SUMMARY
Influenza A virus polymerase subunit PB2 is a major virulence determinant implicated in pathogenicity and host adaptation. During cross-species virus transfer from avian to mammalian cells, PB2 switches specificity from importin α3 to α7. This specificity is not recapitulated in vitro, where PB2 binds all importin α isoforms with comparably high affinity. In this paper, we investigated the structure, conformational dynamics and autoinhibition of importin α isoforms 1, 3 and 7 in complex with PB2. Our data suggest that association of PB2 with α3 and α7 is favored by reduced autoinhibition of these isoforms and by the unique structure of PB2’s NLS-domain. We propose that by recruiting importin α3 or α7 in the absence of importin β, PB2 reduces the complexity of adaptor-mediated import to a pseudo-bimolecular reaction, thereby acquiring a kinetic advantage over classical NLS-cargos that form an import complex only when importin α and β are simultaneously available.
Keywords: nuclear transport, importin α isoforms, importin α3, importin α7, Influenza virus PB2, IBB-autoinhibition, NLS
INTRODUCTION
Trafficking of macromolecules between the cytoplasm and the nucleus occurs through the selectively-permeable barrier of nuclear pore complexes (NPC), large channels that span the nuclear envelope. Although permeable to molecules smaller than ~40 kDa, the majority of proteins are escorted through the NPC in an energy-dependent process mediated by soluble transport factors of the importin β-superfamily (also known as β-karyopherins) (Stewart, 2007). Originally identified on the basis of a conserved N-terminal Ran-binding domain (Mosammaparast and Pemberton, 2004), β-karyopherins share significant structural homology with importin β (Cingolani et al., 1999) and are able to move through the barrier of the NPC by interacting with the ‘FG-repeats’ present in many of the nucleoporins composing the NPC (Bednenko et al., 2003). The classical nuclear import pathway also relies on a second karyopherin, importin α, which acts as an adaptor between importin β and NLS-cargos (Goldfarb et al., 2004). Importin α contains an N-terminal importin β binding (IBB) domain (Lott and Cingolani, 2011) and a C-terminal helical core composed of 10 Armadillo (Arm) repeats that form an extended binding surface for NLSs (Conti et al., 1998; Kobe, 1999). The IBB domain is a functional NLS that, in the context of the full length importin α, folds back to autoinhibit the Arm core, thereby preventing NLS binding in the absence of importin β (Harreman et al., 2003; Kobe, 1999). The classical import complex assembles in the cytoplasm upon recognition of an NLS by importin α, which requires importin β-initiated displacement of the IBB from the Arm core.
Importin α and importin β are built of helical repeats that stack together to form solenoid structures (Cook et al., 2007). Importin β contains 19 HEAT repeats, each of which consists of two α-helices that form a flexible superhelical spring (Forwood et al., 2010; Kappel et al., 2010; Zachariae and Grubmuller, 2008). Arm repeats found in importin α's helical core are similar to HEAT repeats, but the first helix of the HEAT repeat is split into two smaller helices, generating a three-helix motif (Andrade et al., 2001). This subtle change in secondary structure leads to a far more rigid structure (Conti et al., 2006). Importin α's Arm core contains two NLS-binding pockets on its concave surface, between Arms 2–4 and 6–8, that are known as the major and minor NLS-binding pockets, respectively. The classical NLS consists of a short stretch of basic residues that can be clustered consecutively, as in the SV40 monopartite NLS (KKKRK), or separated by 10–12 residues, as in the nucleoplasmin bipartite NLS (KR-10X-KKKK) (Jans et al., 2000). Monopartite NLSs generally bind at the major NLS and, to a lesser extent, at the minor site, while bipartite NLSs span both NLS binding sites (Chen et al., 2005; Conti and Kuriyan, 2000; Conti et al., 1998; Fontes et al., 2003; Fontes et al., 2000; Marfori et al., 2012). Each NLS-binding pocket in importin α contains several sites that accommodate basic residues and are termed P1-P5 for the major and P1'-P4' for the minor NLS-binding site. Strong electrostatic interactions between acidic NLS-binding pockets and most classical NLSs usually result in low nanomolar (~5–50 nM) dissociation constants (Catimel et al., 2001; Hodel et al., 2001), although weaker affinities have also been reported (Lott et al., 2011).
There are seven isoforms of importin α in mammals (Mason et al., 2009), of which importin α1 is thought to function as the generic adaptor for NLS-cargos (Kohler et al., 1999). A growing number of import cargos have been shown to rely on specific importin α isoforms (Pumroy and Cingolani, 2014). An interesting example is the influenza A polymerase subunit PB2, one of the three subunits of the influenza A RNA-dependent RNA polymerase complex (PA, PB1, and PB2) that catalyzes viral RNA replication and transcription in the nuclei of infected host cells (Resa-Infante et al., 2011). PB2 is a major virulence determinant of influenza viruses that has been implicated in pathogenicity and host adaptation (Resa-Infante and Gabriel, 2013). In vivo studies have shown that PB2 found in avian influenza strains is imported into the nucleus by importin α3, but in mammalian adapted strains PB2 switches dependency to importin α7 (Gabriel et al., 2011; Hudjetz and Gabriel, 2012; Resa-Infante et al., 2014). Structural studies, including a crystal structure of PB2 in complex with importin α5 (Tarendeau et al., 2007), revealed that PB2 contains an extended NLS-domain (referred to as NLD) that is connected by a flexible linker to an independently folded N-terminal domain (627-domain) (Tarendeau et al., 2008). Several mutations implicated in cross-species transfer are located on the 627-domain surface and possibly affect PB2 interactions with other viral proteins and/or host factors, including importins (Resa-Infante and Gabriel, 2013). Despite the selectivity for importin α isoforms observed in vivo, PB2-NLD binds the Arm core of all importin α isoforms with equivalent low nanomolar affinity in vitro (Boivin and Hart, 2011). To determine the molecular basis for PB2-discrimination among isoforms of importin α, in this paper we have studied the structure, dynamics and association of PB2-NLD with three representative isoforms of importin α.
RESULTS
Evolution and diversification of importin α isoforms
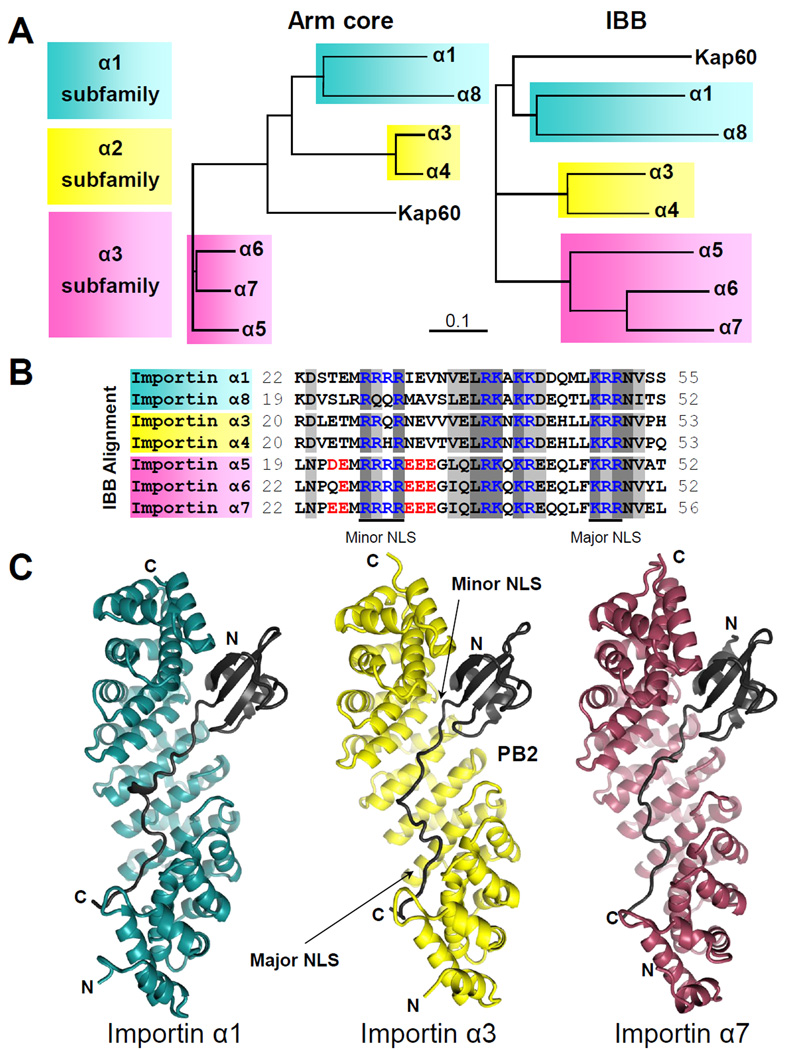
The human genome encodes seven isoforms of importin α grouped into three subfamilies: α1, α2 and α3 (Mason et al., 2009) (Table S1). These isoforms have similar lengths (516 to 539 amino acids) and share as many as 31% identical and 50% conserved residues in their Arm cores (Figure 1A, left). Subfamilies α1 and α2 are more similar to each other than subfamily α3, which diverged earlier in evolution from an ancestral gene, possibly similar to yeast importin α (Kap60) (Mason et al., 2009). Amino acid conservation in the Arm core varies within each subfamily: the α2 and α3 subfamilies are the most conserved, with 94% and 88% sequence identity, respectively, that drops to 59% in the α1 subfamily. Aligning importin α isoforms by the IBB-domain results in a classification consistent with that produced using the Arm core (Figure 1A, right). Within the IBB-domain, three clusters of basic amino-acids are conserved in all isoforms: the ‘KRR’ and ‘RxxR’ motifs that bind major and minor NLS-binding sites of importin α, respectively (Kobe, 1999), and the ‘RKxKK/R’ motif, which interacts with the recycling factor Cse1 (Matsuura and Stewart, 2004) (Figure 1B). The minor NLS-binding motif is the least conserved and shows significant variation at positions P2’ and P3’. Whereas the fully autoinhibited importin α1 has four consecutive Rs (‘RRRR’), the position P3’ is replaced with Q in importin α8/α3 and an H is found in importin α4. Likewise, isoforms of the α3 subfamily have acidic residues immediately flanking each side of the minor NLS binding site. We predict that subtle differences in Arm core sequence, together with differential autoinhibition due to amino acid differences in IBBs, are important determinants to decipher how NLS-cargos discriminate among different isoforms of importin α.
Figure 1. Evolution and structure of the importin α isoforms.
(A) Phylogenetic trees of the Arm cores (left) and IBBs (right) of human importin α isoforms and the yeast importin α (Kap60). (B) ClustalX2 alignment of the IBB region of human importin α isoforms. Basic conserved residues are in blue, with acidic residues of subfamily α3 surrounding the minor NLS in red. Highlighted in dark and light grey are identical and similar residues, respectively. (C) Ribbon diagrams of PB2-NLD, in black, bound to the Arm core of importin α1 (right, in cyan), importin α3 (middle, in yellow) and importin α7 (left, in purple). See also Table S1.
Crystallization of importin α isoforms bound to PB2-NLD
We have determined crystal structures of the PB2-NLD (res. 678–759) in complex with representative isoforms from each subfamily of mammalian importin α, namely α1, α3, and α7. All isoforms used for crystallization lacked the N-terminal IBB-domain that is linked to the Arm core by a protease sensitive flexible linker (Cingolani et al., 2000). High resolution diffracting crystals of isoforms α1 and α7 bound to PB2-NLD were readily obtained, whereas the isoform importin α3 was recalcitrant to crystallization. Controlled dehydration of crystallization droplets against a reservoir solution containing 1M salt gave ~10 µm thick needle-like crystals that diffracted to 2.7 Å resolution. The 3D-structures of subfamily representative isoforms bound to PB2-NLD were solved and refined to an Rfree between 19% and 23% (Table 1). Thus, the NLS-domain of influenza stabilizes the solenoid structure of all importin α isoforms, allowing for crystallization. As dehydration is a classic approach to increasing the resolution of crystals of flexible proteins (Roy et al., 2012), it is possible that importin α3 adopts a more flexible conformation than the other isoforms.
Table 1.
Crystallographic data collection and refinement statistics.
| Importin α1:PB2 | Importin α3:PB2 | Importin α7:PB2 | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P41 | C2 |
| Cell dimensions | |||
| a, b, c (Å) | 77.1, 90.2, 100.7 | 135.68, 135.68, 47.31 | 242.3, 38.8, 67.9 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 98, 90 |
| Resolution limits (Å) | 15-1.70 (1.76-1.70) | 15-2.70 (2.80-2.70) | 15-2.42 (2.51-2.42) |
| Reflections (total/unique) | 344,360 / 76,101 | 83,033 / 23,809 | 566,026 / 21,920 |
| Completeness (%) | 97.5 (97.9) | 98.8 (98.2) | 90.0 (60.4) |
| Rsyma | 4.9 (50) | 22.3 (58.7) | 8.3 (37.5) |
| I / σ(I) | 48.5 (5.6) | 11.1 (2.3) | 24.9 (4.7) |
| Wilson B value (Å2) | 23.5 | 23.8 | 30.5 |
| Refinement | |||
| Resolution (Å) | 15-1.70 | 15-2.70 | 15-2.42 |
| No. reflections | 141,899 | 23,788 | 40,116 |
| Rwork / Rfreeb | 16.29 / 19.08 | 18.48 / 23.86 | 18.89 / 23.10 |
| No. atoms | |||
| Importin α | 3247 | 3219 | 3298 |
| PB2 | 559 | 568 | 569 |
| Water | 475 | 288 | 157 |
| B-factors (Å2) | |||
| Importin α | 31.8 | 28.3 | 36.3 |
| PB2 | 45.5 | 44.7 | 33.4 |
| Water | 45.3 | 31.2 | 28.0 |
| R.m.s deviations | |||
| Bond lengths (Å) | 0.006 | 0.011 | 0.003 |
| Bond angles (°) | 1.06 | 1.08 | 0.73 |
| Ramachandran plot (%) | 99.0 / 0.8 / 0.2 | 95.9 / 4.1 / 0 | 98.8 / 1.2 / 0 |
| Core/Allowed and Generously Allowed/Disallowed |
Values in parentheses are for highest-resolution shells.
Rsym =Σi,h | I(i,h) − <I(h)> | /Σi,h | I(i,h) | where I(i,h) and <I(h)> are the ith and mean measurement of intensity of reflection h.
The Rfree value was calculated using 2,000 reflections selected randomly
Subtle differences in the Arm superhelix among importin α isoforms
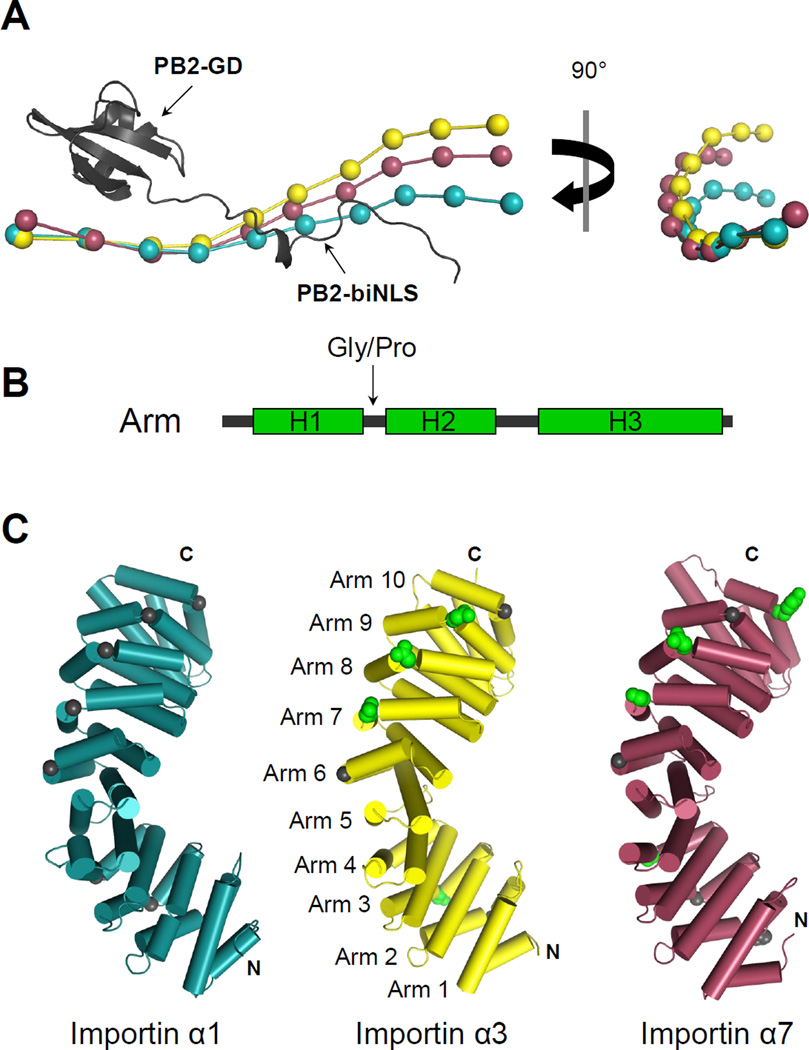
Importin α isoforms bound to PB2-NLD share a similar 3D-organization consisting of 10 stacked Arm repeats (Figure 1C). The highly conserved architecture of the Arm repeats leads to extreme structural conservation, which is also observed in non-karyopherin Arm repeat containing proteins such as β-catenin (Huber et al., 1997). PB2-NLD contains a globular domain (PB2-GD) (res. 678–736) built by 3 β-stands packed against two short α- helices that connect to an extended bipartite-like NLS (PB2-biNLS) (res. 737–759). As seen in complex with importin α5 (Tarendeau et al., 2007), PB2-GD sits above the minor NLSbinding pocket of importin α and makes minimal contacts with C-terminal Arms 7–10. PB2- biNLS runs antiparallel to the importin α Arm core, making extensive contacts that span from minor to major NLS-binding pockets (Figure 1C). Secondary structure matching alignment of the three importin α isoforms reveals a global RMSD of 1.27 Å between importin α1 and α3, 1.33 Å between importin α3 and α7, and 1.82 Å between importin α1 and α7. A beads-on-a-string representation of the three isoforms, aligned with respect to PB2-GD (Figure 2A), reveals significant deviation in position of the importin α N-termini. To break down these differences, the RMSD between pairs of adjacent Arm repeats was measured for each isoform and carefully compared (Figure S1). This analysis suggested that the regions of greatest deviation between isoforms are between Arms 3–4 for importin α3, Arms 4–5 among all importin isoforms, and Arms 8–10 for importin α7. Comparing the amino acid sequence and individual Arm repeats in each isoform, we identified two structural determinants that may contribute to differences in flexibility and possibly curvature: first, importin α3 and α7 have a larger number of bulky hydrophobic residues conserved at the interfaces between Arm repeats in their regions of high RMSD. Second, Arm repeats show different degeneration among isoforms at the break between helix 1 (H1) and helix 2 (H2) of each Arm repeat (Figure 2B). In importin α1, a helix breaking glycine is conserved between helices H1 and H2 (Figure 2C) at 8 of the 10 Arm repeats, with the exception of Arm 1, which lacks a proper helix H1 and Arm 5 that presents two helices fused together into a single helix, analogous to a HEAT repeat (Andrade et al., 2001). In contrast, importin α3 and α7 have several Arm repeats where the helix breaking residue between H1 and H2 is replaced by a helix promoting polar or charged residue. In α3 this is particularly evident at the C-terminus, where Arms 7–9 have D, N, and N respectively between H1 and H2 (Figure 2C). Thus, degenerated Arm repeats may contribute to a higher flexibility of importin α3 and possibly α7 as compared to importin α1.
Figure 2. Structural comparison of human importin α isoforms.
(A) Structural alignment of α1, α3 and α7 (shown as ‘beads-on-a-string’) bound to PB2-NLD (in ribbons). Alignment is based on superimposition of PB2 globular domains using as reference PB2-NLD solved in complex with importin α1. Beads-on-a-string models of importin α isoforms were drawn in PyMol by reducing each Arm repeat to a single point based on the α-carbon of the conserved tryptophan positioned in the middle of helix H3. (B) Schematic diagram of the helices of an Arm repeat. (C) Cartoon representation of the importin α isoforms with helices depicted as cylinders. Residues at the H1/H2 interface are shown as spheres, with glycines in black and polar or charged residues in green. The color coding of importin α isoforms in panels (A) and (C) is same as in (Figure 1C). See also Figure S1.
Importin α isoforms share an invariant NLS-binding groove
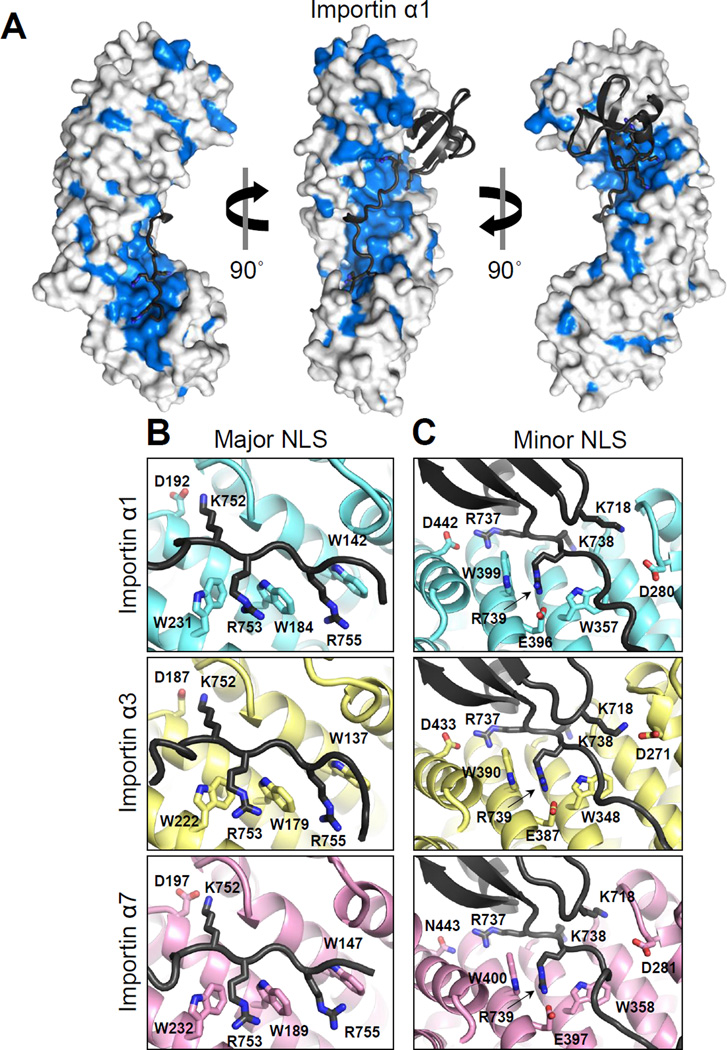
PB2 binding to importin α isoforms buries 9.7%, 9.8% and 9.0% of the total exposed surface of importin α1 (1,818 Å2/18,654 Å2), α3 (1,854 Å2/ 18,831 Å2) and α7 (1,740 Å2/19,247 Å2), respectively. Mapping conserved residues on the concave surface of importin α isoforms occupied by PB2 reveals an essentially identical NLS-binding groove (Figure 3A). In contrast, systematic differences exist at the C-termini (Arm 8–10) and on the convex surface, which are not involved in binding to PB2. Comparing the electrostatic potential molecular surface of importin α isoforms reveals highly conserved patches of acidic residues at the major and minor NLS-binding pockets, as well as scattered acidic residues especially at N- and C-termini of importin α3 (Figure S2). This is consistent with the slightly lower isoelectric point of this isoform as compared to α1 and α7, which, excluding the IBB domain is 4.55 versus 4.91 and 4.62.
Figure 3. Interactions of PB2-NLD at the major and minor NLS-binding sites of importin α isoforms.
(A) Surface representation of importin α1 bound to PB2-NLD (shown as black ribbon): in blue are residues identical in all human isoforms; in light grey are residues which are not conserved in all human isoforms. Magnified view of the major (B) and minor (C) NLSbinding sites of importin α1 (top), importin α3 (middle) and importin α7 (bottom). See also Figure S2.
Recognition of PB2-NLD by importin α isoforms follows similar principles as previously described for importin α5 (Tarendeau et al., 2007), with subtle differences in the three subfamilies. PB2 binding to the major NLS-binding pocket is essentially identical in the three isoforms (Figure 3B): PB2K752 occupies the P2 site and forms a salt bridge with D192/187/197 (in importin α1/α3/α7, respectively), while PB2R753 and PB2R755 are held in place in the P3 and P5 sites, respectively, by hydrogen bonds with the conserved tryptophans on Arm 2–4, namely W142/137/147, W184/179/189, and W231/222/232. Instead, at the minor NLS-binding site (Figure 3C), PB2K738 occupies the P1’ site and makes hydrogen bonds in the cavity made by the loops between Arm 7–8 and 6–7. PB2R739 in the P2’ site forms a salt bridge with E396/387/397 as well as hydrogen bonds with the conserved tryptophans on Arms 7 and 8: W357/348/358 and W399/390/400. Interestingly, PB2-NLD contains additional determinants binding to the minor NLS-binding site outside of the classical bipartite NLS, namely, PB2R737, which binds at a P0’ position, and PB2K718 that in all three isoforms projects from the PB2-GD to bind in trans to the P3’ position (Figure 3C). In α1 and α3, PB2R737 forms a salt bridge with D442/D433, while a hydrogen bond is made with N443 in importin α7. On the other hand, in α1 and α7 PB2K718 inserts into the pocket at the helical interface between ARM6-7 where it makes hydrogen bonds with polar residues. In contrast, for importin α3 only, PB2K718 is found outside this binding pocket, where it forms a salt bridge with nearby D271, a residue conserved in all isoforms. Apart from PB2K718, PB2-GD makes negligible contacts with the Arm core of importin α isoforms mainly due to the conformation of PB2-biNLS at the minor NLS-site, which forms a barrier between GD and the Arm core. Thus, consistent with the comparably high binding affinity measured in vitro (Boivin and Hart, 2011), recognition of PB2-NLD by importin α isoforms reveals a conserved pattern of contacts. This suggests that the molecular determinants responsible for in vivo discrimination of importin α3 and α7 must reside outside the NLS-binding groove.
Importin α3 is more flexible than other isoforms and contains a hinge at the major NLS-binding site
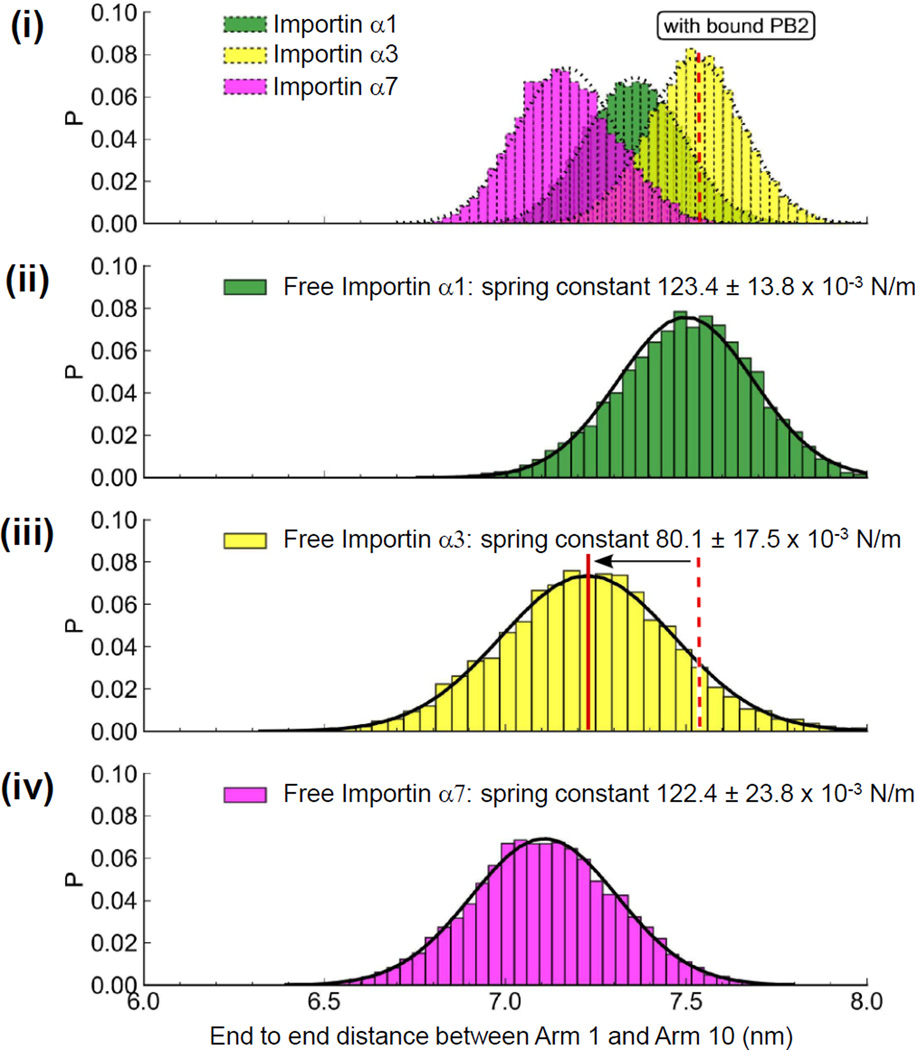
We used equilibrium molecular dynamics (MD) simulations to examine the conformational flexibility of importin α isoforms in the apo- and PB2-bound conformations. Similar to previous analyses of importin β flexibility (Kappel et al., 2010), we measured the distance between the first and last Arm repeat to monitor fluctuations in the overall length of the proteins over the course of 0.4 µs simulations. This analysis revealed that the three isoforms importin α1, α3 and α7 display only small end-to-end distance fluctuations when bound to PB2-NLD (Figure 4, (i)). All isoforms retain a similar equilibrium length and show narrow distributions around their equilibrium conformation. By contrast, the flexibility of the unliganded importin isoforms is increased, and larger differences between α3 and the other two isoforms occur (Figure 4, (ii–iv)). In particular, α3 exhibits a greater degree of flexibility, as evidenced by a broader end-to-end distance distribution, and a somewhat larger shift from its liganded equilibrium length. All fluctuations of importin α3 can be described by a Gaussian function centered at 7.25 nm (Figure 4, (iii)). In analogy to importin β (Kappel et al., 2010), importin α can be thought of as a Hookean spring and a spring constant keq (defined as force/compression (N/m)) can be calculated from the end-to-end distance distribution in Figure 4 (ii–iv) and used to compare the relative flexibility of each isoform. We found that the mean spring constant keq of free importin α1, α3 and α7 is (123.4 ± 13.8) × 10−3 N/m, (80.1 ± 17.5) × 10−3 N/m and (122.4 ± 23.8) × 10−3 N/m, respectively. This suggests that importin α3 is a 65% softer spring than α1 and α7, which allows larger endto- end equilibrium fluctuations, though it is still 8 times stiffer than yeast importin β (spring constant of (10 ± 4) × 10−3 N/m) (Kappel et al., 2010).
Figure 4. Equilibrium molecular dynamics simulations of importin α isoforms.
End-to-end distance measurements of importin α isoforms bound to PB2-NLD (i) or free importin α1 (ii), α3 (iii) and α7 (iv). Shaded bars display equilibrium fluctuations of importin α isoforms elongations obtained from a 0.4 µs equilibrium simulation. On the Y-axis, ‘P’ is the normalized probability distribution. See also Figure S3.
We then examined the interfaces between individual Arm repeats to determine the regions of greatest motion in each isoform. This analysis revealed a hinge region between Arm 3 and 4 of the apo-importin α3 not present in importin α1 or α7 (Figure S3). This hinge is immobilized in an extended position by the binding of PB2-NLD. Additionally, importin α3 has large variability in curvature between Arms 6 and 7 and Arms 8 and 9, regions which were also constrained by binding to PB2-NLD. In contrast, importin α1 and α7 had little difference between their Arm repeats between apo- and PB2-bound forms. In both isoforms, the C-terminal Arm repeats contain the most significant variations, regardless of the presence of PB2-NLD. As the hinge observed in importin α3 occurs right at the binding site for the major NLS, we predict that importin α3 will bind weakly to an NLS that relies primarily on the major NLS-binding pocket.
Reduced autoinhibition of importin α3 for PB2-NLD
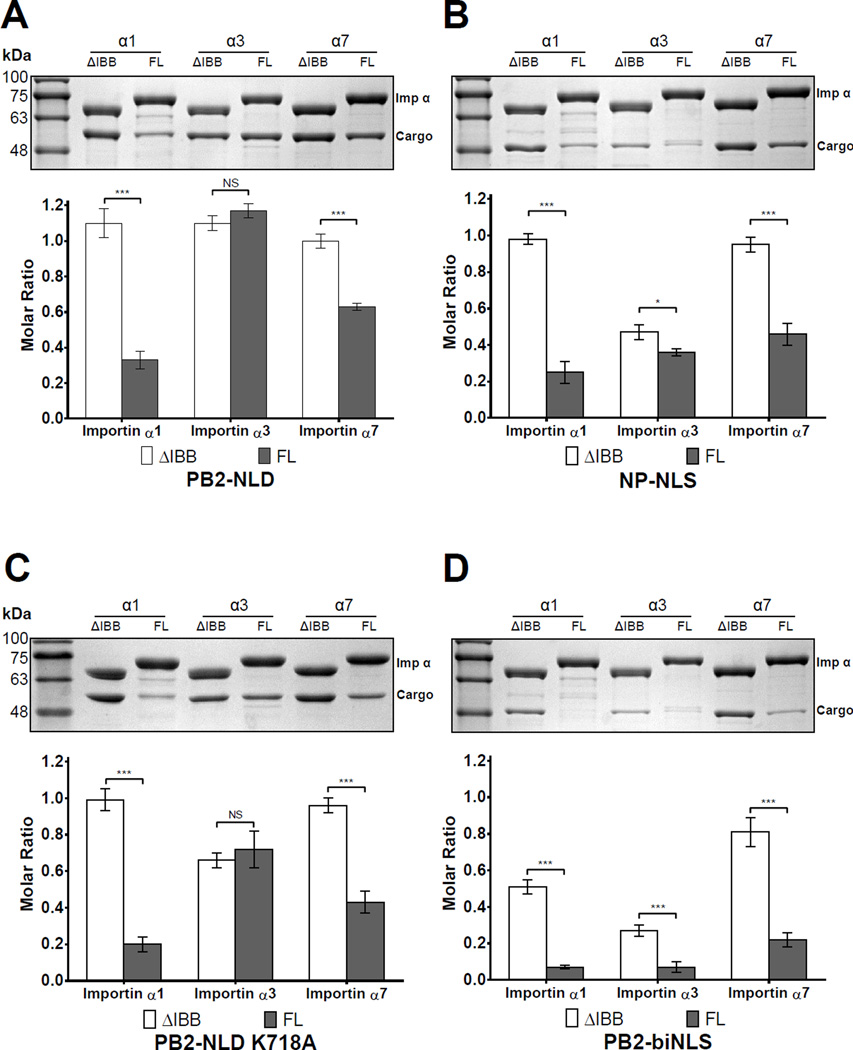
We used a semi-quantitative GST-pulldown assay to examine the interplay between PB2-NLD binding to specific importin α isoforms and autoinhibition by the IBB-domain. GST-tagged importin α isoforms either lacking the IBB-domain (ΔIBB-importin α) or full length (FL-importin α) were incubated with MBP-tagged PB2-NLD and, after extensive washing, bound PB2-NLD was analyzed by SDS-PAGE and quantified. As expected, all ΔIBB-importin α isoforms bound PB2-NLD in a 1:1 molar ratio (or 100% of importin α bound to cargo). Due to autoinhibition, reduced binding of PB2-NLD with FL-importin α1 (32%) and α7 (62%) was observed, whereas a stoichiometric quantity of PB2-NLD was recovered with importin α3, in the presence and absence of IBB (Figure 5A). To control for different levels of autoinhibition between cargos, a MBP-tagged construct of the nucleoplasmin NLS (NP-NLS) was also tested for binding to importin α isoforms (Figure 5B). As observed for PB2-NLD, ΔIBB-importin α1 and α7 bound stoichiometric quantities of NP-NLS, while drastically reduced binding to the FL-importin α1 (25%) and FL-importin α7 (46%) was observed, confirming that both isoforms are autoinhibited by their IBBs, although to a different extent. As reported for influenza A virus nucleoprotein (Melen et al., 2003), we observed significantly reduced binding of NP-NLS to ΔIBB-importin α3, with only 45% of possible binding sites occupied by this cargo (reduced to 35% with FLimportin α3). Likewise, importin α5, which is 81% identical to α7, showed autoinhibition and binding properties comparable to those of importin α7 (Figure S4).
Figure 5. Pulldown analysis of the interaction of importin α isoforms with NLS-cargos.
GST-tagged importin α isoforms lacking the IBB (ΔIBB-) or full length (FL-) were immobilized on glutathione beads and incubated with (A) PB2-NLD, (B) NP-NLS, (C) PB2-NLD-K718A and (D) PB2-biNLS (residues 738–759). Pulldowns are shown as mean ± SD for three experiments. Student's t-test was used to determine significance, where *P<0.05, **P<0.01, ***P<0.001 and NS is not significant. See also Figure S4.
To further dissect the interaction of PB2-NLD and importin α3, two mutants of PB2 were generated and tested: PB2-NLD-K718A, which lacks the PB2K718 binding in trans to the P3’ site (Figure 3C), and PB2-biNLS that contains only the bipartite-like NLS but lacks PB2-GD. Importin α1 and α7 showed similar association to the K718A mutant as with the wild type PB2- NLD (Figure 5C), characterized by equimolar binding to the ΔIBB constructs, and reduced binding to the FL-importin α1 (20%) and α7 (43%) due to autoinhibition. In contrast, this mutation reduced by one third the affinity of PB2-NLD for importin α3 (66% for ΔIBB and 72% for FL), suggesting this isoform is more sensitive than α1 and α7 to mutations that weaken binding to the minor NLS-binding pocket. Strikingly, entirely removing the PB2-GD (Figure 5D) reduced binding to all ΔIBB-constructs (51% to α1, 27% to α3 and 81% to α7) and even more pronouncedly to FL-isoforms (7% to α1 and α3, 21% to α7). This suggests that the PB2 bipartite NLS is weaker than the NP-NLS, and that PB2-GD synergizes with the biNLS to bypass autoinhibition upon binding to importin α3. However, as seen in the crystal structure (Figure 1C), PB2-GD makes negligible contacts with importin α isoforms on its own. Interestingly, a crystal structure of importin α1 bound to PB2-biNLS lacking the GD reveals a frame-shift at the minor NLS-site where the first two basic residues of the PB2 minor NLS-box are shifted by two positions (Figure S5). This suggests that PB2-GD provides a register for the minor NLS-binding boxes to associate with the Arm core. In summary, using a semi-quantitative pulldown assay, we have determined that high affinity binding of PB2-NLD to importin α1 and, in part, α7 is autoinhibited by their IBB-domains, but occurs in an IBBindependent manner for importin α3. The preferential binding of this isoform to PB2-NLD requires an N-terminal GD, which provides a topological advantage to the biNLS.
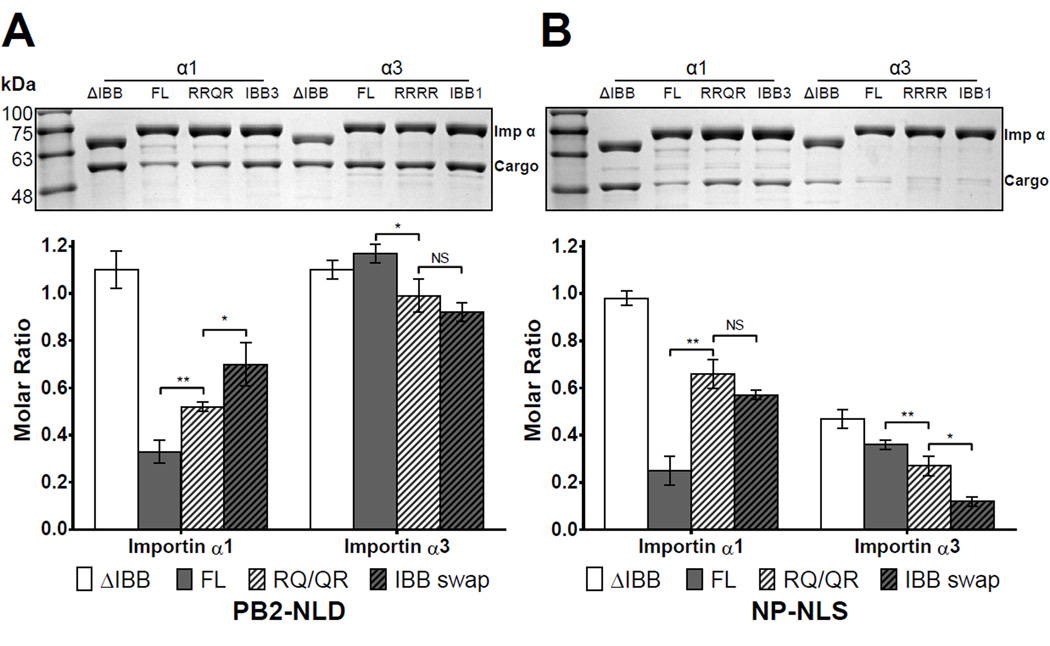
Swapping IBB-domains partially restores autoinhibition
To further examine how differential autoinhibition by IBBs affect isoform specificity for PB2, we focused exclusively on the isoforms α1 and α3, which display the tightest and loosest autoinhibition for PB2-NLD, respectively, and generated two sets of mutants in the IBB-domain. The first set of mutants had a R→Q substitution of position P3’ of α1’s minor NLS-binding cluster ‘RRRR’ (FL-importin α1 (RRQR)) that emulates the equivalent sequence found in importin α3 (Figure 1B); and the reciprocal mutation (Q→R) was also introduced in importin α3 (FL-importin α3(RRRR)). The second set of mutants had a total swap of IBBs between the two isoforms to yield the chimera IBB3-importin α1 and IBB1-importin α3. We tested these mutants for their ability to bind PB2-NLD and, as control, NP-NLS using the pulldown assay previously described. Interestingly, all importin α1 mutants showed increased binding to cargo as compared to wild type FL-importin α1. Binding of PB2-NLD to the FL-importin α1 (RRQR) increased from 32% to 52% and to 70% for IBB3-importin α1 (Figure 6A). Similarly, the NPNLS control showed a comparable trend consistent with loss of autoinhibition, going from 25% of FL-importin α1 to 66% with FL-importin α1(RRQR) and 57% with IBB3-importin α1 (Figure 6B). Thus, the IBB-domain of importin α3 is less autoinhibiting, partially because the presence of a natural Q→R in its minor-NLS-binding box. On the other hand, FL-importin α3 (RRRR) and the chimera IBB1-importin α3 saw a very modest gain of autoinhibition for PB2-NLD (Figure 6A). The control experiment with NP-NLS confirmed this trend: 35% of NP-NLS bound to FL-importin α3, decreased to 27% with FL-importin α3 (RRRR) and only 12% with IBB1- importin α3 (Figure 6B). Thus, natural variation in amino acid sequence at the minor NLSbinding box of the IBB between isoforms modulates autoinhibition.
Figure 6. Pulldown analysis of the interaction of importin α isoform IBB mutants with PB2.
GST-tagged importin α1 and α3 lacking the IBB (ΔIBB-), full length (FL-), Q/R mutants and IBB swaps were immobilized on glutathione beads and incubated with (A) PB2-NLD or (B) control NP-NLS. Pulldowns are shown as mean ± SD for three experiments. No interaction was observed between free MBP, GST-importin and glutathione beads (Figure S5). Student's t-test was used to determine significance, where *P<0.05, **P<0.01, ***P<0.001 and NS is not significant. See also Figure S6.
DISCUSSION
The evolution of an ancestral importin α gene into seven isoforms is part of the regulatory diversification characterizing higher eukaryotes that makes the transport machinery redundant and extraordinarily complex to study. In light of such a plethora of import adaptors, it is not surprising that viruses have learned how to exploit the host import machinery by developing specialized NLS-cargos that target specific isoforms of importin α. In this paper, we have studied how Influenza A Virus PB2 discriminates among importin α isoforms. Our structural and biochemical analysis of importin α isoforms bound to PB2 NLD led us to four novel findings. First, all importin α isoforms share an essentially invariant NLS-binding surface (Figure 3A) but differ greatly in conformational flexibility (Figure 4). A larger range of extension/compression was observed for importin α3 than the other isoforms, with that increased range of motion coming from a hinge between Arms 3 and 4. Second, subtle differences in amino acid sequence in the IBB-domain result in different levels of autoinhibition among importin α isoforms (Figure 5A). We found a gradient of autoinhibition, from importin α1 consistently having the strongest autoinhibition, importin α5/α7 having looser autoinhibition, to α3 with the weakest autoinhibition for classical NLSs and a total loss of autoinhibition for PB2. Third, importin α3 Arm core has an intrinsically reduced affinity for bipartite NLSs as compared to the other isoforms, likely reflecting the increased breathing of the major NLS binding site in this isoform. This reduced affinity is seen for the classical NP-NLS (Figure 5B), for its own IBB (Figure 5A), which is an intramolecular NLS, and for the strongly autoinhibiting IBB domain of importin α1 (Figure 6A). Fourth, PB2 NLS-domain has an unusually strong minor NLS binding box with an additional basic residue at position P0’ and 718K exposed in trans from its GD (Figure 3C). This finding reinforces the biological importance of the so called ‘minor’ NLS-binding site of importin α, which has recently been bolstered by the discovery of import cargos that bind exclusively (Chang et al., 2013; Kosugi et al., 2009; Lott et al., 2011; Pang and Zhou, 2014), or primarily (Giesecke and Stewart, 2010) to the minor NLS-binding site.
PB2 is a specialized viral cargo that is able to hijack specific importin α isoforms
In the attempt to define structural determinants that uniquely define cargos specific to importin α3, we made the interesting observation that a globular domain of PB2, C-terminal of the bipartite NLS, is essential for proper association with importin α3 and to a lesser extent α1 and α7 (Figure 5D). Interestingly, other cargos known to bind importin α3 also share a similar globular domain next to the NLS; for instance, RCC1 has a large β-propeller C-terminal of its NLS (Makde et al., 2010), PARP-2 contains a globular WRG-domain flanking the N-terminal NLS (Haenni et al., 2008), and NF-κB p50/p65 has an immunoglobulin-like domain immediately before its NLS (Jacobs and Harrison, 1998). RCC1 is of particular interest because, like PB2, it is specifically imported by importin α3 despite the ability to bind to the Arm-cores of all isoforms, and its β-propeller domain is a determinant for isoform specificity (Friedrich et al., 2006; Kohler et al., 1999). Thus, we propose that a globular domain flanking the NLS provides a register for high affinity association to importin α3, which may allow certain cargos to stabilize the intrinsically flexible backbone of this isoform, freezing the Armsuperhelix in a more extended conformation (Figure 4(i)).
Evolutionary advantages of having multiple isoforms
Both importin α3 and α5/α7 analyzed in this paper are significantly less autoinhibited for PB2 than the universal α1 (Figure 5A, S4), as also seen previously with importin α5 and STAT1 (Nardozzi et al., 2010a). This suggests that a fundamental distinctive property of isoforms of subfamilies α2 and α3 could be the reduced degree of intramolecular autoinhibition as compared to the universal importin α1. Based on these variations in autoinhibition and import complex assembly dynamics, we propose that importin α isoforms of subfamilies α2 and α3 provide a “fast-track” for nuclear import of important cellular cargos. Viruses may have learned to exploit this fast-track by evolving proteins that bind preferentially to less autoinhibited isoforms of importin α. In support of this hypothesis, a growing number of viral proteins are found to be imported by members of the subfamily α2 and α3 (Nardozzi et al., 2010b; Pumroy and Cingolani, 2014).
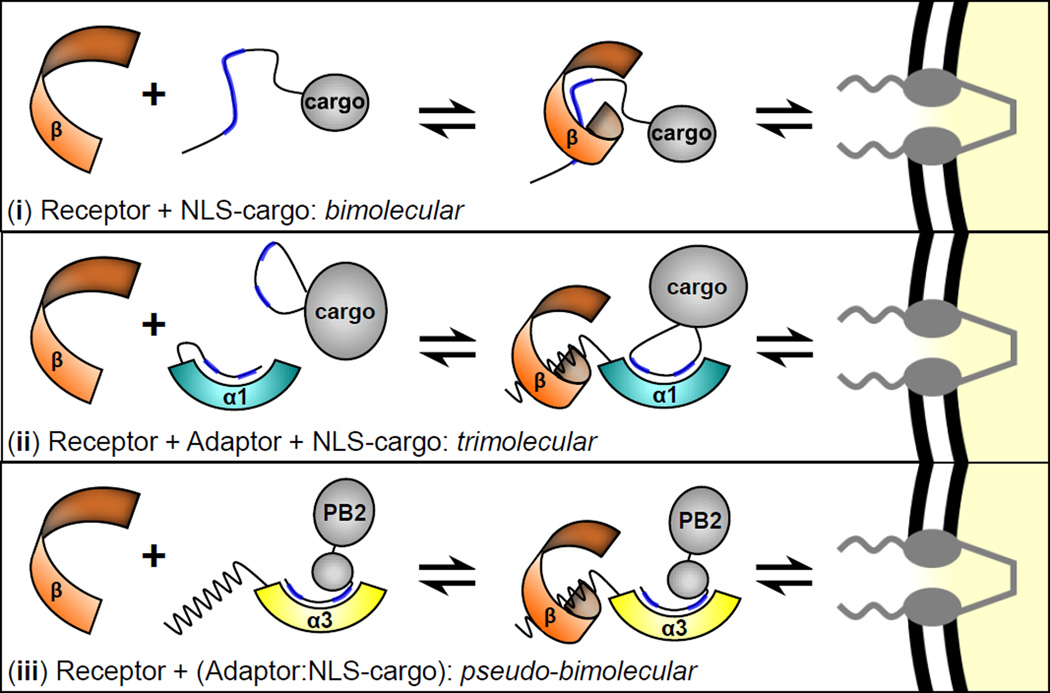
Mechanistically, how does association of an NLS-cargo to a less autoinhibited importin α isoform confer a fast-track to the cell nucleus? Riddick and Macara demonstrated that adaptor-dependent nuclear import that relies on importin α1 (a trimolecular reaction) is significantly slower than adaptor-independent import (a bimolecular reaction) (Figure 7, (i–ii)), mainly because of the kinetic cost of forming a complex of three soluble proteins (e.g. importin β, importin α1 and NLS-cargo), as opposed to two (e.g. importin β and NLS-cargo) (Riddick and Macara, 2007). Import cargos like PB2 and RCC1 (Friedrich et al., 2006) compete off the IBB-domain of importin α3 (or α7) in the absence of importin β, gaining a kinetic advantage over classical NLS-cargos that form an import complex only in the presence of importin β. The ability to pre-complex importin α effectively reduces the complexity of adaptor-mediated import to a pseudo-bimolecular reaction between a preformed cargo-importin α complex and the receptor importin β (Figure 7, (iii)). This may provide a selective advantage over the bulk of classical cargos that compete with each other (Timney et al., 2006), and the IBB for binding to importin α isoforms, and do so productively only in the presence of importin β. This proposed fast-track for nuclear import is expected to be physiologically important especially at low concentrations of importin β, when importin α/β-dependent nuclear transport occurs less efficiently (Yang and Musser, 2006). Though the estimated cellular concentration of importin β is high (~3 µM) relative to importin α (~1 µM) (Riddick and Macara, 2005), free importin β available for binding to importin α/NLS-cargos is likely limiting in a proliferating cell. A significant percentage of importin β is thought to be stuck in the NPC bound to FG-nups (Kapinos et al., 2014; Riddick and Macara, 2007) and much of the remaining soluble importin β also binds RanGTP (Wente and Rout, 2010), snRNPs (via snurportin) (Lott et al., 2010) and adapter-independent cargos like PTHrP (Cingolani et al., 2002). In conclusion, this paper defines novel structural and biochemical properties of importin α isoforms 3 and 7 that help decipher how the nuclear import machinery could function as a determinant for Influenza A virus host adaptation.
Figure 7. Model for PB2 nuclear import by importin α isoform 3 and 7.
Schematic diagrams of (i) adaptor-independent nuclear import; (ii) adaptor and receptor-dependent nuclear import; (iii) proposed nuclear import of PB2.
EXPERIMENTAL PROCEDURES
Molecular biology techniques
The genes encoding mouse α1 (Lott et al., 2011), human importin α3 and α7 (Boivin and Hart, 2011) were cloned as FL and ΔIBB in vector pET28a (Novagen) and pGEX-6P (GE Healthcare). The IBB-swap mutants (IBB3-importin α1 and IBB1-importin α3) were generated by megaprimer PCR and inserted into a pGEX-6P vector. Influenza A PB2-NLD (res. 678–759) from strain (A/Victoria/75/1995(H3N2)) was inserted into the pGEX-6P vector between BamHI and XhoI sites. MBP-tagged PB2-NLD (res. 678–759) and NP-NLS (res. 155–170) were previously described (Pumroy et al., 2012). MBP-tagged PB2-biNLS (res. 738–759) was generated by long PCR. Site directed mutagenesis was used to introduce a stop codon after position 487 of importin α3, point mutations R30Q and Q28R in importin α1 and α3, respectively and K718A mutation in PB2. All constructs generated in this study were entirely sequenced to ensure the correctness of the DNA sequence.
Expression and purification of recombinant proteins
GST-PB2-NLD and untagged or 6xHis tagged importin α isoforms were co-expressed in BL21-AI cells (Invitrogen), induced with 0.1 mM IPTG and 0.1% arabinose, and grown overnight at 20°C. Cell pellets were resuspended in Lysis buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM BME, 1 mM PMSF, and 0.2% Tween-20) and lysed by sonication. Clarified lysates were incubated with GST resin (GenScript), and beads were washed with Complex buffer (20mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA, 5 mM BME, 0.1 mM PMSF). The PB2- NLD:importin α complexes were removed from immobilized GST-resin by the overnight application of PreScission Protease. The 6xHis tag was removed by the overnight application of TEV protease. MBP-cargos were purified as in (Pumroy et al., 2012). GST-importin α isoforms were expressed in BL21-AI cells and purified as in (Nardozzi et al., 2010a). Complexes, MBP-cargos and GST-importin α isoforms were further purified by size exclusion chromatography on a Superdex 200 or 75 16/60 (GE Healthcare) in Complex buffer and concentrated by ultrafiltration.
Pulldown assay and quantification
The pulldown assay was carried out as previously described (Lott et al., 2010). 25 µL of GST-importin α at 24 µM were incubated for 15 minutes with 25 µL of glutathione resin beads equilibrated with Complex buffer. Beads were washed 3 times with 700 µL of complex buffer. 25 µL of MBP-tagged cargo were added at 2.5× molar excess (60 µM), and incubated 15 minutes. Beads were then washed 3 times with 700 µL of Complex buffer; 25 µL of 2× Laemmli buffer were added to the beads and samples boiled for 5 min at 95°C. Samples were then separated by SDS-PAGE (12.5%) electrophoresis and stained with Coomassie Brilliant Blue- G-250 and destained overnight. For quantification, see Supplemental Experimental procedures.
Crystallization and structure determination
Crystals of ΔIBB-importin α1 bound to PB2-NLD were obtained by mixing equal volumes of gel filtration-purified complex at 7.5 mg/mL with 0.49 M Na phosphate monobasic and 0.91 M K phosphate dibasic, pH 6.9 and equilibrating the droplet against 600 µL of the same precipitant. ΔIBB-Importin α7 bound to PB2-NLD was crystallized by mixing together protein complex at 9 mg/mL with an equal volume of 0.2 M magnesium acetate, 12% PEG 8000, 0.1M Na cacodylate at pH 6.5 and equilibrating the droplet against 600 µL of the same precipitant. Crystals of ΔIBB-importin α3 bound to PB2-NLD were obtained mixing equal volumes of protein complex at 20 mg/ml with 0.1 M Na cacodylate, 0.1 M NaCl and 1.1 M ammonium sulfate and equilibrating the droplet over 600 µL of 1 M NaCl. All crystals were harvested in nylon cryo-loops, cryo-protected with 27% ethylene glycol and flash-frozen in liquid nitrogen. Crystals were diffracted at beamlines X6A and X29 at the National Synchrotron Light Source (NSLS) on a Quantum Q270 and a Quantum-315r CCD detector, respectively. Data indexing, integration and scaling were carried out with the HKL2000 (Otwinowski and Minor, 1997). Initial phases were obtained by molecular replacement using Phaser (McCoy, 2007) and PDB entry 3Q5U as a search model. Atomic models were built using Coot (Emsley and Cowtan, 2004) and refined using phenix.refine (Adams et al., 2002). Data collection and refinement statistics are summarized in Table 1.
Molecular Dynamics simulations
All simulations were based on the structures of α1, α3 and α7 bound to PB2-NLD determined in this work and unliganded importin α structures obtained by stripping the atomic coordinates of PB2-NLD. Equilibrium MD simulations were performed for 0.4 µs using GROMACS 3.3 and 4.0 (Hess et al., 2008; Van Der Spoel et al., 2005) as previously described (Kappel et al., 2010).
Supplementary Material
Research Highlights.
All importin α isoforms share an invariant NLS-binding surface
Importin α3 is more flexible than other isoforms and binds weakly to classical NLSs.
Importin α3 and α7 are weakly autoinhibited by their IBBs compared to importin α1
The 3D structure of PB2-NLD allows it to bypass the IBB when binding to importin α3.
ACKNOWLEDGEMENTS
We are thankful to the staff at NSLS beamlines X6A and X29 for beamtime and assistance in data collection. R.P. is supported by NIH Training NIH T32 GM100836. This work was supported in part by NIH grants GM074846 and GM100888 to G.C. Research in this publication includes work carried out at the Kimmel Cancer Center X-ray crystallography and Molecular interaction Facility, which is supported in part by NCI Cancer Center Support Grant P30 CA56036. The coordinates and structure factors for importin α1, α3 and α7 bound to PB2- NLD have been deposited in the protein Data Bank (accession codes 4UAF, 4UAE, 4UAD).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Bednenko J, Cingolani G, Gerace L. Nucleocytoplasmic transport: navigating the channel. Traffic. 2003;4:127–135. doi: 10.1034/j.1600-0854.2003.00109.x. [DOI] [PubMed] [Google Scholar]
- Boivin S, Hart DJ. Interaction of the influenza A virus polymerase PB2 C-terminal region with importin alpha isoforms provides insights into host adaptation and polymerase assembly. J. Biol. Chem. 2011;286:10439–10448. doi: 10.1074/jbc.M110.182964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catimel B, Teh T, Fontes MR, Jennings IG, Jans DA, Howlett GJ, Nice EC, Kobe B. Biophysical characterization of interactions involving importin-alpha during nuclear import. J. Biol. Chem. 2001;276:34189–34198. doi: 10.1074/jbc.M103531200. [DOI] [PubMed] [Google Scholar]
- Chang CW, Counago RM, Williams SJ, Boden M, Kobe B. Distinctive conformation of minor site-specific nuclear localization signals bound to importin-alpha. Traffic. 2013;14:1144–1154. doi: 10.1111/tra.12098. [DOI] [PubMed] [Google Scholar]
- Chen MH, Ben-Efraim I, Mitrousis G, Walker-Kopp N, Sims PJ, Cingolani G. Phospholipid scramblase 1 contains a nonclassical nuclear localization signal with unique binding site in importin alpha. J. Biol. Chem. 2005;280:10599–10606. doi: 10.1074/jbc.M413194200. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol. Cell. 2002;10:1345–1353. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Lashuel HA, Gerace L, Muller CW. Nuclear import factors importin alpha and importin beta undergo mutually induced conformational changes upon association. FEBS Lett. 2000;484:291–298. doi: 10.1016/s0014-5793(00)02154-2. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Conti E, Kuriyan J. Crystallographic analysis of the specific yet versatile recognition of distinct nuclear localization signals by karyopherin alpha. Structure. 2000;8:329–338. doi: 10.1016/s0969-2126(00)00107-6. [DOI] [PubMed] [Google Scholar]
- Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Conti E, Uy M, Leighton L, Blobel G, Kuriyan J. Crystallographic analysis of the recognition of a nuclear localization signal by the nuclear import factor karyopherin alpha. Cell. 1998;94:193–204. doi: 10.1016/s0092-8674(00)81419-1. [DOI] [PubMed] [Google Scholar]
- Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu. Rev. Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Jans D, Brinkworth RI, Kobe B. Structural basis for the specificity of bipartite nuclear localization sequence binding by importin-alpha. J. Biol. Chem. 2003;278:27981–27987. doi: 10.1074/jbc.M303275200. [DOI] [PubMed] [Google Scholar]
- Fontes MR, Teh T, Kobe B. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-alpha. J. Mol. Biol. 2000;297:1183–1194. doi: 10.1006/jmbi.2000.3642. [DOI] [PubMed] [Google Scholar]
- Forwood JK, Lange A, Zachariae U, Marfori M, Preast C, Grubmuller H, Stewart M, Corbett AH, Kobe B. Quantitative structural analysis of importin-beta flexibility: paradigm for solenoid protein structures. Structure. 2010;18:1171–1183. doi: 10.1016/j.str.2010.06.015. [DOI] [PubMed] [Google Scholar]
- Friedrich B, Quensel C, Sommer T, Hartmann E, Kohler M. Nuclear localization signal and protein context both mediate importin alpha specificity of nuclear import substrates. Mol. Cell. Biol. 2006;26:8697–8709. doi: 10.1128/MCB.00708-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Klingel K, Otte A, Thiele S, Hudjetz B, Arman-Kalcek G, Sauter M, Shmidt T, Rother F, Baumgarte S, et al. Differential use of importin-alpha isoforms governs cell tropism and host adaptation of influenza virus. Nat. Commun. 2011;2:156. doi: 10.1038/ncomms1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giesecke A, Stewart M. Novel binding of the mitotic regulator TPX2 (target protein for Xenopus kinesin-like protein 2) to importin-alpha. J. Biol. Chem. 2010;285:17628–17635. doi: 10.1074/jbc.M110.102343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DS, Corbett AH, Mason DA, Harreman MT, Adam SA. Importin alpha: a multipurpose nuclear-transport receptor. Trends Cell Biol. 2004;14:505–514. doi: 10.1016/j.tcb.2004.07.016. [DOI] [PubMed] [Google Scholar]
- Haenni SS, Altmeyer M, Hassa PO, Valovka T, Fey M, Hottiger MO. Importin alpha binding and nuclear localization of PARP-2 is dependent on lysine 36, which is located within a predicted classical NLS. BMC Cell Biol. 2008;9:39. doi: 10.1186/1471-2121-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harreman MT, Hodel MR, Fanara P, Hodel AE, Corbett AH. The auto-inhibitory function of importin alpha is essential in vivo. J. Biol. Chem. 2003;278:5854–5863. doi: 10.1074/jbc.M210951200. [DOI] [PubMed] [Google Scholar]
- Hess B, Kutzner C, Van Der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hodel MR, Corbett AH, Hodel AE. Dissection of a nuclear localization signal. J. Biol. Chem. 2001;276:1317–1325. doi: 10.1074/jbc.M008522200. [DOI] [PubMed] [Google Scholar]
- Huber AH, Nelson WJ, Weis WI. Three-dimensional structure of the armadillo repeat region of beta-catenin. Cell. 1997;90:871–882. doi: 10.1016/s0092-8674(00)80352-9. [DOI] [PubMed] [Google Scholar]
- Hudjetz B, Gabriel G. Human-like PB2 627K influenza virus polymerase activity is regulated by importin-alpha1 and -alpha7. PLoS Pathog. 2012;8:e1002488. doi: 10.1371/journal.ppat.1002488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MD, Harrison SC. Structure of an IkappaBalpha/NF-kappaB complex. Cell. 1998;95:749–758. doi: 10.1016/s0092-8674(00)81698-0. [DOI] [PubMed] [Google Scholar]
- Jans DA, Xiao CY, Lam MH. Nuclear targeting signal recognition: a key control point in nuclear transport? Bioessays. 2000;22:532–544. doi: 10.1002/(SICI)1521-1878(200006)22:6<532::AID-BIES6>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Kapinos LE, Schoch RL, Wagner RS, Schleicher KD, Lim RY. Karyopherin-centric control of nuclear pores based on molecular occupancy and kinetic analysis of multivalent binding with FG nucleoporins. Biophys. J. 2014;106:1751–1762. doi: 10.1016/j.bpj.2014.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappel C, Zachariae U, Dolker N, Grubmuller H. An unusual hydrophobic core confers extreme flexibility to HEAT repeat proteins. Biophys. J. 2010;99:1596–1603. doi: 10.1016/j.bpj.2010.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B. Autoinhibition by an internal nuclear localization signal revealed by the crystal structure of mammalian importin alpha. Nat. Struct. Biol. 1999;6:388–397. doi: 10.1038/7625. [DOI] [PubMed] [Google Scholar]
- Kohler M, Speck C, Christiansen M, Bischoff FR, Prehn S, Haller H, Gorlich D, Hartmann E. Evidence for distinct substrate specificities of importin alpha family members in nuclear protein import. Mol. Cell. Biol. 1999;19:7782–7791. doi: 10.1128/mcb.19.11.7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Matsumura N, Takashima H, Miyamoto-Sato E, Tomita M, Yanagawa H. Six classes of nuclear localization signals specific to different binding grooves of importin alpha. J. Biol. Chem. 2009;284:478–485. doi: 10.1074/jbc.M807017200. [DOI] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Mitrousis G, Pante N, Cingolani G. The importin beta binding domain modulates the avidity of importin beta for the nuclear pore complex. J. Biol. Chem. 2010;285:13769–13780. doi: 10.1074/jbc.M109.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Bhardwaj A, Sims PJ, Cingolani G. A minimal nuclear localization signal (NLS) in human phospholipid scramblase 4 that binds only the minor NLS-binding site of importin alpha1. J. Biol. Chem. 2011;286:28160–28169. doi: 10.1074/jbc.M111.228007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott K, Cingolani G. The importin beta binding domain as a master regulator of nucleocytoplasmic transport. Biochim. Biophys. Acta. 2011;1813:1578–1592. doi: 10.1016/j.bbamcr.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–566. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfori M, Lonhienne TG, Forwood JK, Kobe B. Structural basis of high-affinity nuclear localization signal interactions with importin-alpha. Traffic. 2012;13:532–548. doi: 10.1111/j.1600-0854.2012.01329.x. [DOI] [PubMed] [Google Scholar]
- Mason DA, Stage DE, Goldfarb DS. Evolution of the metazoan-specific importin alpha gene family. J. Mol. Evol. 2009;68:351–365. doi: 10.1007/s00239-009-9215-8. [DOI] [PubMed] [Google Scholar]
- Matsuura Y, Stewart M. Structural basis for the assembly of a nuclear export complex. Nature. 2004;432:872–877. doi: 10.1038/nature03144. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J. Biol. Chem. 2003;278:28193–28200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- Mosammaparast N, Pemberton LF. Karyopherins: from nuclear-transport mediators to nuclear-function regulators. Trends Cell Biol. 2004;14:547–556. doi: 10.1016/j.tcb.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Nardozzi J, Wenta N, Yasuhara N, Vinkemeier U, Cingolani G. Molecular basis for the recognition of phosphorylated STAT1 by importin alpha5. J. Mol. Biol. 2010a;402:83–100. doi: 10.1016/j.jmb.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardozzi JD, Lott K, Cingolani G. Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 2010b;8:32. doi: 10.1186/1478-811X-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Methods in Enzymology 276: Macromolecular Crystallography. 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pang X, Zhou HX. Design rules for selective binding of nuclear localization signals to minor site of importin alpha. PLoS One. 2014;9:e91025. doi: 10.1371/journal.pone.0091025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Cingolani G. Diversification of importin α isoforms in cellular trafficking and disease state. Biochem. J. 2014 doi: 10.1042/BJ20141186. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pumroy RA, Nardozzi JD, Hart DJ, Root MJ, Cingolani G. Nucleoporin Nup50 stabilizes closed conformation of armadillo repeat 10 in importin alpha5. J. Biol. Chem. 2012;287:2022–2031. doi: 10.1074/jbc.M111.315838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Gabriel G. The nuclear import machinery is a determinant of influenza virus host adaptation. Bioessays. 2013;35:23–27. doi: 10.1002/bies.201200138. [DOI] [PubMed] [Google Scholar]
- Resa-Infante P, Jorba N, Coloma R, Ortin J. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol. 2011;8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resa-Infante P, Thieme R, Ernst T, Arck PC, Ittrich H, Reimer R, Gabriel G. Importin-alpha7 is Required for Enhanced Influenza A Virus Replication in the Alveolar Epithelium and Severe Lung Damage in Mice. J. Virol. 2014 doi: 10.1128/JVI.00270-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG. A systems analysis of importin-{alpha}-{beta} mediated nuclear protein import. J. Cell Biol. 2005;168:1027–1038. doi: 10.1083/jcb.200409024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddick G, Macara IG. The adapter importin-alpha provides flexible control of nuclear import at the expense of efficiency. Mol. Sys. Biol. 2007;3:118. doi: 10.1038/msb4100160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Hutcheon ML, Duncan TM, Cingolani G. Improved crystallization of Escherichia coli ATP synthase catalytic complex (F1) by introducing a phosphomimetic mutation in subunit epsilon. Acta Crystallogr. F Struct. Biol. Cryst. Commun. 2012;68:1229–1233. doi: 10.1107/S1744309112036718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- Tarendeau F, Boudet J, Guilligay D, Mas PJ, Bougault CM, Boulo S, Baudin F, Ruigrok RW, Daigle N, Ellenberg J, et al. Structure and nuclear import function of the C-terminal domain of influenza virus polymerase PB2 subunit. Nat. Struct. Mol. Biol. 2007;14:229–233. doi: 10.1038/nsmb1212. [DOI] [PubMed] [Google Scholar]
- Tarendeau F, Crepin T, Guilligay D, Ruigrok RW, Cusack S, Hart DJ. Host determinant residue lysine 627 lies on the surface of a discrete, folded domain of influenza virus polymerase PB2 subunit. PLoS Pathog. 2008;4:e1000136. doi: 10.1371/journal.ppat.1000136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timney BL, Tetenbaum-Novatt J, Agate DS, Williams R, Zhang W, Chait BT, Rout MP. Simple kinetic relationships and nonspecific competition govern nuclear import rates in vivo. J. Cell Biol. 2006;175:579–593. doi: 10.1083/jcb.200608141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJ. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect. Biol. 2010;2:a000562. doi: 10.1101/cshperspect.a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Musser SM. Nuclear import time and transport efficiency depend on importin beta concentration. J. Cell Biol. 2006;174:951–961. doi: 10.1083/jcb.200605053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachariae U, Grubmuller H. Importin-beta: structural and dynamic determinants of a molecular spring. Structure. 2008;16:906–915. doi: 10.1016/j.str.2008.03.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.