Abstract
Background
The inability to quantify sexual exposure to HIV limits the power of HIV prevention trials of vaccines, microbicides and pre-exposure prophylaxis (PrEP) in women. We investigated detection of HIV-1 and Y chromosomal (Yc) DNA in vaginal swabs from 83 participants in the HPTN 035 microbicide trial as biomarkers of HIV exposure and unprotected sexual activity.
Methods
143 vaginal swabs from 85 women were evaluated for the presence of Y chromosomal DNA (Quantifiler Duo DNA quantification kit, Applied Biosystems) and total HIV-1 DNA (single copy in-house qPCR assay). Y DNA detection was paired with self-reported behavioral data with regard to recent coitus (≤ 1 week prior to collection) and condom usage (100% vs. <100% compliance).
Results
Yc DNA was detected in 62/143 (43%) swabs. For the 126 visits at which both behavioral data and swabs were collected, Yc DNA was significantly more frequent in women reporting <100% condom usage (OR 10.69; 95% confidence interval: 2.27 – 50.32; p=0.003). Notably, 27 of 83 (33%) swabs from women reporting 100% condom usage were positive for Yc DNA. HIV DNA was only detected in swabs collected post-seroconversion.
Conclusions
The use of Yc DNA in HIV prevention trials could reliably identify sub-groups of women who have unprotected sexual activity and could provide valuable exposure-based estimates of efficacy.
Keywords: Biomarker, HIV exposure, Y chromosomal detection, Yc, HIV DNA, vaginal swabs, microbicide, pre-exposure prophylaxis, sexual transmission, prevention trial
INTRODUCTION
A major limitation of current HIV prevention trial design is the lack of an accurate and sensitive means to measure exposure to HIV (1, 2). The evaluation of risk behavior using self-reported condom use and frequency of coitus can be unreliable due to miscommunication between interviewer and interviewee, lack of understanding of the questions asked, reporting behavior according to perceived expectations, inability to recall experience, and blatant misreporting (3, 4). Additionally, self-report of condom use frequency does not capture risk associated with improper condom use or accidental condom breakage. Despite >80% reported condom use in VOICE, FEM-PrEP and the TDF2 study, pregnancy and HIV incidence rates remained higher than expected (5–7). A cross-sectional study of 910 women in Zimbabwe found that only 52% of participants who tested positive for prostate-specific antigen (PSA) in vaginal swabs reported unprotected sex during the previous 2 days. Audio computer assisted self-interview (ACASI) technology did not generate significantly different responses about unprotected intercourse compared to face-to-face interview (8).
The detection of semen could provide an unbiased measure of unprotected sex in participants of HIV prevention trials. However, the limited sensitivity of the most commonly used biomarker, PSA, makes it impractical for use in clinical studies where swab collection could occur hours to weeks after intercourse. PSA levels decline 10-fold by 3 hours post-exposure, and are undetectable by 24–48 hours post-exposure (9–11). Rapid stain identification of human semen (RSID) that detects the presence of semenogelin has also been used in trials to indicate that a woman has been exposed to ejaculate in the previous 48 hours, but is 10-fold less sensitive than quantitative PCR methods to detect Y chromosomal (Yc) DNA (12, 13). Depite heterogeneity in both initial deposit of Yc DNA and rate of decline of Yc DNA signal, Yc has the advantage of detectability up to 15 post-coital days from self-collected vaginal swabs without impact from menses on the rate of decay (14, 15). Yc DNA is not detected in women using condoms correctly as demonstrated by a study that showed that only 5/56 women had positive Yc DNA results after condom use following a 14 day abstinence period and the 5 detections were associated with receptive oral sex and digital penetration (16, 17).
The detection of HIV in genital samples from HIV negative women could more directly assess HIV infection risk. In a study of cervical dysplasia in U.S. women, HIV-1 env and gag glycoproteins were identified in cervicovaginal lavage samples from women who were confirmed to be HIV negative by serology (18). However partner HIV status was not known, and the linkage of env and gag detection with future seroconversion was not verified. HIV-1 viral RNA and proviral HIV-1 DNA sequences can be detected in seminal plasma and non-spermatazoal mononuclear cells in HIV infected men throughout successful long-term HAART while plasma HIV-1 viral RNA levels remain undetectable (19–22). The detection of HIV DNA in genital samples from HIV negative women has not been studied as marker of HIV infection risk.
We therefore examined the frequency of Yc DNA and HIV DNA detection in vaginal swabs collected in HIV seroconverters (both pre- and post-seroconversion) and non-seroconverters from the HPTN 035 study, using highly sensitive quantitative PCR assays with detection limits of a single copy.
MATERIALS AND METHODS
Study Population
HPTN 035 was a phase II/IIb safety and effectiveness study of the vaginal microbicides BufferGel and 0.5% PRO2000/5 gel for the prevention of HIV-1 infection in women, conducted from February 2005 through September 2008 (NCT00074425). All participants provided informed consent for swab collection and future testing. The population demographic characteristics, protocol, and trial results are described elsewhere (23). Starting in 2008, vaginal swab specimens were collected from participants during each quarterly pelvic exam by applying a Dacron swab to the posterior fornix of the vagina until the tip was saturated with fluid, then placing the swab in a cryovial containing 400 μL of phosphate-buffered saline. The cryovials were stored at −80°C at the sites and shipped to the MTN Network Laboratory after the primary study results were available. The current study evaluated a case control subset of swabs collected from women at sites in Zimbabwe (Harare and Chitungwiza), South Africa (Hlabisa and Durban) and Malawi (Blantyre and Lilongwe) at a 1:3 ratio (seroconverters: non-seroconverters). Swabs from seroconverters were collected both pre- and post-seroconversion. Swabs from participants post-seroconversion were collected a median of 21 days after detection of seroconversion (range 5 – 124 days). Seroconverters were not taking antiretroviral therapy at the time of swab collection. The operator performing the assays was blinded to the subgroup to which the participant belonged.
Nucleic Acid Extraction
Swabs were processed to isolate the cell pellet as described previously (24). Total nucleic acid was extracted by incubating the vaginal swab cell pellet in 2 mg/ml Proteinase K solution (Applied Biosystems) for 30 min at 55°C. Guanidinium isothiocyanate (Sigma) and glycogen (Roche) were added to final concentrations of 4.58 M and 0.47 mg/ml, respectively, and incubated at room temperature for 30 minutes. Nucleic acids were precipitated by centrifugation at 15,000 × g in the presence of a nearly equal volume of isopropanol. Nucleic acid pellets were washed repetitively with 70% ethanol and air dried before suspending in 5 mM Tris pH 8. One third of the sample was used as template in the Quantifiler Duo assay, one third was used for testing HIV-1 DNA and one third was stored.
Detection of HIV-1 DNA
HIV-1 DNA was detected using a modified version of the single copy assay with primers targeted to a conserved region in the integrase gene (iSCA) (25). Briefly, 10 μl of extracted DNA was diluted with 20 μl of 5 mM Tris pH 8 and run in triplicate in a reaction containing 1X Roche LightCyler 480 probes master mix, 400 nM of primers iSCA-F (5′-TTT GGA AAG GAC CAG CCA A -3′) and iSCA-r (5′-CCT GCC ATC TGT TTT CCA-3′) and 200 nM Taqman probe (5′-6FAM AAA GGT GAA GGG GCA GTA GTA ATA C BHQ_1-3′). DNA was amplified at 95°C for 5 min followed by 45 cycles of 95°C for 15 s and 60°C for 1 min on a LightCycler 480 (Roche). This assay can detect HIV-1 DNA at a single copy per well, as verified by Poisson’s distribution statistics in limiting dilution experiments of purified HIV-1 DNA target in mock swab cell pellets prepared using A431 cells spiked with a known quantity of purified HIV-1 DNA and human semen. False positive results were observed in 0/111 reactions using Tris buffer as a no template control. Each sample was run in triplicate and considered “undetectable” if all three reactions had no amplification, “detected, quantifiable” if at least 2/3 reactions had one or more copies detected per reaction and “detected, not quantifiable” if only 1/3 reactions had one or more copies detected.
Detection of Y Chromosomal DNA
Yc and total human DNA was detected using the commercially available Quantifiler Duo kit (Applied Biosystems). Reactions were prepared according to the manufacturers guidelines with the modification of increasing the total number of PCR cycles from 40 to 55 to allow late amplification (>35 cycles) to reach the plateau phase of PCR, and to observe if any cases of non-specific amplification occurred after 40 cycles. Despite these adjustments, all positive wells had cycle threshold (Ct) values of 40 cycles or below, consistent with the kit manufacturers guidelines. Wells with Ct values of 40–41 that had exponential amplification were included as positive only if the calculated copy number was 1 copy or above. No cases of false amplification between 41 and 55 cycles occurred. The assay can detect Yc DNA at a single copy per well, as verified by Poisson’s distribution statistics in limiting dilution experiments of semen in mock swab cell pellets. False positive Yc DNA was observed in 0/135 reactions using Tris buffer as a no template control and 0/65 reactions using A431 female epithelial cell pellets as a negative control. Positive detection using dilutions of semen was obtained in 89/89 wells with a predicted copy number of 2 or higher. Additional sensitivity and specificity information can be found in the Quantifiler Duo user’s manual. Each test sample was run in replicates of five, and considered “undetectable” if all five reactions had no amplification, “detected, quantifiable” if at least 3/5 reactions had one or more copies detected per reaction, and “detected, not quantifiable” if only one or two reactions had at least one copy detected per reaction. Total Human DNA, measured through the RPPH1 target as part of the Quantifiler Duo kit, was used to calculate approximate total cell numbers in each swab cell pellet and quantified according to kit manufacturer’s instructions.
Behavioral Data Collection and Analysis
Participants were queried on gel and condom use during the last coital act and during all coital acts in the last seven days at each quarterly visit, and data was collected by participant self-report (23). Detected, but not quantifiable Yc DNA samples were excluded from the analysis of Yc DNA detection with self-reported condom use due to the inferior quality of these samples.
Statistical Analysis
SPSS Version 20 (IBM Corp.) was used for all analyses. Conditional logisitic regression was used to compare the odds of being a seroconverter among those with detectable Yc DNA and those with no detectable Yc DNA. The association between Yc DNA detection (versus no detection) and reported sexual activity was assessed using Generalized Estimating Equation models with a binary link, robust errors and independent correlation structure.
RESULTS
Swab collection and cell recovery
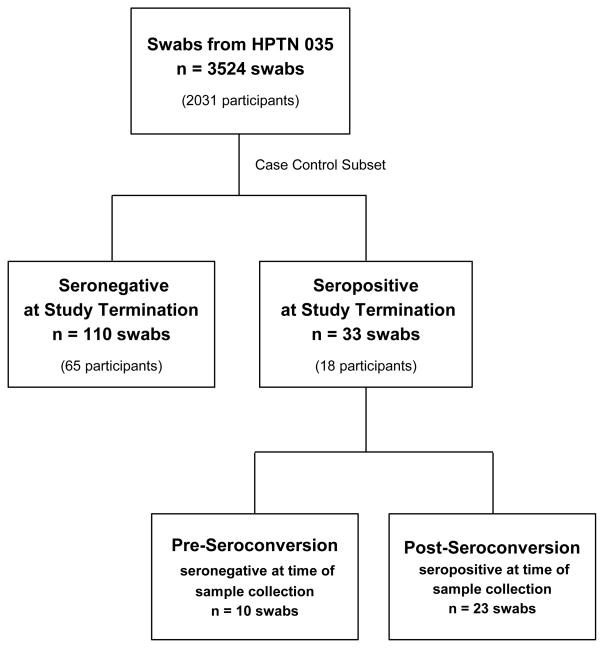
In the HPTN 035 study, 3524 vaginal swab specimens were collected from 2031 women from the African sites. A randomly selected subset of 110 swabs from 65 women who remained HIV negative throughout the study (controls) and 33 swabs from 18 women who seroconverted post-enrollment (cases) were evaluated for the presence of HIV and Y chromosomal DNA (Figure 1). Of 33 swabs from seroconverters, 10 were collected while the women were still HIV negative (prior to seroconversion) and 23 were collected after the participant was confirmed as having seroconverted. Cell recovery from swabs as assessed through total human DNA detection was a median of 3.0 × 105 cells/swab (IQR 1.4 × 105 to 6.1 × 106 cells/swab). ROC curve analysis demonstrated no minimal cell pellet size requirement for the detection of Yc DNA. The minimum cell number needed for HIV DNA recovery could not be determined due to small sample size (data not shown).
Figure 1.
Study Design
HIV-1 DNA detection
HIV-1 DNA was only detected in 10/23 swabs collected post-seroconversion. Five had quantifiable HIV-1 DNA with a median of 141 copies/sample and a range of 55 – 1593 copies/sample from swabs that were collected post-seroconversion, and 5 had HIV-1 DNA that was detected but not quantifiable (Table 1). HIV-1 DNA could not be detected in the 10 samples collected prior to seroconversion or in the 110 samples from women who remained HIV-1 negative throughout the study.
Table 1.
Detection of HIV-1 DNA in a subset of vaginal swabs from HPTN 035
| Seronegativea (n = 110) | Pre-Seroconversionb (n = 10) | Post-Seroconversion (n = 23) | ||
|---|---|---|---|---|
|
| ||||
| HIV-1 DNAc | n (%) | n (%) | n (%) | median (range) copies/sample |
| Undetectable | 110 (100%) | 10 (100%) | 13 (50%) | - |
| Detected, Quantifiable | 0 | 0 | 5 (25%) | 141 (55–1593) |
| Detected, Not Quantifiable | 0 | 5 (25%) | - | |
Swab sample collected from participant who remained seronegative at study termination.
Swab sample collected from participant who was seronegative at time of sample collection, but seropositive at study termination.
Each sample was run in triplicate and considered “undetectable” if all three reactions had no amplification, “detected, quantifiable” if at least 2/3 reactions had one or more copies detected per reaction and “detected, not quantifiable” if only 1/3 reactions had one or more copies detected.
Y Chromosomal DNA detection
All 143 swabs were evaluated for the presence of Yc DNA as a measure of unprotected sexual activity. Yc DNA was detected in 62/143 (43%) swabs. Forty-four of 62 samples had quantifiable Yc DNA with a median of 425 copies/sample and a range of 20 – 11926 copies/sample. Yc DNA was detected but not quantifiable in 18/62 (29%) samples (Table 2). Using conditional logistic regression with serostatus as the outcome, no significant differences in Yc DNA detection were found between the HIV seroconverter cases (5/33 [15%]) and HIV non-seroconverter controls (39/110 [35%]) in samples that had quantifiable Yc DNA (p = 0.1). This difference remained non-significant when samples collected prior to seroconversion were included as cases (OR 0.48; p = 0.3). There was no correlation between Yc and HIV-1 DNA detection in the sample set.
Table 2.
Detection of Y Chromosomal DNA in a subset of vaginal swabs from HPTN 035
| Seronegativea (n = 110) | Seropositiveb (n = 33) | |||
|---|---|---|---|---|
|
| ||||
| Yc DNAc | n (%) | median (range) copies/sample |
n (%) | median (range) copies/sample |
| Undetectable | 62 (56%) | - | 19 (58%) | - |
| Detected, Quantifiable | 39 (35%) | 413 (16–7003) | 5 (15%) | 626 (74–11926) |
| Detected, Not Quantifiable | 9 (8%) | - | 9 (27%) | - |
Swab sample collected from participant who remained seronegative at study termination.
Swab sample collected from participant who was seropositive at study termination. 10 samples were collected pre-seroconversion, and 23 samples were collected post-seroconversion.
Each sample was run in replicates of five, and considered “undetectable” if all five reactions had no amplification, “detected, quantifiable” if at least 3/5 reactions had one or more copies detected per reaction, and “detected, not quantifiable” if only one or two reactions had at least one copy detected per reaction.
Association of Yc DNA Detection with Self-Reported Condom Use
Participant responses to frequency of coitus and condom use were also collected at 126 out of the 143 visits at which a swab sample was collected. There were 27 (21%) reports of no vaginal sex in the week prior to sample collection, 83 (66%) reports of vaginal sex in the past week with 100% condom use, and 16 (13%) reports of vaginal sex in the past week with less than 100% condom use. Excluding samples that were not quantifiable, the proportion of participants with Yc DNA increased as reported condom use decreased. Only 4/27 (15%) samples from participants reporting no coitus had Yc DNA (median 98 copies), compared to 27/83 (33%) samples with Yc DNA (median 518 copies) from participants reporting vaginal sex in the past week with 100% condom use, and 9/16 (56%) samples from participants with Yc DNA (median 568 copies) reporting vaginal sex in the past week with <100% condom use. The proportion of samples with Yc DNA was significantly higher (OR 10.69; p=0.003) among those participants with inconsistent condom use compared to those reporting no coitus in the past week (Table 3). Reporting of feminine hygiene practices did not correlate with Yc DNA detection (data not shown).
Table 3.
Correlation of Y Chromosomal DNA with Self-Reported Condom Usage
| Yc DNA | Univariate Analysis | |||
|---|---|---|---|---|
| Not detected | Detected, Quantifiable (%)a | OR (95% CI) | P-value | |
| No vaginal sex in past week (n = 27) | 19 (70%) | 4 (15%) | 1 | - |
| Vaginal sex in past week with 100% condom use (n = 83) | 44 (53%) | 27 (33%) | 2.92 (0.93, 9.13) | 0.066 |
| Vaginal sex in past week with <100% condom use (n = 16) | 4 (25%) | 9 (56%) | 10.69 (2.27, 50.32) | 0.003 |
Samples that were detected but not quantifiable were excluded from the analysis.
DISCUSSION
The advent of new, highly sensitive quantitative PCR technologies for nucleic acid detection and quantification in genital specimens enabled us to evaluate the feasibility of using HIV and Yc DNA detection as a biomarker for risk behavior in HIV prevention clinical trials. We quantified HIV and Yc DNA from the same vaginal swab and stratified the results against behavioral data on condom use from women both pre- and post-seroconversion, and from women who never seroconverted in the trial period. The operator performing the assays was blinded as to the subgroups to which the participants belonged.
Despite having an assay with sensitivity down to a single copy of HIV-1 DNA per PCR reaction, we did not find evidence of HIV-1 in specimens from seronegative women, including from those women who eventually seroconverted. The inability to detect seminal viral DNA (from an infected male partner) in a vaginal swab sample could have been influenced by sample quality, timing of swab sample collection from last coital act, and lack of frequent exposure to HIV. Further, CD4+ lymphocytes only comprise 2% of the total cell number in an average semen sample and only 0.1% of CD4+ T cells carry provirus in an HIV-infected individual. The number of absolute CD4+ lymphocytes in semen can also depend on the health of the individual and decrease dramatically in viremic patients (26). Of note, the maximum number of Yc DNA copies from any swab in our study was 1000 copies/well, with the majority 134/143 (94%) having Yc copies of 100 or less meaning that only a small fraction of semen was being detected. Because so little of Yc DNA is being recovered from an ejaculate it is very unlikely that rare HIV-infected would have been detected.
We also did not observe a difference in frequency of Yc DNA detection in seroconverters compared to non-seroconverters (15% versus 35%), and the number of copies of Yc detected did not predict risk of seroconversion. Walsh et al. showed that there was a significant difference in levels of PSA and sperm counts with different types of risk exposures such as condom breakage, but a similar analysis using Yc DNA has not been done. There are several explanations for Yc DNA not predicting seroconversion including variation in donor sperm count, variable time between last coitus and sample collection, non-coital exposure (e.g. digital) (27), and limited sampling of seroconverters. Larger observations over longer periods of time may reveal an association of Yc DNA exposure and risk of HIV or other sexually-transmitted infections.
Our data does provide further evidence that Yc chromosomal DNA detection serves as a reliable biomarker to monitor sexual activity (28). A significantly higher proportion of women (p = 0.003) reporting unprotected sex in the past week had detectable Yc DNA compared to women reporting no vaginal sex in past week. Interestingly, 33% of women reporting 100% condom use had detectable Yc DNA, with copy numbers at similar levels to those women reporting <100% condom use (518 versus 568 copies), suggesting that condom usage was over-reported in this study or that exposure occurred prior to the use of a condom. The behavioral questionnaire was administered through face-to-face interviews, which may contribute to inconsistent reporting particularly for sensitive topics (29). Our data shows promise for the use of Yc DNA as an objective measure for condom use. Further study is needed to determine if level of risk or type of risk behavior can be correlated with number of Yc copies detected.
One limitation of this study is that testing for HIV and Yc DNA was done retrospectively on stored swabs from quarterly sampling, where timing of swab collection after coitus and the HIV infection status of the male partner was not known. Testing a larger number of pre-seroconversion swabs or swabs from serodiscordant couples could provide further insight into the feasibility of using HIV or Yc DNA as a biomarker. Self-collected samples could provide the best timing for detecting HIV exposure, but would rely on the ability and willingness of participants to collect high quality samples. More frequent sample collection could provide risk information to statisticians for refined secondary analysis of clinical trial data in populations of highest risk. Modifying the assay for HIV detection to include HIV-1 RNA or total nucleic acid detection may improve sensitivity.
In summary, we demonstrated that by using highly sensitive quantitative PCR assays, Yc DNA and total HIV-1 DNA can be detected down to a single copy in vaginal swab samples. Yc DNA detection is more frequent among women reporting <100% condom use with coitus but can also be detected in a third of women reporting 100% condom usage. These results suggest that Yc DNA detection in vaginal fluids could refine assessments of HIV risk and efficacy of preventive strategies.
Acknowledgments
Financial support. This work was supported by the Bill and Melinda Gates Foundation (grant 48293) and the Microbicide Trials Network, which is cofunded by the National Institute of Allergy and Infectious Diseases (grant UM1 AI068633), the National Institute of Child Health and Development, and the National Institute of Mental Health, all of the National Institutes of Health.
Footnotes
Potential conflicts of interest. JWM is a consultant for Gilead Sciences and owns shares of RFS Pharmaceuticals. No other authors have reported conflicts.
Previous Presentation. This work was partially presented at the 20th Conference on Retroviruses and Opportunistic Infections in Atlanta, Georgia, March 3–7, 2013.
References
- 1.Mauck CK, Doncel GF. Biomarkers of semen in the vagina: applications in clinical trials of contraception and prevention of sexually transmitted pathogens including HIV. Contraception. 2007;75(6):407–19. doi: 10.1016/j.contraception.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Mauck CK. Biomarkers for evaluating vaginal microbicides and contraceptives: discovery and early validation. Sex Transm Dis. 2009;36(3 Suppl):S73–5. doi: 10.1097/OLQ.0b013e3181994155. [DOI] [PubMed] [Google Scholar]
- 3.Anglewicz P, Gourvenec D, Halldorsdottir I, et al. The effect of interview method on self-reported sexual behavior and perceptions of community norms in Botswana. AIDS Behav. 2013;17(2):674–87. doi: 10.1007/s10461-012-0224-z. [DOI] [PubMed] [Google Scholar]
- 4.Pool R, Montgomery CM, Morar NS, et al. Assessing the accuracy of adherence and sexual behaviour data in the MDP301 vaginal microbicides trial using a mixed methods and triangulation model. PLoS One. 2010;5(7):e11632. doi: 10.1371/journal.pone.0011632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marrazzo J, Ramjee G, Nair G, et al. Pre-exposure prophylaxis for HIV in women: daily oral tenofovir, oral tenofovir/emtricitabine or vaginal tenofovir gel in the VOICE study (MTN 003). 20th Conference on Retroviruses and Opportunistic Infections; Atlanta, GA. 2013. [Google Scholar]
- 6.Thigpen MC, Kebaabetswe PM, Paxton LA, et al. Antiretroviral Preexposure Prophylaxis for Heterosexual HIV Transmission in Botswana. N Engl J Med. 2012;367(5):423–34. doi: 10.1056/NEJMoa1110711. [DOI] [PubMed] [Google Scholar]
- 7.Van Damme L, Corneli A, Ahmed K, et al. Preexposure prophylaxis for HIV infection among African women. N Engl J Med. 2012;367(5):411–22. doi: 10.1056/NEJMoa1202614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minnis AM, Steiner MJ, Gallo MF, et al. Biomarker validation of reports of recent sexual activity: results of a randomized controlled study in Zimbabwe. Am J Epidemiol. 2009;170(7):918–24. doi: 10.1093/aje/kwp219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahamondes L, Diaz J, Marchi NM, Castro S, Villarroel M, Macaluso M. Prostate-specific antigen in vaginal fluid after exposure to known amounts of semen and after condom use: comparison of self-collected and nurse-collected samples. Hum Reprod. 2008;23(11):2444–51. doi: 10.1093/humrep/den283. [DOI] [PubMed] [Google Scholar]
- 10.Jamshidi R, Penman-Aguilar A, Wiener J, et al. Detection of two biological markers of intercourse: prostate-specific antigen and Y-chromosomal DNA. Contraception. 2013;88(6):749–57. doi: 10.1016/j.contraception.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Macaluso M, Lawson L, Akers R, et al. Prostate-specific antigen in vaginal fluid as a biologic marker of condom failure. Contraception. 1999;59(3):195–201. doi: 10.1016/s0010-7824(99)00013-x. [DOI] [PubMed] [Google Scholar]
- 12.Abbott SA, Friedland BA, Sarna A, et al. An evaluation of methods to improve the reporting of adherence in a placebo gel trial in Andhra Pradesh, India. AIDS Behav. 2013;17(6):2222–36. doi: 10.1007/s10461-012-0402-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simson Oechsle C, Haddad S, Sgueglia JB, Grgicak CM. Screening Biological Stains with qPCR versus Lateral Flow Immunochromatographic Test Strips: A Quantitative Comparison using Analytical Figures of Merit. J Forensic Sci. 2014;59(1):199–207. doi: 10.1111/1556-4029.12284. [DOI] [PubMed] [Google Scholar]
- 14.Brotman RM, Melendez JH, Smith TD, Galai N, Zenilman JM. Effect of menses on clearance of Y-chromosome in vaginal fluid: implications for a biomarker of recent sexual activity. Sex Transm Dis. 2010;37(1):1–4. doi: 10.1097/OLQ.0b013e3181b5f15d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zenilman JM, Yuenger J, Galai N, Turner CF, Rogers SM. Polymerase chain reaction detection of Y chromosome sequences in vaginal fluid: preliminary studies of a potential biomarker for sexual behavior. Sex Transm Dis. 2005;32(2):90–4. doi: 10.1097/01.olq.0000149668.08740.91. [DOI] [PubMed] [Google Scholar]
- 16.Melendez JH, Giles JA, Yuenger JD, et al. Detection and quantification of Y-chromosomal sequences by real-time PCR using the LightCycler system. Sex Transm Dis. 2007;34(8):617–9. doi: 10.1097/01.olq.0000258336.65285.31. [DOI] [PubMed] [Google Scholar]
- 17.Ghanem KG, Melendez JH, McNeil-Solis C, et al. Condom use and vaginal Y-chromosome detection: the specificity of a potential biomarker. Sex Transm Dis. 2007;34(8):620–3. doi: 10.1097/01.olq.0000258318.99606.d9. [DOI] [PubMed] [Google Scholar]
- 18.Basu J, Romney SL, Angeletti RH, et al. Human immunodeficiency virus (HIV) antigens and RNA in HIV-seronegative women with cervical intraepithelial neoplasia. AIDS Res Hum Retroviruses. 2009;25(3):249–59. doi: 10.1089/aid.2008.0096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mermin JH, Holodniy M, Katzenstein DA, Merigan TC. Detection of human immunodeficiency virus DNA and RNA in semen by the polymerase chain reaction. J Infect Dis. 1991;164(4):769–72. doi: 10.1093/infdis/164.4.769. [DOI] [PubMed] [Google Scholar]
- 20.Ball JK, Curran R, Irving WL, Dearden AA. HIV-1 in semen: determination of proviral and viral titres compared to blood, and quantification of semen leukocyte populations. J Med Virol. 1999;59(3):356–63. doi: 10.1002/(sici)1096-9071(199911)59:3<356::aid-jmv16>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Dornadula G, Beumont M, et al. Human immunodeficiency virus type 1 in the semen of men receiving highly active antiretroviral therapy. N Engl J Med. 1998;339(25):1803–9. doi: 10.1056/NEJM199812173392502. [DOI] [PubMed] [Google Scholar]
- 22.Lambert-Niclot S, Tubiana R, Beaudoux C, et al. Detection of HIV-1 RNA in seminal plasma samples from treated patients with undetectable HIV-1 RNA in blood plasma on a 2002–2011 survey. AIDS. 2012;26(8):971–5. doi: 10.1097/QAD.0b013e328352ae09. [DOI] [PubMed] [Google Scholar]
- 23.Abdool Karim SS, Richardson BA, Ramjee G, et al. Safety and effectiveness of BufferGel and 0.5% PRO2000 gel for the prevention of HIV infection in women. AIDS. 2011;25(7):957–66. doi: 10.1097/QAD.0b013e32834541d9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dezzutti CS, Richardson BA, Marrazzo JM, et al. Mucosal Escherichia coli bactericidal activity and immune mediators are associated with HIV-1 seroconversion in women participating in the HPTN 035 trial. J Infect Dis. 2012;206(12):1931–5. doi: 10.1093/infdis/jis555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cillo AR, Krishnan A, Mitsuyasu RT, et al. Plasma viremia and cellular HIV-1 DNA persist despite autologous hematopoietic stem cell transplantation for HIV-related lymphoma. J Acquir Immune Defic Syndr. 2013;63(4):438–41. doi: 10.1097/QAI.0b013e31828e6163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Politch JA, Mayer KH, Anderson DJ. Depletion of CD4+ T cells in semen during HIV infection and their restoration following antiretroviral therapy. J Acquir Immune Defic Syndr. 2009;50(3):283–9. doi: 10.1097/QAI.0b013e3181989870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walsh TL, Frezieres RG, Peacock K, et al. Use of prostate-specific antigen (PSA) to measure semen exposure resulting from male condom failures: implications for contraceptive efficacy and the prevention of sexually transmitted disease. Contraception. 2003;67(2):139–50. doi: 10.1016/s0010-7824(02)00478-x. [DOI] [PubMed] [Google Scholar]
- 28.Jadack RA, Yuenger J, Ghanem KG, Zenilman J. Polymerase chain reaction detection of Y-chromosome sequences in vaginal fluid of women accessing a sexually transmitted disease clinic. Sex Transm Dis. 2006;33(1):22–5. doi: 10.1097/01.olq.0000194600.83825.81. [DOI] [PubMed] [Google Scholar]
- 29.Phillips AE, Gomez GB, Boily MC, Garnett GP. A systematic review and meta-analysis of quantitative interviewing tools to investigate self-reported HIV and STI associated behaviours in low- and middle-income countries. Int J Epidemiol. 2010;39(6):1541–55. doi: 10.1093/ije/dyq114. [DOI] [PubMed] [Google Scholar]