Abstract
Neurotransmitter:sodium symporters (NSS) terminate synaptic signal transmission by Na+-dependent reuptake of released neurotransmitters, with key conformational states reported for a bacterial homolog LeuT and an inhibitor-bound Drosophila dopamine transporter. However, a coherent mechanism of Na+-driven transport has not been described. Here, we present two crystal structures of MhsT, a NSS member from Bacillus halodurans, in occluded inward-facing states with bound Na+ ions and L-Trp that provide insight into the cytoplasmic release of Na+. The switch from outward- to inward-oriented states is centered on the partial unwinding of transmembrane helix 5, which is facilitated by a conserved GlyX9Pro motif that opens an intracellular pathway for water to access the Na2 site. Based on our structural and functional findings we propose a mechanism according to which solvation through the TM5 pathway facilitates Na+ release from Na2 and the transition to an inward-open state.
Introduction
The neurotransmitter: sodium symporter (NSS) family encompasses a large group of prokaryotic and eukaryotic proteins that exploit the energy stored in the Na+ electrochemical gradient to power the uphill transport of their respective substrates1-3. Whereas bacterial NSS mediate the Na+-coupled uptake of amino acids, many human NSS (also known as the SLC6 family)4 are expressed in presynaptic neurons where they perform the Na+/Cl−-coupled reuptake of neurotransmitters such as serotonin, dopamine, γ -aminobutyric acid (GABA) and glycine from the synaptic cleft2,3. Notably, SLC6 members are therapeutic drug targets for the treatment of a broad range of neuropsychiatric conditions including depression, epilepsy and ADHD5,6 as well as being targets for widely abused psychostimulants such as cocaine and amphetamine6.
Crystal structures of the Aquifex aeolicus amino acid transporter LeuT7-10 have revealed the general structure of NSS showing an inverted pseudo-twofold symmetry relating two transmembrane domains comprising of transmembrane helices (TMs) 1-5 and 6-10, respectively. The Na1 and Na2 binding sites as well as the primary substrate-binding site (S1) are situated between the so-called scaffold (TMs 3-4, 8-9) and bundle domains (TMs 1-2, 6-7)7. The available structures for LeuT have revealed a number of the main states proposed to be involved in the transport cycle of the NSS family: outward-open with bound Na+ (ref. 10), occluded outward-oriented with bound Na+- and L-Leu7, and an inward-open apo state10. The inhibitor-bound, outward-open Drosophila melanogaster dopamine transporter (dDAT) structure11 is similar to LeuT, indicating that the general mechanism of transport is conserved across the NSS family.
Despite the lack of significant sequence identity, the LeuT-fold architecture12,13 is shared by several transporter families that extend beyond the NSS family7,10,14-19. Notably, the Na2 site of LeuT is conserved as a Na+ or H+ binding site in different transporters, highlighting its central role as the essential driver of ion-coupled transport in LeuT-fold transporters in general7,10,14-17.
However, fundamental questions addressing the transition from the substrate-bound, outward-facing occluded state7 to the substrate-free, inward-facing10 state and how it is coupled to the driving Na+ gradient have in fact remained unanswered for the NSS family and other Na+-driven transporters. Here we report the first crystal structures of a NSS member in an occluded inward-facing state with bound Na+ and substrate, namely the Bacillus halodurans multi-hydrophobic amino acid transporter (MhsT) determined at 2.1 Å and 2.6 Å resolution using two lipid-based crystallization methods, i.e., using detergent-solubilized lipids (HiLiDe)20 and lipidic cubic phase (LCP)21, respectively (Table 1; Fig. 1).
Table 1. Data collection and refinement statistics.
| MhsT HiLiDe | MhsT LCP | |
|---|---|---|
| Data collection | ||
| Space group | P2 | C2 |
| Cell dimensions | ||
| a, b, c (Å) | 44.3, 49.9, 110.1 | 44.2, 92.7, 112.5 |
| α, β, γ (°) | 90, 96.8, 90 | 90, 99.5, 90 |
| Resolution (Å) | 44.0−2.10 (2.21−2.10)* |
39.4−2.60 (2.72−2.60)* |
| Rsym (%) | 15.7 (124.3) | 15.6 (92.5) |
| I/σI | 8.9 (1.15) | 5.0 (1.0) |
| CC1/2 (%)# | 99.5 (44.0) | 97.5 (21.2) |
| Completeness (%) | 98.7 (98.5) | 96.8 (97.6) |
| Redundancy | 3.8 (3.8) | 2.0 (2.0) |
| Refinement | ||
| Resolution (Å) | 44.0−2.10 (2.21−2.10) |
39.4−2.60 (2.72− 2.60) |
| No. reflections | 27,894 (3,907) | 13,315 (1,632) |
| Rwork/Rfree | 0.191/0.235 | 0.201/0.255 |
| No. atoms | ||
| Protein | 3320 | 3243 |
| L-tryptophan ligand | 15 | 15 |
| Na+ ions | 2 | 2 |
| Water | 77 | 22 |
| Detergents/lipids | 184 | 66 |
| B-factors | ||
| Protein | 23.7 | 45.9 |
| L-tryptophan ligand | 12.8 | 30.9 |
| Na+ ions | 14.9 | 35.2 |
| Water | 26.1 | 41.2 |
| Detergents/lipids | 41.7 | 64.1 |
| R.m.s. deviations | 24.4 | 46.2 |
| Bond lengths (Å) | 1.73 | 0.80 |
| Bond angles (°) | 0.012 | 0.003 |
Values in parentheses are for highest-resolution shell.
Percentage of correlation between intensities from random half-datasets.
Figure 1. MhsT structure.
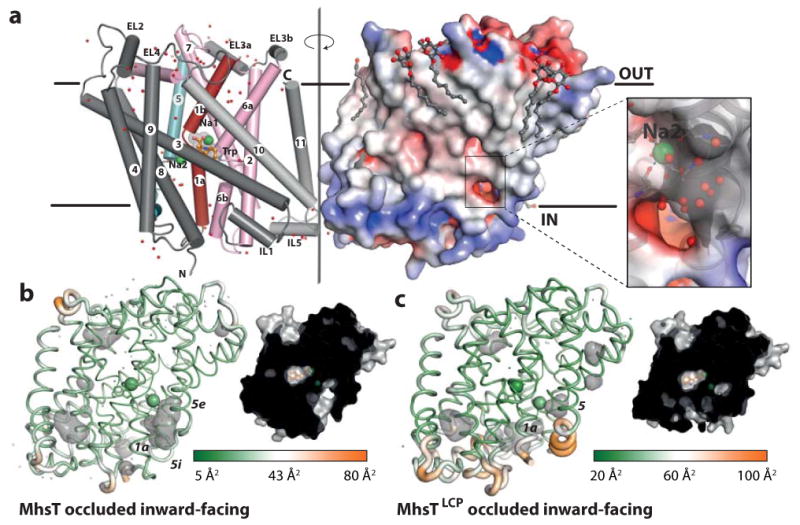
a, MhsT forms a crystallographic dimer through the intracellular half of helix 11 and intracellular loop 5. MhsT is shown in a schematic representation (scaffold helices in dark grey, bundle helices in red and pink, and TM5 in cyan) with associated water molecules (red dots). The electrostatic surface is shown for the symmetry related MhsT (right side, associated detergent molecules as ball-and-sticks, Na+ as green spheres, L-Trp as orange sticks) and the close-up shows a cytoplasmic cavity lined by negatively charged residues that reaches the Na2 site. b-c, Schematic structures based on HiLiDe (b) and LCP crystallization (c) with cavities (grey, left) and cut-through surface representations (right). Structural flexibility is indicated by the atomic displacement coded cartoon putty thickness and color gradient from green (low disorder) to orange (high disorder), with average values 23.7 Å2 and 46.5 Å2 for MhsT and MhsTLCP, respectively
Results
MhsT substrate transport and binding
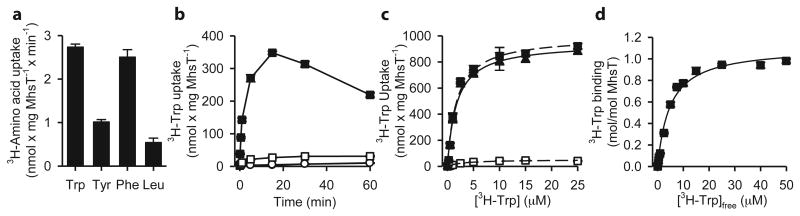
MhsT, a NSS homolog from Bacillus halodurans, is a transporter for hydrophobic L-amino acids22 (Fig. 2a). Transport of L-Trp in MhsT-containing proteoliposomes is time-dependent (Fig. 2b) and saturable with a Kt (substrate concentration at half-maximum transport velocity) of 1.7 ± 0.3 μM and a Vmax (maximum transport velocity) of 0.95 ± 0.04 μmol × mg MhsT-1 × min-1, yielding a catalytic turnover number (kcat) of 0.8 ± 0.03 s-1 (Fig. 2c). This number is similar to many other Na+-coupled transporters22-24 but about two and three orders of magnitude faster than LeuT-mediated transport of L-Ala and L-Leu, respectively7,25. MhsT binds L-Trp with an apparent Kd (dissociation constant) value of 4.8 ± 0.6 μM and a molar binding stoichiometry of 1.1 ± 0.04 as determined by saturation equilibrium binding studies in dodecylmaltoside (DDM), the detergent we used for solubilization and purification of MhsT (Fig. 2d).
Figure 2. MhsT uptake and binding.
The activity of MhsT reconstituted into proteoliposomes was determined by a, 1-minute uptake measurements of 3H-Trp, 3H-Tyr, 3H-Phe, and 3H-Leu at 0.1 μM in the presence of NaCl, b, a time-course of 1 μM 3H-Trp accumulation. c, Kinetics of MhsT-mediated L-tryptophan uptake in proteoliposomes. Initial rates and time course of transport were measured against increasing concentrations of 3H-Trp in the presence of an inwardly-directed Na+ electrochemical gradient (▪) or by dissipation of the gradient with gramicidin (□, reflecting binding). Net transport (▲) is the difference between total accumulation and binding. The control liposomes (without MhsT, ○) served as control in (b). d, Saturation equilibrium binding of 3H-Trp by MhsT was assayed with the SPA of purified, DDM-solubilized MhsT plotted as function of free 3H-Trp. Error bars represent the S.E.M., n=3.
Two MhsT crystal structures
We obtained two independent crystal forms of MhsT using HiLiDe20 (Fig. 1a and b; Table 1) and LCP21 crystallization methods (Fig. 1c; Table 1) with the resulting structures determined at 2.1 and 2.6 Å resolution, respectively. Both structures represent an occluded state with two bound Na+ ions and one L-Trp molecule (Fig. 1; Table 1). The structures are overall similar and superimpose with a 0.65 Å r.m.s.d., but differ at a specific, local region (see below). In the following MhsT and MhsTLCP will refer to the HiLiDe and the LCP structures, respectively.
Functional state of the MhsT structure
The overall fold of MhsT is similar to LeuT7 (Supplementary Fig. 1) and dDAT11, with which it shares 33% and 25% sequence identity, respectively (see Supplementary Discussion). Similar to the inward-open conformation of LeuT10 we observe a tight hydrophobic cluster of TM1b, 2, 6a and 7 that effectively seals the extracellular side of the transporter (Fig. 3b and c). Thus, compared to the occluded outward-facing state of LeuT7, which displays an open and hydrophobic extracellular vestibule, the extracellular vestibule of MhsT is collapsed. Interestingly at the intracellular side, we observe a water access pathway that reaches the Na2 site (Fig. 1a), while the bound substrate and Na1 site remain inaccessible. Thus, the MhsT structure represents an occluded, inward-facing state (see below).
Figure 3. Comparison of MhsT and LeuT in different functional states.
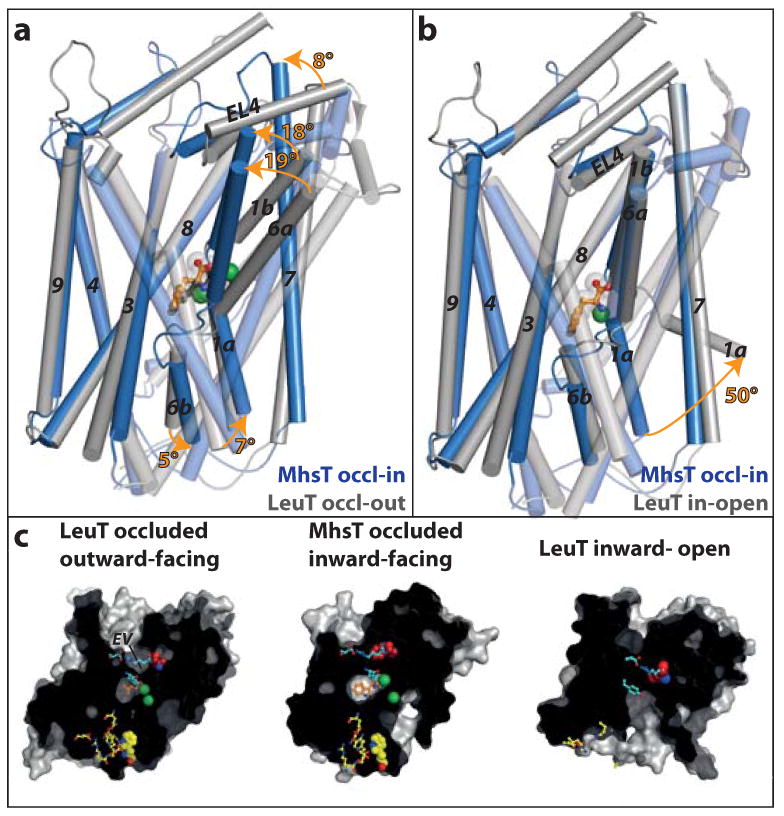
a-b MhsT occluded inward-facing (occl-in, blue) is compared to LeuT (a) occluded outward-facing7 (occl-out, PDB 2A65, grey) and (b) inward-open10 (in-open, PDB 3TT3, grey) based on superpositioning of the scaffold domains (TMs 3-4, 8-9). Key structural changes are marked with orange arrows. a, The switch from outward- to inward-facing occluded states is associated with a movement of the extracellular parts of helices 1b, 6a and 7 and extracellular loop 4 (EL4) towards the scaffold domain that closes the extracellular vestibule. b, The switch from occluded inward-facing to inward-open is linked to the release of Na+ from the Na2 site, which triggers TM1a to swing out and open a wide, intracellular pathway. c, Intracellular and extracellular cavities. Residues forming the extracellular gates are colored in cyan (Arg34MhsT/Arg30LeuT, Tyr108MhsT/Tyr108LeuT, Phe230MhsT/Phe253LeuT, Asp385MhsT/Asp404LeuT). Trp33MhsT is a key residue of the hydrophobic extracellular vestibule (EV) and is colored red (conserved, except unique Leu29LeuT). Intracellular gates (Arg9MhsT/Arg5LeuT, Glu10MhsT/Glu6LeuT, Ser244MhsT/Ser267LeuT, Tyr245MhsT/Tyr268LeuT, Glu330MhsT/Gln361LeuT, Asp369LeuT, Arg344MhsT/Arg375LeuT) are shown in yellow sticks and a hydrophobic plug (Trp12MhsT/Trp8LeuT) in yellow spheres. The substrate is shown in orange and Na+ ions in green spheres.
The collapsed extracellular vestibule forms a hydrophobic cluster centered on Trp33 of TM1b (Fig. 3b and c), which is conserved across the NSS family except for LeuT that has Leu29LeuT at the equivalent position (Supplementary Fig. 2a). The bulky nature of Trp33 leaves no void for an additional ligand, in contrast to the inward-open state of LeuT, which also displays extracellular closure, but with a yet unidentified entity enclosed near Leu29LeuT (ref. 10) and possibly marks a second substrate site (S2) as previously identified by molecular dynamics simulations and functional studies25.
A number of structures of non-NSS LeuT-fold transporters have also been assigned as occluded and inward-facing, including a Na+ -released, galactose-bound vSGLT14,26,27, a Na+ -released betaine-displaced BetP17,28 and a post-substrate released ApcT in a return state16. All of these models therefore represent later intermediates of a transport cycle than the Na+-bound and occluded inward-facing states reported here for the NSS family transporter MhsT.
Substrate and Na1 binding sites
The binding site for L-Trp in MhsT is similar to the L-Leu occupied S1 site in LeuT7 (Supplementary Fig. 3a and b). The carboxyl and amino groups of L-Trp make hydrogen bonds with the side chains of Tyr108 and Ser233 and backbone atoms of Gly30, Ala26, Thr231 and Phe230. A flexible Met236 side chain accommodates the bulky indole group of L-Trp, and the indole nitrogen makes a hydrogen bond with Ser327 (Supplementary Fig. 3a). Comparing MhsT and LeuT, the bulky side chains of Phe259LeuT and Ile359LeuT (Met236MhsT and Leu328MhsT, respectively) cannot accommodate L-Trp8 (Supplementary Fig. 3d), and although a Ile359GlnLeuT mutation supports L-Trp transport and binding29 it imposes a different rotamer configuration of L-Trp (Supplementary Fig. 3c) than in the L-Trp-transporting MhsT (Supplementary Fig. 3a). The coordination of Na1 in MhsT is similar to LeuT7 (Supplementary Table 1), showing octahedral geometry by side chains of Asn31, Thr231 and Asp263, backbone carbonyls of Ala26 and Thr231, as well as the L-Trp carboxyl group (Supplementary Fig. 3e and f).
TM5 unwinding and intracellular access to the Na2 site
Unlike the inward-open conformation of LeuT10, the extracellular closure of MhsT is not paired with an open configuration of the intracellular side (Fig. 3b and c; Fig. 4a). Rather, the intracellular side is reminiscent of the outward-facing states7,10 (Fig. 3a; Fig. 4a and c), where the N-terminal tail seals the cytoplasmic side through specific, polar interactions and with Trp12 (Trp8LeuT, conserved across the NSS family) plugging a hydrophobic pocket (Fig. 4a and b; Supplementary Fig. 4). However, the intracellular side also differs in key details from that of the outward-facing forms of LeuT. Most importantly, we observe unwinding of the intracellular part of TM5 – a feature that creates a solvent pathway that provides water access from the cytoplasm to the Na2 site (Fig. 4a; Supplementary Fig. 4b and see below). We propose that the unwinding results from the strain imposed by the movement of the extracellular part of TM5 (TM5e) together with TM6a in closure of the extracellular vestibule around Trp33, while at the same time the intracellular part of TM5 (TM5i) holds on to a short coiled-coil interaction with TM1a, which in turn is held in place through coordination with Na+ at the Na2 site (Fig. 4a-c). Furthermore, the negative end of TM4 and the positive end of TM5i dipoles30 interact with the salt bridge between the conserved Arg344MhsT (Arg375LeuT) of the TM8-9 loop and Glu10MhsT (Glu6LeuT) of the N-terminal end, respectively (Fig. 4a and c; Supplementary Fig. 4). Thus, the substrate-occluding configuration of the intracellular interface is dependent on the occupied Na2 site, which promotes the TM1a enclosure of the Na1 and L-Trp binding sites (Fig. 4a, see below).
Figure 4. A proposed TM5 “spring mechanism”.
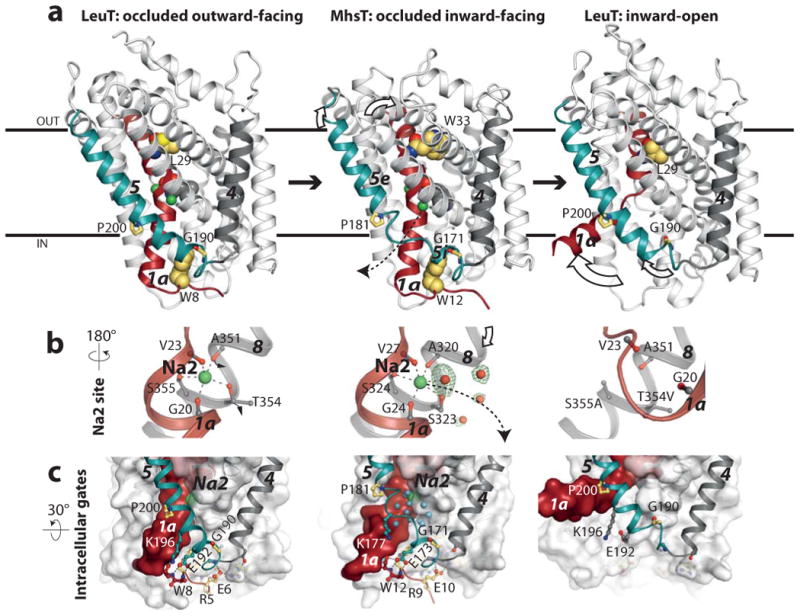
Conformational changes depicted by structures of the occluded outward-facing LeuT7, occluded inward-facing MhsT (this work), and inward-open LeuT10. a, TM5 (cyan) and TM1 (red) movements (white arrows). b, Changes at the Na2 site: trigonal-bipyramidal in the outward-facing LeuT, octahedral in the occluded inward-facing MhsT, and Na+ released in the inward-open LeuT. A Fo-Fc simulated annealing omit map (3 r.m.s.d. contour in green) shows water molecules in a solvent pathway reaching Na2. c, Intracellular gates encompass tight interactions between the N-terminus and TM4-5. Conserved residues in NSS are shown in yellow and waters of the Na2 solvation pathway as cyan spheres. The switch from outward- to inward-facing states occurs by collapse of the extracellular vestibule on the conserved Trp33MhsT that associates with unwinding at the conserved GlyX9Pro motif at the intracellular side of TM5 next to the Na2 site.
Importantly, the unwinding of TM5 is likely facilitated by a conserved GlyX9Pro helix-breaking motif (Gly171X9Pro181MhsT, Gly190X9Pro200LeuT, Fig. 5a). A sequence alignment of NSS for the poorly conserved TM4-TM5 region31 was confirmed and further refined by the recent dDAT structure11. This alignment indicates two populations for the GlyX9Pro motif, where in mammalian NSS it is slightly shifted in position with respect to bacterial NSS (Fig. 5a; Supplementary Fig. 2). However, we predict a preserved ability to promote unravelling of TM5i as observed here. We speculate that the sequence variations fine-tune the TM5 deformation mechanism to the specific physico-chemical conditions of individual species.
Figure 5. Conservation of transmembrane helix 5 GlyX9Pro motif and mutational studies.
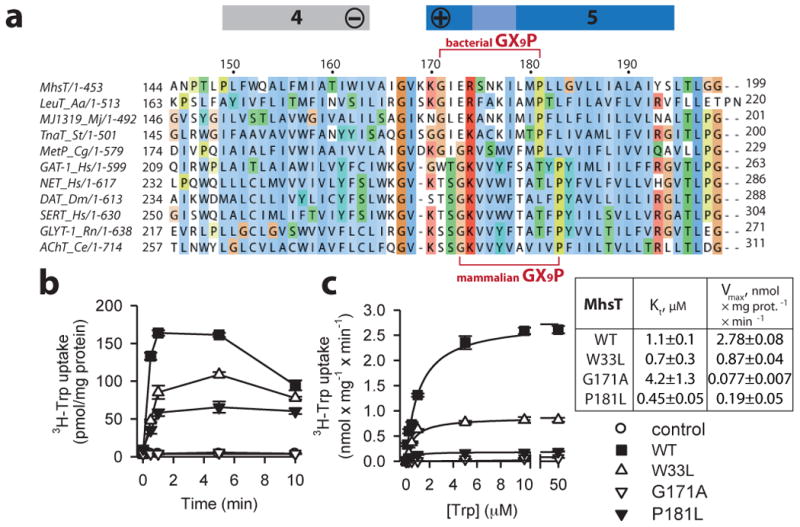
a, Sequence alignment of the NSS family based on a structural alignment of LeuT, MhsT and dDAT highlighting a conserved GlyX9Pro motif in TM5. b, Time course of 0.1 μM 3H-Trp (20 Ci/mmol) accumulation in intact cells of MQ614 expressing WT and mutant MhsT. Uptake was measured in 10 mM Tris/MES pH 8.5 and 50 mM NaCl. c, Kinetics of 3H-Trp uptake in intact MQ614 cells expressing MhsT-WT (▪), W33L (△), G171A (▽), and P181L (▾). Symbols are consistent in b and c (control cells lacking MhsT are shown as O in b) and data represent the mean ± SEM of triplicate determinations of representative experiments that were repeated ≥ three times.
Interestingly, structural alignment of MhsT (NSS family) and Mhp115 (nucleobase:cation symporter-1 (NCS1) family), which is another LeuT-fold transporter, reveals a recurring GlyXnPro pattern in the intracellular part of TM5 (Supplementary Fig. 5), suggesting that the unwinding and cytoplasmic water access to the Na+ binding site in the inward-facing states may also be exploited in other Na+-dependent, LeuT-fold transporters (See Supplementary Discussion for additional details on structural alignment of other LeuT-fold symporters).
Na2 solvation
In the occluded outward-facing state of LeuT the Na+ ion at the Na2 site is buried and has trigonal-bipyramidal coordination by five ligands7. However, for MhsT a cytoplasmic water molecule is added as a sixth ligand, which accesses Na2 through the solvent pathway afforded by TM5 unwinding (Fig. 1 and 4; Supplementary Table 1). TM8 is displaced with respect to TM1, and the Ser323 side chain has shifted, providing space for the water molecule. The equivalent Thr354LeuT has previously been suggested to be important for Na2 solvation32. With this additional water ligand, Na2 has changed coordination chemistry from trigonal-bipyramidal and occluded to octahedral yet distorted (Fig. 4b), and is now in contact with the intracellular environment. The Na2 site is, therefore, effectively primed for further solvation and Na+ release to the cytoplasmic milieu; a process that may be facilitated by the negative membrane potential.
MhsT Trp33 and GlyX9Pro mutational study
Functionally important residues in MhsT were tested by mutagenesis and measurement of L-Trp uptake into intact E. coli (MQ614 strain) cells. The mutation of Trp33MhsT to Leu reduced the Vmax by nearly 70% compared to MhsT-WT with little effect on Kt (Fig. 5b and c). These findings support a role for the conserved Trp33 in enhancing the transition to the inward-facing conformation. Notably, for the mammalian γ-aminobutyric acid (GABA) transporter GAT1 the corresponding Trp68LeuGAT1 mutation shows unaffected ligand binding affinity yet markedly reduced transport due to impaired substrate release33. In addition, mutation of the aligned residue in human DAT (Trp85Leu/AlahDAT) enhanced cocaine binding, consistent with a more outward-open configuration, and reduced transport by slowing inward release of dopamine34,35.
Similarly, the functional role of the Gly171X9Pro181 motif in MhsT was investigated. Mutating Gly171 to Ala or Pro181 to Leu drastically reduced the uptake of 3H-Trp (Fig. 5b and c) with Vmax values that were 97.2±0.2% and 93.2±1.8% lower than the Vmax for WT, respectively, while the Kt was 3.8 times higher for Gly171Ala and 2.4 times lower for the Pro181Leu mutant compared to WT (Fig. 5c). Pro272rDAT of the TM5-GlyX9Pro motif of the rat dopamine transporter was previously shown to be important for the apparent affinity for dopamine and a cocaine analog, and for the turnover of dopamine transport36, all consistent with a central role for this structural motif in transport. Also of note, hSERT is activated by protein kinase G phosphorylation of Thr276, which precedes the Gly278X9Pro288hSERT motif37 hinting at a regulatory interplay.
Thus, mutations of the GlyX9Pro motif primarily reduce the Vmax, and have smaller and diverse effects on the Kt, suggesting that such mutant forms generally allow binding of substrate in the outward open state but are impaired in the forward transition towards substrate release.
Differences between MhsT and MhsTLCP structures
Comparing our two independent crystal forms of the occluded, inward-facing state we note that the MhsTLCP structure exhibits tighter crystal packing (Supplementary Fig. 6) and a displaced N-terminal segment that is disordered in the crystal structure (Fig. 1b and c). Without the N-terminal segment stabilizing the unwound TM5 the latter reverts to a continuous but slightly kinked helix (Fig. 1c; Supplementary Fig. 6e; Supplementary Movie 1), similar to that observed in the LeuT7,10 and dDAT11 structures. The helical TM5 now disconnects the solvent pathway from the cytoplasm to the Na2 site. Importantly, the MhsTLCP structure therefore shows that TM5 of MhsT is capable of forming a helix as observed in LeuT and dDAT while also highlighting that conserved features of the N-terminal segment are important for stabilization of a conformation that opens the Na2 site to the cytoplasm (Supplementary Movie 1). Dynamics of the N-terminal segment have also been observed in single-molecule studies as being integral to function38. Furthermore we note that the continuous TM5 helix thus formed upon N-terminal tail disengagement remains flexible as indicated by high displacement factors for that region in the MhsTLCP structure (Fig. 1c).
Comparison of the MhsT, MhsTLCP and LeuT structures7,10 therefore suggests a “spring mechanism” for Na+ release (Fig. 4a): the TM5 helix is continuous in the occluded outward-facing state7, then exploits unwound conformations as an extended spring (or other flexible conformations) in the inward-facing state observed here, and finally reforms as a continuous helix in the inward-open state upon Na+ and substrate release10.
Discussion
The MhsT structures reported here provide new insights into the coupling of Na+ and substrate release for the NSS family. A coupled mechanism has been proposed for secondary transporters where the driving ion binds before the main substrate (‘first-on’) in the outward-facing state and has to be released before the main substrate (‘first-off’) in the inward-oriented state13,39. Functional studies of the NSS family have shown that Na+ binding stabilizes the outward-open state and precedes substrate binding38,40-42. This model was confirmed by the crystal structure of the Na+-bound outward-open state of LeuT10. Furthermore, molecular dynamics simulations of LeuT for the occluded, outward-facing state proposed that cytoplasmic water access to the Na2 site primes substrate release32. The MhsT structures presented here provides experimental support for this, but through an unanticipated mechanism, namely TM5 unwinding.
A related release mechanism was recently proposed for a Major Facilitator Superfamily transporter, namely the proton-dependent phosphate transporter PiPT43. For PiPT an occluded inward-facing state displays a narrow putative H+ release pathway, suggesting also a two-step intracellular release mechanism: firstly of the driving H+ through the narrow pathway formed upon substrate occlusion, and secondly of the transported substrate as H+ release triggers inward opening.
In conclusion, our observations suggest a general model of NSS function (Fig. 6; Supplementary Movie 2): First, Na+ and substrate binding in an outward-open state initiates occlusion by interactions with TM1b and TM6a7. This occluded, outward-facing state exposes a hydrophobic vestibule, which primes the transporter for further, extracellular closure upon movement of TM1b, TM6a TM7 and EL4 together with TM5e that at the same time drives the transporter towards the inward-facing state presented here. The extracellular closure is centered on the conserved Trp33MhsT of TM1, but may involve binding of a second substrate (S2) in LeuT9,10, which lacks the conserved Trp at this position. In the occluded inward-facing state, TM5e has moved relative to TM5i, which maintains its interaction with the N-terminal segment and TM1a through joint coordination of Na+ at the Na2 site (Fig. 4b); TM5 therefore extends as a spring at the cytoplasmic membrane interface, with the unwinding facilitated by the conserved GlyX9Pro deformation points. TM5 unwinding however provides access of intracellular solvent to the Na2 site, with the Na2 site accepting a water molecule as a sixth ligand, completing octahedral coordination (Fig. 4b; Supplementary Fig. 3g and h). The occluded inward-facing state is therefore primed for full solvation and intracellular release of Na+ from the Na2 site. Due to the low intracellular concentration of Na+, maintained by active Na+ extruding transporters, the intracellular release of Na2 triggers the forward reaction. Once Na2 is released, possibly stimulated by the structural flexibility of TM5 and a negative membrane potential, TM1a becomes liberated and can swing out while TM5i reassociates with TM5e in reformation of the TM5 helix, which in combination provides the opening for substrate and Na1 release to the intracellular environment10.
Figure 6. Transport mechanism of the NSS family.
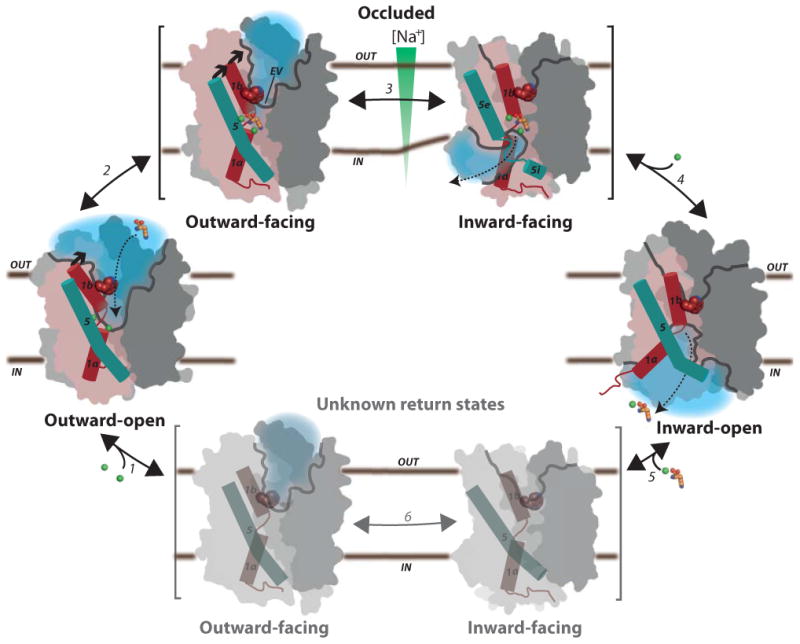
Na+ binding at the Na1 and Na2 sites stabilizes the outward-open state (1) and allows the substrate to bind (dashed arrow), triggering substrate site occlusion (2). Closure of the hydrophobic extracellular vestibule (EV) on Trp33 facilitates formation of an inward-facing state where TM5 unwinding provides a solvation pathway for the Na2 site (3) and an opportunity for Na+ to escape to the intracellular low-Na+ environment (4). Na+ release from Na2 allows TM5 to reform and TM1a to swing out to release the substrate with Na+ from Na1 in the inward-open state. Finally, the transporter switches back to the outward-open state through yet unknown occluded return states (5-6). Scaffold is shown in dark grey, bundle domain in pink, cavities opening in grey line, and solvated regions in light blue, TM1 as red and TM5 as cyan cylinders, Na+ ions as green, and amino acid substrate in orange spheres.
Thus the MhsT structures provide a missing link that depicts in concrete structural terms how an intimate sequence of intermediate steps during the transport cycle of a secondary transporter is connected to the downhill movement of Na+ along its transmembrane electrochemical gradient. Furthermore, these structures suggest that the dynamics of such individual states play another key role, with further studies of structure, function and dynamics required to establish a comprehensive understanding of how membrane transporter molecules work in a biomembrane.
Methods
Protein expression and purification
The mhsT gene flanked by a sequence that encodes an N-terminal 10×His tag and TEV protease cleavage site (in plasmid pNZ8048) was expressed in Lactococcus lactis NZ9000 and purified as previously described22.
Transport in proteoliposomes and intact cells
Transport of 3H-Trp in intact E. coli MQ614 [aroP mtr tnaB tyrP1 pheP::cat] was performed as described44. For uptake studies in proteoliposomes purified MhsT was reconstituted at a 1:150 (w/w) ratio in preformed liposomes made from E. coli polar lipid extract (Avanti Polar Lipids) that were destabilized with 1.7 mM Triton X-10022,24. Detergent was removed by stepwise addition of a total of 240 mg/mL Bio-Beads SM-2 (Biorad) and the proteoliposomes (or control liposomes) were collected by ultracentifugation at 320,000 × g for 45 min, resuspended (to ∼100 mg lipids/mL) in 100 mM potassium phosphate pH 6.5, 1 mM TCEP and stored in liquid N2. Prior to uptake measurements the proteoliposomes were thawed at 23 °C and extruded through a 400 nm filter (Avanti). The accumulation of 3H-Trp (20 Ci/mmol; American Radiolabeled Chemicals, Inc.) was measured with increasing concentrations of 3H-Trp (2 Ci/mmol) for the indicated periods of time at 22 °C in assay buffer composed of 10 mM Tris/MES pH 8.5, 50 mM NaCl in the presence of 1 μM valinomycin. To determine 3H-Trp binding to the MhsT-containing proteoliposomes or to control liposomes, the time course of 3H-Trp uptake was performed by dissipating the electrochemical NaCl gradient with gramicidin (25 μg/mL) for 5 min prior to the start of the reaction. Reactions were stopped by ice-cold assay buffer followed by rapid filtration through 0.22 μm nitrocellulose filters (Millipore) and used for scintillation counting. Counts per minute (cpm) were transformed into pmol using known amounts of 3H-Trp. Protein incorporated in proteoliposomes (as well as in detergent-solubilized form) was assayed using the amidoblack protein assay45.
Binding studies
Saturation binding of 3H-Trp to MhsT purified in n-dodecyl-β,D-maltopyranoside (DDM, Anagrade, Anatrace) was performed as described22 by means of the scintillation proximity assay (SPA) with 4.1 pmol of purified protein per assay in buffer composed of 150 mM Tris/MES pH 7.5, 50 mM NaCl, 1 mM TCEP, 20% glycerol, and 2 mM DDM. Samples were incubated with increasing concentrations of radioligand and measured in the SPA cpm mode of the MicroBeta™ counter. The efficiency of detection was calculated with a standard curve of known concentration to transform cpm into pmol. Specific binding was determined by subtracting the non-proximity signal (as determined in the presence of 800 mM imidazole) from the total binding, and was plotted as a function of free radioligand. Nonlinear regression fitting of the data was performed to obtain the Kd (concentration at 50% maximum binding) and molar ratio of Trp-to-MhsT binding (Graphpad Prism5).
Crystallization, data collection and processing
For crystallization experiments MhsT purified in DDM (SOL-GRADE, Anatrace) was digested with TEV protease to remove the His-tag and loaded on a second Ni-column (Ni-NTA, Qiagen) collecting the MhsT-containing flow-through. The protein was concentrated using a Viva-spin concentration device with molecular weight (MW) cut-off of 50 kDa to 7 mg/mL for size exclusion chromatography on a TSKgel G3000SW (Tosoh Bioscience) column with a running buffer consisting of 10 mM Tris-HCl pH 7.0, 100 mM NaCl, 10% (v/v) glycerol, 0.5 mM L-tryptophan (Sigma), 0.35 mM DDM. Peak fractions were collected and concentrated to 9-10 mg/mL using a Viva-spin concentration device with a MW cut-off of 50 kDa. MhsT was re-lipidated by adding 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Sigma) lipids according to the HiLiDe method20 with a 6:1 protein to lipids weight ratio.
A reservoir buffer containing 100 mM Tris-HCl pH 7.0, 400 mM NaCl, 20-24% w/w PEG400 and 5% trimethylamine-N-oxide (Sigma), supplemented with 11 mM n-nonyl-β-D-glucoside (NG, Anagrade, Anatrace) was mixed with the protein solution as 1 + 1 μL drops and subjected to vapor diffusion at 19 °C in hanging drops sealed with immersion oil20. Mounted crystals were subjected to controlled dehydration46 with a HC1c humidity control device at BESSY II beamline BL14.347, reducing humidity from 93% to 89% in 1% steps with 2 min incubation prior to flash cooling in liquid N2. X-ray diffraction data were collected at 0.8726 Å wavelength at the ESRF ID23-2 microfocus beamline and processed in space group P2 using the XDS package48 and TRUNCATE49 yielding a final dataset at 2.1 Å resolution as judged from the Wilson plot (with Wilson B-factor 27.8), the CC1/2 coefficient and model refinement statistics (see Table 1).
The MhsT reconstituted lipidic cubic phase (LCP) was prepared by the two syringe method21,50 by mixing 7.8 MAG (1-(7Z-pentadecenoyl)-rac-glycerol, Avanti Polar Lipids) lipid with protein (10 mg/mL) in a 1:1 weight ratio of protein solution to lipid. Crystallization trials were set up using an LCP dispensing robot (LCP Gryphon, Art Robbins Instruments) with the LCP dispensed on to homemade silanized 96-well glass sandwich crystallization plates with well diameter of 6 mm and well depth of 140 μm21. Each well comprised of 50 nL of LCP with 800 nL crystallization solution. Crystals of the MhsT-Trp complex (30 μm plates) were obtained at 19 °C in a crystallization solution containing 20% PEG400, 400 mM NaCl, 100 mM HEPES pH 7.0. Crystals were harvested directly from the LCP and flash cooled in liquid N2. X-ray diffraction data was collected at 0.9686 Å wavelength at I24 microfocus beamline (Diamond Light Source) and processed in the C2 space group using the XDS package48 and TRUNCATE49 yielding a final data set at 2.6 Å resolution as judged from the Wilson plot (with Wilson B-factor 27.5 Å2), the CC1/2 coefficient and model refinement statistics. The quality of the datasets was checked with PHENIX XTRIAGE51 (see Table 1).
Structure determination and analysis
For MhsT, initial phases were obtained by molecular replacement using the PHENIX MR_ROSETTA51 program. A search model was prepared with PHENIX SCULPTOR52 combining the MhsT sequence to the LeuT structure of the occluded outward-facing state (PDB 2A65)7 using a sequence alignment of MhsT and LeuT prepared with Clustal Omega53. The model obtained from PHENIX AUTOBUILD51 yielded R/Rfree of 0.31/0.43 using a 2.8 Å resolution preliminary data set of the HiLiDe crystal form. The model was rebuilt manually in COOT54 using density modification and prime-and-switch maps55, and refinement of coordinates and individual B-factors using PHENIX REFINE51. The intermediate model was reintroduced to PHENIX MR_ROSETTA51 using the 2.1 Å resolution data set (R/Rfree 0.31/0.36) followed by manual rebuilding, which led to a final model with Rwork/Rfree of 0.191/0.235 and Ramachandran plot favored/outliers 97.5/0.0%. Although waters have been modeled at the Na2 solvent access pathway, there were also indications of continuous density that could be interpreted as PEG400 or glycerol molecules, both of which were present in the crystallization buffer. However, each of these assignments led to unfavorable clashes with the protein structure. It was concluded that the electron density maps reveal water, PEG400 and glycerol in the solvent pathway at overlapping, partial sites, but that water molecules represent an adequate, single model at the available resolution. Initial phases for the LCP model were obtained by MR using the final HiLiDe model. Iterative rounds of structure refinement were performed in PHENIX REFINE51 with the models revised in real space with the program COOT54 leading to Rwork/Rfree of 0.201/0.255 and Ramachandran plot favored/outliers 96.5/0.2%. The final refinement statistics are summarized for MhsT HiLiDe and LCP structures in Table 1 (PDB code 4US3 and 4US4, respectively).
Figures were created using PYMOL56. The movie was generated from a morph between structures as created using PYMOL56 with geometry minimization for each iteration performed using PHENIX51.
A multiple sequence alignment of the NSS family was created using Muscle57 taking 200 randomly selected scores from a BLAST search on the human serotonin transporter. Using a Blosum30 scoring matrix and a gap open penalty of -45, the sequence alignment reproduced the structural alignment of MhsT, LeuT7,10 and dDAT11, in particular for regions showing low sequence identity. This alignment was used for the TM5 analysis of the GlyX9Pro motif and is essentially identical to a previously published alignment31. The structural alignments were performed with DaliLite58, converted to sequence alignment with Chimera59 and visualized with Jalview60. Electrostatic surface representations were prepared with the Adaptive Poisson-Boltzmann Solver61.
Supplementary Material
Acknowledgments
We thank A. M. Winther for her initial work on crystallization of detergent-solubilized MhsT, A.M. Nielsen for technical assistance and J.L. Karlsen for support with scientific computing. We are grateful to L. Shi and H. Weinstein for valuable discussion. We thank S.G. Rasmussen for access to a LCP dispensing robot. We are thankful to D. Flot and S. Russi at the European Synchrotron Radiation Facility ID23-2, and U. Müller and M.S. Weiss at the Helmholtz-Zentrum Berlin synchrotron radiation source BESSY II BL 14.1 and 14.3, and R. Owen and D. Axford at the Diamond Light Source 124, for help with X-ray diffraction data collection. Access to synchrotron facilities were supported by the Danscatt program of the Danish Council for Independent Research and the EU-FP7 infrastructure program Biostruct-X (grants 860 and 5624). This work was supported by research grants of the Lundbeck Foundation and NIH grants DA17293 and DA022413. L.M. was supported by a Boehringer-Ingelheim Fonds fellowship. L.R. was supported by the Danish Council for Independent Research in Medical Sciences and J. A.L. by the Danish Council for Independent Research in Natural Sciences.
Footnotes
Author Contributions: The functional characterization was performed by M.Q., H.Y. and L.M. HiLiDe crystallization, data collection and structure determination was performed by L.M. and assisted by L.R. LCP crystallization and data collection was performed by L.M. and J.A.L. and structure determination by L.M. The manuscript was written by L.M., M.Q., J.A.J, and P.N. and all authors commented on the manuscript.
Contributor Information
Matthias Quick, Email: mq2102@columbia.edu.
Poul Nissen, Email: pn@mb.au.dk.
References
- 1.Kristensen AS, et al. SLC6 neurotransmitter transporters: structure, function, and regulation. Pharmacol Rev. 2011;63:585–640. doi: 10.1124/pr.108.000869. [DOI] [PubMed] [Google Scholar]
- 2.Rudnick G. Contemporary Neuroscience: Neurotransmitter Transporters: Structure, Function, and Regulation. 2nd. Humana Press Inc.; Totowa, NJ: 2002. Mechanisms of Biogenic Amine Neurotransmitter Transporters; pp. 25–51. [Google Scholar]
- 3.Torres GE, Gainetdinov RR, Caron MG. Plasma membrane monoamine transporters: structure, regulation and function. Nat Rev Neurosci. 2003;4:13–25. doi: 10.1038/nrn1008. [DOI] [PubMed] [Google Scholar]
- 4.Hediger MA, et al. The ABCs of solute carriers: physiological, pathological and therapeutic implications of human membrane transport proteins Introduction. Pflugers Arch. 2004;447:465–8. doi: 10.1007/s00424-003-1192-y. [DOI] [PubMed] [Google Scholar]
- 5.Singh SK. LeuT: a prokaryotic stepping stone on the way to a eukaryotic neurotransmitter transporter structure. Channels (Austin) 2008;2:380–9. doi: 10.4161/chan.2.5.6904. [DOI] [PubMed] [Google Scholar]
- 6.Gether U, Andersen PH, Larsson OM, Schousboe A. Neurotransmitter transporters: molecular function of important drug targets. Trends Pharmacol Sci. 2006;27:375–83. doi: 10.1016/j.tips.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437:215–23. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 8.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322:1655–61. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quick M, et al. Binding of an octylglucoside detergent molecule in the second substrate (S2) site of LeuT establishes an inhibitor-bound conformation. Proc Natl Acad Sci USA. 2009;106:5563–8. doi: 10.1073/pnas.0811322106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481:469–74. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013 doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abramson J, Wright EM. Structure and function of Na(+)-symporters with inverted repeats. Curr Opin Struct Biol. 2009;19:425–32. doi: 10.1016/j.sbi.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forrest LR, Kramer R, Ziegler C. The structural basis of secondary active transport mechanisms. Biochim Biophys Acta. 2011;1807:167–88. doi: 10.1016/j.bbabio.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 14.Faham S, et al. The crystal structure of a sodium galactose transporter reveals mechanistic insights into Na+/sugar symport. Science. 2008;321:810–4. doi: 10.1126/science.1160406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weyand S, et al. Structure and molecular mechanism of a nucleobase-cation-symport-1 family transporter. Science. 2008;322:709–13. doi: 10.1126/science.1164440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shaffer PL, Goehring A, Shankaranarayanan A, Gouaux E. Structure and mechanism of a Na+-independent amino acid transporter. Science. 2009;325:1010–4. doi: 10.1126/science.1176088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez C, Koshy C, Yildiz O, Ziegler C. Alternating-access mechanism in conformationally asymmetric trimers of the betaine transporter BetP. Nature. 2012;490:126–30. doi: 10.1038/nature11403. [DOI] [PubMed] [Google Scholar]
- 18.Tang L, Bai L, Wang WH, Jiang T. Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol. 2010;17:492–6. doi: 10.1038/nsmb.1788. [DOI] [PubMed] [Google Scholar]
- 19.Fang Y, et al. Structure of a prokaryotic virtual proton pump at 3.2 A resolution. Nature. 2009;460:1040–3. doi: 10.1038/nature08201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gourdon P, et al. HiLiDe—Systematic Approach to Membrane Protein Crystallization in Lipid and Detergent. Crystal Growth & Design. 2011;11:2098–2106. [Google Scholar]
- 21.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–31. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quick M, Javitch JA. Monitoring the function of membrane transport proteins in detergent-solubilized form. Proc Natl Acad Sci USA. 2007;104:3603–8. doi: 10.1073/pnas.0609573104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quick M, Wright EM. Employing Escherichia coli to functionally express, purify, and characterize a human transporter. Proc Natl Acad Sci USA. 2002;99:8597–601. doi: 10.1073/pnas.132266599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jung H, Tebbe S, Schmid R, Jung K. Unidirectional reconstitution and characterization of purified Na+/proline transporter of Escherichia coli. Biochemistry. 1998;37:11083–8. doi: 10.1021/bi980684b. [DOI] [PubMed] [Google Scholar]
- 25.Shi L, Quick M, Zhao Y, Weinstein H, Javitch JA. The mechanism of a neurotransmitter:sodium symporter—inward release of Na+ and substrate is triggered by substrate in a second binding site. Mol Cell. 2008;30:667–77. doi: 10.1016/j.molcel.2008.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Tajkhorshid E. Ion-releasing state of a secondary membrane transporter. Biophys J. 2009;97:L29–31. doi: 10.1016/j.bpj.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Watanabe A, et al. The mechanism of sodium and substrate release from the binding pocket of vSGLT. Nature. 2010;468:988–91. doi: 10.1038/nature09580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ressl S, Terwisscha van Scheltinga AC, Vonrhein C, Ott V, Ziegler C. Molecular basis of transport and regulation in the Na(+)/betaine symporter BetP. Nature. 2009;458:47–52. doi: 10.1038/nature07819. [DOI] [PubMed] [Google Scholar]
- 29.Piscitelli CL, Gouaux E. Insights into transport mechanism from LeuT engineered to transport tryptophan. EMBO J. 2012;31:228–35. doi: 10.1038/emboj.2011.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sengupta D, Behera RN, Smith JC, Ullmann GM. The alpha helix dipole: screened out? Structure. 2005;13:849–55. doi: 10.1016/j.str.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70:1630–42. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 32.Zhao C, Noskov SY. The role of local hydration and hydrogen-bonding dynamics in ion and solute release from ion-coupled secondary transporters. Biochemistry. 2011;50:1848–56. doi: 10.1021/bi101454f. [DOI] [PubMed] [Google Scholar]
- 33.Mager S, et al. Ion binding and permeation at the GABA transporter GAT1. J Neurosci. 1996;16:5405–14. doi: 10.1523/JNEUROSCI.16-17-05405.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Z, Wang W, Uhl GR. Dopamine transporter tryptophan mutants highlight candidate dopamine- and cocaine-selective domains. Mol Pharmacol. 2000;58:1581–92. doi: 10.1124/mol.58.6.1581. [DOI] [PubMed] [Google Scholar]
- 35.Chen N, Zhen J, Reith ME. Mutation of Trp84 and Asp313 of the dopamine transporter reveals similar mode of binding interaction for GBR12909 and benztropine as opposed to cocaine. J Neurochem. 2004;89:853–64. doi: 10.1111/j.1471-4159.2004.02386.x. [DOI] [PubMed] [Google Scholar]
- 36.Lin Z, Itokawa M, Uhl GR. Dopamine transporter proline mutations influence dopamine uptake, cocaine analog recognition, and expression. FASEB J. 2000;14:715–28. doi: 10.1096/fasebj.14.5.715. [DOI] [PubMed] [Google Scholar]
- 37.Ramamoorthy S, Samuvel DJ, Buck ER, Rudnick G, Jayanthi LD. Phosphorylation of threonine residue 276 is required for acute regulation of serotonin transporter by cyclic GMP. J Biol Chem. 2007;282:11639–47. doi: 10.1074/jbc.M611353200. [DOI] [PubMed] [Google Scholar]
- 38.Zhao Y, et al. Substrate-modulated gating dynamics in a Na+-coupled neurotransmitter transporter homologue. Nature. 2011;474:109–13. doi: 10.1038/nature09971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krupka RM. Coupling mechanisms in active transport. Biochim Biophys Acta. 1993;1183:105–13. doi: 10.1016/0005-2728(93)90009-5. [DOI] [PubMed] [Google Scholar]
- 40.Claxton DP, et al. Ion/substrate-dependent conformational dynamics of a bacterial homolog of neurotransmitter: sodium symporters. Nat Struct Mol Biol. 2010;17:822–9. doi: 10.1038/nsmb.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhao Y, et al. Single-molecule dynamics of gating in a neurotransmitter transporter homologue. Nature. 2010;465:188–93. doi: 10.1038/nature09057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kazmier K, et al. Conformational dynamics of ligand-dependent alternating access in LeuT. Nat Struct Mol Biol. 2014;21:472–9. doi: 10.1038/nsmb.2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen BP, et al. Crystal structure of a eukaryotic phosphate transporter. Nature. 2013;496:533–6. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Quick M, et al. State-dependent conformations of the translocation pathway in the tyrosine transporter Tyt1, a novel neurotransmitter: sodium symporter from Fusobacterium nucleatum. J Biol Chem. 2006;281:26444–54. doi: 10.1074/jbc.M602438200. [DOI] [PubMed] [Google Scholar]
- 45.Schaffner W, Weissmann C. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal Biochem. 1973;56:502–14. doi: 10.1016/0003-2697(73)90217-0. [DOI] [PubMed] [Google Scholar]
- 46.Russi S, et al. Inducing phase changes in crystals of macromolecules: status and perspectives for controlled crystal dehydration. J Struct Biol. 2011;175:236–43. doi: 10.1016/j.jsb.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 47.Mueller U, et al. Facilities for macromolecular crystallography at the Helmholtz-Zentrum Berlin. J Synchrotron Radiat. 2012;19:442–9. doi: 10.1107/S0909049512006395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–32. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Collaborative Computational Project, N. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 50.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem Phys Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–21. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bunkoczi G, Read RJ. Improvement of molecular-replacement models with Sculptor. Acta Crystallogr D Biol Crystallogr. 2011;67:303–12. doi: 10.1107/S0907444910051218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Terwilliger TC. Using prime-and-switch phasing to reduce model bias in molecular replacement. Acta Crystallogr D Biol Crystallogr. 2004;60:2144–9. doi: 10.1107/S0907444904019535. [DOI] [PubMed] [Google Scholar]
- 56.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 57.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holm L, Park J. DaliLite workbench for protein structure comparison. Bioinformatics. 2000;16:566–7. doi: 10.1093/bioinformatics/16.6.566. [DOI] [PubMed] [Google Scholar]
- 59.Pettersen EF, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 60.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci USA. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Forrest LR, Rudnick G. The rocking bundle: a mechanism for ion-coupled solute flux by symmetrical transporters. Physiology (Bethesda) 2009;24:377–86. doi: 10.1152/physiol.00030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao Y, et al. Substrate-dependent proton antiport in neurotransmitter:sodium symporters. Nat Chem Biol. 2010;6:109–16. doi: 10.1038/nchembio.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wohlers I, Andonov R, Klau GW. DALIX: optimal DALI protein structure alignment. IEEE/ACM Trans Comput BiolBioinform. 2013;10:26–36. doi: 10.1109/TCBB.2012.143. [DOI] [PubMed] [Google Scholar]
- 65.Hasegawa H, Holm L. Advances and pitfalls of protein structural alignment. Curr Opin Struct Biol. 2009;19:341–8. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.