SUMMARY
TOR Complex 1 (TORC1) is a potent anabolic regulator of cellular growth and metabolism. When cells have sufficient amino acids, TORC1 is active due to its lysosomal localization mediated via the Rag GTPases. Upon amino acid removal, the Rag GTPases release TORC1, causing it to become cytoplasmic and inactive. We show here that upon amino acid removal, the Rag GTPases also recruit TSC2 to the lysosome, where it can act on Rheb. Only when both the Rag GTPases and Rheb are inactive is TORC1 fully released from the lysosome. Upon amino acid withdrawal, cells lacking TSC2 fail to completely release TORC1 from the lysosome, fail to completely inactivate TORC1, and fail to adjust physiologically to amino acid starvation. These data suggest that regulation of TSC2 subcellular localization may be a general mechanism to control its activity, and places TSC2 in the amino acid sensing pathway to TORC1.
Keywords: mTORC1, TSC2, Drosophila, amino acids, Rag GTPases
INTRODUCTION
Small GTPases act as molecular switches that alternate between GTP-bound and GDP-bound states, thereby regulating a vast array of cellular parameters including mitochondrial activity, cell growth, cell metabolism and cell morphology. Small GTPases are activated or inactivated by guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs) respectively, which regulate the GDP/GTP load of the GTPase. Hence, understanding how GEFs and GAPs are regulated is an important aspect of understanding GTPase function. Compared to the regulation of GEFs, relatively little is known about how the activity of GAPs is regulated (Cherfils and Zeghouf, 2013). GAPs acting on members of the Rho and Arf GTPase superfamilies are activated via membrane recruitment, causing rearrangements in the structure of the GAPs upon membrane binding (Cherfils and Zeghouf, 2013). Regulation of GAPs acting on members of the Ras superfamily of GTPases is less well understood. One such GAP is comprised of the TSC1/TSC2/TBC1D7 trimeric tumor suppressor complex (Dibble et al., 2012), which acts on the small GTPase Rheb. Although it is known that activity of this complex is regulated by phosphorylation on multiple sites, it is not yet clear how these phosphorylations affect TSC1/2 activity at the molecular level.
The kinase TOR complex 1 (TORC1) is a potent anabolic regulator of cellular growth and metabolism that is often hyperactivated in human cancers (Guertin and Sabatini, 2007; Heitman et al., 1991; Huang and Manning, 2008; Laplante and Sabatini, 2012; Proud, 2011). To be active, TORC1 needs to bind a molecule of Rheb in the active, GTP-bound state (Inoki et al., 2003; Tee et al., 2003). By inactivating Rheb, the TSC1/2 complex is therefore a critical upstream inhibitor of TORC1. The TSC1/2 complex acts as a central point of integration of almost all known inputs regulating TORC1, including cellular stresses such as low oxygen or low ATP, and various growth-promoting signals, such as PI3K, Ras, TNF and Wnt signaling (reviewed in (Huang and Manning, 2008)). The importance of the TSC complex on TORC1 signaling and growth is highlighed by the fact that TSC2-inactivating mutations have been found in various human growth-related diseases (Huang and Manning, 2008). One other important input regulating TORC1 activity is the availability of amino acids (Blommaart et al., 1995; Efeyan et al., 2012; Hara et al., 1998; Jewell et al., 2013). Whether TSC2 is also involved in regulating TORC1 in response to amino acids, however, is unclear since various studies have come to differing conclusions (Gao et al., 2002; Roccio et al., 2006; Smith et al., 2005).
Unlike all the inputs that regulate TORC1 via TSC1/2 and Rheb, amino acids regulate TORC1 via a separate set of small GTPases, the Rag GTPases (Kim et al., 2008; Sancak et al., 2008). The Rag GTPases form hetero-dimeric complexes consisting of RagA or RagB bound to RagC or RagD. These complexes are stably anchored to lysosomal membranes via the LAMTOR/Ragulator complex (Sancak et al., 2010). In the presence of amino acids, the Rag dimers are in an ‘active’ conformation with RagA or RagB bound to GTP and RagC or RagD bound to GDP. The active Rag dimers recruit TORC1 to the lysosomal surface, where it binds Rheb to form an active holoenzyme. In the absence of amino acids, the Rag GAP complex termed GATOR1 causes the Rag dimers to switch into an inactive conformation containing GDP-bound RagA/B, thereby releasing TORC1 from the lysosomal surface (Bar-Peled et al., 2013; Panchaud et al., 2013). This causes TORC1 to become inactive, presumably because it no longer binds active Rheb on the lysosomal surface. Hence, TORC1 activation can currently be viewed as consisting of two aspects – the activation of Rheb in response to a plethora of regulatory inputs, and the localization of TORC1 to lysosomal membranes, in response to amino acids, which allows it to meet Rheb.
In this study, we uncover subcellular localization as a mechanism regulating activity of the TSC1/2 GAP complex. We find that upon amino acid removal, TSC1/2 is recruited to lysosomes via binding to the Rag proteins, thereby bringing TSC1/2 in close proximity of its target, Rheb. This suggests that relocalization of GAPs to the vicinity of their substrates is one mechanism for their regulation. Unexpectedly, we find that regulation of Rheb by TSC1/2 upon amino acid starvation is required for TORC1 to be released from lysosomal membranes. This suggests a ‘dual anchoring’ mechanism of TORC1 at the lysosome, perhaps with the Rag proteins playing a crucial role in recruiting TORC1 to the lysosomal membrane, and Rheb helping to retain it there. We find that cells lacking TSC2 are impaired in their response to amino acid starvation, failing to efficiently turn off TORC1. As a result, cells lacking TSC2 are very sensitive to amino acid starvation and die under conditions that control cells can cope with. In sum, our data indicate that the TSC1/2 complex is responsive to amino acids starvation and participates in amino acid signaling to TORC1. Hence, the TSC1/2 complex appears to play a role in regulating TORC1 in response to all regulatory inputs known to date.
RESULTS
Rag GTPases bind TSC2
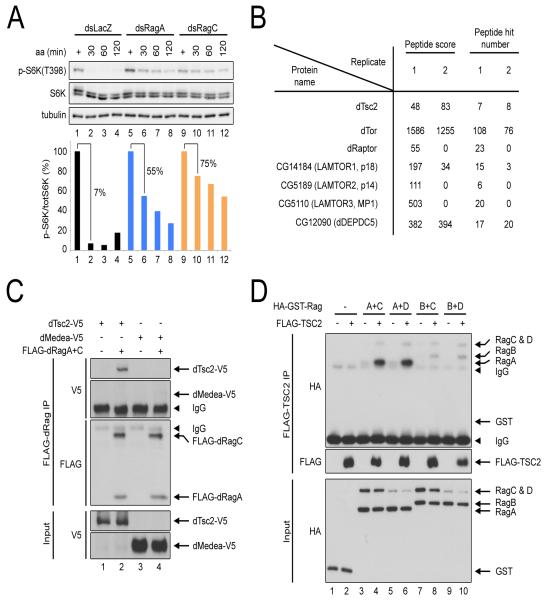
While studying regulation of TORC1 by the Rag GTPases, we noticed that Drosophila cells with reduced Rag protein levels are unable to completely shut off TORC1 in the absence of amino acids. When treated with medium lacking amino acids, control S2 cells shut off TORC1 activity, assayed via S6 Kinase (S6K) phosphorylation, to roughly 7% the level of fully-fed cells (Figure 1A). In contrast, S2 cells with RagA or RagC knock-down are significantly impaired in their response to amino acid removal, retaining circa 50% the TORC1 activity levels of fully fed cells (Figure 1A). In this and all subsequent experiments, amino acid removal is performed in the presence of dialyzed serum, thereby specifically removing amino acids but not growth factors from cell culture media. Furthermore, immunoblot quantifications are performed on a LI-COR imaging system, providing a means to quantitatively study TORC1 activity (see Experimental Procedures). This phenotype was reiterated with independent dsRNAs targeting non-overlapping regions of RagA and RagC (Figure S1A), proving specificity of the phenotype. Furthermore, knock-down of LAMTOR3 also led to similar, impaired TORC1 inactivation upon amino acid withdrawal (not shown). Similar effects can also be observed in human HEK293FT cells (Figure S1C) and in previous reports (see Figure 3H in (Sancak et al., 2008)). As previously shown (Kim et al., 2008; Sancak et al., 2008), S2 cells and HEK293FT cells with Rag protein knock-down also show severely compromised reactivation of TORC1 upon amino acid re-addition (Figures S1B and S1D). Together, these data indicate that the Rag GTPases not only activate TORC1 in the presence of amino acids, but also actively repress TORC1 in the absence of amino acids. Since the Rag proteins ‘let go’ of TORC1 in the absence of amino acids (Sancak et al., 2008), it is not easy to explain how they could also be actively repressing TORC1. Thus, the Rag GTPases appear to have an additional activity besides their ability to reversibly bind TORC1.
Figure 1. Conserved binding of TSC2 to the Rag GTPase complex.
(A) Drosophila Rag proteins are required for complete TORC1 inactivation upon amino acid withdrawal. Immunoblots of S2 cells treated with dsRNAs against LacZ (control), RagA or RagC, incubated with media lacking only amino acids in the presence of dialyzed serum for indicated times.
(B) Shot-gun mass spectroscopy analysis identifies Tsc2 as a Rag binding protein. Two statistics of peptide abundance are shown for Tsc2 and control proteins from two independent FLAG-RagA + FLAG-RagC immunoprecipitates (IP) from S2 cells.
(C) Confirmation by coIP that Rag proteins bind Tsc2 but not an unrelated protein, Medea. Immunoblots of anti-FLAG IPs from Drosophila S2 cells expressing FLAG-tagged RagA + RagC and V5-tagged Tsc2 or Medea, as indicated.
(D) Human TSC2 binds human Rag dimeric complexes. Immunoblots of anti-FLAG IPs from HEK293FT cells expressing FLAG-tagged TSC2 and HA-GST-tagged Rag proteins or an HA-GST control, as indicated.
We hypothesized that the Rag proteins might be recruiting inhibitory factors to the lysosome upon amino acid starvation. To investigate this, we immunoprecipitated FLAG-RagA and FLAG-RagC from Drosophila S2 cells and performed shot-gun mass spectrometry analysis to identify interacting partners. Amongst the identified proteins, as expected, were known components of the Ragulator complex (Sancak et al., 2010) such as p14, p18 and MP1 as well as TOR and Raptor (Figure 1B). This analysis also found significant amounts of Tsc2 as a Rag binding protein (Figure 1B). One possibility could be that binding of Tsc2 to the Rag GTPases is indirect, due to the Rag GTPases binding TORC1, which binds Rheb, which binds Tsc2. However, as shown below, binding between Tsc2 and the Rag GTPases increases when the Rag GTPases are in the inactive state, and hence bind less TORC1, arguing against this possible explanation. Since Tsc2 is a negative component of the TOR signaling pathway, we decided to study this interaction in more detail.
We first aimed to confirm the interaction between Tsc2 and the Rag GTPases by co-immunoprecipitation (coIP). Indeed, FLAG-tagged Drosophila RagA and RagC were able to coIP epitope-tagged Tsc2 but not an unrelated protein, Medea (Figure 1C). Likewise, epitope-tagged human TSC2 was able to coIP human Rag GTPases in HEK293FT cells (Figures 1D). As discussed below, a complex consisting of human RagA and RagC can also coIP endogenous TSC2 (Figure 2E). In sum, the interaction between the Rag GTPases and TSC2 appears to be specific and evolutionarily conserved from flies to humans.
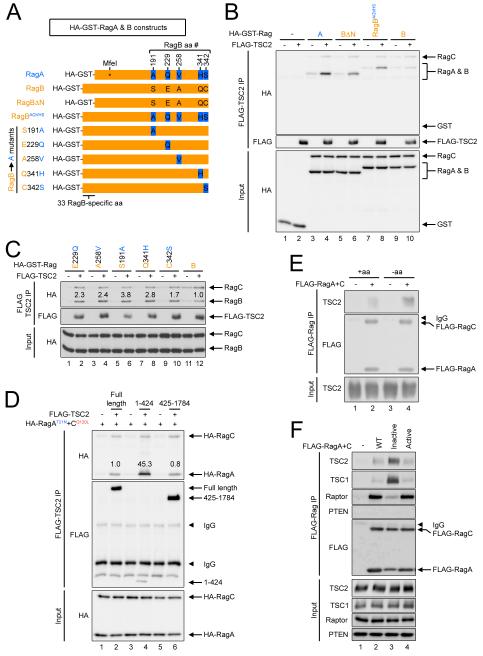
Figure 2. TSC2 binding to the Rag proteins depends on cellular amino acid signaling.
(A) Schematic diagram of RagA, RagB, and various mutants used in panels B-D to delineate the binding interfaces between TSC2 and RagA.
(B) Both N-terminal and C-terminal regions of RagA are involved in TSC2 binding. CoIP of FLAG-TSC2 with RagA, RagB, and hybrid versions of the two proteins as shown in panel A, showing that both the N-terminal 33 amino acids as well as the 5 C-terminal amino acids specific for RagA impact TSC2 binding.
(C) All five C-terminal amino acids specific for RagA, as shown in panel A, contribute towards TSC2 binding, as each one improves the binding of RagB to TSC2 when introduced singly. CoIP experiments were performed as in (B).
(D) The N-terminal 424 amino acids of TSC2 bind strongly to Rag proteins, although the remaining amino acids 425-1784 also contribute towards Rag binding. CoIP in HEK293FT cells of truncated versions of FLAG-TSC2 with HA-RagA+C. ‘Inactive’-locked RagAlow-nucleotide and RagCGTP were used as this complex binds TSC2 most strongly, as in panel F.
(E) Amino acid starvation increases binding of endogenous TSC2 to the Rag GTPases. Immunoblots of anti-FLAG IPs from HEK293FT cells expressing FLAG-tagged RagA + RagC, treated +/− amino acids in the presence of dialyzed FBS for 1h.
(F) Endogenous TSC2 and TSC1 bind most strongly to Rag dimers in the ‘inactive’ state, which antiparallels Raptor binding. Immunoblots of anti-FLAG IPs from HEK293FT cells expressing FLAG-tagged WT RagA + RagC and mutants locked into the active RagAGTP/RagClow-nucleotide or inactive RagAlow-nucleotide/RagCGTP states.
Delineation of interacting regions of TSC2 and the Rag proteins
We next aimed to characterize in more detail the binding between TSC1/2 and the Rag complex. We first asked which component of the Rag complex is binding TSC1/2. Immunoprecipitation of tagged TSC2 showed that it binds RagA significantly more strongly than the other Rag proteins (Figure 1D), suggesting RagA is a likely binding partner for the TSC1/2 complex. To study the residues in RagA involved in TSC2 binding, we exploited the fact that RagA has significantly stronger binding to TSC2 compared to RagB despite the two proteins being almost identical. Compared to RagB, RagA is lacking an N-terminal extension of 33 amino acids, and has 5 amino acid substitutions at the C-terminus of the protein (Figure 2A). We asked which of these differences are important for RagA binding to TSC2. Both removal of the N-terminal extension of RagB, as well as introduction of the five C-terminal amino acid changes into RagB caused improved binding to TSC2 (Figures 2A-B) indicating that both the N-terminal and the C-terminal residues affect TSC2 binding. We then introduced the five RagA-specific amino acid substitutions individually into RagB and found that each of them improves the binding of RagB to TSC2 (Figures 2A and 2C). Hence, each of these five amino acids of RagA contributes towards TSC2 binding, and indeed these 5 amino acid differences between RagA and RagB are evolutionarily conserved.
To identify the binding partner in the TSC1/2 complex, we coIPed epitope tagged TSC1 or TSC2 individually with RagA+C, revealing that TSC2 but not TSC1 binds the Rag complex (Figure S2A). To study which residues in TSC2 are responsible for Rag binding, we generated a series of C-terminal truncations of TSC2 and tested them by coIP (Figure S2B). This analysis revealed that the N-terminal 424 amino acids of TSC2 bind very strongly the Rag proteins, but that nonetheless the remaining amino acids 425-1784 also have Rag binding capacity (Figure 2D). In sum, binding between the two complexes appears to be mediated via TSC2 binding to RagA. Although the interaction surfaces are complex, binding depends strongly on five individual amino acids in RagA identified in this analysis.
Binding between Rag proteins and TSC2 increases upon amino acid removal
We asked whether binding between the Rag proteins and TSC2 depends on cellular amino acid signaling, since our initial findings suggested that the Rag complex might be recruiting a TOR inhibitor upon amino acid removal. Indeed, a complex consisting of human RagA and RagC (“RagA+C”) was able to coIP endogenous TSC2 (lanes 1 and 2, Figure 2E) and this interaction became stronger upon amino acid removal (lanes 2 versus 4, Figure 2E). One mechanistic explanation could be that binding between TSC2 and the Rag GTPases depends on the state of activation of the Rag proteins, which switch to the ‘inactive’ RagAlow-nucleotide/RagCGTP conformation upon amino acid removal. To test this, we assayed binding between endogenous TSC2 and mutant variants of the RagA/RagC complex, locked into either the ‘active’ (RagAGTP/RagClow-nucleotide) or ‘inactive’ (RagAlow-nucleotide/RagCGTP) states (Sancak et al., 2008), and found that indeed TSC2 binds the Rag proteins much more strongly when they are in the ‘inactive’ state (Figure 2F). Binding of endogenous TSC1 to the Rag proteins closely paralleled binding of TSC2 (Figure 2F), indicating that TSC2 is mediating binding of the entire TSC1/2 complex since TSC1 cannot bind the Rag proteins by itself (Figure S2A). As expected (Sancak et al., 2008), Raptor binding was strongest when the Rag proteins were in the active state (Figure 2F), therefore anti-correlating with TSC2 binding. As a negative control endogenous PTEN did not show any binding to the Rag proteins (Figure 2F). This preference for binding the inactive Rag complex cannot be ascribed to the state of activation of any one of the two Rags, but rather appears to depend on the conformation of both RagA and RagC, since states of intermediate activation (RagAGTP/RagCGTP or RagAlow-nucleotide/RagClow-nucleotide) also showed intermediate levels of TSC2 binding (Figure S2C). In sum, binding of TSC2 to the Rag complex appears to anti-correlate with binding of mTORC1 to the Rag complex, with mTORC1 binding predominantly when the Rag complex is active, in the presence of amino acids, and TSC2 binding predominantly when the Rag complex is inactive in the absence of amino acids.
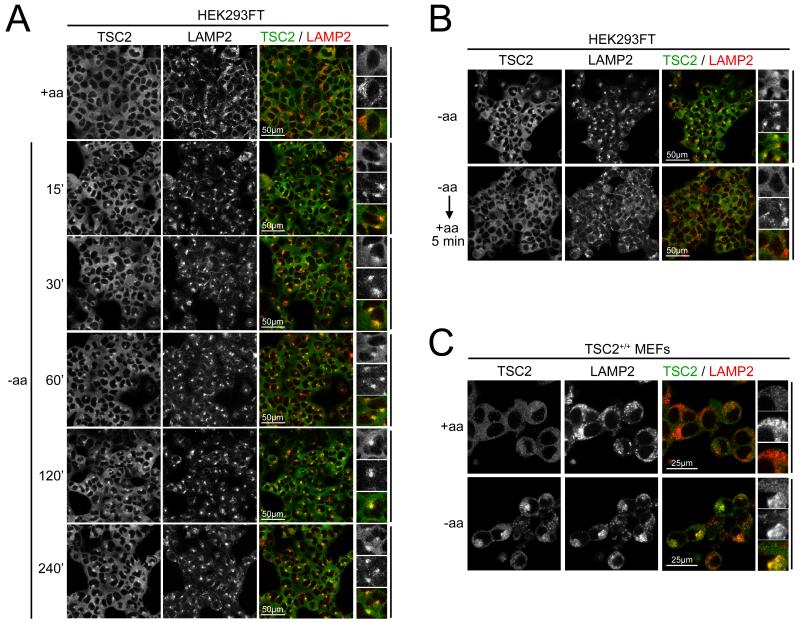
TSC2 is recruited to the lysosome in a Rag-dependent manner upon amino acid removal
Preferential binding of TSC2 for the ‘inactive’ Rag complex, which is localized on the lysosome, raised the possibility that TSC2 might be recruited to lysosomes upon amino acid removal. Using an antibody that specifically recognizes endogenous TSC2 ((Dibble et al., 2012) and Figure S3A), we found that in the presence of amino acids TSC2 is diffusely cytoplasmic in HEK293FT cells, in a pattern that is not concentrated on lysosomes/late-endosomes (marked by LAMP2, henceforward referred to as ‘lysosomes’) relative to elsewhere in the cytoplasm (+aa, Figure 3A). Upon removal of amino acids, however, TSC2 quickly accumulates on lysosomes (15min −aa, Figure 3A), and remains associated with lysosomes for hours (240min −aa, Figure 3A). Upon re-addition of amino acids, TSC2 quickly becomes diffusely cytoplasmic again (Figure 3B). Similar relocalization of TSC2 to lysosomes could also be observed in mouse embryonic fibroblasts (MEFs) (Figure 3C), and it was confirmed at the ultrastructural level by anti-TSC2 immuno-electron microscopy (Figure S3B). Furthermore, TSC1 also relocalizes to lysosomes upon amino acid removal, suggesting that the whole TSC1/2 complex relocalizes (Figure S3C).
Figure 3. Amino acid starvation causes TSC2 lysosomal localization in a Rag-dependent manner.
(A,C) TSC2 accumulates on lysosomes upon amino acid deprivation. Confocal micrograph of HEK293FT cells (A) or MEFs (C) treated +/− all amino acids in the presence of dialyzed FBS for the indicated times, stained for TSC2 (green) and the lysosomal marker LAMP2 (red). LAMP2 aggregates do not show TSC2 accumulation in the +aa condition.
(B) TSC2 quickly relocalizes away from lysosomes upon amino acid readdition. HEK293FT cells, starved for amino acids in the presence of dialyzed serum for 4h, then re-supplied with amino acid-containing media for 5 minutes. Cells stained and imaged as in (A).
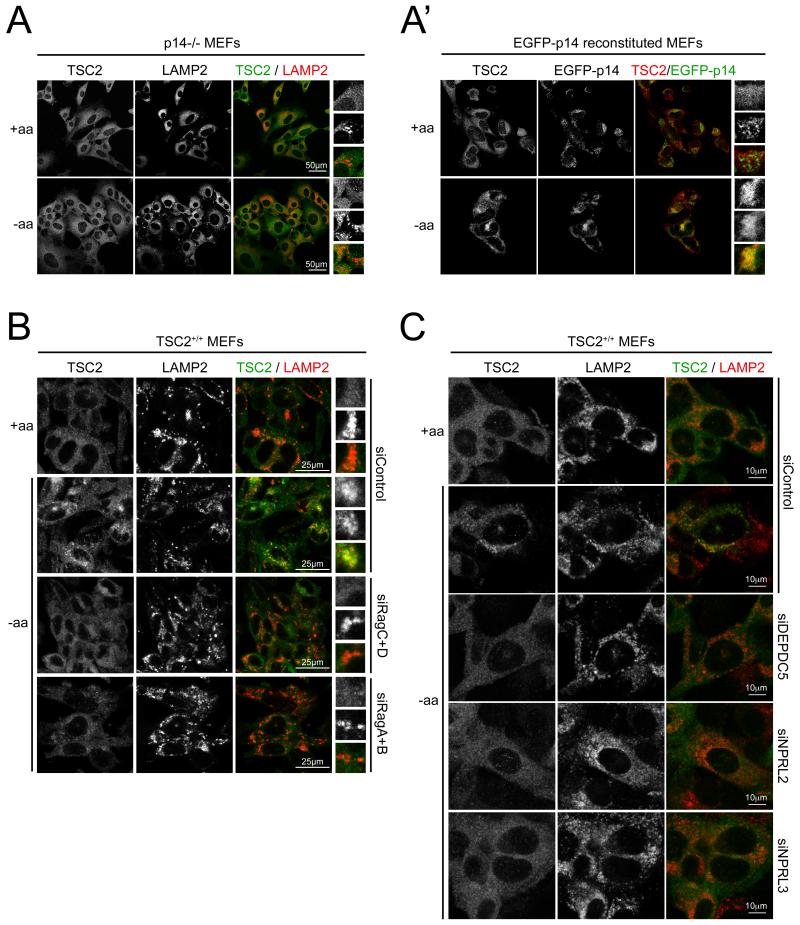
In agreement with the biochemical data presented above, lysosomal accumulation of TSC2 upon amino acid withdrawal is mediated via the Rag proteins, since it is strongly blunted upon amino acid starvation in MEFs lacking p14/LAMTOR2 in which the Rag proteins are not lysosomally localized (Sancak et al., 2010) (Figure 4A-A’), in MEFs or HEK293FT cells transiently lacking expression of LAMTOR components due to siRNA-mediated knock-down (data not shown), or in MEFs in which the Rag proteins have been knocked-down (Figure 4B). As expected (Sancak et al., 2008), Rag knock-down caused reduced TORC1 activation upon amino acid re-addition (Figure S4A) as well as reduced TOR lysosomal localization (Figure S4B), confirming successful knock-down of the Rag proteins. In agreement with the biochemical data showing preferential binding of TSC2 for the inactive Rag complex, recruitment of TSC2 to lysosomes in the absence of amino acids was also abrogated upon knock-down of the Rag GAP GATOR1 complex components DEPDC5, NPRL2 and NPRL3 (Figures 4C and S4E). In this setup, the Rag complex is present on lysosomes but remains in the activated state (Bar-Peled et al., 2013; Panchaud et al., 2013), leading, as previously shown, to increased TORC1 activity and lysosomal localization in the absence of amino acids (Figures S4C-D). These data suggest that recruitment of TSC2 to the lysosome upon amino acid withdrawal is due to the Rag proteins changing to the inactive conformation.
Figure 4. Lysosomal recruitment of TSC2 depends on the Rag proteins.
(A-A’) Lysosomal localization of TSC2 upon amino acid removal requires the LAMTOR complex. Confocal micrograph of p14/LAMTOR2 knock-out MEFs (A), or control p14 knock-out MEFs reconstituted to express an EGFP-p14 fusion (A’), treated +/− amino acids for 1h in the presence of dialyzed FBS. Lysosomes marked either with anti-LAMP2 antibody or with the lysosomaly localized EGFP-p14.
(B) Lysosomal localization of TSC2 upon amino acid removal requires the Rag proteins. MEFs transfected with control siRNA against Renilla luciferase or siRNAs targeting the various Rag proteins, treated +/− amino acids for 1h in the presence of dialyzed FBS. Cells stained and imaged as in (A).
(C) Lysosomal localization of TSC2 upon amino acid removal requires that the Rag proteins change to the inactive conformation. MEFs transfected with control siRNA or siRNAs targeting components of the GATOR1 complex DEPDC5, NPRL2, or NPRL3 required for the Rag proteins to become inactive upon amino acid removal. Cells stained and imaged as in (A).
TSC2 is required for complete inactivation of TORC1 upon amino acid removal
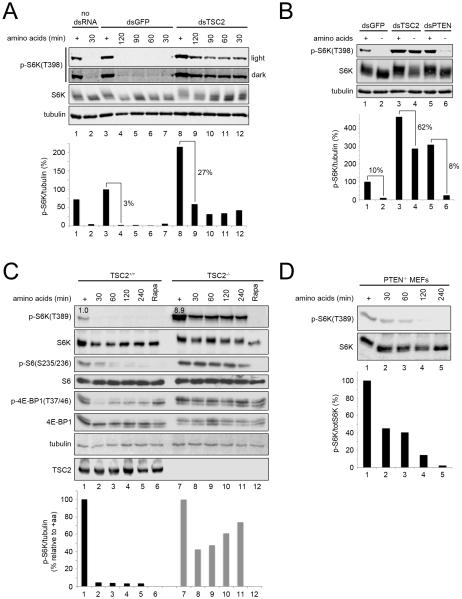
Having seen that TSC2 is recruited to lysosomes upon amino acid removal, we asked whether this has a functional consequence in terms of TORC1 activity. We tested if cells lacking TSC2 are impaired in their ability to turn off TORC1 upon amino acid removal. Unlike control Drosophila S2 cells which shut off TORC1 almost completely upon amino acid starvation, S2 cells with decreased Tsc2 levels show only a partial, initial drop in TORC1 activity, after which they retain a significantly elevated amount of TORC1 activity stably for up to 2 hours (Figure 5A). This cannot be explained simply by elevated starting levels of TORC1 activity in the Tsc2 knock-down cells, because S2 cells lacking PTEN have similarly elevated starting levels of TORC1 activity, but nonetheless efficiently shut off TORC1 upon amino acid removal (Figure 5B). These data are in agreement with previous observations (Gao et al., 2002). As in Drosophila cells, mouse embryonic fibroblasts lacking TSC2 or TSC1 are also unable to completely inactivate mTORC1 upon amino acid starvation (Figures 5C and S5A). Also in mouse cells this cannot be explained by elevated starting levels of mTORC1 activity because PTEN knock-out MEFs also start with elevated mTORC1 activity but efficiently shut off mTORC1 upon amino acid withdrawal (Figure 5D). Treatment of TSC1−/− or TSC2−/− MEFs with the TORC1-specific inhibitor rapamycin eliminated the aberrantly elevated S6K phosphorylation, showing that it is reflecting elevated activity of TORC1 and not another kinase (Figures 5C and S5A). An alternate explanation could be that lack of the TSC1/2 complex leads to reduced phosphatase activity towards S6K. To test this, we performed a rapamycin treatment timecourse to compare the rate of S6K dephosphorylation in control and TSC2 knock-down conditions. In both MEFs and Drosophila S2 cells, however, rapamycin treatment caused rapid dephosphorylation of S6K with similar kinetics in cells containing or lacking TSC2 (Figures S5F-G, see quantification), indicating that phosphatase activity is unchanged. We wondered whether cells lacking TSC2 might simply have a delayed response to amino acid starvation, in which case extending the time of incubation in medium lacking amino acids would allow them to more completely shut off mTORC1. Astoundingly, however, TSC2−/− MEFs retained elevated levels of mTORC1 activity in the absence of amino acids for up to 1.5-2 days, at which point they started dying, leading to concomitant drops in both phospho-S6K and total S6K signals on the immunoblots (Figure S5B). When phospho-S6K signals are normalized to total S6K levels, one can see that TSC2−/− MEFs retain a consistent level of mTORC1 activity up to their death (lower panel Figure S5B). In sum, TSC2 seems to be intimately linked to amino acid sensing by cells, in that 1) TSC2 localization changes upon amino acid removal, and 2) TSC2 is required for mTORC1 to fully turn off in the absence of amino acids.
Figure 5. TSC2 loss causes insensitivity to amino acid removal in Drosophila and mammalian cells.
(A-B) Tsc2 but not PTEN knock-down makes Drosophila cells largely insensitive to amino acid removal. (A) Immunoblots of Drosophila S2 cells treated with no dsRNA, dsRNA against GFP (control), or dsRNA against Tsc2, treated with media containing (+) or lacking amino acids for the indicated times. (B) PTEN knock-down S2 cells efficiently inactivate TORC1 upon amino acid removal, despite starting with elevated TORC1 activity levels.
(C-D) TSC2 knock-out MEFs do not completely shut off TORC1 upon amino acid withdrawal, whereas PTEN knock-out MEFs do. Immunoblots from (C) TSC2−/− and the respective control TSC2+/+ MEFs, or (D) PTEN knock-out MEFs, treated with medium +/− amino acids for the indicated times. Lanes 6 and 12 (C), cells treated +20nM Rapamycin for 1h in the presence of amino acids (‘Rapa’). Quantified with a LI-COR imaging system.
TSC2 is required to completely release mTORC1 from the lysosome upon amino acid starvation
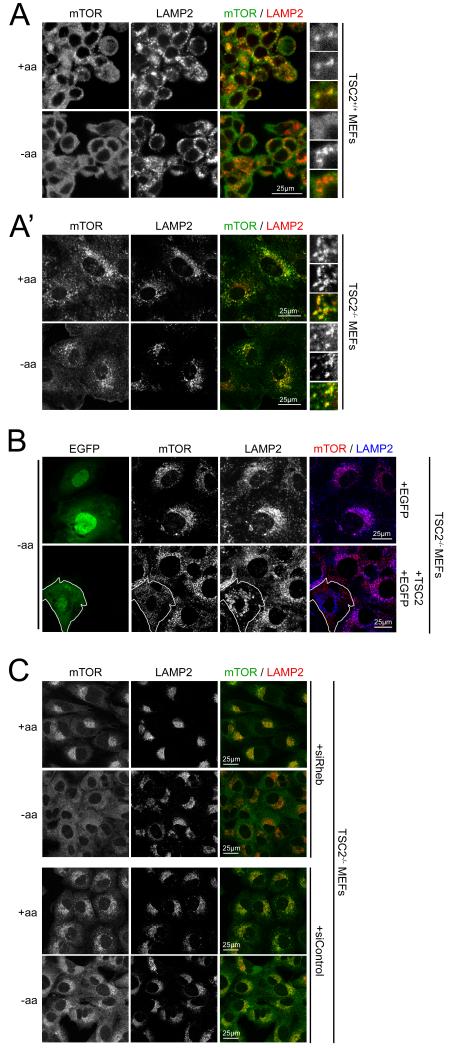
mTORC1 activation requires two regulated steps – one is activation of Rheb, and the other is localization of mTORC1 to the lysosomal surface where it meets Rheb, thereby forming an active complex (Inoki et al., 2003; Tee et al., 2003). In particular, the presence or absence of amino acids is known to regulate mTORC1 localization (Kim et al., 2008; Sancak et al., 2008). Since TSC2 appears to be involved in amino acid signaling to mTORC1, we asked whether TSC2 is required for the proper subcellular localization of mTORC1. As previously described (Kim et al., 2008; Sancak et al., 2008), in control cells mTOR accumulates on lysosomes in the presence of amino acids, and becomes diffusely cytoplasmic in the absence of amino acids (Figure 6A). In contrast, in TSC2-null MEFs (which, as expected, are larger than control cells) mTOR localization is very strongly lysosomal, both in the presence and in the absence of amino acids (Figure 6A’ and top panels Figure 6B). This defect can be rescued by transfecting them with a TSC2 expression plasmid, confirming specificity of the phenotype (lower panels in Figure 6B, note lysosomally localized mTOR in the starved non-transfected TSC2−/− cells, which is rescued in the transfected cell expressing TSC2 and EGFP). As expected from the known effect of TSC1 loss on TSC2 stability, this defect in mTOR localization could also be observed in TSC1-null MEFs (Figure S6A). Therefore, unexpectedly, the TSC1/2 complex is required for mTORC1 to be released completely from lysosomes upon amino acid withdrawal. Consistent with this phenotype being specific for loss of the TSC1/2 complex, and in agreement with the data presented in Figure 5, mTOR was readily released upon amino acid removal from lysosomes in PTEN-mutant MEFs, which also start with elevated mTORC1 activation levels (Figure S6B).
Figure 6. TSC2 is required for mTOR to localize away from lysosomes upon amino acid removal in a Rheb dependent manner.
(A-B) mTOR is released from lysosomes upon amino acid removal in control (A) but not TSC2-null MEFs (A’). This is rescued by re-expressing TSC2 + EGFP (to mark transfected cells) but not EGFP alone in the TSC-null MEFs (B). Note that the cell expressing TSC2 and GFP no longer has mTOR accumulated on lysosomes, whereas the surrounding, non-transfected cells retain lysosomally localized mTOR.
(C) Defective release of mTOR from lysosomes upon amino acid withdrawal in TSC2-null MEFs is rescued by knocking down Rheb. TSC2-null MEFs transfected with either Rheb siRNAs (upper panels) or control siRNAs (lower panels) treated 1h with +/− amino acid containing medium in the presence of dialyzed FBS.
Since mTORC1 localization is known to be regulated via the GTP/GDP load of the Rag GTPases (Sancak et al., 2008), and TSC2 has been described as a GAP for other small GTPases (Inoki et al., 2003; Wienecke et al., 1995; Xiao et al., 1997), we hypothesized TSC2 might regulate mTORC1 localization by acting as a Rag GAP. However, we radioactively quantified the relative GTP/GDP load of overexpressed Rag proteins in cells in the presence or absence of co-expressed TSC2, and could find no evidence to support this (data not shown). Another possible explanation could be that TSC2 affects the level of intracellular amino acids, thereby affecting mTORC1 localization. However, this does not seem to be the case: The intracellular levels of 10 amino acids drop upon amino acid starvation. For these, the intracellular levels drop equivalently, or even more dramatically, in the TSC2-null MEFs compared to control cells (Figure S5C). The intracellular levels of 8 other amino acids, unexpectedly, either remain constant or increase in control cells upon amino acid removal from the medium (Figures S5D-E). For these, the intracellular levels either increase less dramatically, or actually decrease, in TSC2-null MEFs compared to controls (Figures S5D-E). Since TSC2 acts via Rheb (Inoki et al., 2003), and Rheb is known to bind the mTORC1 complex (Long et al., 2005), this raised the possibility that Rheb might mediate the effect of TSC2 on mTOR localization by helping to anchor mTOR at the lysosomal membrane in collaboration with the Rag proteins. Indeed, in TSC2-null MEFs, knock-down of Rheb by siRNA allowed mTOR to be released from lysosomes upon amino acid removal (Figure 6C, controls in Figures S6C-D). Equivalent results could be observed in TSC2-null cells stably transfected with inducible shRNA constructs targeting an independent region of Rheb, confirming specificity of the effect (Figure S6E). Furthermore, overexpression of active, but not inactive, Rheb was sufficient to localize mTOR to the lysosome even in the absence of amino acids (data not shown). In sum, these results identify Rheb as the TORC1-anchoring activity in the TSC2-null MEFs.
TSC2 is required for cells to respond physiologically to amino acid starvation
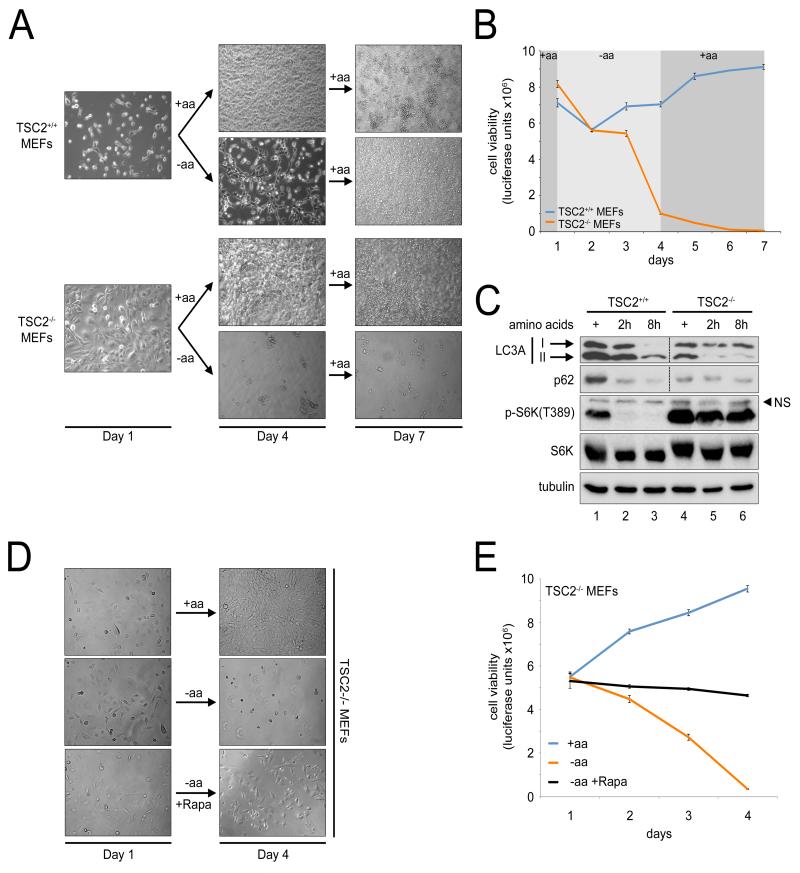
The data presented above support a model (detailed below, and Figure S7C) whereby TSC2 is required for cells to respond correctly to the removal of amino acids, allowing mTORC1 to be fully displaced from the lysosomal surface, thereby allowing it to become fully inactivated. We asked whether the inability of TSC2-null cells to respond correctly to amino acid starvation at the molecular level is paralleled by an equivalent defect at the physiological level. One might expect that if cells do not fully shut off mTORC1 upon amino acid starvation, this may lead to a metabolic catastrophe, as the demand for nutrients by cellular processes is not correctly reduced to match the reduced supply. To this end, we deprived control and TSC2-null MEFs of amino acids in the presence of dialyzed serum. Surprisingly, control MEFs were able to survive these conditions for several days; the cells were still visibly present after the treatment, and they recommenced proliferating when given complete medium (Figure 7A), suggesting they were in a quiescent state. This could also be quantified using a standard viability assay that quantifies ATP levels (blue trace, Figure 7B). In contrast (and in agreement with data in Figure S5B), TSC2-null MEFs died within 3 days in response to amino acid removal, indicating a physiologically defective response to amino acid starvation (Figures 7A-B). If TSC2-null MEFs are dying upon amino acid withdrawal due to an incomplete inhibition of mTORC1 activity, then blocking mTORC1 pharmacologically in these cells should improve their survival. Indeed, treating TSC2-null MEFs with rapamycin rescued their death upon amino acid starvation (Figures 7D-E). One catabolic process that is activated in response to low mTORC1 to promote cell survival upon amino acid starvation is autophagy. Control MEFs efficiently activated autophagy upon amino acid starvation (seen as accumulation of a faster migrating form of LC3A and degradation of p62, Figure 7C). In contrast, TSC2-null MEFs were impaired in activating autophagy (Figure 7C), thereby identifying one of probably multiple mTORC1-dependent physiological responses to amino acid starvation that are defective in TSC2-null cells. In sum, these data support the notion that TSC2 is required physiologically for cells to respond to amino acid starvation.
Figure 7. TSC2 null cells are impaired in their response to amino acid starvation, leading to their death.
(A-B) Amino acid removal causes cell death in TSC2 null cells. TSC2−/− and the respective TSC2+/+ control MEFs were treated on day 1 with medium +/− amino acids in the presence of dialyzed FBS. On day 4, all samples were given fresh medium containing amino acids. (A) DIC images captured on days 1, 4, and 7. (B) Cell viability determined using the Cell-Titer Glo kit (Promega).
(C) TSC2-null MEFs show defective autophagy induction upon amino acid withdrawal. TSC2−/− and the respective TSC2+/+ control MEFs were treated with medium containing (+) or lacking amino acids in the presence of dialyzed FBS for the indicated times. Activation of autophagy visualized on an immunoblot via increased conversion of LC3A to the faster migrating lipidated form II and degradation of p62 protein.
(D-E) Pharmacological inhibition of TORC1 rescues survival of TSC2-null MEFs upon amino acid withdrawal. TSC2−/− mouse embryonic fibroblasts were treated with medium containing (+aa) or lacking (−aa) amino acids with or without 20nM Rapamycin (−aa/+Rapa) for 3 days in the presence of dialyzed FBS. DIC images (D) and respective cell viability titers determined using the Cell-Titer Glo kit (Promega) (E).
Error bars: std. dev.
DISCUSSION
The data presented here suggest a model whereby in the presence of amino acids mTORC1 accumulates on lysosomes due to a dual anchoring activity composed primarily of the Rag proteins, but supported by Rheb (Figure S7C). While amino acids are present, binding between the Rag proteins and TSC1/2 is low, causing the TSC1/2 complex to remain cytoplasmic. Upon amino acid removal, the Rag proteins cause mTORC1 to be released from lysosomes via two independent activities, both of which result from the Rag proteins shifting to an ‘inactive’ conformation. Firstly, the Rag proteins reduce their binding for mTORC1, thereby releasing one of the two activities tethering mTORC1 at the lysosome. Secondly, the Rag proteins actively recruit TSC2 to the lysosome. This allows TSC2 to act on Rheb, thereby releasing the second tethering activity keeping mTORC1 on the lysosome. In the absence of TSC2, this second activity is unaffected, causing mTORC1 to remain lysosomally localized (Figure S7C, right panel).
The LAMTOR complex (composed of the p18, p14 and MP1) has been shown to serve as a docking point for mTORC1 (Sancak et al., 2010) and MEK/ERK complexes (Schaeffer et al., 1998; Teis et al., 2002), regulating their recruitment to late endosomes/lysosomes and their activation status. Our data demonstrate that integrity of the LAMTOR complex is critical for proper TSC2 subcellular localization upon amino acid withdrawal, therefore highlighting the importance of this scaffold complex for endomembrane-mediated activation/inactivation of signaling pathways.
The data presented here show that the TSC1/2 complex is part of the molecular machinery required for mTORC1 to respond properly to the absence of amino acids. We find that the TSC1/2 complex responds to amino acid starvation by changing its subcellular localization (Figure 3-4, S3B, S4E), and that TSC2 is required for mTORC1 to be fully released from lysosomes and fully inactivated upon amino acid removal (Figures 5-6). That said, however, in cells lacking TSC2, there is nonetheless a clear initial drop in TORC1 activity upon amino acid removal (Figures 5A,C). The remaining activity is then sustained indefinitely (Figure S5B). Hence, mTORC1 appears to consist of two pools or two degrees of activation, one of which requires TSC2 to become inactive upon amino acid withdrawal, and one of which responds independently of TSC2. This might explain why previous studies arrived at differing interpretations of their data (Gao et al., 2002; Roccio et al., 2006; Smith et al., 2005), since there is some response of TORC1 to amino acid removal in TSC2-null cells, however the response is severely blunted compared to controls. Further work will hopefully shed light on these two pools of activity. Although the impairment in mTORC1 response to amino acids in TSC2-null cells is partial, it is nonetheless of critical physiological relevance because TSC2-null MEFs die upon amino acid removal in sharp contrast to control MEFs (Figures 7 and S5B).
Looking at the model in Figure S7C from several perspectives yields various insights. (1) Regulation of mTORC1 activation could previously be rationalized as consisting of two independent, parallel steps: first, regulation of Rheb via TSC1/2 in response to a plethora of signals including stresses and growth factor signaling, and second, regulation of mTORC1 subcellular localization to lysosomal membranes in response to amino acids. Only when mTORC1 is properly localized to meet active Rheb would an active holoenzyme form. The data presented here blur the distinction between these two steps, since Rheb also affects mTORC1 localization, and amino acids also signal through TSC1/2. Instead, the two sets of regulatory inputs into mTORC1 appear to be more integrated. (2) Amino acid removal is ‘dominant’ over growth factor signaling, causing mTORC1 to shut off despite the presence of growth factors (Blommaart et al., 1995; Hara et al., 1998). This was previously explained by the fact that in the absence of amino acids, mTORC1 could not localize near active Rheb to form an active complex. Our dual tethering model is also consistent with this notion, but for a slightly modified reason, which is that amino acid starvation acts to sever both the Rag and Rheb lysosomal tethering activities. (3) Seen from the perspective of the Rag proteins, they swap binding partners depending on the state of amino acid signaling, binding preferentially to mTORC1 in the presence of amino acids, and binding preferentially to the TSC1/2 complex in the absence of amino acids. Consequently, mTORC1 and TSC1/2 also swap subcellular localizations.
We noticed that in our hands, knock-down of Rag proteins did not result in as strong a reduction in TORC1 activity in the presence of amino acids as was previously reported (Kim et al., 2008; Sancak et al., 2008). The most simple explanation is technical – that our Rag knock-downs are not strong enough to fully abrogate Rag recruitment of mTORC1 in the presence of amino acids, but are sufficient to impair Rag recruitment of TSC2 in the absence of amino acids. In that case, optimizing the Rag knock-downs might lead to even stronger effects than the ones we present here. Two alternate biological explanations, however, might be worth investigating in the future. The first is that our data suggest a dual anchoring mechanism of mTORC1 at the lysosomal membrane – one by the Rag proteins and one by Rheb. It is possible that the relative contribution of lysosomal tethering of mTORC1 by the Rag proteins and by Rheb might depend on their relative levels of expression and activation in the system being studied. This balance will likely depend on the cell line and on cell culture conditions. A second possible explanation could be one of biological kinetics, influenced by treatment strategy. The outcome might be quantitatively different if one looks at acute amino acid removal from cells adapted to complete medium (which we do here), or if one looks at amino acid add-back to cells that have equilibrated their signaling to the absence of amino acids. Indeed, we see that if we knock-down the Rag proteins in HEK293FT cells, we do not see a dramatic reduction in mTORC1 activity in the presence of amino acids (lanes 1-3 Figure S1D). However, if we remove amino acids for 1 hour and then re-add amino acids for 30 minutes, the same degree of Rag knock-down causes an obvious reduction in mTORC1 activity (lanes 7-9 Figure S1D). Likewise, in Drosophila S2 cells, RagC knock-down only had a mild effect on TORC1 activity in untreated cells (Figures 1A and S1A), but severely blunted the ability of cells to respond to amino acid add-back (Figure S1B). This difference between amino acid removal and amino acid add-back raises the interesting possibility that the Rag proteins are key in recruiting mTORC1 to the lysosome, a process that happens upon amino acid re-addition, and that both the Rag proteins and Rheb work together to keep mTORC1 on the lysosome once it is there. Indeed, in agreement with this model, mTOR is able to be recruited to the lysosome upon amino acid re-addition in TSC2-null MEFs in which Rheb is knocked down (Figure S7A-B), indicating that although Rheb tethers mTOR to the lysosome upon amino acid removal (Figure 6C), it is not required for de-novo recruitment of mTOR to the lysosome upon amino-acid add-back.
Previous reports showed that hyperactive mTORC1 signaling or dysregulated translation can lead to a metabolic mismatch in supply and demand, leading to cellular or organismal death (Choo et al., 2010; Efeyan et al., 2013; Kim et al., 2008; Leprivier et al., 2013; Teleman et al., 2005). We hypothesized that if TSC2 is required for mTORC1 activity to respond to amino acid starvation, then TSC2 might also be necessary for cells to respond physiologically to this stress. Indeed, TSC2 knock-out MEFs die upon removal of amino acids whereas control cells do not (Figure 7). The fact that cells with elevated mTORC1 activity due to impaired nutrient sensing die when deprived of amino acids raises the interesting hypothesis that limiting nutrient supply to tumors of certain genotypes might have a beneficial effect on their treatment. Consistent with this effect being due to elevated mTORC1 activity, the death of TSC2 knock-out MEFs is rescued by rapamycin treatment (Figure 7). This leads to the unexpected finding that rapamycin can actually promote cell survival under nutrient deprivation conditions, which might have therapeutic implications in mTOR-related malignancies.
We identify here the subcellular localization of TSC1/2 as one mechanism regulating this GAP holoenzyme. In an accompanying manuscript, Manning and colleagues show that TSC2 subcellular localization is also regulated by growth factor signaling. They show that in the absence of growth factor signaling, TSC2 is lysosomally localized even in the presence of amino acids. We show here that in the absence of amino acids, TSC2 is lysosomally localized, even in the presence of growth factor signaling (FBS). Hence, the presence of both amino acids and growth factor signaling are required to keep TSC2 in the cytoplasm and as long as one of the two is missing, TSC2 becomes lysosomally localized. Combined, our findings raise the possibility that TSC2 subcellular localization is a general mechanism for regulating this complex. It would be interesting to study whether the other inputs known to regulate TSC2 also affect its subcellular localization.
EXPERIMENTAL PROCEDURES
Immunoblot quantifications
Immunoblots were imaged and quantified with a LI-COR Odyssey FC imaging system, which has a larger dynamic range than what can be shown on the figure images. Data analysis was performed on the raw data, and not the image displayed, hence bands that look saturated on the figures for visualization purposes do not impact the data analysis. Quantifications of CoIP efficiency (Figures 2C, 2D and S2B) were calculated as the amount of Rag in the FLAG-TSC2 IP minus the negative control IP, normalized to the amount of TSC2 in the IP and the amount of Rag in the input.
Cell Imaging
All cell images within one panel were acquired and displayed with the same settings. Details of confocal microscopy in Supplemental Material.
Cell culture
Drosophila S2 cells were cultured in Schneider’s medium, supplemented with 10% FBS (PAA). HEK293FT cells (Invitrogen) and mouse embryonic fibroblasts were cultured in high glucose DMEM, supplemented with 10% FBS (Biochrom), except PTEN−/− MEFS which were also supplemented with 2mM Glutamine. TSC1−/−, TSC2+/+p53−/− and TSC2−/−p53−/− MEFs were a kind gift by David Kwiatkowski and Michael Hall and were described previously (Kwiatkowski et al., 2002; Zhang et al., 2003). p14−/− and EGFP-p14 reconstituted MEFs were a kind gift of Lukas Huber, described in (Teis et al., 2006). PTEN−/− MEFs were provided by Hong Wu, described in (Lesche et al., 2002).
Detailed experimental procedures are provided in the Supplemental Material.
Supplementary Material
HIGHLIGHTS.
TSC2 is recruited by the Rag GTPases to lysosomes upon amino acid starvation
TSC2 is required for complete inactivation of TORC1 upon amino acid starvation
Loss of TSC2 causes Rheb-mediated lysosomal retention of mTOR upon a.a. starvation
TSC2 is required for the correct physiological response of cells to a.a. starvation
ACKNOWLEDGEMENTS
A.T. would like to dedicate this paper to Rich Losick and Steve Cohen, who taught him how to do research. We thank David Kwiatkowski and Michael Hall for the TSC2 mutant MEFs, Lukas Huber for the p14−/− MEFs, and Hong Wu for the PTEN-mutant MEFs. We would also like to aknowledge David Sabatini for supplying the pRK5-HA-GST-Rag plasmids (Adgene #19298, 19301, 19304, 19307), Kun-Liang Guan for the pcDNA3-HA-RagA and RagC point mutants, Brendan Manning for the pcDNA3-FLAG-TSC2 and pRK7-FLAF-TSC1 expression vectors, and DJ Pan for the pAc5.1-Tsc2-V5-His expression vector. Several antibodies were obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD and maintained by The University of Iowa. We thank the Metabolomics Core Technology Platform of the Excellence cluster ‘CellNetworks’ (University of Heidelberg), and the Deutsche Forschungsgemeinschaft (grant ZUK 40/2010-3009262) for support with the HPLC-based intracellular amino acid measurements, the DKFZ Mass Spec core facility for protein identifications, Androniki Kolovou and Jacomine Krijnse Locker and the University of Heidelberg EM Core Facility for support with electron microscopy experiments, and Sibylle Schleich and Katrine Weischenfeldt for technical support. This work was supported by the European Research Council (ERC) under the European Union’s 7th Framework Programme.
REFERENCES
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. A Tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science. 2013;340:1100–1106. doi: 10.1126/science.1232044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blommaart EF, Luiken JJ, Blommaart PJ, van Woerkom GM, Meijer AJ. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. The Journal of biological chemistry. 1995;270:2320–2326. doi: 10.1074/jbc.270.5.2320. [DOI] [PubMed] [Google Scholar]
- Cherfils J, Zeghouf M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiological reviews. 2013;93:269–309. doi: 10.1152/physrev.00003.2012. [DOI] [PubMed] [Google Scholar]
- Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, Cantley LC, Blenis J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol Cell. 2010;38:487–499. doi: 10.1016/j.molcel.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Chang S, Gumper I, Snitkin H, Wolfson RL, Kirak O, Sabatini DD, Sabatini DM. Regulation of mTORC1 by the Rag GTPases is necessary for neonatal autophagy and survival. Nature. 2013;493:679–683. doi: 10.1038/nature11745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Zoncu R, Sabatini DM. Amino acids and mTORC1: from lysosomes to disease. Trends in molecular medicine. 2012;18:524–533. doi: 10.1016/j.molmed.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Zhang Y, Arrazola P, Hino O, Kobayashi T, Yeung RS, Ru B, Pan D. Tsc tumour suppressor proteins antagonize amino-acid-TOR signalling. Nature cell biology. 2002;4:699–704. doi: 10.1038/ncb847. [DOI] [PubMed] [Google Scholar]
- Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- Hara K, Yonezawa K, Weng QP, Kozlowski MT, Belham C, Avruch J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. The Journal of biological chemistry. 1998;273:14484–14494. doi: 10.1074/jbc.273.23.14484. [DOI] [PubMed] [Google Scholar]
- Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905–909. doi: 10.1126/science.1715094. [DOI] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. The Biochemical journal. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–1834. doi: 10.1101/gad.1110003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewell JL, Russell RC, Guan KL. Amino acid signalling upstream of mTOR. Nature reviews Molecular cell biology. 2013;14:133–139. doi: 10.1038/nrm3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nature cell biology. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski DJ, Zhang H, Bandura JL, Heiberger KM, Glogauer M, el-Hashemite N, Onda H. A mouse model of TSC1 reveals sex-dependent lethality from liver hemangiomas, and up-regulation of p70S6 kinase activity in Tsc1 null cells. Human molecular genetics. 2002;11:525–534. doi: 10.1093/hmg/11.5.525. [DOI] [PubMed] [Google Scholar]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, et al. The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell. 2013;153:1064–1079. doi: 10.1016/j.cell.2013.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesche R, Groszer M, Gao J, Wang Y, Messing A, Sun H, Liu X, Wu H. Cre/loxP-mediated inactivation of the murine Pten tumor suppressor gene. Genesis. 2002;32:148–149. doi: 10.1002/gene.10036. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Current biology: CB. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Panchaud N, Peli-Gulli MP, De Virgilio C. Amino acid deprivation inhibits TORC1 through a GTPase-activating protein complex for the Rag family GTPase Gtr1. Science signaling. 2013;6:ra42. doi: 10.1126/scisignal.2004112. [DOI] [PubMed] [Google Scholar]
- Proud CG. mTOR Signalling in Health and Disease. Biochemical Society transactions. 2011;39:431–436. doi: 10.1042/BST0390431. [DOI] [PubMed] [Google Scholar]
- Roccio M, Bos JL, Zwartkruis FJ. Regulation of the small GTPase Rheb by amino acids. Oncogene. 2006;25:657–664. doi: 10.1038/sj.onc.1209106. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer HJ, Catling AD, Eblen ST, Collier LS, Krauss A, Weber MJ. MP1: a MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science. 1998;281:1668–1671. doi: 10.1126/science.281.5383.1668. [DOI] [PubMed] [Google Scholar]
- Smith EM, Finn SG, Tee AR, Browne GJ, Proud CG. The tuberous sclerosis protein TSC2 is not required for the regulation of the mammalian target of rapamycin by amino acids and certain cellular stresses. The Journal of biological chemistry. 2005;280:18717–18727. doi: 10.1074/jbc.M414499200. [DOI] [PubMed] [Google Scholar]
- Tee AR, Manning BD, Roux PP, Cantley LC, Blenis J. Tuberous sclerosis complex gene products, Tuberin and Hamartin, control mTOR signaling by acting as a GTPase-activating protein complex toward Rheb. Current biology: CB. 2003;13:1259–1268. doi: 10.1016/s0960-9822(03)00506-2. [DOI] [PubMed] [Google Scholar]
- Teis D, Taub N, Kurzbauer R, Hilber D, de Araujo ME, Erlacher M, Offterdinger M, Villunger A, Geley S, Bohn G, et al. p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. The Journal of cell biology. 2006;175:861–868. doi: 10.1083/jcb.200607025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teis D, Wunderlich W, Huber LA. Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Developmental cell. 2002;3:803–814. doi: 10.1016/s1534-5807(02)00364-7. [DOI] [PubMed] [Google Scholar]
- Teleman AA, Chen YW, Cohen SM. 4E-BP functions as a metabolic brake used under stress conditions but not during normal growth. Genes Dev. 2005;19:1844–1848. doi: 10.1101/gad.341505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienecke R, Konig A, DeClue JE. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. The Journal of biological chemistry. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- Xiao GH, Shoarinejad F, Jin F, Golemis EA, Yeung RS. The tuberous sclerosis 2 gene product, tuberin, functions as a Rab5 GTPase activating protein (GAP) in modulating endocytosis. The Journal of biological chemistry. 1997;272:6097–6100. doi: 10.1074/jbc.272.10.6097. [DOI] [PubMed] [Google Scholar]
- Zhang H, Cicchetti G, Onda H, Koon HB, Asrican K, Bajraszewski N, Vazquez F, Carpenter CL, Kwiatkowski DJ. Loss of Tsc1/Tsc2 activates mTOR and disrupts PI3K-Akt signaling through downregulation of PDGFR. The Journal of clinical investigation. 2003;112:1223–1233. doi: 10.1172/JCI17222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.