SUMMARY
Hematopoietic stem and progenitor cells (HSPCs) can reconstitute and sustain the entire blood system. We generated a highly specific transgenic reporter of HSPCs in zebrafish. This allowed us to perform high-resolution live imaging on endogenous HSPCs not currently possible in mammalian bone marrow. Using this system we have uncovered distinct interactions between single HSPCs and their niche. When an HSPC arrives in the perivascular niche, a group of endothelial cells remodel to form a surrounding pocket. This structure appears conserved in mouse fetal liver. Correlative light and electron microscopy revealed that endothelial cells surround a single HSPC attached to a single mesenchymal stromal cell. Live imaging showed mesenchymal stromal cells anchor HSPCs and orient their divisions. A chemical genetic screen found the compound lycorine promotes HSPC-niche interactions during development and ultimately expands the stem cell pool into adulthood. Our studies provide evidence for dynamic niche interactions upon stem cell colonization.
INTRODUCTION
Hematopoietic stem and progenitor cells (HSPCs) self-renew and give rise to all blood cell types. Definitive HSPCs arise from the hemogenic endothelium of the dorsal aorta (DA) (Bertrand et al., 2010; Boisset et al., 2010; Kissa and Herbomel, 2010), are released into circulation, and then seed an intermediate hematopoietic niche before colonizing the adult marrow. In mammals, this intermediate tissue is the fetal liver (FL), and in zebrafish it is the caudal hematopoietic tissue (CHT), a vascular plexus in the ventral tail of the embryo (Murayama et al., 2006; Orkin and Zon, 2008). After rapid expansion in the intermediate niche, HSPCs will leave and go on to seed the adult marrow, which in mammals is bone and in zebrafish is kidney (Traver et al., 2003).
The adult niche is a complex microenvironment that maintains and regulates HSPCs throughout life. The bone marrow contains a complex network of sinusoidal vessels that act as an interface between circulation and the niche. Most HSPCs are proximal to these vessels and are considered to be in a perivascular niche (Kiel et al., 2005; Nombela-Arrieta et al., 2013). Studies have shown that endothelial cells (ECs) have distinct properties that enable them to support and expand associated HSPCs (Butler et al., 2010; Hooper et al., 2009). However, the perivascular niche is not limited to ECs and many other cell types also play a role, including mesenchymal stromal cells, osteoblasts, arterioles and sympathetic nerves (Morrison and Scadden, 2014). Stromal cells are likely heterogenous throughout the bone marrow and provide HSPC maintenance factors such as CXCL12 and KITLG (Ding and Morrison, 2013; Ding et al., 2012; Greenbaum et al., 2013; Méndez-Ferrer et al., 2010; Sugiyama et al., 2006). HSPCs in the bone marrow have been observed in a number of elegant studies (Köhler et al., 2009; Lo Celso et al., 2009; Xie et al., 2009). Primarily these studies used multiphoton intravital microscopy to locate transplanted HSPCs in surgically accessed bone or bone explants. A high resolution and dynamic live view of the physical interactions between endogenous cell types in the niche has not been achieved.
We have developed a transgenic zebrafish line to observe the migration and behavior of endogenous HSPCs. Conserved hematopoietic regulatory genes have led to the development of HSPC transgenic reporter lines, although none of these are entirely specific (Lin et al., 2005; North et al., 2007). To establish a more specific HSPC line, we utilized a regulatory element from the first intron of the mouse Runx1 locus (+23 kb downstream of the P1 promoter) to drive expression of a marker (Nottingham et al., 2007). The Runx1+23 enhancer from mouse marks definitive HSPC in the zebrafish in all sites of definitive hematopoiesis and has been confirmed by long-term transplantation.
The ability to track endogenous HSPCs in the live embryo allowed us to observe dynamic interactions with the niche. We discovered a cellular behavior that involves triggered remodeling of perivascular ECs upon arrival of an HSPC in a new site of hematopoiesis. We also show that mesenchymal stromal cells can anchor and orient the division plane of an HSPC. Using correlative light and electron microscopy, we reveal the high-resolution ultrastructure of a single HSPC in the perivascular niche after live tracking and lodgement. Lastly, we used a chemical genetic approach to modulate the HSPC-niche interactions observed in the embryo. These interactions lead to long-term changes in the size of the stem cell pool into adulthood. Our studies suggest that the niche reacts to the arrival of stem cells.
RESULTS
Establishment of a highly specific HSPC transgenic zebrafish line
To observe and study endogenous HSPCs, we established transgenic zebrafish lines with the mouse Runx1+23 enhancer driving either cytoplasmic EGFP (Runx:GFP) or nuclear localized NLS-mCherry (Runx:mCherry). These two lines were crossed to demonstrate that the green and red fluorescent proteins marked mostly the same cell populations, with the Runx:mCherry line showing broader expression in progenitors (Figure S1A,B). Time-lapse live imaging showed that Runx+ cells emerge from the hemogenic endothelium of the DA (Figure 1A; Movie S1). Intercrossing the Runx:mCherry line with cmyb:EGFP and cd41:EGFP lines showed that Runx+ cells marked an overlapping population in all major hematopoietic sites of the embryo, including the DA, CHT, thymus and kidney (Figure S1C-I, data not shown). We detected Runx+ cells in the adult kidney marrow using flow cytometry and immunohistochemistry (Figure S1J-L).
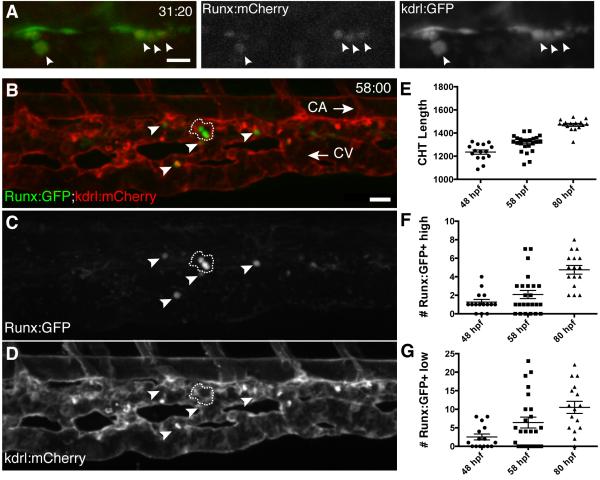
Figure 1. A transgenic zebrafish line that specifically marks HSPCs.
(A) Single frame of time-lapse (hours post fertilization:minutes) showing Runx:mCherry+ HSPCs (arrowheads; red nuclei) budding from kdrl:GFP+ hemogenic endothelium of DA (green). See Movie S1. (B-D) CHT is colonized by Runx:GFP+ HSPCs (green) that are closely associated with kdrl:mCherry+ ECs (red). Caudal artery (CA) is dorsal to CHT, caudal vein (CV) is ventral, circulation runs posterior (right arrow) and anterior (left arrow), respectively. Cluster of 3 Runx:GFP+ high cells outlined with dashed line and 4 Runx:GFP+ low cells indicated with arrowheads. 58 hpf embryo. (E-G) Runx:GFP+ high and low cells quantified, with CHT length as indicator of stage (anterior from cloaca to posterior limit of CA). Note: Confocal images are 3D rendered depth or max projections of 20-30 μm z-stacks. Scale bars: (A) 15 μm; (B) 25 μm. Error bars show mean ± SEM. See also Figure S1.
Next, we followed newly born HSPCs as they colonized the CHT niche. We quantified the number of Runx:GFP+ cells in the CHT at different developmental stages. We found there were an average of 1, 2, or 5 Runx:GFP+ high cells (and 2, 6, or 10 Runx:GFP+ low cells) in the CHT at 48, 58, and 80 hours post fertilization (hpf), respectively (Figure 1B-G). Cell numbers in the Runx:mCherry line were comparable, except for greater expansion of Runx:mCherry+ low cells from 72 hpf (Figure S1A-B). Looking at Runx:GFP+ cells in the CHT together with a vascular reporter, we found that HSPCs were closely associated with ECs of the caudal vein plexus (Figure 1B-D). Both cell numbers and localization of Runx:GFP+ cells are consistent with previous lineage tracing experiments of HSPC from the hemogenic endothelium to the CHT (Kissa et al., 2008).
The functional stem cell characteristics of Runx+ HSPCs in the zebrafish
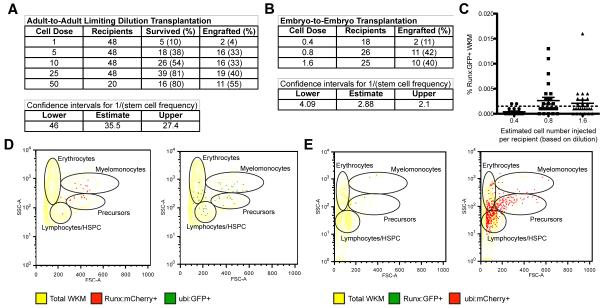
To assess the functional characteristics of cells in the Runx+ pool, we performed limiting dilution transplantation as a standard assay to determine hematopoietic stem cell content. We purified Runx:mCherry+ cells from the kidney marrow of adult transgenic donors. These donors also carried the ubiquitous ubi:GFP transgene (Mosimann et al., 2011) so that multilineage contribution could be assessed if the Runx:mCherry HSPC enhancer was down-regulated upon differentiation. Sorted double positive cells were diluted in a range between 1 and 50 then transplanted into irradiated recipients (Figure 2A). Survival rates improved in recipients that received a greater number of donor HSPCs, and at 3 months post-transplantation the kidney marrow of recipients was dissected for flow cytometry analysis. Runx:mCherry+ cells were present in the kidney marrow and had contributed ubi:GFP+ progeny to all lineages (Figure 2A,D). Statistical analysis of engraftment over a range of cell doses estimated a stem cell frequency of approximately 1 in 35 (Figure 2A) (Hu and Smyth, 2009). We consider this to be an underestimate because the donors and recipients were not immune-matched; previous work from our lab showed that immune matching can improve hematopoietic transplantation in zebrafish (de Jong et al., 2011). The Runx+ cell pool can be sorted with a single transgenic marker, and no additional labeling of cell surface markers, to a purity that is within the range of the well-characterized KSL (c-Kit+ Sca-1+ Lin−) population in mouse (Osawa et al., 1996). Based on the ability of a small number of adult Runx+ cells to engraft long-term, self-renew, and produce all lineages, we have demonstrated there is substantial stem cell content within this pool of cells.
Figure 2. Functional HSPCs are highly purified using a single transgenic marker.
Summary of results from (A) adult-to-adult and (B) embryo-to-embryo limiting dilution transplantation experiments. (C) Embryo-to-embryo transplantation recipients with engraftment of Runx:GFP+ cells in kidney marrow at 3 months (above background; >0.001%). Representative kidney marrow flow cytometry analysis of (D) adult-to-adult transplantation recipient with Runx:mCherry+ HSPCs and ubi:GFP+ lineages, and (E) embryo-to-embryo transplantation recipient with Runx:GFP+ HSPCs and ubi:mCherry+ lineages. Error bars show mean ± SEM. See also Figure S2.
We also wanted to evaluate the stem cell characteristics of the Runx+ population in the embryo. Studies have shown that hematopoietic stem cells isolated from different tissues of the mouse embryo have the capacity to contribute to hematopoiesis when transplanted into recipient embryos (Fleischman and Mintz, 1979) or adults (Medvinsky and Dzierzak, 1996; Müller et al., 1994). Based on a previous approach (Traver et al., 2003), we have further developed an HSPC embryo-to-embryo transplantation assay in zebrafish. Runx+ cells were sorted from a pool of 3 days post fertilization (dpf) double transgenic Runx+ and ubi+ donor embryos because sufficient cell numbers could be collected at that stage. Only 1-2 cells were injected into the circulation of a wild-type recipient embryo at 2 dpf. A dilution series established the number of cells injected for each experiment. At 2 dpf the CHT is being colonized by endogenous HSPCs but the thymus has not formed, allowing introduction of exogenous cells without the possibility of immune rejection. Recipient embryos are then raised to adulthood and their kidney marrow is analyzed by flow cytometry for engraftment at 3-5 months. We scored engraftment as any detectable Runx+ cells above background (Figure 2B,C). This rationale was chosen because approximately one donor HSPC will be competing with endogenous stem cells in an unconditioned wild-type recipient embryo—there is no precedent to predict chimerism in this scenario. The transplanted cells must seed the CHT, migrate to the kidney, and persist into adulthood where they will self-renew and contribute to all lineages.
We identified recipients with Runx+ HSPCs and ubi+ progeny in the kidney marrow, as well as some that had ubi+ cells in the peripheral blood (Figure 2E, data not shown). Statistical analysis estimated the stem cell frequency of the Runx+ population in the 3 dpf embryo to be approximately 1/2.88 cells (Figure 2B). These results were representative of three independent experiments—two with Runx:GFP;ubi:mCherry double transgenic lines and a third with the opposite Runx:mCherry;ubi:GFP transgenic combination (Figure S2). Together, our limiting dilution adult and embryo transplantation assays demonstrate the Runx+ populations contain functional stem cells based on long-term multilineage engraftment. Our data also suggests that some Runx+ cells are progenitors, based on: 1) overlap with transgenic reporters that mark both stem and progenitor cells (cmyb:EGFP and cd41:EGFP; Figure S1); 2) a Runx+ low population that appears to expand from Runx+ high cells (Figures 1, S1, data not shown); 3) flow cytometry analysis of adult kidney marrow that shows Runx+ cells in gates that contain progenitors (Figures 2D,E and S2C). There are no antibodies available in zebrafish to further purify these populations, or to distinguish between a stem and progenitor cell, so we will operationally define engrafting Runx+ cells as HSPCs in this study.
Dynamic visualization of stem cell colonization of the CHT niche
To directly observe the interaction of HSPCs with surrounding ECs during CHT colonization, we performed time-lapse live imaging with spinning-disk confocal microscopy. We were able to routinely acquire image series with high temporal resolution (1-2 minutes per 2-channel confocal z-stack) for up to 16 continuous hours. A widefield image was acquired using a 20x objective because the rare lodgement of a single HSPC could occur anywhere in the length of the CHT (Figure 1B). We observed HSPC arrival in the CHT via circulation, followed by adherence to endothelial walls (Figure S3A; Movie S2). Next, cells underwent rapid extravasation to the abluminal side of the endothelial wall (<5 minutes; Figure S3A). Once HSPCs lodged in the CHT, we made a striking observation: a small group of EC actually remodeled around a single HSPC to form a stem cell pocket, which we call “endothelial cuddling” (Figures 3A, S3B; Movie S2). Within the 12-16 hour limit of time-lapse acquisition, an HSPC would make one of three cell division decisions: 1) symmetric (defined as two daughter cells that remain in the pocket); 2) asymmetric (defined as one daughter cell remaining in the pocket and the other migrating out); 3) or no division. In this example the division is asymmetrical, with one daughter cell crawling out of the endothelial niche and the other remaining (Figure S3C; Movie S2).
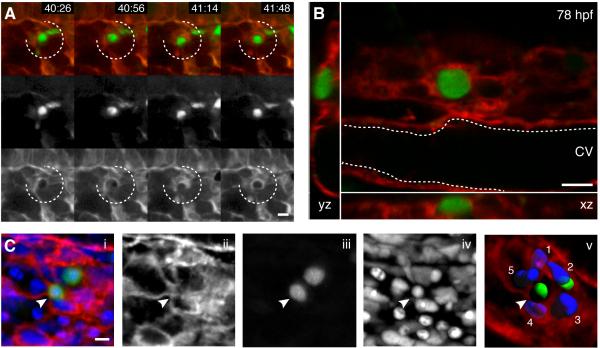
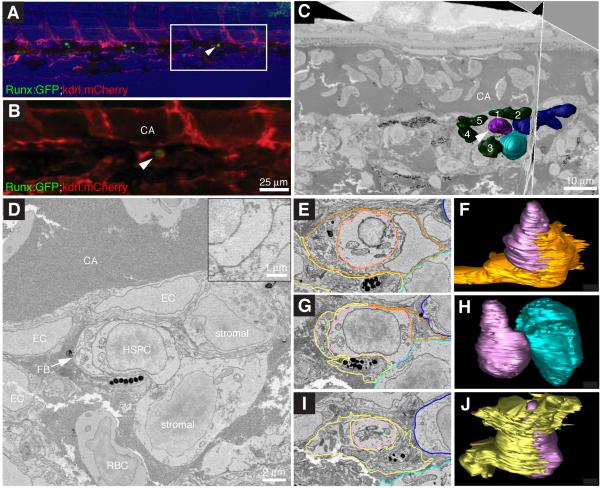
Figure 3. Endothelial cells in the perivascular niche remodel to surround a single HSPC.
(A) 4 frames from time-lapse Movie S2 between 40-42 hpf (hours post fertilization:minutes). Upper row is merge of Runx:GFP+ HSPC (green, middle row) and kdrl:RFP ECs (red, lower row). A group of surrounding ECs (broken circle) remodel around a single HSPC soon after its arrival. (B) Higher magnification (60x) live image of single Runx:GFP+ HSPC surrounded by kdrl:mCherry ECs at 78 hpf (orthogonal views). (C) 3D rendered projection of scanning confocal image from fixed 80 hpf embryo. (i) merge, (ii) kdrl:mCherry ECs, (iii) Runx:GFP+ HSPCs, (iv) DRAQ5 nuclei, (v) kdrl:mCherry projection with 3D modeled green HSPCs and 5 blue surrounding EC nuclei (arrowhead indicates HSPC in EC surround). All views: dorsal up, ventral down. Scale bars: 10 μm. See also Figure S3.
After HSPC lodgement in the CHT, we used higher magnification (40-60x) to better detail the close association and contact of surrounding ECs (Figure 3B). To quantify the number of surrounding ECs, we briefly fixed and stained embryos with a nuclear dye, then imaged using scanning confocal microscopy (Figure 3C). The ECs in contact with a single HSPC were outlined with membrane-bound mCherry and their nuclei were counted. In each structure we typically observed 5-6 ECs surrounding a single HSPC. Time-lapse live imaging of endogenous Runx+ HSPCs in the embryo has revealed striking interactions with perivascular ECs in the niche microenvironment.
Endothelial niche remodeling is conserved in mammalian fetal liver
We sought to establish if the distinct endothelial niche structure identified in the zebrafish CHT was also found in mammals. The equivalent tissue is the FL as it is the first tissue colonized by definitive HSPCs from the DA. The FL is an intermediate site of hematopoiesis where HSPCs expand before leaving to colonize the adult marrow and it produces the majority of blood during development (Morrison et al., 1995; Sánchez et al., 1996). To examine the earliest stages of FL colonization, we dissected E11.5 FLs from Ly6a-GFP (Sca-1) mice, which have GFP+ HSPC (Ma et al., 2002). Together with HSPC marker Runx1, and EC marker VE-cadherin (Cdh5), we found Ly6a-GFP+/Runx1+ HSPCs in one of 3 compartments in the FL (Figure 4A-C; Movie S3A-C): 1) an abluminal space with no EC contact; 2) adherent to ECs on one side; 3) inside an EC pocket. The similarity of a single HSPC surrounded by a small group of ECs in both the mouse FL and zebrafish CHT (compare Figure 3B and 4C), suggests that this is a conserved cellular structure that forms during stem cell lodgement in hematopoietic niches.
Figure 4. Endothelial cells surround HSPCs in the fetal liver microenvironment.
(A-C) FLs from E11.5 Ly6a-GFP mice were fixed and stained for immunofluorescence with anti-VE-Cadherin (red), anti-Runx1 (blue), and anti-GFP (green) antibodies. We scored 59 Ly6a-GFP+/Runx1+ cells from 3 FLs and identified three different HSPC-EC configurations: (A) abluminal with no contact between HSPC and ECs (18/59; 30%); (B) EC contact on one side of the HSPC (27/59; 46%); (C) HSPC surrounded on all sides with ECs (14/59; 24%). See Movie S3. (D) c-kit+ cell (magenta) adhered to CD31+ ECs (green) in one lobe of an E11.5 FL (arrowhead). White box marks details below. Time-lapse frames show in <90 minutes the HSPC migrates into a field of ECs. Soon after, ECs surround HSPC to form niche. See Movie S4. (E) c-kit+(blue)/Ly6a-GFP+(green) HSPC adhered to abluminal side of CD31+ EC (red). Following this cell >2 hours (1 frame/5 minutes) shows a division with distal and proximal daughters relative to sinusoid, the latter remains in an endothelial surround. See Movie S5. Confocal images: 3D rendered depth projection (A,B,C,E), orthogonal view (A,B,C below), maximum projection (D) of z-stack. Scale bars: 10 μm. See also Figure S4.
The identification of a potentially conserved HSPC-endothelial niche structure raised the possibility that HSPCs also trigger a dynamic remodeling of ECs during colonization of the FL. The FL tissue in mouse is not directly accessible to confocal microscopy, so we instead applied a protocol for live imaging of embryo explants (Boisset et al., 2010). We dissected wild-type and Ly6a-GFP E11.5 FLs, soaked the explant FL in fluorescently conjugated Kit (c-kit) and Pecam1 (CD31) antibodies then immediately performed live imaging for up to 4 hours. With the caveat that the marker profile of early E11.5 FL HSPCs has not been well defined, and that many c-kit+ cells in the FL are progenitors, we have observed hematopoietic and EC interactions that are strikingly similar to those in the zebrafish CHT (compare Figure 3A and 4D). We observed c-kit+ hematopoietic cells adhered to the sinusoidal network of CD31+ ECs (Figure 4D-E and S4). We followed a c-kit+ cell attached to the sinusoid as it migrated into and was surrounded by a small group of ECs (Figure 4D; Movie S4). Tracking a Ly6a-GFP+/c-kit+ cell in another explant, we observed one cell division (Figure 4E). Intriguingly, the daughter cell proximal to the sinusoid remained surrounded by ECs, while the daughter cell distal to the sinusoid migrated away into the abluminal space (Figure 4E; Movie S5). In another time-lapse sequence, a c-kit+ cell was observed undergoing discrete steps towards lodgement: adherence, extravasation, abluminal migration, and endothelial niche remodeling (Figure S4A-C). Even though FL explants have been removed from circulation and are imaged ex vivo, our live imaging data strongly suggest an evolutionary conserved process of dynamic EC remodeling around a single hematopoietic cell in a site of hematopoiesis.
Stem cell divisions are oriented relative to mesenchymal stromal cells
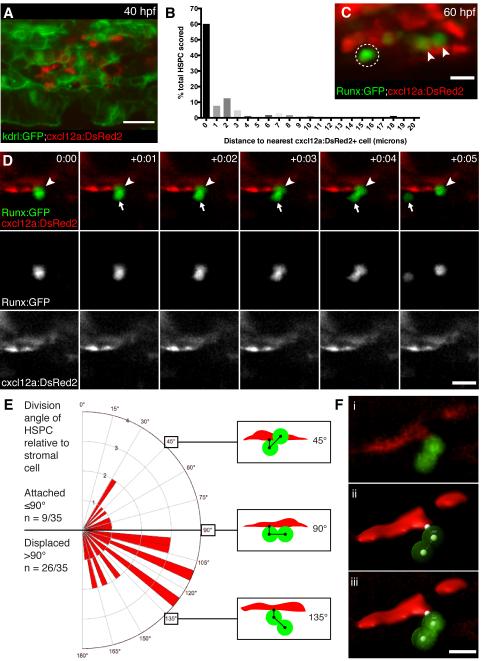
To further understand the interaction of HSPCs with the perivascular niche, we identified a cxcl12a:DsRed2 transgenic line that allowed us to observe mesenchymal stromal cells in the CHT (Glass et al., 2011). These are likely the “fibroblastic reticular cells” that were previously described in the CHT (Murayama et al., 2006) and similar to the “CXCL12-abundant reticular” cells in the perivascular bone marrow niche (Sugiyama et al., 2006). By combining cxcl12a:DsRed2 and kdrl:GFP transgenic lines, we could see that mesenchymal stromal and endothelial cells are closely associated in the CHT, with cxcl12a+ cells distributed in abluminal spaces that underlie kdrl+ ECs (Figure 5A). We then crossed the Runx:GFP and cxcl12a:DsRed2 transgenic lines to observe HSPCs together with mesenchymal stromal cells. Most of the HSPCs in the CHT were lodged next to a stromal cell (Figure 5C). To quantify the proximity of these two cell types within 3D confocal z-stacks, the distance between a Runx+ HSPC and its nearest cxcl12a+ stromal cell was measured (Figure 5B,C). This analysis showed that 60% of HSPCs are in direct contact with a cxcl12a+ cell and 85% are ≤3 microns away. We performed time-lapse live imaging of CHT colonization in these double transgenic embryos and similarly found that arrival and expansion of most HSPCs occurred in close proximity to stromal cells (data not shown). When we tracked HSPCs attached to stromal cells over time, we observed that a significant majority of divisions had a distinct orientation: one daughter cell remained joined and was proximal to the stromal cell and the other daughter cell was distal and displaced away from the niche (Figure 5D-F; Movie S6). These data suggest that the cxcl12a+ stromal cell might orient the division plane of the HSPC by providing a polarizing signal. Together, this work shows that cxcl12a+ stromal cells dynamically interact with HSPCs in the CHT niche.
Figure 5. HSPCs are anchored to perivascular stromal cells during cell divisions.
(A) cxcl12a:DsRed2+ stromal cells (red) underlie kdrl:GFP+ ECs (green). 40 hpf embryo. (B) Percentage of HSPCs in CHT scored by distance to nearest stromal cell (n=168 total cells from 25 embryos). (C) Detail of Runx:GFP+ HSPCs (green) in proximity to cxcl12a:DsRed2+ stromal cells (red). Arrowheads mark HSPCs in contact with stromal cell. Circle marks HSPC with 2 μm gap between it and stromal cell. 60 hpf embryo. (D) 6 frames selected from time-lapse Movie S6 (hours:minutes). Upper row is merge of Runx:GFP+ HSPC (green, middle row) and cxcl12a:DsRed2+ stromal cells (red, lower row). Proximal HSPC anchored to stromal cell (arrowhead) divides and releases distal daughter cell into circulation (arrow). (E) Rose diagram showing division plane of HSPC oriented relative to stromal cell. Majority of divisions result in displaced daughter cell (n = 26/35 cell divisions from 22 embryos; 95% confidence interval 0.567—0.875; mean division angle of 110°). Diagrams show HSPCs dividing over stromal cell surface (45°), perpendicular to stromal cell (90°), or displaced away from stromal cell (135°). Angles may be affected by release into flow of circulation. (F) 3D models used to measure HSPC divisions relative to stromal cells. (i) Volume rendered confocal image. (ii) 3D model showing angle measurement between attachment point on stromal cell surface and center points of proximal and distal HSPCs (example shown is 110°). (iii) Overlay of confocal image and 3D model. Scale bars: (A) 25 μm; (C,F) 10 μm; (D) 15 μm.
High-resolution ultrastructure of an endogenous HSPC in the perivascular niche
We wanted to reveal the high-resolution architecture and cellular interactions of a single rare HSPC in the perivascular niche. To do this we used correlative light and electron microscopy (Mironov and Beznoussenko, 2009). First, to confirm that an HSPC was lodged in the CHT, we performed time-lapse live imaging of Runx:GFP;kdrl:mCherry embryos as above (Figure 3A). Imaging multiple stage-matched embryos in parallel, we confirmed that 1-2 HSPCs per embryo had not only migrated to the CHT, but had also lodged and triggered an EC remodeling event that was stable for more than 6 hours (Figure 6A-B). These same embryos were then fixed and embedded for electron microscopy (EM). Serial block face scanning EM captured large sections of the CHT at high resolution (Figure 6C; XY: 10 nm/pixel, Z: 100 nm/slice). Based on a number of cellular and anatomical markers (e.g. vessels, melanocytes, somites), the position of the lodged HSPC in time-lapse could be correlated with EM serial sections (Figure S5A,B, data not shown). In summary, we tracked a single cell in an embryo by time-lapse live imaging, processed that same embryo for serial block face scanning EM, and then identified the single cell in a high-resolution 3D reconstruction of the EM sections.
Figure 6. High resolution electron microscopy of endogenous HSPC in the perivascular niche.
(A) Last frame of CHT time-lapse (60 hpf). Arrowhead marks HSPC lodged >6 hours. Runx:GFP (green), kdrl:mCherry (red), brightfield (blue). Anterior left, posterior right, dorsal top, ventral bottom. (B) Detail of region in (A) marked by box. (C) Single section and orthogonal slice from serial block face EM scans. Lodged HSPC (purple, arrowhead), surrounding EC nuclei (green, numbered), stromal cells (dark and light blue). (D) High resolution EM of HSPC lodged in perivascular niche ventral to DA. The HSPC is in direct contact with one stromal cell (see higher magnification inset). (E-J) Selected sections (left) through niche with cell membrane traces used to build 3D models (right). (E) In this section, the HSPC (purple) is mostly surrounded by EC (orange). Portions of fibroblastic (yellow) and stromal (light and dark blue) cells are visible. (F) About half of the HSPC surface is wrapped by EC. (G) The HSPC directly contacts the stromal cell. Portions of the fibroblastic cell, EC, and second stromal cell are visible. (H) Only the midsection of the HSPC contacts the stromal cell. (I) The fibroblastic cell surrounds the HSPC. Portions of two stromal cells are visible. (J) Most of the HSPC surface is wrapped by the fibroblastic cell. Scale bars: (B) 25 μm; (C) 10 μm; (D) 2 μm and inset 1 μm. See also Figure S5 and Movie S7.
3D reconstruction of EM scans shows that the HSPC lodges in a region just adjacent to the caudal artery (Figures 6C, S5E). As predicted from our confocal microscopy analysis (Figure 3C), a pocket of at least 5 ECs surrounds the HSPC (Figures 6C, S5E). Our EM scans showed the details of cellular interactions between HSPCs and ECs. Regarding the endothelial cells, one example showed the HSPC partially wrapped by an EC (Figure 6E,F). In another example only one region of the HSPC was attached to an EC (Figure S5G,H). In other sections an EC would only contact the HSPC with a small protrusion, or not contact it all, lying opposite an extracellular space (Figures 6D, S5F, data not shown). Regarding the mesenchymal stromal cells, in one case we found two stromal cells in proximity to the HSPC, but only one is in direct contact in a narrow region near the mid-point of the cell (Figure 6D,G,H). This reveals the actual anchored attachment of HSPC to stromal cell that was suggested by our confocal microscopy (Figure 5B-F). In addition, a previously unidentified cell type, a fibroblastic cell with melanophore inclusions, was found within the perivascular niche and tightly wrapped around the HSPC (Figure 6D,I,J; Movie S7). The melanophore inclusions suggest a cell of neural crest origin, as such cells have been found in the adult marrow (Isern et al., 2014; Méndez-Ferrer et al., 2010). The combined technology of time-lapse confocal imaging and electron microscopy has shown that perivascular niche cells tightly wrap, abut, or lie opposite the extracellular space surrounding a single stem cell.
A chemical genetic screen to discover small molecule regulators of CHT niche colonization
Next we wanted to modulate the interactions between HSPC and perivascular niche to test their functional significance. We decided to use a chemical genetic approach so that we could perturb the embryo with small molecules during a discrete window of CHT colonization (48-72 hpf; Figure S6A). Hits were scored if they increased or decreased hematopoietic progenitor markers cmyb and runx1 with activities that were similar to control compounds (Figure S6B-E, data not shown). We used (±)11,12-epoxyeicosatrienoic acid (EET) as a control because it was identified by our lab as a strong positive regulator of HSPC (P.L., L.I.Z., unpublished). We tested the CXCR4 antagonist, AMD3100, as a potential inhibitor of CHT colonization because cxcr4a/b and cxcl12a/b genes are expressed in the CHT (Figure S6K), as is the cxcl12a transgenic reporter (Figure 5). After treating embryos with a range of AMD3100 doses, we observed dose-dependent reduction of HSPC markers in the CHT (Figure S6F). Following these results, we screened ~2400 known bioactive compounds, and identified 40 individual compounds that increased and 107 that decreased CHT hematopoiesis.
We tested a subset of our chemical screen hits by adding individual compounds to the media of Runx:GFP;kdrl:mCherry embryos during time-lapse imaging to directly observe the behavior of HSPCs as they colonize the CHT. Validation of our approach came from one of our positive hits for increased CHT hematopoiesis, SB-431542 (Figure S6D,G-J), a selective inhibitor of transforming growth factor (TGF)-β type I receptors, and most potently ALK5/TGFBR1 (Inman et al., 2002). When added during time-lapse imaging, a small number of newly arrived HSPCs in the CHT would undergo a greater number of divisions (Figure S6H-J), which was never observed in control treated embryos. We measured these differences by performing lineage tree analysis of HSPCs during parallel time-lapse movies (i.e. control and treated stage-matched embryos imaged side-by-side). HSPCs in control treated embryos would undergo 0-1 divisions, while rare HSPCs in SB-431542 treated embryos would divide 2-3 times. These data illustrate that inhibition of TGF-β receptor signaling using the chemical SB-431542 expands HSPC populations by increasing cell divisions. This is consistent with previously published in vitro and in vivo data that showed TGF-β signal negatively regulates HSPC proliferation (Soma et al., 1996; Yamazaki et al., 2011). Directly visualizing the effect of SB-431542 on HSPCs has demonstrated that we can rapidly identify cellular response to a specific signal in the endogenous niche.
HSPC-niche interactions in the embryo have a long-term effect on the stem cell pool
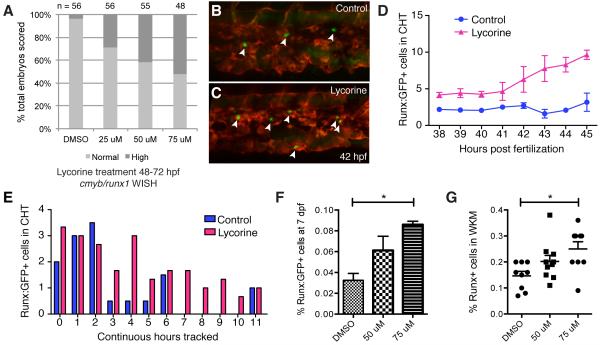
A second compound we identified in our screen was lycorine, a natural alkaloid extracted from the Amaryllidaceae plant family that dose-dependently increased cmyb and runx1 in the CHT (Figures 7A, S6E). This drug does not have a defined target or mechanism of action, but is a candidate anti-inflammatory and anti-cancer drug (Kang et al., 2012; Lamoral-Theys et al., 2009). Running parallel time-lapse movies, we observed more HSPCs lodged in the CHT in lycorine-treated embryos compared to controls (Figure 7B,C). Over time, lycorine treatment dramatically increased HSPC number in the CHT (Figure 7D). We also scored the total amount of time each HSPC was resident in the CHT during the time-lapse, and found lycorine treatment produced a significant shift towards longer durations spent in the niche (Figure 7E).
Figure 7. Modulating HSPC niche colonization with lycorine has long-term effects on the stem cell pool.
(A) Lycorine treatment dose-dependently increases the percentage of embryos with high cmyb/runx1 expression levels, as shown in Figure S6E (linear regression of Y (response) on X (dose) is significant: r2=0.146607, P=6.46E-9). (B,C) Stage-matched frames from parallel time-lapse movies show more HSPCs (arrowheads) with (C) 25 μM lycorine treatment than (B) DMSO-treated controls (42 hpf, Runx:GFP;kdrl:RFP). (D) HSPCs counted in each frame then averaged each hour. 5 movies imaged in parallel: n=2 controls and n=3 lycorine-treated. (E) HSPCs in the same embryos were scored for continuous hours tracked in CHT. Longer median time HSPCs spent in CHT of lycorine-treated embryos was significant (1.67 hours; Wilcoxon signed-rank test, p=0.01; HSPC counts normalized because treatment group was n=3 and control group was n=2). (F,G) Pools of Runx:GFP+ embryos treated with lycorine from 2-3 dpf and washed off had significantly increased HSPC at (F) 7 dpf (DMSO vs 75 μM, p=0.0004; flow cytometry analysis of 4 independent pools per dose) and (G) 4 months (DMSO vs 75 μM, p=0.006). Error bars show mean ± s.e.m. See also Figure S6.
To better understand the molecular changes that occur during lycorine treatment and how this positively regulates HSPC-niche interactions in the embryo, we treated Runx:GFP;kdrl:RFP embryos from 2-3 dpf, then sorted Runx+ HSPCs and kdrl+ ECs for whole genome microarray analysis (GEO accession number: GSE56015). Lycorine treatment induces significant gene expression changes that suggest altered adhesive properties of ECs and a changed activation state of HSPCs (Table S1A). These changes are possibly related to inflammatory pathways (e.g. TNF, IL1B, INFG) that could alter the interaction between ECs and HSPCs within the CHT niche (Table S1B).
Lastly, we wanted to determine if there was long-term functional significance for these changes in embryonic HSPC-niche interactions. Pools of Runx:GFP+ embryos were treated with lycorine from 2-3 dpf then washed off and examined at later timepoints. Strikingly, transient lycorine treatment during CHT colonization had a sustained effect on the total number of HSPCs. At 7 dpf, after colonization and early expansion in the kidney, lycorine treated embryos had a significantly higher number of Runx:GFP+ HSPCs (Figure 7F). We then followed treated embryos into adulthood. After four months, analysis of whole kidney marrow (WKM) showed a significant increase in the percentage of Runx:GFP+ HSPCs (Figure 7G). Using a chemical genetic screening approach we found a single compound, lycorine, that increased HSPC lodgement during development and ultimately led to a sustained increase in the size of the stem cell pool into adulthood.
DISCUSSION
The ability to follow endogenous HSPCs in the live zebrafish embryo has allowed us to directly observe cellular behaviors during lodgement in the perivascular niche. Current technology has not visualized similar events in the mammalian bone marrow. We watched as HSPCs attached to the endothelial wall of a vessel, underwent extravasation, migrated into the abluminal space, then triggered a striking endothelial remodeling event (Figure S3D). Live imaging of mouse FL explants revealed comparable interactions between hematopoietic cells and sinusoidal ECs, suggesting these steps towards HSPC lodgement are conserved (Figure S4D). Correlative light and electron microscopy allowed us to identify a single lodged HSPC in the perivascular niche that we could then resolve at very high resolution. This confirmed our initial observations made by confocal microscopy and also revealed new detailed cellular interactions. Perivascular niche cells can contact a single stem cell by wrapping, firm attachment, and extension of projections. Other niche cells stay close to the stem cell but keep their distance across small extracellular spaces. Together our data is beginning to reveal the spatial relationships between a stem cell and its niche.
Our finding that HSPC arrival triggers cellular changes in the local perivascular niche raises interesting questions about what constitutes a hematopoietic stem cell niche. Rather than a static number of niches that can be either cleared or filled (Schofield, 1978), our results are suggestive of basic niche components that create a permissive environment for an arriving stem cell. Once attracted to these general locations in the marrow, the stem cell will move out of circulation and lodge in its new surroundings. The rapid remodeling of endothelial cells around a stem cell may provide a mechanism to retain and protect these new arrivals. The formation of a pocket by endothelial cells could effectively increase the concentration of local growth factors and signaling molecules, and maintain more productive stem cell interactions of HSPCs with stromal cells. We observed that the division plane of HSPCs is often oriented relative to attached stromal cells. Future studies should investigate the potential for plasticity in terms of location and the number of sites where stem cells can reside. Our study focused on the hematopoietic microenvironment in the zebrafish CHT and mouse FL. These are intermediate niches required to rapidly expand the stem cell pool. With continual advancements in imaging technology we hope to soon be able to resolve similar cellular dynamics in the adult marrow.
EXPERIMENTAL PROCEDURES
All animals were handled according to approved Institutional Animal Care and Use Committee (IACUC) of Boston Children's Hospital protocols.
Transgenic zebrafish lines
To generate the Runx transgenic lines, the Runx1 +23 enhancer (Nottingham et al., 2007) and mouse β-globin minimal promoter were PCR amplified from C57/BL6 mouse genomic DNA. MultiSite Gateway Cloning (Invitrogen) was used to assemble constructs for injection into embryos using Tol2 transgenesis. See also Extended Experimental Procedures.
Imaging
Transgenic zebrafish lines were crossed and staged embryos were selected by fluorescence microscopy. Zebrafish embryos were mounted for live imaging in glass bottom dishes or multi-well plates with 1% LMP agarose and covered with E3 media and tricaine as described (Bertrand et al., 2010). Zebrafish embryos were imaged in an incubated chamber at 28.5°C. Mouse embryos were dissected and staged at E11-E11.5 by counting >42 somite pairs. FL explants were imaged at 37°C with humidified CO2 and in culture media as previously described (Boisset et al., 2010). Live confocal microscopy was performed using a Yokogawa spinning disk and Nikon inverted Ti microscope. Multiple embryos were imaged within a 1-5 minute interval using a moving XY stage, as well as acquisition of z-stacks through the entire CHT in multiple fluorescent channels. Fixed transgenic zebrafish embryos were scanned using a Nikon C2si confocal NiE upright microscope. Fixed mouse FLs were scanned using a Zeiss LSM 710 confocal microscope. See also Extended Experimental Procedures.
Image analysis
Image processing and rendering was done using Fluorender, Imaris (Bitplane), NIS-elements (Nikon), Volocity (PerkinElmer) and ImageJ/Fiji. See also Extended Experimental Procedures.
Adult-to-Adult HSPC Transplantation
WKM from 3-month Runx:mCherry;ubi:GFP fish was isolated and sorted by fluorescence-activated cell sorting (FACS). Double-positive cells were transplanted into irradiated casper recipients (n=20-48 recipients per cell dose) along with untreated helper marrow at the following ratios: 1:20,000; 5:20,000; 10:20,000; 25:20,000; 50:20,000. At 3 months post-transplant, WKM from recipient fish was collected and analyzed by flow cytometry to detect chimerism levels of mCherry and GFP-positive cells in the marrow. We confirmed multi-lineage reconstitution by observing differentiated GFP-positive cells in multiple cell populations, as determined by forward and side scatter profiles (Traver et al., 2003). Stem cell frequency was determined using ELDA software (confidence interval=0.95; (Hu and Smyth, 2009)).
Embryo-to-Embryo HSPC Transplantation
Double positive transgenic embryos were finely chopped and dissociated using Liberase (Roche). Cell suspensions were filtered and mCherry+/GFP+ cells were collected using a FACSAria cell sorter (BD Biosciences). Collected cells were resuspended in PBS at an estimated concentration of 400 cells/microliter and 1, 2, or 4 nanoliters were injected directly into the circulation of wild-type 2 dpf embryos—based on these dilutions, the estimated cell numbers were 0.4, 0.8, and 1.6, respectively. Approximately 30 embryos were injected per dose and 12-26 embryos per group survived to adulthood (3-5 months). WKM was analyzed for percentage of engrafted Runx+ cells using a LSR II flow cytometer (BD Biosciences). Any recipients with positive cells detected above background (>0.001% of WKM) were scored as engrafted. Flow cytometry data was analyzed using FACSDiva and FlowJo software. See also Extended Experimental Procedures.
Serial block face scanning electron microscopy and 3D reconstructions
Immediately at the end of live imaging time-lapse acquisition 60 hpf embryos were fixed in 2.5% glutaraldehyde and 4% paraformaldehyde in a 0.1 M sodium cacodylate buffer. Samples were submitted to Renovo Neural Inc (Cleveland, USA) for further processing and serial block face scanning electron microscopy. Images were imported into the program IMOD 4.5 then aligned and reconstructed using dual-axis tomography. Cells were manually outlined and 3D reconstructions were generated. See also Extended Experimental Procedures.
Chemical screening
Embryos were treated from 48-72 hpf with individual library chemicals (~2400 bioactives) by placement directly in 96-well receiver plates containing small molecules diluted in E3/1% DMSO (average final concentration of library compounds was ~30 μM). After treatment and before fixation we checked for secondary defects (e.g. no circulation, toxicity, developmental delay). We performed whole mount in situ hybridization with HSPC markers cmyb and runx1 and scored for expression levels in the CHT. See also Extended Experimental Procedures.
Microarrays
Runx:GFP;kdrl:RFP embryos were treated from 48-72 hpf with E3/1% DMSO or E3/1%DMSO/75 uM Lycorine (~100-150 embryos per group). Embryos were dissociated and sorted as above. Three populations were collected: GFP+ HSPC (~1-2k cells/experiment), RFP+ EC (~10-20k cells/experiment), and negative cells (~100k cells/experiment; total embryo as a comparator population). In total, 18 samples were collected: 3 biological replicates × 2 treatment conditions × 3 cell populations. Cells were sorted directly in Trizol LS. Total RNA was amplified and hybridized to Affymetrix Zebrafish GeneChip ZebGene-1_0-st microarrays. See also Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank the following for their support and contributions: L. Cameron, confocal imaging (Dana Farber Cancer Institute); T. Schlaeger, S. Datta, & P.D. Manos, chemical screening (Boston Children’s Hospital (BCH)); R. Mathieu, flow cytometry (BCH); L. Bu & A. Hill, confocal imaging (BCH); H.A. Feldman & W. London, statistical analysis (BCH); T.P. Ward & Y. Zhou, computer support (BCH); J. Hutchinson, O.M. Hofmann, & W.A. Hide, microarray analysis (Harvard School of Public Health); M. Ericsson & E.J. Benecchi, conventional electron microscopy (Harvard Medical School); A. Roholt, serial block face scanning EM (Renovo Neural, Cleveland, USA); T. Fuerstenhaupt, guidance in EM tomographic reconstructions (University of Calgary); T. Bowman, immunohistochemistry (Brigham & Women’s Hospital). We thank T.V. Bowman, J.R. Perlin, and C.K. Kaufman for critical reading of this manuscript. This work was supported by HHMI and NIH grants: R01 HL04880, 5P30 DK49216, 5R01 DK53298, 5U01 HL10001-05, R24 DK092760 (to L.I.Z.); R01 HL091724, U01HL100405 (to N.A.S.); 1F31HL120615 (to A.D.Y.). O.J.T. was supported by the American Society of Hematology, Canadian Institutes of Health Research (CIHR), and Heart and Stroke Foundation of Canada. S.J.C. was supported by CIHR (MOP-97787) and Alberta Innovates Health Solutions. E.J.H. was supported by a Helen Hay Whitney Foundation Fellowship. L.I.Z. is a founder and stockholder of Fate Therapeutics, Inc. and Scholar Rock, and a scientific advisor for Stemgent.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bertrand JY, Chi NC, Santoso B, Teng S, Stainier DYR, Traver D. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature. 2010;464:108–111. doi: 10.1038/nature08738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boisset J-C, Van Cappellen W, Andrieu-Soler C, Galjart N, Dzierzak E, Robin C. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature. 2010;464:116–120. doi: 10.1038/nature08764. [DOI] [PubMed] [Google Scholar]
- Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, Seandel M, Shido K, White IA, Kobayashi M, et al. Endothelial cells are essential for the self-renewal and repopulation of Notch-dependent hematopoietic stem cells. Cell stem cell. 2010;6:251–264. doi: 10.1016/j.stem.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong JLO, Burns CE, Chen AT, Pugach E, Mayhall EA, Smith ACH, Feldman HA, Zhou Y, Zon LI. Characterization of immune-matched hematopoietic transplantation in zebrafish. Blood. 2011;117:4234–4242. doi: 10.1182/blood-2010-09-307488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Morrison SJ. Haematopoietic stem cells and early lymphoid progenitors occupy distinct bone marrow niches. Nature. 2013;495:231–235. doi: 10.1038/nature11885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischman RA, Mintz B. Prevention of genetic anemias in mice by microinjection of normal hematopoietic stem cells into the fetal placenta. Proc Natl Acad Sci U S A. 1979;76:5736–5740. doi: 10.1073/pnas.76.11.5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass TJ, Lund TC, Patrinostro X, Tolar J, Bowman TV, Zon LI, Blazar BR. Stromal cell-derived factor-1 and hematopoietic cell homing in an adult zebrafish model of hematopoietic cell transplantation. Blood. 2011;118:766–774. doi: 10.1182/blood-2011-01-328476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Hsu Y-MS, Day RB, Schuettpelz LG, Christopher MJ, Borgerding JN, Nagasawa T, Link DC. CXCL12 in early mesenchymal progenitors is required for haematopoietic stem-cell maintenance. Nature. 2013;495:227–230. doi: 10.1038/nature11926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooper AT, Butler JM, Nolan DJ, Kranz A, Iida K, Kobayashi M, Kopp H-G, Shido K, Petit I, Yanger K, et al. Engraftment and reconstitution of hematopoiesis is dependent on VEGFR2-mediated regeneration of sinusoidal endothelial cells. Cell stem cell. 2009;4:263–274. doi: 10.1016/j.stem.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. Journal of Immunological Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Inman GJ, Nicolas FJ, Callahan JF, Harling JD, Gaster LM, Reith AD, Laping NJ, Hill CS. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol Pharmacol. 2002;62:65–74. doi: 10.1124/mol.62.1.65. [DOI] [PubMed] [Google Scholar]
- Isern J, García-García A, Martín AM, Arranz L, Martín-Pérez D, Torroja C, Sánchez-Cabo F, Méndez-Ferrer S, Cossu G. The neural crest is a source of mesenchymal stem cells with specialized hematopoietic stem-cell-niche function. eLife. 2014;3:e03696. doi: 10.7554/eLife.03696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Zhang Y, Cao X, Fan J, Li G, Wang Q, Diao Y, Zhao Z, Luo L, Yin Z. Lycorine inhibits lipopolysaccharide-induced iNOS and COX-2 up-regulation in RAW264.7 cells through suppressing P38 and STATs activation and increases the survival rate of mice after LPS challenge. International immunopharmacology. 2012;12:249–256. doi: 10.1016/j.intimp.2011.11.018. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz OH, Iwashita T, Yilmaz OH, Terhorst C, Morrison SJ. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121:1109–1121. doi: 10.1016/j.cell.2005.05.026. [DOI] [PubMed] [Google Scholar]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature. 2010;464:112–115. doi: 10.1038/nature08761. [DOI] [PubMed] [Google Scholar]
- Kissa K, Murayama E, Zapata A, Cortés A, Perret E, Machu C, Herbomel P. Live imaging of emerging hematopoietic stem cells and early thymus colonization. Blood. 2008;111:1147–1156. doi: 10.1182/blood-2007-07-099499. [DOI] [PubMed] [Google Scholar]
- Köhler A, Schmithorst V, Filippi M-D, Ryan MA, Daria D, Gunzer M, Geiger H. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009;114:290–298. doi: 10.1182/blood-2008-12-195644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamoral-Theys D, Andolfi A, Van Goietsenoven G, Cimmino A, Le Calvé B, Wauthoz N, Mégalizzi V.r., Gras T, Bruyére C.l., Dubois J, et al. Lycorine, the Main Phenanthridine Amaryllidaceae Alkaloid, Exhibits Significant Antitumor Activity in Cancer Cells That Display Resistance to Proapoptotic Stimuli: An Investigation of Structure–Activity Relationship and Mechanistic Insight. Journal of medicinal chemistry. 2009;52:6244–6256. doi: 10.1021/jm901031h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H-F, Traver D, Zhu H, Dooley K, Paw BH, Zon LI, Handin RI. Analysis of thrombocyte development in CD41-GFP transgenic zebrafish. Blood. 2005;106:3803–3810. doi: 10.1182/blood-2005-01-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo Celso C, Fleming HE, Wu JW, Zhao CX, Miake-Lye S, Fujisaki J, Côté D, Rowe DW, Lin CP, Scadden DT. Live-animal tracking of individual haematopoietic stem/progenitor cells in their niche. Nature. 2009;457:92–96. doi: 10.1038/nature07434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Robin C, Ottersbach K, Dzierzak E. The Ly-6A (Sca-1) GFP transgene is expressed in all adult mouse hematopoietic stem cells. Stem cells (Dayton, Ohio) 2002;20:514–521. doi: 10.1634/stemcells.20-6-514. [DOI] [PubMed] [Google Scholar]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell. 1996;86:897–906. doi: 10.1016/s0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- Méndez-Ferrer S, Michurina TV, Ferraro F, Mazloom AR, Macarthur BD, Lira SA, Scadden DT, Ma'ayan A, Enikolopov GN, Frenette PS. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AA, Beznoussenko GV. Correlative microscopy: a potent tool for the study of rare or unique cellular and tissue events. Journal of microscopy. 2009;235:308–321. doi: 10.1111/j.1365-2818.2009.03222.x. [DOI] [PubMed] [Google Scholar]
- Morrison SJ, Hemmati HD, Wandycz AM, Weissman IL. The purification and characterization of fetal liver hematopoietic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:10302–10306. doi: 10.1073/pnas.92.22.10302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison SJ, Scadden DT. The bone marrow niche for haematopoietic stem cells. Nature. 2014;505:327–334. doi: 10.1038/nature12984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosimann C, Kaufman CK, Li P, Pugach EK, Tamplin OJ, Zon LI. Ubiquitous transgene expression and Cre-based recombination driven by the ubiquitin promoter in zebrafish. Development (Cambridge, England) 2011;138:169–177. doi: 10.1242/dev.059345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. Development of hematopoietic stem cell activity in the mouse embryo. Immunity. 1994;1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin H-F, Handin RI, Herbomel P. Tracing Hematopoietic Precursor Migration to Successive Hematopoietic Organs during Zebrafish Development. Immunity. 2006;25:963–975. doi: 10.1016/j.immuni.2006.10.015. [DOI] [PubMed] [Google Scholar]
- Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, Park SY, Lu J, Protopopov A, Silberstein LE. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol. 2013;15:533–543. doi: 10.1038/ncb2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North TE, Goessling W, Walkley CR, Lengerke C, Kopani KR, Lord AM, Weber GJ, Bowman TV, Jang I-H, Grosser T, et al. Prostaglandin E2 regulates vertebrate haematopoietic stem cell homeostasis. Nature. 2007;447:1007–1011. doi: 10.1038/nature05883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng J-F, Prabhakar S, Rubin EM, Li P-S, Sloane-Stanley J, Kong-A-San J, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood. 2007;110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osawa M, Nakamura K, Nishi N, Takahasi N, Tokuomoto Y, Inoue H, Nakauchi H. In vivo self-renewal of c-Kit+ Sca-1+ Lin(low/-) hemopoietic stem cells. Journal of immunology (Baltimore, Md : 1950) 1996;156:3207–3214. [PubMed] [Google Scholar]
- Sánchez MJ, Holmes A, Miles C, Dzierzak E. Characterization of the first definitive hematopoietic stem cells in the AGM and liver of the mouse embryo. Immunity. 1996;5:513–525. doi: 10.1016/s1074-7613(00)80267-8. [DOI] [PubMed] [Google Scholar]
- Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood cells. 1978;4:7–25. [PubMed] [Google Scholar]
- Soma T, Yu JM, Dunbar CE. Maintenance of murine long-term repopulating stem cells in ex vivo culture is affected by modulation of transforming growth factor-beta but not macrophage inflammatory protein-1 alpha activities. Blood. 1996;87:4561–4567. [PubMed] [Google Scholar]
- Sugiyama T, Kohara H, Noda M, Nagasawa T. Maintenance of the Hematopoietic Stem Cell Pool by CXCL12-CXCR4 Chemokine Signaling in Bone Marrow Stromal Cell Niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Traver D, Paw BH, Poss KD, Penberthy WT, Lin S, Zon LI. Transplantation and in vivo imaging of multilineage engraftment in zebrafish bloodless mutants. Nature Immunology. 2003;4:1238–1246. doi: 10.1038/ni1007. [DOI] [PubMed] [Google Scholar]
- Xie Y, Yin T, Wiegraebe W, He XC, Miller D, Stark D, Perko K, Alexander R, Schwartz J, Grindley JC, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009;457:97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
- Yamazaki S, Ema H, Karlsson G, Yamaguchi T, Miyoshi H, Shioda S, Taketo MM, Karlsson S, Iwama A, Nakauchi H. Nonmyelinating Schwann cells maintain hematopoietic stem cell hibernation in the bone marrow niche. Cell. 2011;147:1146–1158. doi: 10.1016/j.cell.2011.09.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.