macrophages and cultured human endometrial stromal cells and that this communication, translated to the in vivo state, may contribute to the pathology of endometriosis.
The current studies used cultured cells as a model system. These experiments were designed to model potential interactions between peritoneal macrophages and endometrial stromal cells that have mislocated to the peritoneal cavity, as well as the effect of decidualization on those interactions. Two cell lines were used, telomerase-immortalized human endometrial stromal cells (T-HESC)13 and U-937 macrophages14. Cell culture simplifies study of the interactions among cells by limiting the numbers of cell types in the system. Moreover, cell culture allows examination of cell interactions in a controlled environment. These experiments analyzed the effect of factors secreted by macrophages on gene expression in T-HESC cells, and the effects of factors secreted by T-HESC cells on gene expression in macrophages. Genes that were differentially expressed in this study were compared to genes that were differentially expressed in endometriosis tissue15 and in cultured endometrial cells exposed to factors secreted by monocytes12.
Methods and Materials
Experimental design
In experiment 1, T-HESC cells were treated with vehicle or estradiol plus medroxyprogesterone acetate in the presence and absence of macrophage conditioned medium. In experiment 2, macrophages were treated under control conditions in the absence or presence of T-HESC conditioned medium. Total RNA was extracted from the cells and gene expression was measured by DNA microarray and by real time RT-PCR for both experiments.
Cell Lines
American Type Culture Collection was the source for human endometrial stromal cells immortalized by telomerase (T-HESC) and the U-937 monocytic cell line. Phenol red-free DMEM/F12 (Sigma, St. Louis, MO) supplemented with 1% ITS+ (insulin, transferrin, selenious acid, bovine serum albumin, and linoleic acid) (Becton Dickinson, Franklin Lakes, NJ), 10% charcoal/dextran treated fetal bovine serum albumin (FBS) (HyClone, South Logan, UT), 1% penicillin/streptomycin (Sigma), and 500 ng/ml puromycin (Sigma) was used for routine maintenance of T-HESC cells. RPMI-1640 media (ATCC) supplemented with 10% charcoal/dextran treated FBS and 1% penicillin/streptomycin was used for routine maintenance of the U-937 cell line.
Preparation of the U-937 macrophage conditioned medium
For maintenance of the cell line, U-937 cells were grown as monocytes in suspension in 10% FBS RPMI-1640 with pen/strep media until the cells covered 80% of the flask surface. The cells were treated with 32 nM phorbol myristate acetate (PMA) for 48 hours to stimulate differentiation into macrophages16. The PMA- and FBS-containing medium was removed from the macrophages and the cells were rinsed with phosphate buffered saline (PBS). Fresh serum-free, phenol red-free DMEM/F12 media supplemented with 1% penicillin/streptomycin was placed on the cells for 24 hours. The conditioned medium was collected, centrifuged, and filtered through a 0.2 micron filter and stored at −70°C until used as macrophage conditioned medium (CM). The U-937 macrophage CM was supplemented with 1% ITS+ and puromycin before use. The U-937 cells differentiated from the monocytic phenotype (suspension culture) to the macrophage phenotype (adherent and containing inclusion bodies) in response to PMA16 (data not shown).
Preparation of T-HESC conditioned medium
T-HESC cells were maintained in their routine media (phenol red-free DMEM/F-12 with 10% charcoal/dextran treated FBS supplemented as described above) until 80% confluent. The cells were rinsed with PBS and the DMEM/F-12 media was replaced with serum-free phenol red-free RPMI-1640 supplemented with 16.4 mM L-glutamine, 10 mM HEPES, and 1% penicillin/streptomycin for 24 hours, at which time the medium was removed from the T-HESC cells, centrifuged, filtered through a 0.2 micron filter, and stored at −70°C until used as T-HESC CM.
Hormone and Conditioned Medium Treatments and RNA extraction12
For experiment 1, 4 flasks each of three separate passages of T-HESC cells were cultured for 8 days. Two flasks of each passage were grown in the presence of 0.1% ethanol (vehicle control) and the remaining 2 flasks were treated with 10−8 M estradiol + 10−7 M medroxyprogesterone acetate (E+P) (Sigma, St. Louis, MO)12, 13. Vehicle or E+P was added to the cells every 48 h when the media was changed. This dose and duration of E+P treatment induces a decidual-like reaction in cultured endometrial stromal cells.13 Cells were serum starved beginning on day 6 of the experiment. U-937 macrophage CM was added to one flask of each treatment group (vehicle and E+P) on day 7. The remaining two flasks of cells were treated with non-conditioned control medium (serum-free, phenol red-free DMEM/F12 supplemented with 1% penicillin/streptomycin). To collect total RNA on day 8, the medium was removed from the T-HESC cells, and 1 ml TRI reagent (Molecular Research Center, Cincinnati, OH) was added to each flask of rinsed cells. A cell scraper was used to suspend and homogenize the cells. Bromochloropropane (200 μl) and 3M sodium acetate (60 μl) were added to the homogenate and the mixture was centrifuged to elicit phase separation. The aqueous phase of each sample containing the RNA was collected and purified on an RNeasy column (Qiagen, Valencia, CA). The possibility of contamination of the RNA with DNA was eliminated by with an on-column RNase-free DNase. The RNA quality and quantity were assessed using the RNA 6000 Nano LabChip in an Agilent Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
For experiment 2, 2 flasks each of 3 separate passages of U-937 cells were grown in their routine RPMI-1640 media (ATCC) supplemented with 10% charcoal/dextran treated FBS and 1% penicillin/streptomycin until the cells covered 80% of the flask surface. The cells were treated with 32 nM PMA for 48 hours to stimulate differentiation of the monocytes into macrophages. The macrophages were serum-starved for 24 hours, then the medium was replaced with T-HESC CM or with non-conditioned control medium for 24 hours. Total RNA extraction and assessment of RNA were carried out as described above for T-HESC cells.
DNA Microarrays
Applied Microarrays (Tempe, AZ) was the source of the CodeLink Whole Human Genome Bioarrays12, 15, 17, 18 used for the analysis of gene expression in experiments 1 and 2. In experiment 1, T-HESC cells were treated with vehicle, E+P, vehicle + macrophage CM, or E + P + macrophage CM (n=3 passages/group, no samples were pooled). In experiment 2, U-937 macrophages were treated with nonconditioned medium or T-HESC conditioned medium (n=3 passages/group, no samples were pooled). CodeLink microarrays are spotted with single-stranded 30-mer oligonucleotide probes for 53,000 human genes/transcribed sequences. Total RNA was the starting material for the synthesis of biotinylated complementary RNA (cRNA) as described15. Biotinylated cRNA was hybridized with the CodeLink microarrays for 18 h, and hybridized slides were processed and scanned as described15. GenePix Pro software (MDS, Inc., Toronto, Canada) was used to acquire the microarray images. Microarray data were aligned, background correction applied, and individual spots were associated with their gene identifiers using CodeLink software (Applied Microarrays, Tempe, AZ). Acuity software (MDS, Inc., Toronto, Canada) was used to obtain fold expression values. Statistical analysis of the microarray data, normalization of the expression of each gene to the median gene expression and of each slide to the 50th percentile of gene expression were carried out using GeneSpring 7.3 software (Agilent, Santa Clara, CA). Comparisons of differential gene expression in the current study were made to lists of differentially expressed genes in endometriosis tissue15 and to cultured T-HESC cells that had been treated with monocyte conditioned medium12.
Real time RT-PCR
Real time reverse transcription-polymerase chain reaction (real time RT-PCR) was used to confirm differential expression of selected genes identified by DNA microarray. Pre-designed primers and minor groove binding probes and TaqMan Gold reagents were obtained from Applied Biosystems (Foster City, CA) as described12. Differences in expression of each gene of interest were calculated relative to the expression of an endogenous control, GAPDH (housekeeping gene). The qBase software19 package was used to analyze real time RT-PCR data.
Statistical analyses
The GeneSpring 7.3 (Agilent, Santa Clara, CA) software package was used to analyze the DNA microarray data by analysis of variance. The multiple testing correction used the Benjamini and Hochberg false discovery rate. Approximately 5% of the identified genes would be expected to pass this restriction by chance. Real time RT-PCR data were analyzed by ANOVA followed by Newman-Keuls Multiple Comparison Test using GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA).
Results
An increase in prolactin expression is considered a universal marker for decidualization13. Assessment of prolactin expression by real time RT-PCR showed an increase from 3.04 ± 0.7 in control conditions to 60.34 ± 7.2 in the presence of E+P. The cells also underwent phenotypic changes in response to E+P as described with decidualization13 (data not shown). The increase in prolactin expression and phenotypic changes in the presence of E+P confirm the endometrial identity of the T-HESC cell line. The presence of macrophage CM reduced prolactin expression to 1.0 ± 0.3 in control conditions and 6.77 ± 1.1 in the presence of E+P.
Statistical analysis demonstrated that treatment of T-HESC cells with macrophage CM resulted in two-fold up- or down-regulation of 716 named genes with p values of 0.05 or less and with expression values greater than 1. Of those genes, 644 responded to macrophage CM in the absence or presence of E+P (Supplemental Table 1). The remaining 72 genes responded to E+P but not to macrophage CM (Supplemental Table 2). The genes were sorted into 23 gene ontologies based on the categories of differential expression in response to macrophage CM (Table 1); some of the ontologies were empty in the E+P-only responding genes. The entire data set for all DNA microarrays in this study has been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO; www.ncbi.nlm.nih.gov/geo) as recommended by Minimum Information About a Microarray Experiment (MIAME) standards20. The data can be accessed through GEO Series accession number GSE19834.
Table 1.
Ontologies of genes that were differentially expressed in endometrial stromal cells (T-HESC cell line) in the presence of macrophage conditioned medium (Mph CM) ± estrogen plus progesterone (E+P) compared to ontologies of genes differentially expressed in T-HESC cells in response to E+P only (no response to Mph CM) compared to control conditions.
| Mph CM ± E+P | E+P only | |
|---|---|---|
| Signal transduction | 189 | 27 |
| Proteases and related | 24 | 2 |
| Steroid metabolism | 3 | 0 |
| Transcriptional regulation | 92 | 9 |
| Cytoskeleton | 25 | 6 |
| Defense/Immunity | 23 | 2 |
| Cell cycle/Cell death | 28 | 1 |
| Adhesion | 18 | 3 |
| Extracellular matrix | 19 | 2 |
| Angiogenesis | 4 | 0 |
| Translation | 3 | 0 |
| Oxidation-Reduction | 7 | 1 |
| Proteoglycans | 4 | 0 |
| Chaperonins | 9 | 0 |
| Ubiquitin system | 10 | 3 |
| Lipid metabolism | 14 | 1 |
| Membrane proteins | 21 | 2 |
| Protein-protein interactions | 6 | 0 |
| Vesicle trafficking | 4 | 0 |
| Nucleic acid regulation | 13 | 2 |
| Enzymes/metabolism | 83 | 6 |
| Channels/transporters | 34 | 3 |
| Unknown function | 11 | 2 |
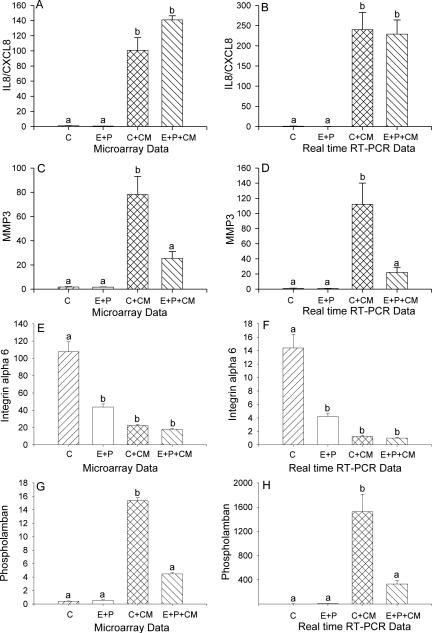
Real time RT-PCR was used to confirm the differential expression of selected genes. Treatment of cultured T-HESC cells with macrophage conditioned medium (CM) significantly increased the expression of interleukin 8, also known as chemokine CXCL8 (IL8/CXCL8); the presence of E+P did not affect IL8/CXCL8 expression in the presence or absence of macrophage CM (Figure 1A-B). Treatment of cultured T-HESC cells with macrophage conditioned medium (CM) significantly increased the expression of matrix metalloproteinase 3 (MMP3) as shown by both DNA microarray and by real time RT-PCR. The presence of E+P significantly reduced the up-regulation of MMP3 expression caused by treatment with macrophage CM (Figure 1C-D). Matrix metalloproteinase 1 (MMP1) showed a similar pattern of expression as MMP3 (Supplemental Table 1). In contrast, the expression of integrin alpha 6 (ITGA6) was significantly reduced by treatment with both E+P and macrophage CM (Figure 1E-F). The presence of macrophage CM stimulated a significant increase in the expression of phospholamban (PLN). The addition of E+P reduced the expression of phospholamban to control levels (Figure 1G-H).
Figure 1.
Differential expression of interleukin 8/CXCL8 (IL8/CXCL8, A, B), matrix metalloproteinase 3 (MMP3, C, D), integrin alpha 6 (E, F), and phospholamban (G, H) in cultured human endometrial stromal cells treated with vehicle (control, C) or estradiol plus medroxyprogesterone acetate (E+P) in the absence or presence of macrophage CM as measured by DNA microarray (A, C, E, G) and by real time RT-PCR (B, D, F, H). Significance was determined by ANOVA using GeneSpring software for microarray data and GraphPad 4 for real time RT-PCR data, p<0.05, n=3. Bars with different letter superscripts denote that the data for those groups were significantly different from each other.
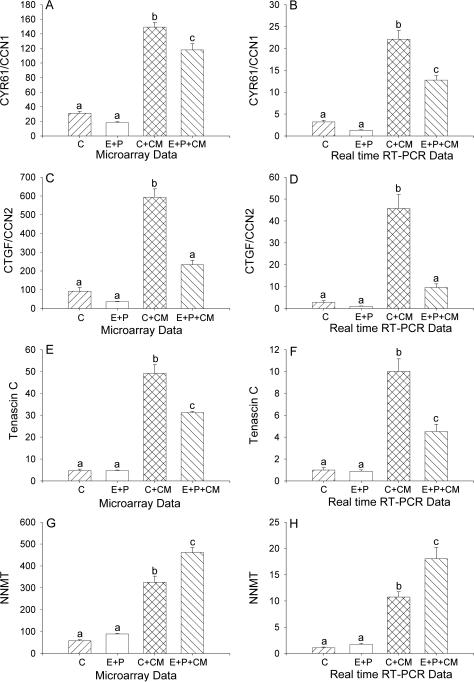
The expression of cysteine-rich angiogenic inducer 61, also known as CCN1 (CYR61/CCN1) was significantly increased in T-HESC cells in the presence of macrophage CM. The addition of E+P had no effect on control expression of CYR61/CCN1 but down-regulated its expression in the presence of macrophage CM (Figure 2A-B). The presence of macrophage CM stimulated a significant increase in the expression of connective tissue growth factor, also known as CCN2 (CTGF/CCN2). The addition of E+P reduced the expression of CTGF/CCN2 to control levels (Figure 2C-D). Treatment of cultured T-HESC cells with macrophage conditioned medium (CM) significantly increased the expression of tenascin C (TNC) as shown by both DNA microarray and real time RT-PCR. For tenascin, the presence of E+P significantly reduced the up-regulation of expression caused by treatment with macrophage CM (Figure 2E-F). Nicotinamide N-methyltransferase (NNMT) expression was significantly increased in T-HESC cells treated with macrophage CM. The presence of E+P caused a further increase in expression of NNMT in cultured T-HESC cells in the presence of macrophage CM (Figure 2G-H).
Figure 2.
Differential expression of cysteine rich angiogenic inducer 61/CCN1 (CYR61/CCN1, A, B), connective tissue growth factor/CCN2 (CTGF/CCN2, C, D), tenascin C (E, F), and nicotinamide N-methyltransferase (NNMT, G, H) in cultured human endometrial stromal cells treated with vehicle (control, C), estradiol plus medroxyprogesterone acetate (E+P) in the absence or presence of macrophage CM as measured by DNA microarray (A, C, E) and by real time RT-PCR (B, D, F). Significance was determined by ANOVA using GeneSpring software for microarray data and GraphPad 4 for real time RT-PCR data, p<0.05, n=3. Bars with different letter superscripts denote that the data for those groups were significantly different from each other.
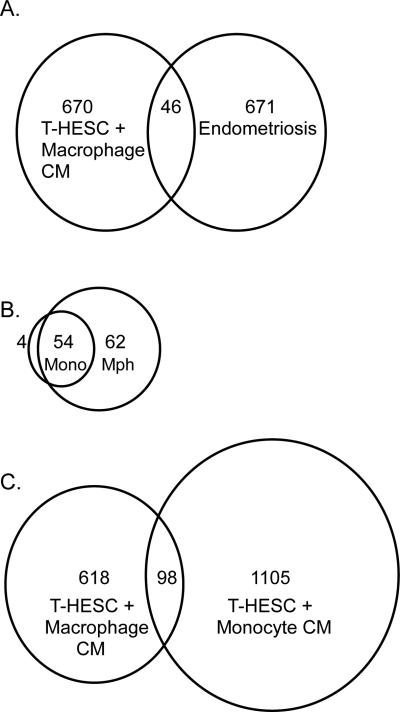
The genes that were differentially expressed in T-HESC cells in response to treatment with macrophage CM (Supplemental Table 1) were compared to the genes that were shown to be differentially expressed in endometriosis in a previous study from our laboratory15 using Access database and Excel to identify duplicates in the two gene lists. Forty-six genes appeared in both lists (Table 2). The list of overlapping genes included 6% of each individual gene list (Figure 3A). Five genes from the group that appeared on both lists were in the group chosen for follow-up with real time RT-PCR in T-HESC cells described above (integrin alpha 6, phospholamban, CYR61/CCN1, CTGF/CCN2, and NNMT).
Table 2.
| Fold Eosisb | Fold T-HESCc | ||
|---|---|---|---|
| NM_004098 | empty spiracles homolog 2 (EMX2) | 0.50 | 0.35 |
| NM_032638 | GATA binding protein 2 (GATA2) | 0.40 | 0.44 |
| NM_133443 | glutamic pyruvate transaminase 2 (GPT2) | 0.40 | 0.51 |
| NM_002147 | homeo box B5 (HOXB5) | 0.40 | 0.52 |
| NM_000210 | integrin, alpha 6 (ITGA6) | 0.40 | 0.21 |
| NM_181354 | oxidation resistance 1 (OXR1) | 0.50 | 0.54 |
| NM_007169 | phosphatidylethanolamine N-methyltransferase (PEMT) | 0.50 | 0.64 |
| NM_019012 | phosphoinositol 3-phosphate-binding protein-2 (PEPP2) | 0.40 | 0.56 |
| NM_020809 | Rho GTPase activating protein 20 (ARHGAP20) | 0.50 | 0.38 |
| NM_004755 | ribosomal protein S6 kinase, 90kDa, polypeptide 5 (RPS6KA5) | 0.50 | 0.22 |
| M77810 | transcription factor GATA-2 (GATA-2) | 0.40 | 0.42 |
| NM_015833 | adenosine deaminase, RNA-specific, B1 (ADARB1) | 2.00 | 2.25 |
| NM_000682 | adrenergic, alpha-2B-, receptor (ADRA2B) | 3.10 | 4.81 |
| NM_170732 | brain-derived neurotrophic factor (BDNF) | 4.00 | 1.74 |
| NM_203416 | CD163 antigen (CD163) | 9.20 | 1.91 |
| NM_005623 | chemokine (C-C motif) ligand 8 (CCL8) | 6.20 | 3.2 |
| NM_002089 | chemokine (C-X-C motif) ligand 2 (CXCL2) | 10.70 | 3.23 |
| NM_001276 | chitinase 3-like 1 (cartilage glycoprotein-39) (CHI3L1) | 9.90 | 1.72 |
| NM_016206 | colon carcinoma related protein | 2.70 | 1.68 |
| BC070085 | colony stimulating factor 2 receptor, beta, low-affinity | 2.80 | 13.02 |
| NM_000760 | colony stimulating factor 3 receptor (granulocyte) (CSF3R) | 5.10 | 1.78 |
| NM_001901 | connective tissue growth factor (CTGF) | 3.10 | 6.46 |
| NM_001554 | cysteine-rich, angiogenic inducer, 61 (CYR61) | 4.60 | 4.78 |
| NM_004946 | dedicator of cytokinesis 2 (DOCK2) | 2.80 | 1.8 |
| NM_003507 | frizzled homolog 7 (FZD7) | 8.10 | 1.34 |
| NM_005512 | glycoprotein A repetitions predominant (GARP) | 2.30 | 2.66 |
| NM_139211 | homeodomain-only protein (HOP) | 3.20 | 7.93 |
| NM_005525 | hydroxysteroid (11-beta) dehydrogenase 1 (HSD11B1) | 10.60 | 25.38 |
| NM_181501 | integrin, alpha 1 (ITGA1) | 2.50 | 3.43 |
| NM_006332 | interferon, gamma-inducible protein 30 (IFI30) | 3.30 | 2.21 |
| NM_002185 | interleukin 7 receptor (IL7R) | 7.40 | 3.34 |
| NM_001557 | interleukin 8 receptor, beta (IL8RB) | 2.70 | 1.63 |
| NM_020169 | latexin (LXN) | 3.70 | 3.35 |
| NM_012320 | lysophospholipase 3 (lysosomal phospholipase A2) (LYPLA3) | 4.00 | 6.19 |
| NM_002317 | lysyl oxidase (LOX) | 2.90 | 1.99 |
| NM_000900 | matrix Gla protein (MGP) | 3.90 | 6.95 |
| NM_005952 | metallothionein 1X (MT1X) | 2.30 | 1.57 |
| NM_006169 | nicotinamide N-methyltransferase (NNMT) | 7.40 | 5.63 |
| NM_002667 | phospholamban (PLN) | 13.30 | 38.4 |
| NM_004675 | ras homolog gene family, member I (ARHI) | 10.60 | 4.65 |
| NM_002928 | regulator of G-protein signalling 16 (RGS16) | 2.80 | 3.72 |
| NM_206963 | retinoic acid receptor responder 1 (RARRES1) | 4.40 | 1.81 |
| NM_002889 | retinoic acid receptor responder 2 (RARRES2) | 2.90 | 1.66 |
| NM_001175 | Rho GDP dissociation inhibitor (GDI) beta (ARHGDIB) | 2.30 | 1.99 |
Statistical differences in gene expression were calculated with GeneSpring 7.3 software.
Fold decrease or increase in expression in ectopic endometriosis (eosis) tissue compared to eutopic endometrial tissue15.
Fold decrease or increase in expression in T-HESC cells treated with macrophage CM compared to control.
Figure 3.
A. Overlap of genes in cultured endometrial cells that were differentially expressed in response to macrophage CM and genes that were differentially expressed in endometriosis15. B. Overlap of factors secreted by monocytes12 versus factors secreted by macrophages. C. Overlap of genes in cultured endometrial cells that were differentially expressed in response to macrophage CM versus monocyte CM12.
The genes that were differentially expressed in T-HESC cells in response to macrophage CM (Supplemental Table 1) were also compared to the genes that were differentially expressed in T-HESC cells in response to monocyte CM as described in a previous study from our laboratory12 using Access database and Excel to identify duplicates in the two gene lists. Under control conditions, 98 genes were differentially expressed in T-HESC cells in response to treatment with either macrophage CM or monocyte CM (Supplemental Table 3).
Secreted factors synthesized by macrophages grown under control conditions that are expected to be responsible for stimulating differential gene expression in T-HESC cells were identified by DNA microarray (Table 3). The group of factors secreted by macrophages was compared to the list of factors secreted by monocytes reported previously12. There was substantial overlap of the factors secreted by monocytes with the factors secreted by macrophages (Figure 3B). Of the 58 factors secreted by monocytes, 54 were also secreted by macrophages. In contrast, there was less overlap of factors secreted by macrophages in that 54 of the 116 macrophage-secreted factors matched those of monocytes (Figure 3B). However, the overlap of differentially expressed genes in response to macrophage CM versus monocyte CM was not as great as the overlap of factors secreted by the two cell types (Figure 3C).
Table 3.
Factors secreted by macrophages. Fold expression compares macrophage secretion to monocyte secretion of the same factor.a
| Genbank Accession # | Gene Name | Macrophage Expression | Fold Exp Mph/Monoa |
|---|---|---|---|
| Cytokines/Chemokines | |||
| NM_138284 | interleukin 17D (IL17D)* | 2.77 | 0.2 |
| NM_001252 | tumor necrosis factor (ligand) superfamily 7 (TNFSF7)* | 2.37 | 0.3 |
| NM_001562 | interleukin 18 (interferon-gamma-inducing factor) (IL18)* | 15.16 | 0.4 |
| NM_006573 | tumor necrosis factor (ligand) superfamily 13b (TNFSF13B)* | 32.57 | 0.4 |
| NM_000606 | complement component 8, gamma polypeptide (C8G)* | 4.27 | 0.5 |
| NM_002415 | macrophage migration inhibitory factor (MIF)* | 7.11 | 0.5 |
| NM_022059 | chemokine (C-X-C motif) ligand 16 (CXCL16)* | 12.98 | 2.2 |
| NM_009588 | lymphotoxin beta (TNF superfamily, member 3) (LTB)* | 7.50 | 2.3 |
| NM_002988 | chemokine (C-C motif) ligand 18 (CCL18)* | 10.39 | 2.3 |
| NM_000660 | transforming growth factor, beta 1 (TGFB1)* | 24.62 | 2.7 |
| AK023341 | similar to Pre-B cell enhancing factor precursor | 6.25 | 5.0 |
| NM_005746 | pre-B-cell colony enhancing factor 1 (PBEF1) | 14.15 | 5.2 |
| NM_006072 | chemokine (C-C motif) ligand 26 (CCL26) | 3.70 | 7.3 |
| NM_005409 | chemokine (C-X-C motif) ligand 11 (CXCL11) | 3.80 | 8.1 |
| NM_003807 | tumor necrosis factor (ligand) superfamily 14 (TNFSF14) | 5.63 | 8.9 |
| NM_172369 | complement component 1, q subcomponent, gamma polypeptide (C1QG) | 3.28 | 10.6 |
| NM_000064 | complement component 3 (C3) | 10.73 | 15.1 |
| NM_002089 | chemokine (C-X-C motif) ligand 2 (CXCL2) | 10.67 | 17.3 |
| NM_002982 | chemokine (C-C motif) ligand 2 (CCL2) | 36.98 | 23.2 |
| NM_004591 | chemokine (C-C motif) ligand 20 (CCL20) | 8.89 | 23.6 |
| NM_001565 | chemokine (C-X-C motif) ligand 10 (CXCL10) | 9.05 | 24.2 |
| NM_000572 | interleukin 10 (IL10) | 17.92 | 25.8 |
| NM_000576 | interleukin 1, beta (IL1B)* | 328.10 | 26.2 |
| NM_002984 | chemokine (C-C motif) ligand 4 (CCL4) | 40.08 | 28.0 |
| NM_002983 | chemokine (C-C motif) ligand 3 (CCL3)* | 124.58 | 40.1 |
| NM_006273 | chemokine (C-C motif) ligand 7 (CCL7) | 82.37 | 55.2 |
| NM_000584 | interleukin 8 (IL8)* | 353.04 | 76.5 |
| NM_005623 | chemokine (C-C motif) ligand 8 (CCL8) | 31.88 | 103.2 |
| NM_002704 | pro-platelet basic protein (chemokine (C-X-C motif) ligand 7) (PPBP) | 42.78 | 114.4 |
| NM_002981 | chemokine (C-C motif) ligand 1 (CCL1) | 99.19 | 274.0 |
| NM_182486 | C1q and tumor necrosis factor related protein 6 (C1QTNF6)* | 2.49 | nd |
| NM_002995 | chemokine (C motif) ligand 1 (XCL1)* | 9.51 | nd |
| NM_002990 | chemokine (C-C motif) ligand 22 (CCL22)* | 56.91 | nd |
| NM_005064 | chemokine (C-C motif) ligand 23 (CCL23)* | 4.15 | nd |
| NM_002991 | chemokine (C-C motif) ligand 24 (CCL24)* | 8.72 | nd |
| NM_002985 | chemokine (C-C motif) ligand 5 (CCL5)* | 98.65 | nd |
| NM_002996 | chemokine (C-X3-C motif) ligand 1 (CX3CL1) | 3.72 | nd |
| NM_001511 | chemokine (C-X-C motif) ligand 1 (CXCL1)* | 6.37 | nd |
| NM_002090 | chemokine (C-X-C motif) ligand 3 (CXCL3) | 3.93 | nd |
| NM_016951 | chemokine-like factor (CKLF)* | 89.05 | nd |
| NM_144601 | chemokine-like factor super family 3 (CKLFSF3)* | 28.39 | nd |
| NM_138460 | chemokine-like factor super family 5 (CKLFSF5)* | 8.62 | nd |
| NM_017801 | chemokine-like factor super family 6 (CKLFSF6)* | 34.22 | nd |
| NM_138410 | chemokine-like factor super family 7 (CKLFSF7)* | 20.92 | nd |
| NM_178868 | chemokine-like factor super family 8 (CKLFSF8)* | 3.58 | nd |
| NM_015991 | complement component 1, q subcomponent, alpha (C1QA) | 9.50 | nd |
| NM_182528 | complement component 1, q subcomponent-like 2 (C1QL2) | 11.93 | nd |
| NM_001734 | complement component 1, s subcomponent (C1S) | 12.66 | nd |
| NM_000063 | complement component 2 (C2) | 51.29 | nd |
| NM_001928 | D component of complement (adipsin) (DF) | 14.32 | nd |
| NM_021268 | interferon, alpha 17 (IFNA17) | 4.47 | nd |
| NM_000605 | interferon, alpha 2 (IFNA2) | 3.12 | nd |
| NM_014440 | interleukin 1 family, member 6 (epsilon) (IL1F6) | 4.56 | nd |
| NM_002187 | interleukin 12B (IL12B)* | 8.20 | nd |
| NM_004513 | interleukin 16 (IL16)* | 15.92 | nd |
| NM_013278 | interleukin 17C (IL17C) | 4.09 | nd |
| NM_013371 | interleukin 19 (IL19)* | 3.40 | nd |
| NM_020525 | interleukin 22 (IL22)* | 3.74 | nd |
| NM_016584 | interleukin 23, alpha subunit p19 (IL23A)* | 2.89 | nd |
| NM_181339 | interleukin 24 (IL24)* | 6.63 | nd |
| NM_000589 | interleukin 4 (IL4)* | 5.81 | nd |
| NM_000600 | interleukin 6 (interferon, beta 2) (IL6)* | 3.24 | nd |
| NM_004221 | natural killer cell transcript 4 (NK4) | 167.69 | nd |
| NM_002975 | stem cell growth factor; lymphocyte secreted C-type lectin (SCGF)* | 10.55 | nd |
| NM_003326 | tumor necrosis factor (ligand) superfamily 4 (TNFSF4)* | 3.74 | nd |
| NM_000074 | tumor necrosis factor (ligand) superfamily 5 (TNFSF5)* | 5.27 | nd |
| NM_000594 | tumor necrosis factor (TNF superfamily 2) (TNF)* | 5.44 | nd |
| Ligands/Growth factors | |||
| NM_005860 | follistatin-like 3 (secreted glycoprotein) (FSTL3) | 3.91 | 0.3 |
| NM_032536 | netrin G2 (NTNG2)* | 4.05 | 0.3 |
| NM_020415 | resistin (RETN) | 28.54 | 0.4 |
| NM_015973 | galanin (GAL) | 13.15 | 0.5 |
| NM_003393 | wingless-type MMTV integration site family 8B (WNT8B)* | 22.50 | 0.5 |
| NM_003395 | wingless-type MMTV integration site family 9A (WNT9A) | 3.74 | 2.0 |
| NM_138573 | neuregulin 4* | 14.85 | 2.6 |
| NM_001719 | bone morphogenetic protein 7 (BMP7) | 6.04 | 4.1 |
| NM_016362 | ghrelin precursor (GHRL) | 11.90 | 4.2 |
| NM_001953 | endothelial cell growth factor 1 (ECGF1) | 8.43 | 5.7 |
| NM_020530 | oncostatin M (OSM) | 6.27 | 8.1 |
| NM_002192 | inhibin, beta A (INHBA) | 18.69 | 11.6 |
| NM_024866 | adrenomedullin 2 (ADM2) | 4.35 | nd |
| NM_015985 | angiopoietin 4 (ANGPT4) | 9.44 | nd |
| NM_006129 | bone morphogenetic protein 1 (BMP1) | 9.97 | nd |
| NM_014482 | bone morphogenetic protein 10 (BMP10)* | 6.50 | nd |
| NM_005448 | bone morphogenetic protein 15 (BMP15)* | 5.63 | nd |
| NM_022640 | chorionic somatomammotropin hormone 1 (CSH1) | 3.78 | nd |
| NM_000759 | colony stimulating factor 3 (granulocyte) (CSF3)* | 10.62 | nd |
| NM_005618 | delta-like 1 (DLL1) | 7.64 | nd |
| NM_020892 | deltex homolog 2 (DTX2) | 5.15 | nd |
| NM_032890 | dispatched homolog 1 (DISP1) | 4.76 | nd |
| NM_003792 | endothelial differentiation-related factor 1 (EDF1) | 68.19 | nd |
| NM_004952 | ephrin-A3 (EFNA3) | 9.97 | nd |
| NM_005227 | ephrin-A4 (EFNA4) | 3.93 | nd |
| NM_001962 | ephrin-A5 (EFNA5) | 9.31 | nd |
| NM_001963 | epidermal growth factor (beta-urogastrone) (EGF)* | 43.71 | nd |
| NM_002087 | granulin (GRN) | 95.45 | nd |
| NM_004494 | hepatoma-derived growth factor (HDGF)* | 105.76 | nd |
| NM_016073 | hepatoma-derived growth factor, related protein 3 (HDGFRP3) | 3.44 | nd |
| NM_032631 | hepatoma-derived growth factor-related protein 2 (HDGF2) | 20.19 | nd |
| NM_031479 | inhibin, beta E (INHBE) | 3.95 | nd |
| NM_199349 | kielin-like | 7.75 | nd |
| NM_002309 | leukemia inhibitory factor (LIF)* | 6.52 | nd |
| NM_002405 | manic fringe homolog (MFNG) | 27.22 | nd |
| NM_002514 | nephroblastoma overexpressed gene (NOV) | 43.87 | nd |
| NM_006181 | netrin 2-like (chicken) (NTN2L)* | 3.96 | nd |
| NM_006179 | neurotrophin 5 (neurotrophin 4/5) (NTF5)* | 16.82 | nd |
| NM_000266 | Norrie disease (pseudoglioma) (NDP) | 5.55 | nd |
| AK055132 | similar to bone morphogenetic protein 1 | 5.21 | nd |
| AI583741 | similar to Neuregulin-3* | 5.57 | nd |
| AA461028 | similar to protein FGF-5 precursor | 20.60 | nd |
| NM_003377 | vascular endothelial growth factor B (VEGFB)* | 2.19 | nd |
| BC058855 | vascular endothelial growth factor | 13.88 | nd |
| NM_005430 | wingless-type MMTV integration site family 1 (WNT1)* | 51.07 | nd |
| NM_057168 | wingless-type MMTV integration site family16 (WNT16) | 3.59 | nd |
| NM_033131 | wingless-type MMTV integration site family 3A (WNT3A)* | 19.24 | nd |
| NM_030761 | wingless-type MMTV integration site family 4 (WNT4) | 3.10 | nd |
| NM_058238 | wingless-type MMTV integration site family 7B (WNT7B) | 3.19 | nd |
| Secreted proteinasesc | |||
| NM_002658 | plasminogen activator, urokinase (PLAU) | 30.38 | 0.5 |
| NM_004995 | matrix metalloproteinase 14 (membrane-inserted) (MMP14) | 13.77 | 2.4 |
| NM_002424 | matrix metalloproteinase 8 (neutrophil collagenase) (MMP8) | 4.21 | 16.1 |
| NM_004994 | matrix metalloproteinase 9 (gelatinase B) (MMP9) | 48.50 | 17.7 |
| NM_022792 | matrix metalloproteinase 19 (MMP19) | 22.23 | 22.4 |
| NM_002423 | matrix metalloproteinase 7 (matrilysin, uterine) (MMP7) | 109.09 | 95.3 |
| NM_002426 | matrix metalloproteinase 12 (macrophage elastase) (MMP12) | 10.31 | 104.2 |
| NM_002421 | matrix metalloproteinase 1 (interstitial collagenase) (MMP1) | 752.87 | 118.6 |
| NM_004132 | hyaluronan binding protein 2 (HABP2) | 3.73 | nd |
| NM_002776 | kallikrein 10 (KLK10) | 4.75 | nd |
| NM_002425 | matrix metalloproteinase 10 (stromelysin 2) (MMP10) | 50.42 | nd |
| NM_004530 | matrix metalloproteinase 2 (gelatinase A) (MMP2) | 5.43 | nd |
| BC047614 | matrix metalloproteinase 24 (membrane-inserted) | 7.20 | nd |
| NM_022468 | matrix metalloproteinase 25 (MMP25) | 14.22 | nd |
| NM_021801 | matrix metalloproteinase 26 (MMP26) | 5.14 | nd |
| NM_002422 | matrix metalloproteinase 3 (stromelysin 1) (MMP3) | 52.72 | nd |
| NM_004142 | matrix metalloproteinase-like 1 (MMPL1) | 7.43 | nd |
| NM_173462 | papilin, proteoglycan-like sulfated glycoprotein (PAPLN) | 5.22 | nd |
Statistical differences in gene expression between macrophages and monocytes were calculated by GeneSpring 7.3 software.
b Expression of secreted factor in macrophages divided by expression of the secreted factor in monocytes12. nd = monocyte data not significantly different from macrophage data.
Expression values for secreted proteinases were not reported previously for monocytes12.
= data for monocyte-secreted factors has been reported previously12
Treatment of U-937 macrophages with T-HESC CM resulted in the differential expression of 15 named genes that exhibited more than 2-fold difference (increase or decrease) compared to control and with p values of 0.05 or less (Table 4). Five genes that were up-regulated in U-937 macrophages in response to treatment with T-HESC CM were also up-regulated in T-HESC cells treated with macrophage CM. These five genes were matrix metalloproteinase 3 (MMP3), metallothionein 1X, oxidised low density lipoprotein receptor 1, tumor necrosis factor receptor superfamily member 21 (TNFR21), and inhibin beta A.
Table 4.
Differentially expressed genes in macrophages in response to T-HESC conditioned medium (CM).a
| GenBank Accession # | Gene name | Control | T-HESC CM |
|---|---|---|---|
| NM_001657 | amphiregulin (AREG) | 3.43 | 7.47 |
| NM_004591 | chemokine (C-C motif) ligand 20 (CCL20) | 8.89 | 34.84 |
| NM_006072 | chemokine (C-C motif) ligand 26 (CCL26) | 3.70 | 7.67 |
| NM_002984 | chemokine (C-C motif) ligand 4 (CCL4) | 40.08 | 94.61 |
| NM_004369 | collagen, type VI, alpha 3 (COL6A3) | 3.10 | 6.43 |
| NM_004091 | E2F transcription factor 2 (E2F2) | 6.23 | 1.71 |
| NM_005143 | haptoglobin (HP) | 2.65 | 11.32 |
| NM_002192 | inhibin, beta A (INHBA) | 18.69 | 39.81 |
| NM_000627 | latent transforming growth factor p binding protein (LTBP1) | 1.99 | 8.07 |
| NM_002422 | matrix metalloproteinase 3 (stromelysin 1) (MMP3) | 52.72 | 108.74 |
| NM_005952 | metallothionein 1X (MT1X) | 17.64 | 42.22 |
| NM_002543 | oxidised low density lipoprotein receptor 1 (OLR1) | 1.61 | 5.82 |
| NM_006573 | tumor necrosis factor (ligand) 13b (TNFSF13B) | 32.57 | 14.75 |
| NM_014452 | tumor necrosis factor receptor 21 (TNFRSF21) | 7.13 | 14.80 |
| NM_007115 | tumor necrosis factor, alpha-induced protein 6 (TNFAIP6) | 2.77 | 7.12 |
Statistical differences in gene expression in macrophages (Mph) under control conditions and in response to T-HESC CM were calculated by GeneSpring 7.3 software.
Discussion
The most important finding of this study is that reciprocal communication occurs between cultured endometrial stromal cells and macrophages. First, treatment of cultured endometrial stromal cells with macrophage CM resulted in changes in expression of a large number of genes which indicates that macrophages secrete factors that act on endometrial stromal cells. Secretory activity increases dramatically when monocytes differentiate into macrophages16 and we identified a number of secreted factors from macrophages using DNA microarray. These secreted factors include cytokines, chemokines, growth factors, and other ligands, all of which may contribute to the changes in gene expression observed in cultured endometrial stromal cells. Endometrial stromal cells up-regulate the expression of cytokines, chemokines and other ligands in response to macrophage CM. These factors may act on macrophages and other cell types of an endometriosis lesion, and they may act in a paracrine manner on neighboring endometrial stromal cells. Genes in ontologies associated with cellular functions associated with the establishment of endometriosis such as cell survival (cell cycle/cell death ontology), adhesion, neovascularization (angiogenesis ontology), and invasion (ontologies: proteases and related, extracellular matrix, signal transduction, and cytoskeleton) were found to be differentially expressed in response to macrophage CM. Second, treatment of macrophages with CM from cultured endometrial stromal cells resulted in changes in expression of genes in the macrophages, albeit a smaller number than with the converse treatment. Together, these data confirm our hypothesis that macrophages and endometrial stromal cells communicate with each other and that this communication, translated to the in vivo state, may contribute to the pathology of endometriosis.
Although the number of differentially expressed genes in macrophages in response to treatment with endometrial stromal cell CM was not large, it is interesting to note that 5 genes were reciprocally up-regulated in both cell types in response to conditioned medium from the other cell type. These data suggest the potential for positive feedback between the two cell types. Of the 5 genes, MMP3 has been proposed to be involved in the invasive processes of endometriosis21. A role for tumor necrosis factor α (TNFα) in endometriosis has been proposed22; one of its receptors, TNFR21, is another of the 5 genes for which positive feedback may occur between endometrial stromal cells and macrophages.
The list of genes that was differentially expressed in cultured endometrial cells treated with macrophage CM overlapped with the list of genes that was differentially expressed in endometriosis lesions reported previously15. The overlap of the two gene lists supports our hypothesis that communication between macrophages and endometrial stromal cells contributes to the pathology of endometriosis. As described further below, many of the genes that appear in both lists have been associated with endometriosis. However, it is also important to note that the list of overlapping genes included only 6% of each individual gene list. The endometriosis tissue samples were obtained from well-established lesions which had been chronically exposed to the inflammatory conditions of the lesion, whereas the cultured endometrial stromal cells were newly exposed to the factors secreted by macrophages, so the cellular responses may be different based on time of exposure. Moreover, endometriosis lesions are composed of multiple cell types including endometrial stromal and epithelial cells, vascular cells, and various types of leukocytes, whereas the cultured cells are composed of a single cell type, so the cultured cells can only express a partial response compared to the multicellular lesion. In addition, it has been reported that the peritoneal mesothelial cells lining the peritoneal cavity interact with endometrial cells in a model of early development of endometriosis, and that this interaction results in changes in gene expression in both cell types23. This interaction most likely also contributes to the gene expression changes in endometriosis that do not overlap with the genes reported here for cultured endometrial stromal cells.
Cysteine-rich angiogenic inducer 61 (CYR61)/CCN1, connective tissue growth factor (CTGF)/CCN2, and tenascin C are all matricellular proteins which are proteins of the extracellular matrix (ECM) that do not have a structural role24. Rather, matricellular proteins are adaptors and modulators of cell-ECM interactions. They interact with and integrate subsets of the actions of cytokines, growth factors, proteases, ECM structural proteins, and cell adhesion receptors in a cell type-specific manner24. CYR61/CCN1 and CTGF/CCN2 support a number of functions associated with endometriosis including cell adhesion, cell migration, and angiogenesis24. CTGF/CCN2 is known to synergize with transforming growth factor β (TGFβ) in the synthesis of ECM proteins during normal wound healing and in pathological fibrosis25,26. An interaction between CTGF/CCN2 and TGFβ may be responsible for the fibrosis that occurs in endometriotic lesions1. Both CTGF/CCN215 and CYR61/CCN127, 28 have been associated with endometriosis. Up-regulation of tenascin C has also been reported in endometriosis15, 31, 32. Tenascin C is associated with cellular functions associated with endometriosis such as migration and invasion, angiogenesis29, 30, and cell proliferation30.
Cleavage of components of the extracellular matrix such as collagen by MMPs is believed to play an important role in the remodeling of the extracellular matrix33, 34 that allows endometrial cells to invade the peritoneum during the establishment of endometriosis lesions21. In addition, MMPs may release active growth factors and cytokines from inactive precursors that are bound to the extracellular matrix34 and which may also contribute to the development of endometriosis. Moreover, suppression of MMP expression with E+P reduced the formation of endometriosis-like lesions in the nude mouse model of endometriosis35. Thus, our finding that macrophage CM stimulates the expression of MMPs 1 and 3 in cultured endometrial cells, and that MMP 1 and 3 expression was decreased by treatment with E+P, support the hypothesis that communication between macrophages and endometrial cells may contribute to the pathology of endometriosis.
IL8, also known as chemokine CXCL8, is a chemotactic factor primarily for neutrophils and lymphocytes36, but has effects on other cell types as well37, 38. In addition, IL8/CXCL8 is a potent angiogenic factor39. Increases in IL8/CXCL8 expression have been reported in endometriosis40 and IL8/CXCL8 is significantly increased in peritoneal fluid of women with endometriosis41. Moreover, IL8/CXCL8 expression by cultured endometrial stromal cells has been reported42. Thus our demonstration of increased IL8/CXCL8 expression in endometrial stromal cells in response to macrophage CM corroborates information in the literature. Our data also support the hypothesis that communication between macrophages and endometrial cells contributes to the pathology of endometriosis through stimulating neovascularization. The recruitment of neutrophils to the endometrial stromal cells by IL8/CXCL8 brings on board an additional cell type that is likely to contribute to the developing intercellular communication network and may further exacerbate the pathology of endometriosis.
Phospholamban was shown to be increased in endometriosis in a microarray study15, and in the current study, macrophage CM increased phospholamban in cultured endometrial stromal cells. Phospholamban suppresses the activity of the Ca2+-ATPase pump that returns cytoplasmic calcium levels to basal concentrations by pumping calcium into the endoplasmic reticulum (ER)43. The primary focus of phospholamban research has been on cardiac muscle contractility because of the importance of returning calcium to the sarcoendoplasmic reticulum after each muscle contraction44. Recently, a new role for phospholamban has been proposed in cell survival. A decrease in calcium levels in the ER reduces the sensitivity of a cell to apoptosis, whereas an increase in ER calcium increases the sensitivity of the cell to apoptosis44-46. We can speculate that an increase in expression of phospholamban in endometrial stromal cells in response to factors secreted by macrophages may reduce cellular sensitivity to apoptosis and support endometrial cell survival outside the uterus.
Nicotinamide N-methyltransferase (NNMT) is involved in the metabolism of xenobiotics47 and its expression has been shown to be a prognostic indicator in cancer48. NNMT is up-regulated in endometriosis tissue15 and is expressed in normal endometrium49. Recently, NNMT expression has been associated with increased cell migration and tumor invasion in bladder cancer cell lines50. The increased expression of NNMT in endometrial stromal cells treated with macrophage CM may be related to the migration and invasion of endometrial cells that occurs during the establishment of endometriosis.
Adhesion of endometrial cells to the peritoneum is an integral step in the establishment of endometriosis4, 51. Integrins are one of the large families of cell adhesive molecules51, 52 and are considered to be important players in adhesion of endometrial cells in the peritoneal cavity during the development of endometriosis. In addition to adhesion, integrins act as mediators of signal transduction52 as well as cell motility53. The expression of integrin alpha 6 was shown to decrease in endometriosis15, 54 and its expression is depolarized in the endometrium of women with endometriosis55. The reduction in expression of integrin alpha 6 in cultured endometrial cells in response to macrophage CM in the current study mimics the data from endometriosis tissue. This change in expression may reflect a shift in the migratory and/or adhesive properties of the endometrial stromal cells that ultimately contribute to the establishment of endometriosis.
A comparison of the list of factors secreted by macrophages to that of monocytes demonstrated that only 4 factors secreted by monocytes were not secreted by macrophages as well. On the other hand, macrophages secreted 62 factors not present in monocytes. The pattern of overlap of the gene expression responses of cultured endometrial stromal cells to macrophage versus monocyte CM was different from the pattern of overlap of secreted factors. Although we had expected that macrophage CM would elicit the expression of many genes that did not respond to monocyte CM, we were surprised to observe the large number of genes that were expressed in response to monocyte CM but not macrophage CM. A possible explanation for this discrepancy lies in the fact that the expression of many genes is regulated by multiple factors. The presence of a single factor, ie, a monocyte secreted factor, may up-regulate the expression of a gene, whereas the presence of a second factor in addition to the first may prevent the expression of that gene. Although gene expression responses of endometrial stromal cells to monocyte CM and macrophage CM were different, they do not appear to oppose each other. Rather, the responses to both seem to support development of endometriosis. Since both monocytes and macrophages are present in endometriosis lesions and have different effects on endometrial stromal cells, it is likely that the effects of both cell types influence the development and maintenance of endometriosis and may even synergize in their effects on endometriosis.
In summary, this report documents reciprocal communication between macrophages and endometrial stromal cells. The data lead us to propose that this intercellular dialogue contributes to the establishment of endometriosis
Supplementary Material
Acknowledgments
This work was supported in part by internal funds from the Sanford School of Medicine Department of Obstetrics & Gynecology. The authors would like to thank Sandy Bradley for technical assistance. The Genomics Core Facility at the University of South Dakota is supported by NIH INBRE 2 P20 RR016479.
References cited
- 1.Schenken RS. Endometriosis. In: Danforth's Obstetrics and Gynecology. In: James R, Scott, Philip J, Di Sain, Charles B, Hammond, William N, Spellacy, editors. Lippincott. Eighth Edition Williams & Wilkins; Baltimore: 1999. pp. 669–675. [Google Scholar]
- 2.Bulun SE, Adashi EY. The physiology and pathology of the female reproductive axis. In: Kronenberg HM, Melmed S, Polonsky KS, Larsen PR, editors. Williams Textbook of Endocrinology. 11th edition. Saunders Elsevier; Philadelphia, PA: 2008. pp. 541–614. [Google Scholar]
- 3.Christodoulakos G, Augoulea A, Lambrinoudaki I, Sioulas V, Creatsas G. Pathogenesis of endometriosis: the role of defective ‘immunosurveillance’. Eur J Contracep Reprod Health Care. 2007;12:194–202. doi: 10.1080/13625180701387266. [DOI] [PubMed] [Google Scholar]
- 4.Khan KN, Kitajima M, Hiraki K, Fujishita A, Sekine I, Ishimaru T, et al. Immunopathogenesis of pelvic endometriosis: role of hepatocyte growth factor, macrophages and ovarian steroids. Am J Reprod Immunol. 2008;60:383–404. doi: 10.1111/j.1600-0897.2008.00643.x. [DOI] [PubMed] [Google Scholar]
- 5.Halme J, Becker S, Wing R. Accentuated cyclic activation of peritoneal macrophages in patients with endometriosis. Am J Obstet Gynecol. 1984;148:85–90. doi: 10.1016/s0002-9378(84)80037-x. [DOI] [PubMed] [Google Scholar]
- 6.Oosterlynck DJ, Cornillie FJ, Waer M, Koninckx PR. Immunohistochemical characterization of leucocyte subpopulations in endometriotic lesions. Arch Gynecol Obstet. 1993;253:197–206. doi: 10.1007/BF02766646. [DOI] [PubMed] [Google Scholar]
- 7.Cakmak H, Guzeloglu-Kayisli O, Kayisli UA, Arici A. Immune-endocrine interactions in endometriosis. Front Biosci. 2009;E1:429–443. doi: 10.2741/E39. [DOI] [PubMed] [Google Scholar]
- 8.Gazvani R, Templeton A. Peritoneal environment, cytokines and angiogenesis in the pathophysiology of endometriosis. Reproduction. 2002;123:217–226. doi: 10.1530/rep.0.1230217. [DOI] [PubMed] [Google Scholar]
- 9.Bedaiwy MA, Falcone T. Peritoneal fluid environment in endometriosis. Clinicopathological implications. Minerva Ginecol. 2003;55:333–345. [PubMed] [Google Scholar]
- 10.Wu M-Y, Ho H-N. The role of cytokines in endometriosis. Am J Reprod Immunol. 2003;49:285–296. doi: 10.1034/j.1600-0897.2003.01207.x. [DOI] [PubMed] [Google Scholar]
- 11.Minici F, Tiberi F, Tropea A, Fiorella M, Orlando M, Gangale MF, et al. Paracrine regulation of endometriotic tissue. Gynecol Endocrinol. 2007;23:574–580. doi: 10.1080/09513590701581721. [DOI] [PubMed] [Google Scholar]
- 12.Klinkova O, Hansen KA, Winterton E, Mark CJ, Eyster KM. Two-way communication between endometrial stromal cells and monocytes. Reprod Sci. 2010;17(2):125–135. doi: 10.1177/1933719109348922. [DOI] [PubMed] [Google Scholar]
- 13.Krikun G, Mor G, Alvero A, Guller S, Schatz F, Sapi E, et al. A novel immortalized human endometrial stromal cell line with normal progestational response. Endocrinology. 2004;145:2291–2296. doi: 10.1210/en.2003-1606. [DOI] [PubMed] [Google Scholar]
- 14.Sundstrom C, Nilsson K. Establishment and characterization of a human histiocytic lymphoma cell line (U-937). Int J Cancer. 1976;17:565–577. doi: 10.1002/ijc.2910170504. [DOI] [PubMed] [Google Scholar]
- 15.Eyster KM, Klinkova O, Kennedy V, Hansen KA. Whole genome deoxyribonucleic acid microarray analysis of gene expression in ectopic versus eutopic endometrium. Fertil Steril. 2007;88:1505–1533. doi: 10.1016/j.fertnstert.2007.01.056. [DOI] [PubMed] [Google Scholar]
- 16.Radzun HJ, Parwaresch MR, Sundström C, Nilsson K, Eissner M. Monocytic origin of the human hematopoietic cell line U-937 and its convertibility to macrophages evidenced by isoenzyme mapping. Int J Cancer. 1983;31:181–186. doi: 10.1002/ijc.2910310208. [DOI] [PubMed] [Google Scholar]
- 17.Yauk CL, Berndt ML, Williams A, Douglas GR. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shippy R, Sendera TJ, Lockner R, Palaniappan C, Kaysser-Kranich T, Watts G, et al. Performance evaluation of commercial short-oligonucleotide microarrays and the impact of noise in making cross-platform correlations. BMC Genomics. 2004;5:61. doi: 10.1186/1471-2164-5-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hellemans J, Mortier G, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoecker C, et al. Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 21.Cox KE, Piva M, Sharpe-Timms KL. Differential regulation of matrix metalloproteinase-3 gene expression in endometriotic lesions compared with endometrium. Biol Reprod. 2001;65:1297–1303. doi: 10.1095/biolreprod65.4.1297. [DOI] [PubMed] [Google Scholar]
- 22.Harada T, Yoshioka H, Yoshida S, Iwabe T, Onohara Y, Tanikawa M, et al. Increased interleukin-6 levels in peritoneal fluid of infertile patients with active endometriosis. Am J Obstet Gynecol. 1997;176:593–597. doi: 10.1016/s0002-9378(97)70553-2. [DOI] [PubMed] [Google Scholar]
- 23.Nair AS, Nair HB, Lucidi RS, Kirchner AJ, Schenken RS, Tekmal RR, et al. Modeling the early endometriotic lesion: mesothelium-endometrial cell co-culture increases endometrial invasion and alters mesothelial and endometrial gene transcription. Fertil Steril. 2008;90(Suppl 2):1487–1495. doi: 10.1016/j.fertnstert.2007.09.047. [DOI] [PubMed] [Google Scholar]
- 24.Chen C-C, Lau LF. Functions and mechanisms of action of CCN matricellular proteins. Int J Biochem Cell Biol. 2009;41:771–783. doi: 10.1016/j.biocel.2008.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leask A, Abraham DJ. The role of connective tissue growth factor, a multifunctional matricellular protein, in fibroblast biology. Biochem Cell Biol. 2003;81:355–363. doi: 10.1139/o03-069. [DOI] [PubMed] [Google Scholar]
- 26.Gressner OA, Gressner AM. Connective tissue growth factor: a fibrogenic master switch in fibrotic liver diseases. Liver Int. 2008;28:1065–1079. doi: 10.1111/j.1478-3231.2008.01826.x. [DOI] [PubMed] [Google Scholar]
- 27.Absenger Y, Hess-Stumpp H, Kreft B, Kratzschmar J, Haendler B, Schutze N, et al. Cyr61, a deregulated gene in endometriosis. Mol Hum Reprod. 2004;10:399–407. doi: 10.1093/molehr/gah053. [DOI] [PubMed] [Google Scholar]
- 28.Gashaw I, Hastings JM, Jackson KS, Winterhager E, Fazleabas AT. Induced endometriosis in the baboon (Papio anubis) increases the expression of the proangiogenic factor CYR61 (CCN1) in eutopic and ectopic endometria. Biol Reprod. 2006;74:1060–1066. doi: 10.1095/biolreprod.105.049320. [DOI] [PubMed] [Google Scholar]
- 29.Chiquet-Ehrismann R. Molecules in focus: Tenascins. Int J Biochem Cell Biol. 2004;36:986–990. doi: 10.1016/j.biocel.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 30.Orend G, Chiquet-Ehrismann R. Tenascin-C induced signaling in cancer. Cancer Lett. 2006;244:143–163. doi: 10.1016/j.canlet.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Tan O, Ornek T, Seval Y, Sati L, Arici A. Tenascin is highly expressed in endometriosis and its expression is upregulated by estrogen. Fertil Steril. 2008;89:1082–1089. doi: 10.1016/j.fertnstert.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 32.Harrington DJ, Lessey BA, Rai V, Bergqvist A, Kennedy S, Manek S, et al. Tenascin is differentially expressed in endometrium and endometriosis. J Pathol. 1999;187:242–248. doi: 10.1002/(SICI)1096-9896(199901)187:2<242::AID-PATH221>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 33.Hulboy DL, Rudolph LA, Matrisian LM. Matrix metalloproteinases as mediators of reproductive function. Mol Hum Reprod. 1997;3:27–45. doi: 10.1093/molehr/3.1.27. [DOI] [PubMed] [Google Scholar]
- 34.McCawley LJ, Matrisian LM. Matrix metalloproteinases: they're not just for matrix anymore! Curr Opin Cell Biol. 2001;13:534–540. doi: 10.1016/s0955-0674(00)00248-9. [DOI] [PubMed] [Google Scholar]
- 35.Bruner KL, Matrisian LM, Rodgers WH, Gorstein F, Osteen KG. Suppression of matrix metalloproteinases inhibits establishment of ectopic lesions by human endometrium in nude mice. J Clin Invest. 1997;12:2851–2857. doi: 10.1172/JCI119478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raman D, Baugher PJ, Thu YM, Richmond A. Role of chemokines in tumor growth. Canc Lett. 2007;256:137–165. doi: 10.1016/j.canlet.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am J Physiol Lung Cell Mol Physiol. 2003;284:L566–L577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 38.Zhu YM, Woll PJ. Mitogenic effects of interleukin-8/CXCL8 on cancer cells. Future Oncol. 2005;1:699–704. doi: 10.2217/14796694.1.5.699. [DOI] [PubMed] [Google Scholar]
- 39.Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of neovascularization. Arterioscler Thromb Vasc Biol. 2008;28:1928–1936. doi: 10.1161/ATVBAHA.108.162925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ulukus M, Ulukus EC, Tavmergen Goker EN, Tavmergen E, Zheng W, Arici A. Expression of interleukin-8 and monocyte chemotactic protein 1 in women with endometriosis. Fertil Steril. 2009;91:687–693. doi: 10.1016/j.fertnstert.2007.12.067. [DOI] [PubMed] [Google Scholar]
- 41.Iwabe T, Harada T, Tsudo T, Tanikawa M, Onohara Y, Terakawa N. Pathogenetic significance of increased levels of interleukin-8 in the peritoneal fluid of patients with endometriosis. Fertil Steril. 1998;69:924–930. doi: 10.1016/s0015-0282(98)00049-1. [DOI] [PubMed] [Google Scholar]
- 42.Arici A, Head JR, MacDonald PC, Casey ML. Regulation of interleukin-8 gene expression in human endometrial cells in culture. Mol Cell Endocrinol. 1993;94:195–204. doi: 10.1016/0303-7207(93)90168-j. [DOI] [PubMed] [Google Scholar]
- 43.Stokes DL. Keeping calcium in its place: Ca2+-ATPase and phospholamban. Curr Opin Struc Biol. 1997;7:550–556. doi: 10.1016/s0959-440x(97)80121-2. [DOI] [PubMed] [Google Scholar]
- 44.Vafiadaki E, Papalouka V, Arvanitis DA, Kranias EG, Sanoudou D. The role of SERCA2a/PLN complex, Ca2+ homeostasis, and anti-apoptotic proteins in determining cell fate. Eur J Physiol. 2009;457:687–700. doi: 10.1007/s00424-008-0506-5. [DOI] [PubMed] [Google Scholar]
- 45.Pinton P, Ferrari D, Rapizzi E, Di Virgilio F, Pozzan T, Tizzuto R. The Ca2+ concentration of the endoplasmic reticulum is a key determinant of ceramide-induced apoptosis: significance for the molecular mechanism of Bcl-2 action. 2001;20:2690–2701. doi: 10.1093/emboj/20.11.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rizzuto R, Pinton P, Ferrari D, Chami M, Szabadkai G, Magalhaes PJ, et al. Calcium and apoptosis: facts and hypotheses. Oncogene. 2003;22:8619–8627. doi: 10.1038/sj.onc.1207105. [DOI] [PubMed] [Google Scholar]
- 47.Aksoy S, Szumlanski CL, Weinshilboum RM. Human liver nicotinamide N-methyltransferase cDNA cloning, expression, and biochemical characterization. J Biol Chem. 1994;269:14835–14840. [PubMed] [Google Scholar]
- 48.Kim J, Hong SJ, Lim EK, Yu YS, Kim SW, Roh JH, et al. Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis. J Exp Clin Cancer Res. 2009;28:20. doi: 10.1186/1756-9966-28-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Allegra A, Marino A, Coffaro F, Lama A, Rizza G, Scaglione P, et al. Is there a uniform basal endometrial gene expression profile during the implantation window in women who became pregnant in a subsequent ICSI cycle? Hum Reprod. 2009;24:2549–2557. doi: 10.1093/humrep/dep222. [DOI] [PubMed] [Google Scholar]
- 50.Wu Y, Siadaty MS, Berens ME, Hampton GM, Theodorescu D. Overlapping gene expression profiles of cell migration and tumor invasion in human bladder cancer identify metallothionein 1E and nicotinamide N-methyltransferase as novel regulators of cell migration. Oncogene. 2008;27:6679–6689. doi: 10.1038/onc.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lessey BA, Young SL. Integrins and other cell adhesion molecules in endometrium and endometriosis. Semin Reprod Endocrinol. 1997;15:291–299. doi: 10.1055/s-2008-1068759. [DOI] [PubMed] [Google Scholar]
- 52.Juliano RL. Signal transduction by cell adhesion receptors and the cytoskeleton: functions of integrins, cadherins, selectins, and immunoglobulin-superfamily members. Annu Rev Pharmacol Toxicol. 2002;42:283–323. doi: 10.1146/annurev.pharmtox.42.090401.151133. [DOI] [PubMed] [Google Scholar]
- 53.Ramsay AG, Marshall JF, Hart IR. Integrin trafficking and its role in cancer metastasis. Cancer Metastasis Rev. 2007;26:567–578. doi: 10.1007/s10555-007-9078-7. [DOI] [PubMed] [Google Scholar]
- 54.Rai V, Hopkisson J, Kennedy S, Bergqvist A, Barlow DH, Mardon HJ. Integrins alpha 3 and alpha 6 are differentially expressed in endometrium and endometriosis. J Path. 1996;180:181–187. doi: 10.1002/(SICI)1096-9896(199610)180:2<181::AID-PATH620>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 55.Vernet-Tomas Mdel M, Perez-Ares CT, Verdu N, Fernandez-Figueras MT, Carreras R. The depolarized expression of the alpha-6 integrin subunit in the endometria of women with endometriosis. J Soc Gynecol Investig. 2006;13:292–296. doi: 10.1016/j.jsgi.2006.03.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.