Abstract
Comparison of the nature of hydride transfer in wild-type and active site mutant (I14A) of dihydrofolate reductase suggests that the size of this side chain at position 14 modulates H-tunneling.
Understanding the nature of the hydride-transfer and how the protein active site structure and dynamics‡ contribute to these reactions is of contemporary interest. Numerous experimental and theoretical data suggest that quantum mechanical tunneling of a nucleus has a role in a large number of enzyme catalyzed hydrogen transfer reactions.1–7 Owing to the multidimensional and quantum-mechanical nature of a hydrogen transfer reaction, the rate of the transfer is sensitive to the protein’s motions that modulate the height and the width of the reaction’s barrier. Subtle changes in the protein structure, some of which take place on the time scale of the hydride-transfer step, might lead to more efficient barrier penetration and H-tunneling. Here we present kinetic data and molecular modeling studies suggesting that an active site mutation modulates the nature of the hydride-transfer in the enzyme dihydrofolate reductase (DHFR; EC 1.5.1.3), from E. coli. The intrinsic kinetic isotope effects (KIEs) for the hydride transfer, and their temperature dependence were evaluated. Our findings implicate the hydrophobic side chain of Ile14 residue, positioned behind the hydride-donor (reduced nicotinamide), in the reorganization of the active site during the DHFR-catalyzed reaction.
DHFR has been a model system for examination of the physical nature of enzymatic hydride transfers and of the contribution of protein dynamics to the hydride transfer.4, 8, 9 DHFR catalyzes the reduction of 7,8-dihydrofolate (H2folate) to 5,6,7,8-tetrahydrofolate (H4folate), an essential co-factor in several biosynthetic pathways. Previous measurements of intrinsic KIEs and their temperature dependences for wild-type EcDHFR (wt DHFR) inferred quantum mechanical tunneling of the hydride in the DHFR catalyzed reaction.10 Kinetic studies of several distal mutants of EcDHFR addressed the role of remote residues in modulating the hydride-tunneling process.4, 8, 9 The latter studies supported the hypothesis that a long range dynamically coupled network acts in the chemical transformation (C-H→C transfer), as predicted by hybrid quantum/classical molecular dynamics simulations.11, 12
In the present study, Ile14, that holds the nicotinamide ring of the cofactor (NADPH) close to the hydride acceptor (H2folate), has been replaced with alanine by side-directed mutagenesis. The presence of a conserved hydrophobic residue behind the nicotinamide ring is a common feature of nicotinamide-dependent enzymes (refs 13, 14 and the PDB) suggesting its critical role in this type of reaction. This residue holds the nicotinamide ring in close proximity to the hydride acceptor (e.g. H2folate), and participates in the organization of the donor and acceptor during the catalyzed hydride-transfer reaction. NMR studies indicated that this residue exists as two rotamers in solution, despite the well-defined orientation in crystal structure (gauche rotamer).15 The trans rotamer, not observed in either closed or occluded crystal structure, is significantly populated in the solution. Modeling studies suggested that in trans rotameric state, the side chain of the I14 clashes with nicotinamide ring forcing it towards the pterin ring. MD simulation confirmed that the position of trans population along the reaction coordinate overlaps with a decrease in the donor-acceptor distance (DAD).16 Therefore, decrease in the size of the side chain, like in the I14A DHFR mutant, alters the DAD and orientation, with minimal effect on the active site’s electrostatics. By comparing the intrinsic primary (1°) KIEs and their temperature dependencies for the mutant I14A DHFR with those previously reported for wtDHFR,10 we can comment on the nature of the hydride transfer and the contribution of protein dynamics to the enzyme catalyzed reaction.
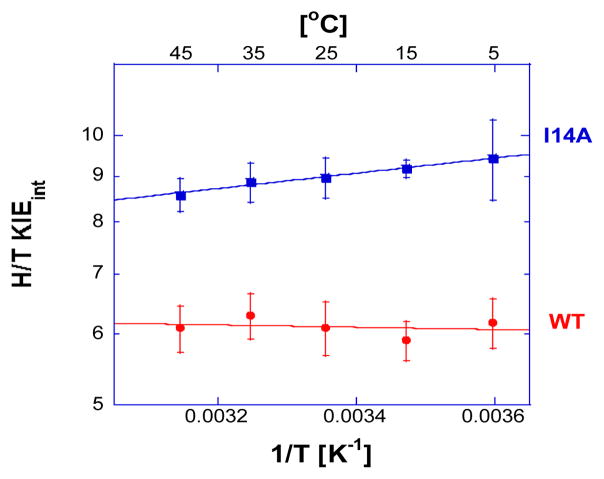
Competitive H/T and D/T KIEs on the second order rate constants (kcat/KM) were measured with the I14A-DHFR mutant using mixtures of [4(R)-3H]-NADPH with [Ad-14C]-NADPH for 1° H/T, and [4(R)-3H]-NADPH and [Ad-14C, 4(R)-2H]-NADPH for 1° D/T KIEs. The synthesis, purification and storage of these labeled cofactors have been presented elsewhere.17 Observed KIEs were used to calculate intrinsic KIEs following the Northrop method as described before (Figure 1, also see Figure 1S and Table 1S in the ESI†).4, 8–10, 18 Table 1 compares the isotope effects on the Arrhenius activation parameters for the wtDHFR and I14A. The values for the isotope effects on the Arrhenius prefactors differ significantly between the two enzymes suggesting noticeable differences in the nature of the hydride tunneling. Moreover, all the isotope effects on the Arrhenius preexponential factors are larger than their semi-classical limits (0.3–1.7).19, 20 Similar findings with the E. coli DHFR have been simulated by high level QM/MM calculations.21–23 A simple phenomenological way to rationalize the data is through the Marcus-like models, also referred to as environmentally coupled tunneling models, tunneling promoting vibrations, etc. In these models, heavy atom motion controls the probability of hydrogen transfer. From this perspective, larger than unity isotope effects on Arrhenius prefactors and small temperature dependencies in KIEs indicate a system with a well reorganized donor and acceptor. A significant temperature dependency in KIEs, on the other hand, suggests poor reorganization, where the average DAD is too long for efficient tunneling. In those cases, thermally activated fluctuation of the DAD allows the system to sample more conformations to enable tunneling at higher temperatures. Most Marcus-like models follow the general expression:2, 3, 24, 25
Fig. 1.
Arrhenius plot of intrinsic H/T KIEs for wild-type (red)10 and mutant (blue) DHFR. The lines represent the nonlinear regression to an exponential equation. More details are presented in the Electronic Supplementary Information (ESI).†
Table 1.
Activation Parameters of Comparative KIEs
| wtDHFRa | I14A | Semiclassical range | |
|---|---|---|---|
| AH/ATb | 7.0±1.5 | 4.7±0.5 | 0.3–1.7 |
| ΔEaT-H*, kcal/mol | −0.1±0.2 | 0.39±0.06 |
From Ref 10
Similar trends were observed for H/D and D/T (data not shown since this is trivial, as they follow the Swain Schaad relationship)
| (1) |
In eq. 1, the rate of hydride-transfer is determined by the isotope independent pre-exponential constant (C), which represents the fraction of reactive complexes; a Marcus term (first exponential); a Franck-Condon term (F(m)) and a “gating term” (EF(m)) that describes the temperature and mass dependence of the DAD fluctuations.2, 3, 24, 25 The Marcus term is almost isotopically insensitive, and represents the heavy-atom reorganization that is required to occur before significant tunneling can take place (i.e., the reorganization of the donor and acceptor toward a “tunneling ready” conformation).3, 26 The Franck-Condon term describes the nuclear wavefunction overlap along the hydrogen coordinate, and is isotopically sensitive. The temperature dependence in KIEs results from the gating term. The last two terms are integrated for all the conformations sampled by the DAD.
Like many highly evolved enzymes under physiological conditions, the wtDHFR exhibited temperature independent KIEs (Figure 1 and Table 1),10 consistent with a well reorganized donor and acceptor with an average DAD that is adequate for tunneling. The active site mutant I14A, on the other hand, exhibits temperature dependent intrinsic KIEs. This implies that at the tunneling-ready conformation, the average DAD is too long for efficient tunneling at low temperature, but at higher temperatures more conformations with a shorter DAD are sampled. This affects D more than H because D can only tunnel from shorter distances, thus leading to temperature dependent KIEs.
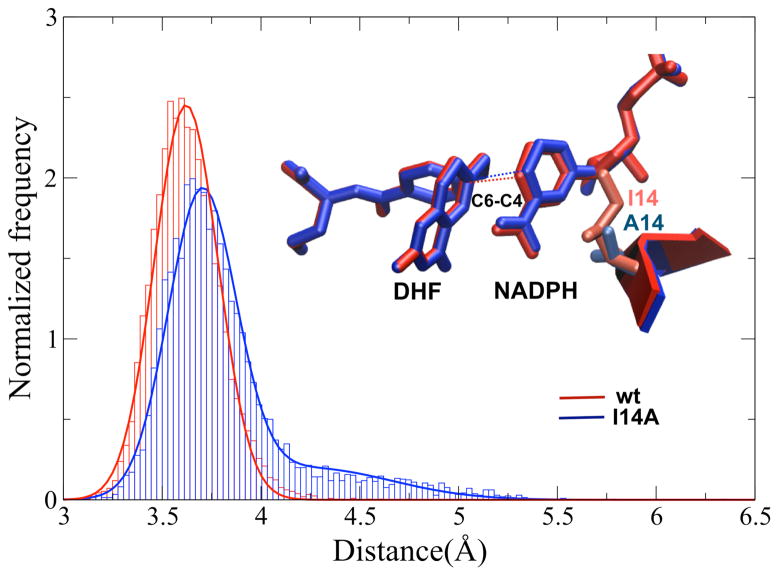
Due to the very different volumes of isoleucine and alanine (reflecting deletion of ethyl and methyl on the β-carbon of residue 14), one would expect that in I14A DHFR mutant the nicotinamide ring (the donor of the hydride) would be less sterically restricted to be close to the hydride acceptor. To better assess the structural aspects of this effect, we modeled the relative orientation of the donor and acceptor in the wild-type and mutant starting from the coordinates of the ternary complex wtDHFR-Folate-NADP+ (PDB entry: 1RX2).27 This ternary complex was chosen because it is the best structural representation of the reactive complex27, 28 The ligands were substituted for the reactants (NADPH and H2folate) and an active site mutation was performed in silico. A 15 ns molecular dynamics simulation (MD) was performed using the Amber9 package29 (see ESI† for details). The results indicate that the mutation leads to an increase in the ground state DADs as well as broader distribution of DADs with smaller fraction of short distances (Figure 2, and Figure 4S in ESI†), which is in accordance with the kinetic findings (Figure 1). The elevated flexibility in the mutant, together with the longer average DAD suggest that the side chain of residue I14 in the wtDHFR compresses the donor toward the acceptor as the system rearranges toward the tunneling-ready state (Figure 5S in ESI†). In support of the less restricted active site, we carefully examined MD trajectories and found that during the timescale of the simulation, on average 1–2 water molecules enter the active site cavity filling the void space between the nicotinamide ring and Ala14. The MD simulation assists in rationalizing the kinetic data and further supports the interpretation of these data by the Marcus-like model. We trust that the findings presented here will lead to rigorous QM/MM calculations that will more thoroughly examine the role of this residue along the reaction coordinate.
Fig. 2.
Histogram representation of DAD distribution for wtDHFR (red) and I14A DHFR (blue). The average structure of reactive complex, together with residue 14, for each enzyme is presented. The lines show the fitting of the data to one and two Gaussians, for the wild type and mutant DHFRs, respectively. For the wt DHFR the C4 (donor) to C6 (acceptor) have an average distance is 3.62 ± 0.16 Å. For I14A DHFR, on the other hand, there are two populations, one with an average DAD of 3.69 ± 0.17 Å, and one 4.24±0.41 Å. These distributions are presented in details at the ESI.† Significantly, I14A DHFR exhibits larger DAD distribution suggesting the increase in the flexibility of the nicotinamide ring for the mutant, and smaller population at short DADs.
The reduction in the size of residue 14 appears to correlate with reorganization of the reacting complex that is less suitable for H-tunneling than with the wild type. The change in the nature of the hydride transfer relative to wtDHFR is also reflected in the reduced rate of hydride transfer. The hydride transfer rate for the protonated ternary complex of EcDHFR at 25 °C was estimated to be 450 s−1 (pH=6.5),30 whereas for I14A DHFR the rate is decreased ~ 14 fold (khyd=33±3 s−1, pH=6.8).31 This decrease in rate further supports the hypothesis that the reorganization to the tunneling-ready conformation is altered by the alanine mutation in such a way that H-tunneling is less efficient.
A previous study that suggested that the longer DAD and distorted geometry diminished tunneling in an enzymatic system, was done on horse liver ADH (HLADH), where a valine residue, analogous to I14 in DHFR, was mutated into alanine. That mutation increased the ground state DAD, as suggested by X-ray crystallography with folate and NADP+ ligands. While no temperature dependent studies were conducted for this mutant of HLADH, the mutation resulted in a reduced coupled motion and tunneling, as suggested by mixed labeling 2° Swain-Schaad exponent (SSE) experiments at a single temperature.13, 14 The current study, on the other hand, allowed for the measurement of the temperature dependence of KIEs that likewise resulted from a change in the orientation between the pterin and nicotinamide rings and changes in the DAD reorganization step. The current findings also suggest that the active site becomes more flexible, which is in accord with a larger temperature dependence of the intrinsic KIEs with I14A DHFR.
In conclusion, a mutation that decreased the restriction on location and motion of the hydride-donor (NADPH) appears to perturb the dynamic behavior at the ground state. Future studies will expand the series of mutations, structural studies, and high level simulations in order to obtain a more rigorous correlation between the size of the side chain and the nature of the hydride-transfer. Of particular interest would be analysis similar to that preformed in ref 21, where the DAD’s distribution is examined along the reaction coordinate.
Acknowledgments
This work was supported by NSF CHE-0133117 and BSF-2007256 (for AK) and 1RO1GM092946 (for SJB). LLP acknowledge CONICET and Dario A. Estrin.
Footnotes
Electronic Supplementary Information (ESI) available: The material and methods used in these studies and more detailed data analysis. See DOI: 10.1039/b000000x/
The term dynamics is used here for atomic motion, structural changes, and alteration of conformational populations.
References
- 1.Basran J, Masgrau L, Sutcliffe MJ, Scrutton NS. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 671–689. [Google Scholar]
- 2.Kohen A. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 743–764. [Google Scholar]
- 3.Nagel ZD, Klinman JP. Chem Rev. 2006;106:3095–3118. doi: 10.1021/cr050301x. [DOI] [PubMed] [Google Scholar]
- 4.Wang L, Goodey NM, Benkovic SJ, Kohen A. Proc Nat Acad Sci USA. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truhlar DG. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 579–620. [Google Scholar]
- 6.Hammes-Schiffer S. Acc Chem Res. 2006;39:93–100. doi: 10.1021/ar040199a. [DOI] [PubMed] [Google Scholar]
- 7.Warshel A, Olsson MHM, Villá-Freixa J. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 621–644. [Google Scholar]
- 8.Wang L, Tharp S, Selzer T, Benkovic SJ, Kohen A. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Goodey NM, Benkovic SJ, Kohen A. Phil Trans R Soc B. 2006;361:1307–1315. doi: 10.1098/rstb.2006.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sikorski RS, Wang L, Markham KA, Rajagopalan PTR, Benkovic SJ, Kohen A. J Am Chem Soc. 2004;126:4778–4779. doi: 10.1021/ja031683w. [DOI] [PubMed] [Google Scholar]
- 11.Radkiewicz J, Brooks C. J Am Chem Soc. 2000;122:225–231. [Google Scholar]
- 12.Wong KF, Selzer T, Benkovic SJ, Hammes-Schiffer S. Proc Natl Acad Sci USA. 2005;102:6807–6812. doi: 10.1073/pnas.0408343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bahnson BJ, Colby TD, Chin JK, Goldstein BM, Klinman JP. Proc Nat Acad Sci USA. 1997;94:12797–12802. doi: 10.1073/pnas.94.24.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Colby TD, Bahnson BJ, Chin JK, Klinman JP, Goldstein BM. Biochemistry. 1998;37:9295–9304. doi: 10.1021/bi973184b. [DOI] [PubMed] [Google Scholar]
- 15.Schnell JR, Dyson HJ, Wright PE. Biochemistry. 2004;43:374–383. doi: 10.1021/bi035464z. [DOI] [PubMed] [Google Scholar]
- 16.Arora K, Brooks CLI. J Am Chem Soc. 2009;131:5642–5647. doi: 10.1021/ja9000135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Markham KA, Kohen A. Curr Anal Chem. 2006;2:379–388. [Google Scholar]
- 18.Northrop DB. In: Enzyme mechanism from isotope effects. Cook PF, editor. CRC Press; Boca Raton, Fl: 1991. pp. 181–202. [Google Scholar]
- 19.Melander L, Saunders WH. Reaction rates of isotopic molecules. Krieger, R.E; Malabar, FL: 1987. [Google Scholar]
- 20.Bell RP. The tunnel effect in chemistry. Chapman & Hall; London & New York: 1980. [Google Scholar]
- 21.Liu H, Warshel A. J Phys Chem B. 2007;111:7852–7861. doi: 10.1021/jp070938f. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal PK, Billeter SR, Hammes-Schiffer S. J Phys Chem B. 2002;106:3283–3293. [Google Scholar]
- 23.Garcia-Viloca M, Truhlar DG, Gao J. Biochemistry. 2003;42:13558–13575. doi: 10.1021/bi034824f. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz SD. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 475–498. [Google Scholar]
- 25.Kuznetsov A, Ulstrup J. Can J Chem. 1999;77:1085–1096. [Google Scholar]
- 26.Kiefer PM, Hynes JT. In: Isotope effects in chemistry and biology. Kohen A, Limbach HH, editors. Taylor & Francis, CRC Press; Boca Raton, FL: 2006. pp. 549–578. [Google Scholar]
- 27.Sawaya MR, Kraut J. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 28.Boehr DD, McElheny D, Dyson HJ, Wright PE. Science. 2006;313:1638–1642. doi: 10.1126/science.1130258. [DOI] [PubMed] [Google Scholar]
- 29.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Crowley M, Walker RC, Zhang W, Merz KM, Wang B, Hayik S, Roitberg A, Seabra G, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Mathews DH, Seetin MG, Sagui C, Babin V, Kollman PA. University of California; San Francisco: p. 200x. [Google Scholar]
- 30.Fierke CA, Johnson KA, Benkovic SJ. Biochemistry. 1987;26:4085–4092. doi: 10.1021/bi00387a052. [DOI] [PubMed] [Google Scholar]
- 31.Adams JA, Fierke CA, Benkovic SJ. Biochemistry. 1991;30:11046–11054. doi: 10.1021/bi00110a006. [DOI] [PubMed] [Google Scholar]