Abstract
Genetic susceptibility to multiple sclerosis (MS) is associated with the MHC located on chromosome 6p21. This signal maps primarily to a 1-Mb region encompassing the HLA class II loci, and it segregates often with the HLA-DQB1*0602, -DQA1*0102, -DRB1*1501, -DRB5*0101 haplotype. However, the identification of the true predisposing gene or genes within the susceptibility haplotype has been handicapped by the strong linkage disequilibrium across the locus. African Americans have greater MHC haplotypic diversity and distinct patterns of linkage disequilibrium, which make this population particularly informative for fine mapping efforts. The purpose of this study was to establish the telomeric boundary of the HLA class II region affecting susceptibility to MS by assessing genetic association with the neighboring HLA-DRB5 gene as well as seven telomeric single nucleotide polymorphisms in a large, well-characterized African American dataset. Rare DRB5*null individuals were previously described in African populations. Although significant associations with both HLA-DRB1 and HLA-DRB5 loci were present, HLA-DRB1*1503 was associated with MS in the absence of HLA-DRB5, providing evidence for HLA-DRB1 as the primary susceptibility gene. Interestingly, the HLA-DRB5*null subjects appear to be at increased risk for developing secondary progressive MS. Thus, HLA-DRB5 attenuates MS severity, a finding consistent with HLA-DRB5’s proposed role as a modifier in experimental autoimmune encephalomyelitis. Additionally, conditional haplotype analysis revealed a susceptibility signal at the class III AGER locus independent of DRB1. The data underscore the power of the African American MS dataset to identify disease genes by association in a region of high linkage disequilibrium.
Multiple sclerosis (MS)3 is a common, disabling disease of the CNS characterized by moderate, but complex risk heritability. In an attempt to map the full array of susceptibility loci and identify the genes that predispose to MS, whole-genome screens for linkage and/or associations were performed with different levels of resolution in multiple datasets. Altogether, these studies detected a number of genomic regions and genes of interest, consistent with the long-held view that MS is a polygenic disorder (1–4). Without exception, the MHC region, located in the short arm of chromosome 6, yielded association scores exceeding stringent thresholds of genome-wide statistical significance, indicating the presence of a major susceptibility gene or genes. This signal maps to a 1-Mb region enclosing the HLA class II segment and segregates primarily with the HLA-DQB1*0602, DQA1*0102, DRB1*1501, DRB5*0101 haplotype (henceforth, all HLA genes will be referred to without the HLA prefix) (5, 6). There is debate, however, whether the DRB1 association explains the entire MHC class II genetic signal (7–12).
The extensive linkage disequilibrium (LD) across the region hindered the identification of the true predisposing factor(s) within the disease susceptibility haplotypes (13). Because LD patterns can differ between populations, the most direct and practical approach to distinguish between primary and secondary effects due to LD is to scrutinize a large number of haplotypes in datasets with different ancestral histories. African Americans are at a lower risk for MS when compared with northern Europeans and white Americans, with recent studies finding a relative risk of 0.64 for developing MS (14). In our early study of MHC class II alleles and haplotypes in an African American MS cohort, selective associations with DRB1*1501 and *1503 independent of DQB1*0602 were revealed (15), indicating that the DRB1 gene constitutes the centromeric edge of the class II association in MS and confirming the power of this approach to fine-map susceptibility genes. However, the telomeric border of the susceptibility locus remains uncertain.
The present study was designed to firmly establish the telomeric boundary of the HLA class II region affecting disease vulnerability by assessing genetic association with the DRB5 gene and seven additional informative markers in a well-characterized African American MS dataset. The results are consistent with a primary role for the DRB1 gene in conferring susceptibility to MS, whereas DRB5 may act as a modifier of progression. Additionally, results suggest an independent effect within or near the class III locus AGER, the gene coding the receptor of advanced glycation end-products, a member of the Ig superfamily and mediator of chronic inflammatory reactions (16).
Materials and Methods
Subjects
The primary dataset studied consisted of 1635 African American individuals, including 769 MS cases, 124 parents, and 742 unrelated control individuals (Table I). All MS subjects met established diagnostic criteria (17). MS phenotypes were characterized by systematic chart review as described (18). Ascertainment protocols and clinical and demographic characteristics were summarized elsewhere (15, 18). All study participants are self-reported African Americans, but European ancestry was documented in most individuals based on genotyping of 186 single nucleotide polymorphisms (SNPs) highly informative for African vs European ancestry as previously described (19). Global estimation of European ancestry using these markers indicated similar mean admixture proportions in cases and controls (see Table I), thereby indicating that both groups were well matched. A second dataset, consisting of 487 white/non-Hispanic MS patients and 434 unrelated controls matched for ethnicity and age, was also included in the study (Table I). STRUCTURE-based analysis with 971 unlinked SNPs distributed across the autosomal genome confirmed the lack of population stratification in this data set (data not shown). Written informed consent was obtained from all participating subjects.
Table I.
Clinical and demographic features of the dataset
| African American MS Cases |
African American Controlsd |
White MS Cases | White Controls | |
|---|---|---|---|---|
| Total N | 769 | 866 | 487 | 434 |
| Female/male ratioa | 3.8:1 | 1.27:1 | 2.2:1 | 2.0:1 |
| % European ancestryb | 22 ± 11.5 | 23 ± 15 | ||
| Mean age of onset in years | 32.6 ± 9.5 | 33.6 ± 9.3 | ||
| Mean disease duration in years | 9.82 ± 7.9 | 8.82 ± 9.0 | ||
| Relapsing remitting cases (n, %)c | 431, 58.5% | 340, 69.8% |
For all markers, including HLA, allele frequencies were similar between female and males study participants (data not shown).
European ancestry in African Americans was documented based on genotyping of 186 informative SNPs in 713 (92.7%) MS cases and 500 (67.4%) controls (19). Mean European ancestry proportions in African American MS cases and controls were not statistically different (p > 0.10).
χ2 test was performed to compare the differences of disease subtype of relapsing remitting cases between the two cohorts (χ2 = 23.84, p < 0.001).
Family controls consist of 124 parents (62 nontransmitted chromosomes); 742 unrelated controls were also used.
Genotyping
DRB1 genotyping
For DRB1, a PCR locus-specific amplification generates a template for DNA sequencing of the relevant polymorphic sites at exon 2 in both the forward and reverse directions, as well as for primer-specific sequencing of the valine motif (GTG) of codon 86 for ambiguity resolution. Analysis is performed using ASSIGN software from Conexio Genomics. Of the African American dataset, 71.1% was characterized for DRB1 variation using this DNA sequencing protocol. Samples without sequence-based DRB1 typing (28.9% of the African American MS dataset and all white MS cases and controls) were genotyped with a validated gene-specific TaqMan assay designed to identify, specifically, the presence or absence of DRB1*1501 and/or *1503 alleles. An internal positive control (β-globin) was included in each well to confirm that the reaction amplified successfully. PCR was conducted in a total volume of 10 µl, containing 20 ng DNA, 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 0.6 µM DRB1*1501/1503-specific primers (forward 5′-ACG TTT CCT GTG GCA GCC TAA-3′, reverse 5′-TGC ACT GTG AAG CTC TCC ACA A-3′), 0.3 µM control primers (forward 5′-ACT GGG CAT GTG GAG ACA GAG AA-3′, reverse 5′-AGG TGA GCC AGG CCA TCA CTA AA-3′), 0.225 µM VIC-labeled DRB1*1501/1503-specific probe (5′-AAC AGC CAG AAG GAC ATC CTG GAG CA-3′), and 0.025 µM 6FAM-labeled control probe (5′-TCT ACC CTT GGA CCC AGA GGT TCT TTG AGT-3′). Amplification was conducted in an ABI Prism 7900HT sequence detection system (Applied Biosystems) with an initial 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 62°C for 1 min. To determine copy number, fluorescence intensity values were analyzed using logistic regression modeling and standardized distribution cutoff statistics. The rs2187668 SNP located within DQA1 shows strong correlation with DRB1*0301 in populations of northern European descent (20), and it was therefore genotyped in white MS cases and controls as a tagging SNP for this DRB1 allele.
DRB5
All study participants were screened for the presence of DRB5 using a validated gene-specific TaqMan assay. An internal positive control (β-globin) was included in each well to confirm that the reaction amplified successfully. PCR was conducted in a total volume of 10 µl, containing 20 ng DNA, 1× TaqMan Universal PCR Master Mix, 0.45 µM DRB5-specific primers (forward 5′-ACGTTTCCTGTGGCAGCCTAA-3′, reverse 5′-TGCACTGTGAAGCTCTCCACAA-3′), 0.45 µM control primers (forward 5′-ACTGGGCATGTGGAGACAGAGAA-3′, reverse 5′-AGGTGAGCCAGGCCATCACTAAA-3′), 0.125 µM VIC-labeled DRB5-specific probe (5′-ACCAGCCAGAAGGACATCCTGGAGCA-3′), and 0.125 µM 6FAM-labeled control probe (5′-TCTACCCTTGGACCCAGAGGTTCTTTGAGT-3′). Amplification was conducted in an ABI Prism 7900HT Sequence Detection System with an initial 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Samples are considered to contain at least one copy of the DRB5 gene if the respective Ct exceeds a preestablished threshold. The second exon for DRB5 was then sequenced for allele determination.
SNP genotyping
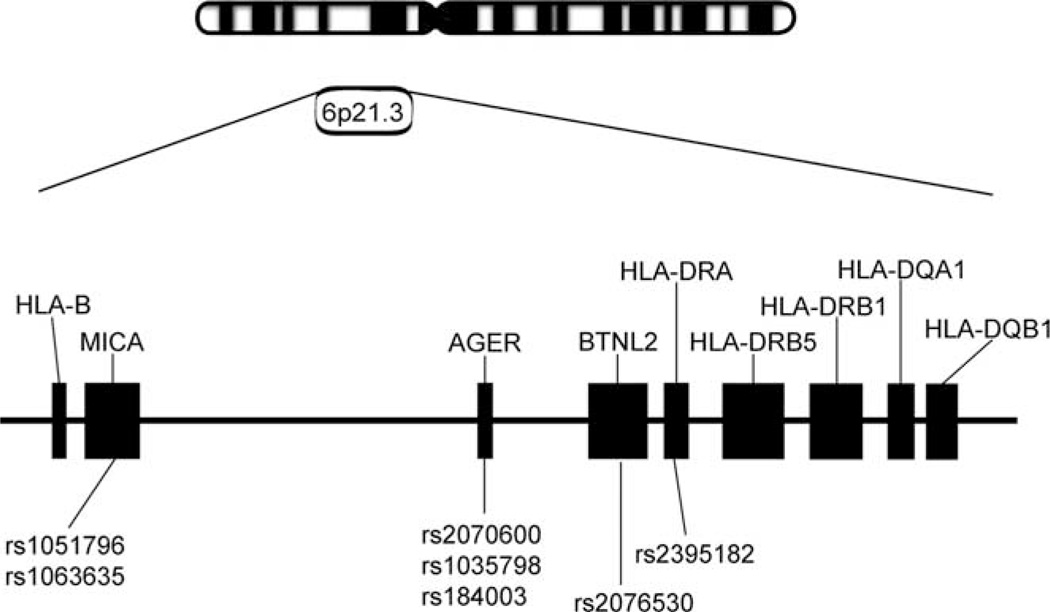
DRA (rs2395182), BTNL2 (rs2076530), AGER (rs2070600, rs1035798, rs184003), and MICA (rs1051796, rs1063635) SNP genotyping (Fig. 1) was completed in the African American dataset (n = 1635 individuals) using ABI custom TaqMan assays designed on File Builder 2.0 software. TaqMan SNP genotyping assays are conducted in 384-well plates using TaqMan Universal PCR Master Mix on an ABI 7900HT Sequence Detection System using SDS 2.0 software. Similarly, two AGER SNPs (rs2070600, rs1035798) were genotyped in white MS cases and controls for confirmatory analyses. The entire AGER gene was sequenced in 10 African American MS patients and 10 African American controls in an effort to locate any causative SNPs in the gene, but no novel SNPs were found. Additional genotype data for CEPH (CEU) and Yoruban (YRI) International HapMap project samples (60 unrelated individuals from each group or 240 total chromosomes) was available for 13,787 (CEU) and 13,820 (YRI) extended MHC region SNPs (827,220 and 829,200 genotypes, respectively) spanning 7.8 Mb (www.hapmap.org, and also, additional SNP data provided kindly by Illumina) for comprehensive LD analyses between the AGER locus SNPs and other surrounding MHC loci.
FIGURE 1.
Genomic organization of the MHC region of human chromosome 6p21.3. Location of DRB1, DRB5, and the seven SNPs in four genes covering a 1.2-Mb segment telomeric to the DRB1 gene. Each filled box represents the relative size and location of each gene in relation to each other. dbSNP rs numbers are listed below each gene.
Ager RNA expression
Transcriptional activity of Ager in lymph nodes and spinal cord of experimental allergic encephalomyelitis (EAE) mice was determined as part of a genome-wide longitudinal expression study previously reported (21, 22). Here we mined the expression of Ager and that of its ligands S100a1–13 and S100b at each of the clinical stages of the induced disease (BE, before EAE; EE, early EAE; PE, peak EAE, ER, early recovery; LR, late recovery). Normalized expression values for these genes were subjected to hierarchical clustering using Euclidean distance and average linkage as the distance metrics.
Statistical analysis
All genotypes were tested for deviation from Hardy-Weinberg expectations in African American and white MS cases and controls using PyPop (version 0.6) (23) or Haploview (version 4.0) (24). Affected family-based controls (nontransmitted parental alleles or “AFBAC”) were derived for MHC SNPs and class II HLA loci in the African American dataset as previously described (25) and combined with data from unrelated controls, when possible, to increase statistical power for association tests. p-values, odds ratios, and confidence intervals for allele or genotype heterogeneity tests in African American and white MS cases and controls were derived using the Fisher’s exact test implemented in Stata (version 9.2, StataCorp). Two sample t tests were used to compare mean European ancestry proportions in African American MS cases and controls. Pairwise LD measures (r2) and significance for MHC region SNP and HLA-DRB1 data were calculated using Haploview (version 4.0) (24). Global (omnibus) haplotype tests of association were performed using the weighted haplotype program (WHAP) (version 2.09) (26). For N haplotypes, the omnibus test is a N − 1 degree-of-freedom test, jointly testing all haplotypes. DRB1 alleles were coded as DRB1*03, DRB1*1501/3 or other for haplotype assignment. Assessment of MHC SNP associations conditioned on DRB1 genotypes was performed using both WHAP and the conditional haplotype method (CHM) (27, 28) to identify specific effects. Significance for association tests using WHAP was determined using 1000 permutations. Only permuted p-values are reported. For CHM, haplotypes were assigned and frequencies summarized for analysis using UNPHASED (version 3.0.7) (29, 30) or PyPop (version 0.6) (23). p-values, odds ratios, and confidence intervals for CHM heterogeneity tests were derived using the Fisher’s exact test. Genotype-phenotype correlations used Kaplan-Meier survival estimates and a Cox proportional hazard model.
Results
Table I lists the clinical and demographic features of the study participants. An increased disease risk associated with the DRB1*15 (both DRB1*1501 and *1503) and DRB1*03 alleles was observed (Table II), as previously reported for a subset of this dataset (15). After accounting for DRB1*15 and *03 effects, no other DRB1 alleles demonstrated evidence for association (data not shown). DRB1*15 haplotypes carry two functional DRβ-chain genes, DRB1 and DRB5, and two different DR dimers can thus be formed by pairing with the nonpolymorphic DRα-chain (31). Since the DRB5 locus is carried exclusively on DRB1*15 and *16 haplotypes, as expected, a strong association with MS was observed with this locus as well (odds ratio (OR) = 1.40, p = 0.0002, Table II). However, DRB5*null individuals were previously described in populations with African ancestry (32) and offer the opportunity to distinguish between independent effects of DRB1 and DRB5. To address this hypothesis, DRB1-DRB5 two-locus haplotypes were assigned in MS cases and controls (Table II). While 100% of observed DRB1*1501 haplotypes in this dataset included the DRB5*0101 gene (overall frequency = 6.4% and 2.9% in cases and controls, respectively), heterogeneity was observed for DRB5 on DRB1*1503 haplotypes: 18 (1.2%) MS cases and 7 (0.4%) controls carried the DRB1*1503-DRB5*null haplotype. Similar to DRB1*1501, the most common DRB5 allele on DRB1*1503 haplotypes was *0101 (>90% of haplotypes). Interestingly, DRB1*1503 was associated with MS in the absence of DRB5 (OR = 2.89, 95% CI = 1.15– 8.22, p = 0.015), thus excluding variation within DRB5 as a major susceptibility factor.
Table II.
DRB1 and DRB5 frequencies in African American MS cases and controlsa
| Cases | Controls | ||||||
|---|---|---|---|---|---|---|---|
| Frequency | Count | Frequency | Count | Odds Ratioc | 95% CI | p Value | |
| DRB1-DRB5 alleles | |||||||
| DRB1*15b | 0.209 | 322 | 0.147 | 237 | 1.53 | 1.27–1.85 | <0.0001 |
| DRB1*1501 | 0.064 | 98 | 0.029 | 47 | 2.35 | 1.63–3.43 | <0.0001 |
| DRB1*1503 | 0.146 | 224 | 0.118 | 190 | 1.33 | 1.07–1.65 | 0.008 |
| DRB1*03 | 0.162 | 216 | 0.133 | 99 | 1.35 | 1.03–1.78 | 0.02 |
| DRB5 | 0.214 | 330 | 0.163 | 262 | 1.40 | 1.17–1.69 | 0.0002 |
| DRB1-DRB5 haplotypes | |||||||
| 1501-0101 | 0.064 | 98 | 0.029 | 47 | 2.35 | 1.62–3.43 | <0.0001 |
| 1502-0101 | 0.003 | 4 | 0 | 0 | |||
| 1502-0102 | 0.001 | 1 | 0 | 0 | |||
| 1503-0101 | 0.133 | 205 | 0.114 | 183 | 1.26 | 1.01–1.57 | <0.04 |
| 1503-0104 | 0.001 | 1 | 0 | 0 | |||
| 1503-null | 0.012 | 18 | 0.004 | 7 | 2.89 | 1.15–8.22 | 0.015 |
| 16-0101 | 0 | 0 | 0.001 | 2 | |||
| 16-0102 | 0 | 0 | 0.001 | 1 | |||
| 16-0202 | 0.014 | 21 | 0.017 | 28 | |||
| 16-0203 | 0 | 0 | 0.001 | 1 | |||
| X-null | 0.774 | 1,190 | 0.833 | 1,339 | |||
PYPOP (version 0.6.0) was used to determine haplotypes for cases and controls (including nontransmitted family-based controls; see Materials and Methods).
DRB1*15 consists of all DRB1*15 alleles, including DRB1*1501 and DRB1*1503.
Odds ratios for DRB1*15/1501/1503 were determined using DRB1*X as the reference group, where DRB1*X are non-DRB1*1501/1503 alleles (1216 case and 1371 control chromosomes). Odds ratios for DRB1*03 were restricted to individuals with complete DRB1 genotype characterization (see Materials and Methods); the reference group consisted of non-DRB1*1501/1503/03 alleles (830 cases and 515 control chromosomes). Odds ratio for DRB1*1501/1503-DRB5 haplotypes were determined using DRB1*X-DRB5*null (X-null) as the reference group, where DRB1*X are non-DRB1*1501/1503 alleles.
HLA-DRβ5DRα heterodimers appear to be effective myelin Ag-presenting molecules (11), and recently published experiments using triple DRB1-DRB5-hTCR transgenics support functional epistasis between DRB1 and DRB5 genes whereby DRβ5 modifies the T cell response activated by DRβ1 thorough activation-induced cell death, resulting in a milder and relapsing form of autoimmune demyelinating experimental disease (33). On the basis of these notable observations, we hypothesized that DRB5*null-affected individuals would manifest a more aggressive and/or progressive disease. Using survival analysis, we found that DRB5*null subjects were at greater risk for developing secondary progressive multiple sclerosis (log rank test p = 0.036) (Fig. 2). The hazard ratio for the DRB5*null subjects is 2.17, p = 0.045 (SE = 0.84, 95% CI = 1.02– 4.64).
FIGURE 2.
Kaplan-Meier curve for time to development of secondary progressive MS according to genotype. The black line is wild-type individuals and the gray line is for DRB5*null individuals.
To determine whether DRB1 is the single susceptibility determinant within the HLA class II–III boundary region, seven additional SNPs in four genes covering a 1.2-Mb segment telomeric to the DRB1 gene (Fig. 1) were genotyped in African American MS cases and controls (Table III). No deviations from Hardy-Weinberg equilibrium were observed for any of the SNPs in control individuals (all p values were >0.01). Using single locus testing, significant associations (p < 0.05) were observed for three of the seven SNPs interrogated; two in AGER and one in BTNL2. Four-loci haplotypes comprised of DRB1, AGER, and BTNL2 SNPs were then tested for association in African American MS cases and controls. The overall (omnibus) haplotype analysis yielded a p-value of 0.007.
Table III.
Results for MHC region SNPs in African American MS cases and controlsa
| Case | Control | ||||||
|---|---|---|---|---|---|---|---|
| Locus/Allele | Allele 1 (Frequency) |
Allele 2 (Frequency) |
Allele 1 (Frequency) |
Allele 2 (Frequency) |
Odds Ratio | 95% CI | p Value |
| rs1051796, MICAex4 1 = C, 2 = T | 890 (0.579) | 648 (0.421) | 914 (0.569) | 692 (0.431) | 1.04 | 0.90–1.20 | 0.59 |
| rs1063635, MICAex4 1 = A, 2 = G | 1020 (0.663) | 518 (0.337) | 1094 (0.680) | 514 (0.320) | 0.93 | 0.79–1.08 | 0.32 |
| rs184003, AGERint7/8 1 = G, 2 = T | 1209 (0.786) | 329 (0.214) | 1289 (0.803) | 317 (0.197) | 0.90 | 0.76–1.08 | 0.27 |
| rs1035798, AGERint3/4 1 = C, 2 = T | 1484 (0.965) | 54 (0.035) | 1498 (0.932) | 110 (0.068) | 2.02 | 1.43–2.87 | <0.0001 |
| rs2070600, AGERex3 1 = G, 2 = A | 1526 (0.992) | 12 (0.008) | 1582 (0.984) | 26 (0.016) | 2.09 | 1.01–4.56 | <0.05 |
| rs2076530, BTNL2 1 = A, 2 = G | 1093 (0.711) | 445 (0.289) | 1082 (0.673) | 526 (0.327) | 1.19 | 1.02–1.39 | 0.023 |
| rs2395182, HLA-DRA 1 = T, 2 = G | 1090 (0.709) | 448 (0.291) | 1181 (0.735) | 425 (0.265) | 0.88 | 0.75–1.03 | 0.10 |
p-values, odds ratios, and 95% CI derived using Fisher’s exact test, two-sided. All analyses were performed in STATA (version 9.2).
To distinguish primary (DRB1) from secondary associations due to LD, extended haplotypes were then utilized for conditional analyses; results are shown in Table IV. When the MS-associated DRB1*15- and DRB1*03-bearing haplotypes were removed from the analyses, strong evidence for association with AGER (rs1035798 SNP allele 1) persisted (OR = 1.85, 95% CI = 1.15–2.95, p = 0.008). The independent association with AGER was confirmed in a white MS dataset (Tables V and VI). Analyses for rs1035798 in African Americans and rs2070600 in whites conditioned on DRB1 using the independent effect test implemented in WHAP yielded very similar results. Both AGER SNPs showed evidence for association when conditioned on the DRB1 genotype using WHAP (p < 0.01 for rs1035798 in African Americans and p < 0.0001 for rs2070600 in whites, data not shown), which were similar to results shown for the conditional haplotype analysis (Tables IV and VI). This is compatible with independent contributions from both AGER and DRB1 to MS susceptibility. The entire AGER gene was sequenced in 10 African American MS patients and 10 African American controls in an effort to locate suggestive causative SNPs in the gene, but no novel SNPs were found.
Table IV.
AGER/BTLN2 SNP allele associations with MS in African Americans in the absence of DRB1*15 and DRB1*15/*03a
|
DRB1*15 Negative Case and Control Haplotypes (Total 2N = 2587) |
DRB1*15 and *03 Negative Case and Control Haplotypes (Total 2N = 1345) |
|||||
|---|---|---|---|---|---|---|
| Locus/Allele | p Value | Odds Ratio | 95% CI | p Value | Odds Ratio | 95% CI |
| rs1035798, AGERint3/4 1 = C, 2 = T | 0.0003 | 1.84 | 1.30–2.62 | 0.008 | 1.85 | 1.15–2.95 |
| rs2070600, AGERex3 1 = G, 2 = A | 0.13 | 1.79 | 0.85–3.94 | 0.20 | 1.78 | 0.71–4.48 |
| rs2076530, BTNL2 1 = A, 2 = G | 0.32 | 1.09 | 0.92–1.29 | 0.87 | 0.98 | 0.78–1.23 |
PYPOP (version 0.6.0) was used to assign haplotypes for cases and controls (including nontransmitted family-based controls; see Materials and Methods). Evaluation of positive MHC SNP allele associations conditional on DRB1 was performed using the conditional haplotype method (see Materials and Methods). p-values, odds ratios, and 95% CI were derived using Fisher’s exact test, two-sided. All analyses shown above were performed in STATA (version 9.2). Overall (omnibus) four-locus haplotype test (rs1035798–rs2070600–rs2076530-DRB1) was performed using WHAP; p = 0.004.
Table V.
MHC region AGER SNPs and HLA-DRB1 in white MS cases and controlsa
| Case | Control | ||||||
|---|---|---|---|---|---|---|---|
| Locus/Allele | Allele 1 (Frequency) |
Allele 2 (Frequency) |
Allele 1 (Frequency) |
Allele 2 (Frequency) |
p Valuec | Odds Ratiod | 95% CI |
| DRB1*15a 1 = *1501, 2 = other | 259 (0.266) | 715 (0.734) | 91 (0.105) | 777 (0.895) | <0.0001 | 3.09 | 2.37–4.05 |
| DRB1*03b 1 = *03, 2 = other | 126 (0.130) | 844 (0.870) | 84 (0.097) | 780 (0.903) | 0.0330 | 1.39 | 1.02–1.88 |
| rs1035798 AGERint3/4, 1 = C, 2 = T | 752 (0.772) | 222 (0.228) | 615 (0.709) | 253 (0.291) | 0.0020 | 1.29 | 1.12–1.73 |
| rs2070600 AGERex3, 1 = G, 2 = A | 959 (0.985) | 15 (0.015) | 824 (0.949) | 44 (0.051) | <0.0001 | 3.41 | 1.85–6.65 |
Total number of MS cases (N = 487); MS controls (N = 434).
See Materials and Methods for DRB1 allele designations.
Fisher’s exact test, two sided.
Odds ratios (95% CI; Fisher’s exact test p-values).
Table VI.
AGER SNP allele associations with MS in whites in the absence of DRB1*15 and DRB1*15/*03a
|
DRB1*15 Negative Case and Control Haplotypes (Total 2N = 1492) |
DRB1*15 and *03 Negative Case and Control Haplotypes (Total 2N = 1274) |
|||||
|---|---|---|---|---|---|---|
| Locus/Allele | p Value | Odds Ratio | 95% CI | p Value | Odds Ratio | 95% CI |
| rs1035798, AGERint3/4 1 = C, 2 = T | 0.4649 | 1.09 | 0.87–1.37 | 0.9531 | 0.99 | 0.78–1.25 |
| rs2070600, AGERex3 1 = G, 2 = A | <0.0001 | 3.75 | 1.88–8.12 | <0.0001 | 4.37 | 2.08–10.25 |
Total number of MS cases (N = 487); MS controls (N = 434); see Materials and Methods for DRB1 allele designations. UNPHASED (version 3.0.7) was used to assign haplotypes for cases and controls; see Materials and Methods). Evaluation of positive MHC SNP allele associations conditional on DRB1 was performed using the conditional haplotype method (CHM). p-values, odds ratios, and 95% CI derived using Fisher’s exact test, two-sided. Overall (omnibus) three-locus haplotype test (rs1035798–rs2070600–DRB1) was performed using WHAP; p < 0.0001.
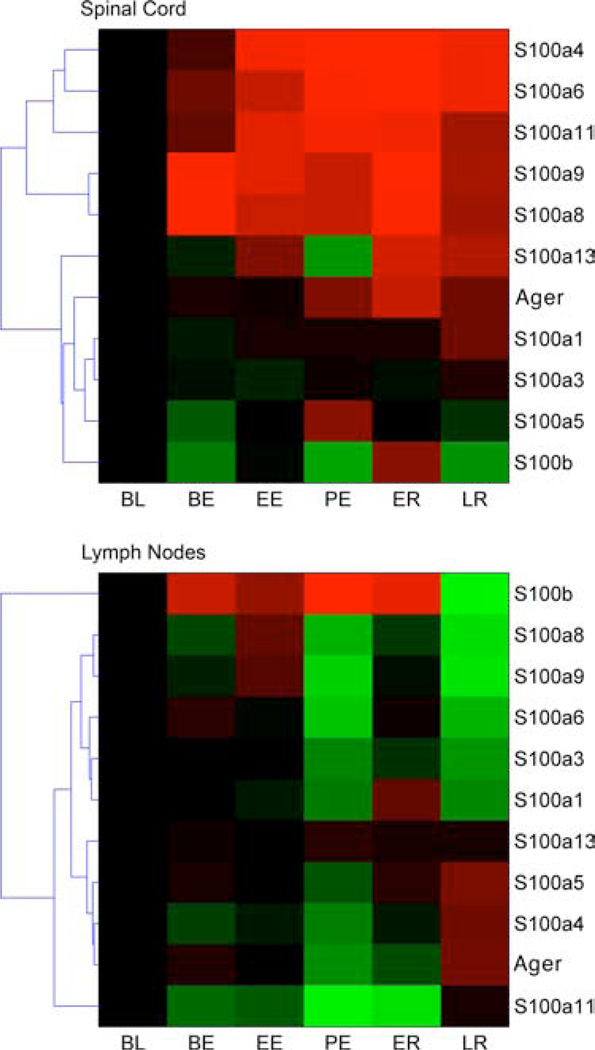
To further assess the involvement of AGER in neuroinflammation, we interrogated a large longitudinal gene expression study of relapsing EAE (21, 22). In that study, microarrays were used to monitor the expression of 22,000 genes in spinal cord and lymph nodes of NOD mice at several stages after immunization with the encephalitogenic peptide myelin oligodendrocyte glycoprotein (MOG)35–55. We thus mined the transcriptional dataset to examine the expression of Ager and its ligands, the S100-calgranulins. Progressive increase in gene expression that correlated with disease symptoms was observed for Ager as well as for all the tested calgranulins except S100b. Notably, S100a8 and S100a9 showed marked up-regulation even before symptoms of EAE were evident (Fig. 3), followed by decreased expression during the recovery phase. Interestingly, a reverse pattern of expression (with concomitant down-regulation as disease progressed) of Ager and S100-calgranulins was observed in the lymph nodes of the same animals. This mirror-like pattern also applies to S100b, whose expression in lymph nodes peaks at the time of maximal disability.
FIGURE 3.
Longitudinal gene expression of Ager and the calgranulins ligands. The expression of Ager and calgranulins was measured longitudinally in EAE by microarray analysis. The expression of each gene was tested at several stages of the disease. Red indicates increased expression, and green indicates reduced expression compared with controls (mice injected with CFA). BL, baseline; BE, before EAE; EE, early EAE; PE peak EAE; ER early recovery; LR, late recovery. Microarray data accession GEO number: GSE7461 (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?accGSE7461).
Discussion
The HLA locus on chromosome 6p21 is the strongest genetic factor identified as influencing MS susceptibility. However, previous attempts to isolate the susceptibility gene in this region did not provide consensus. The discovery of the causal variant was impeded by the high degree of LD that characterizes the DRB1*1501 haplotypes in the high-susceptibility northern European populations (13). The rigidity of this haplotype is the result of recent population history and may indicate selection events (34). Because LD patterns differ between populations, the analysis of African Americans, who have substantially smaller blocks of disequilibrium, is an attractive strategy to identify recombination events that will assist in the identification of disease genes. In a previous study of DRB1 and DQB1 alleles and haplotypes in an African American MS cohort, a selective association with DRB1*15 was revealed, establishing the centromeric boundary of the HLA class II DR-DQ association in MS and suggesting a primary role for the DRB1 gene in MS independent of DQB1*0602 (15). Conversely, the introduction of DQB1*0601 into DRB1*1502 transgenic mice reduced EAE severity, suggesting modulatory effects on disease progression (35). African American patients also exhibited a high degree of DRB1 allelic heterogenity as disease association was found for DRB1*1501, DRB1*1503, and DRB1*0301 alleles. The HLA-DRB1*0301 association with MS confirmed here in African-Americans has been previously demonstrated in Sardinian patients (36), whereas HLA-DRB1*0301 transgenic mice are susceptible to proteolipid protein-induced EAE (37).
Altogether, the haplotypic features of the DRB1*1501-DQB1*X (X = non-0602) and DRB1*1503-positive chromosomes indicated an older African origin for the HLA-associated MS susceptibility genes, predating the divergence of human ethnic groups (15). The present analysis further narrows the susceptibility locus within the class II region to DRB1. A primary role for DRB1 in susceptibility to MS is consistent with a pathogenesis model that involves a T cell-mediated autoimmune response. Susceptibility may be then related to the known function of the encoded molecules in the normal immune response, Ag binding and presentation and T cell repertoire determination.
The crystal structure resolution of a DRα/DRβ5*0101-EBV peptide complex revealed a marked structural equivalence to the DRβ1*1501-myelin basic protein (MBP) peptide complex at the surface presented for TCR recognition (38), suggesting that EBV peptides with only limited sequence identity with a myelin peptide could activate autoreactive T cells and initiate an autoimmune response. Indeed, HLA-DRαDRβ5 heterodimers appear to be effective MBP Ag-presenting molecules (11). However, herein we have demonstrated that ~8% of African American MS DRB1*1503 haplotypes are null for the DRB5 gene, thus excluding DRB5 as an obligatory risk gene. Interestingly, a comparison of DRB1*1503-DRB5*0101 with DRB1*1503-DRB5*null haplotypes in MS cases and controls suggests that DRB5*0101 may modify the risk of DRB1*1503 (OR = 0.43, 95% CI = 0.15–1.11, p = 0.07), although this result did not reach statistical significance. An additive or redundant functional role for DRB5 in disease susceptibility, only applying to DRB1*1501 individuals, is unlikely. On the other hand, animal data using triple DRB1/DRB5/hTCR transgenics strongly favor a disease-modifying effect for DRB5 (33). Although based on a small number of individuals with the rare DRB5*null mutation, we found that these subjects were at higher risk for development of secondary progressive MS. The convergence of findings obtained from these HLA-humanized EAE mice with the emerging human MS genetic data is indeed remarkable, supporting a modulatory role of DRB5 gene products on the progression of human demyelinating disease.
The absence of DRB5 was observed only in the DRB1*1503 haplotypes (~74% of the DRB1*15 samples in the African American cohort contain the *1503 allele vs 0% of the white population). Although structural features of DRB1*1503 have not been described, the two DRB1*15 alleles differ only at position 30 (Tyr in *1501, His in *1503), away from the critical pockets anchoring the peptides. Immunological studies showed that both alleles are equally efficient in presenting the immunodominant epitope MBP 85–99 to specific T cell lines (39), suggesting that β*1501 and β*1503 molecules act similarly in MS development.
A second important observation emerging from this study is the potential independent role for the class III gene AGER, encoding an important member of the Ig superfamily (16). The AGER receptor is present on several cell types, including lymphocytes, mononuclear phagocytes, and vascular endothelial cells (40, 41), and it was first identified as a signal transduction receptor for advanced glycation end products (AGEs). AGEs result from nonenzymatic glycation of proteins, lipids, and nucleic acids, particularly at sites of oxidative stress, and are known to accumulate in a number of chronic inflammatory and neurodegenerative diseases such as diabetes, Alzheimer disease, and amyotrophic lateral sclerosis (42, 43). AGE-AGER ligation induces multiple signal transduction pathways including p21ras, MAP kinase, and the proinflammatory NF-κB pathway (44). Non-AGE ligands for AGER also were identified, including: 1) the S100/calgranulins, which were associated with several chronic inflammatory and systemic autoimmune diseases and are involved in immune cell and vascular endothelium activation (45, 46); 2) amyloid-β peptide, whose interaction with microglia within the CNS is linked to sustained inflammation and neuronal toxicity and cell death (47); 3) amphoterin (high mobility group box chromosomal protein 1 or HMGB-1), a molecule with implications for neurite outgrowth (48); and 4) other uncharacterized cell surface molecules on bacteria and prions (49, 50). While a role for AGER in MS has not yet been established, there is strong evidence for its involvement in the activation of MBP-reactive CD4+ T cells in EAE models (51). Furthermore, blocking AGER ameliorates the model disease by preventing the infiltration of encephalitogenic T cells into the CNS (51). Finally, a correlation between serum AGER levels and disease progression in MS was recently reported (52). Using transcriptional information from the CNS and lymph nodes of mice with EAE and controls, we show differential expression of Ager and its ligands, thus providing additional evidence for a potential role of Ager in EAE/MS. Previous results based on a small study sample also suggest that variation in AGER may influence inflammatory responses (53); therefore, it is a plausible disease candidate for autoimmune conditions such as MS.
The intronic variant of AGER (rs1035798) that was found associated with MS in African Americans is unlikely to be functional by itself. While two other polymorphisms in AGER were also examined in this study (rs2070600, a rare missense variant located in exon 3, and rs184003, another intronic polymorphism), neither demonstrated evidence of association with MS in African Americans. On the other hand, the AGER rs2070600 variant was strongly associated in the white MS case-control dataset. Neither of the associated AGER SNPs are in strong LD with each other, in either dataset (r2 < 0.02 for pairwise correlation in African Americans and whites), which suggests a role for other rare variants within AGER or at nearby loci. Importantly, the class III region within the MHC is the most gene-dense region of the human genome (54), and a comprehensive evaluation of all available MHC SNP data in CEU and YRI populations (see Materials and Methods) shows that the associated AGER SNPs from this study (rs1035798 and rs2070600) are linked with several other class III region genes. These include AGPAT1, PBX2, and NOTCH4 (r2 > 0.6) in CEU, EGFL8, and CREBL1 loci (r2 > 0.6) in both CEU and YRI and, finally, TNXB, CYP21A2, RDBP, HSPA1L, and MSH5 loci (r2 > 0.9) in YRI only, providing a long list of strong candidates for comprehensive mapping efforts.
The current data underscore the power of ethnically defined cohorts to identify disease genes by association for complex diseases. The data demonstrate that, in contrast to the prevailing single locus model, the MHC associations with MS result from complex, multilocus effects that span the entire region. The full characterization of the association range in informative datasets is important to understand MS susceptibility, as well as the role of genetics in progression and response to therapeutics.
Acknowledgments
We are grateful to the MS patients and their families for participating in this study. We thank Robin Lincoln, Wendy Chin, Hourieh Mousavi, and Rosa Guerrero for expert specimen management and Refugia Gomez for database management. We also acknowledge the contribution on non-MS African American samples from John Kane (University of California, San Francisco) and John B. Harley (Oklahoma Medical Research Foundation).
Footnotes
This work was funded by grants from the National Institutes of Health (RO1 NS046297, U19AI067152, K23 NS048869-01, and R01NS049510) and the National Multiple Sclerosis Society (RG3060C8).
Abbreviations used in this paper: MS, multiple sclerosis; AGE, advanced glycation end product; CHM, conditional haplotype method; EAE, experimental autoimmune encephalomyelitis; LD, linkage disequilibrium; MBP, myelin basic protein; OR, odds ratio; SNP, single nucleotide polymorphisms.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Fernald GH, Yeh RF, Hauser SL, Oksenberg JR, Baranzini SE. Mapping gene activity in complex disorders: integration of expression and genomic scans for multiple sclerosis. J. Neuroimmunol. 2005;167:157–169. doi: 10.1016/j.jneuroim.2005.06.032. [DOI] [PubMed] [Google Scholar]
- 2.Abdeen H, Heggarty S, Hawkins SA, Hutchinson M, McDonnell GV, Graham CA. Mapping candidate non-MHC susceptibility regions to multiple sclerosis. Genes Immun. 2006;7:494–502. doi: 10.1038/sj.gene.6364320. [DOI] [PubMed] [Google Scholar]
- 3.Hermanowski J, Bouzigon E, Forabosco P, Ng MY, Fisher SA, Lewis CM. Meta-analysis of genome-wide linkage studies for multiple sclerosis, using an extended GSMA method. Eur. J. Hum. Genet. 2007;15:703–710. doi: 10.1038/sj.ejhg.5201818. [DOI] [PubMed] [Google Scholar]
- 4.International Multiple Sclerosis Genetics Consortium. Hafler DA, Compston A, Sawcer S, Lander ES, Daly MJ, De Jager PL, de Bakker PI, Gabriel SB, Mirel DB, et al. Risk alleles for multiple sclerosis identified by a genomewide study. N. Engl. J. Med. 2007;357:851–862. doi: 10.1056/NEJMoa073493. [DOI] [PubMed] [Google Scholar]
- 5.Barcellos LF, Oksenberg JR, Begovich AB, Martin ER, Schmidt S, Vittinghoff E, Goodin DS, Pelletier D, Lincoln RR, Bucher P, et al. HLA-DR2 dose effect on susceptibility to multiple sclerosis and influence on disease course. Am. J. Hum. Genet. 2003;72:710–716. doi: 10.1086/367781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lincoln MR, Montpetit A, Cader MZ, Saarela J, Dyment DA, Tiislar M, Ferretti V, Tienari PJ, Sadovnick AD, Peltonen L, et al. A predominant role for the HLA class II region in the association of the MHC region with multiple sclerosis. Nat. Genet. 2005;37:1108–1112. doi: 10.1038/ng1647. [DOI] [PubMed] [Google Scholar]
- 7.Fukazawa T, Kikuchi S, Sasaki H, Yabe I, Miyagishi R, Hamada T, Tashiro K. Genomic HLA profiles of MS in Hokkaido, Japan: important role of DPB1*0501 allele. J. Neurol. 2000;247:175–178. doi: 10.1007/s004150050558. [DOI] [PubMed] [Google Scholar]
- 8.Fernandez O, Fernandez V, Alonso A, Caballero A, Luque G, Bravo M, Leon A, Mayorga C, Leyva L, de Ramon E. DQB1*0602 allele shows a strong association with multiple sclerosis in patients in Malaga, Spain. J. Neurol. 2004;251:440–444. doi: 10.1007/s00415-004-0350-2. [DOI] [PubMed] [Google Scholar]
- 9.Khare M, Mangalam A, Rodriguez M, David CS. HLA DR and DQ interaction in myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in HLA class II transgenic mice. J. Neuroimmunol. 2005;169:1–12. doi: 10.1016/j.jneuroim.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 10.Prat E, Tomaru U, Sabater L, Park DM, Granger R, Kruse N, Ohayon JM, Bettinotti MP, Martin R. HLA-DRB5*0101 and -DRB1*1501 expression in the multiple sclerosis-associated HLA-DR15 haplotype. J. Neuroimmunol. 2005;167:108–119. doi: 10.1016/j.jneuroim.2005.04.027. [DOI] [PubMed] [Google Scholar]
- 11.Sospedra M, Muraro PA, Stefanova I, Zhao Y, Chung K, Li Y, Giulianotti M, Simon R, Mariuzza R, Pinilla C, Martin R. Redundancy in antigen-presenting function of the HLA-DR and -DQ molecules in the multiple sclerosis-associated HLA-DR2 haplotype. J. Immunol. 2006;176:1951–1961. doi: 10.4049/jimmunol.176.3.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chao MJ, Barnardo MC, Lui GZ, Lincoln MR, Ramagopalan SV, Herrera BM, Dyment DA, Sadovnick AD, Ebers GC. Transmission of class I/II multi-locus MHC haplotypes and multiple sclerosis susceptibility: accounting for linkage disequilibrium. Hum. Mol. Genet. 2007;16:1951–1958. doi: 10.1093/hmg/ddm142. [DOI] [PubMed] [Google Scholar]
- 13.Miretti MM, Walsh EC, Ke X, Delgado M, Griffiths M, Hunt S, Morrison J, Whittaker P, Lander ES, Cardon LR, et al. A high-resolution linkage-disequilibrium map of the human major histocompatibility complex and first generation of tag single-nucleotide polymorphisms. Am. J. Hum. Genet. 2005;76:634–646. doi: 10.1086/429393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wallin MT, Page WF, Kurtzke JF. Multiple sclerosis in US veterans of the Vietnam era and later military service: race, sex, and geography. Ann. Neurol. 2004;55:65–71. doi: 10.1002/ana.10788. [DOI] [PubMed] [Google Scholar]
- 15.Oksenberg JR, Barcellos LF, Cree BA, Baranzini SE, Bugawan TL, Khan O, Lincoln RR, Swerdlin A, Mignot E, Lin L, et al. Mapping multiple sclerosis susceptibility to the HLA-DR locus in African Americans. Am. J. Hum. Genet. 2004;74:160–167. doi: 10.1086/380997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neeper M, Schmidt AM, Brett J, Yan SD, Wang F, Pan YC, Elliston K, Stern D, Shaw A. Cloning and expression of a cell surface receptor for advanced glycosylation end products of proteins. J. Biol. Chem. 1992;267:14998–15004. [PubMed] [Google Scholar]
- 17.McDonald WI, Compston A, Edan G, Goodkin D, Hartung HP, Lublin FD, McFarland HF, Paty DW, Polman CH, Reingold SC, et al. Recommended diagnostic criteria for multiple sclerosis: guidelines from the international panel on the diagnosis of multiple sclerosis. Ann. Neurol. 2001;50:121–127. doi: 10.1002/ana.1032. [DOI] [PubMed] [Google Scholar]
- 18.Cree BA, Khan O, Bourdette D, Goodin DS, Cohen JA, Marrie RA, Glidden D, Weinstock-Guttman B, Reich D, Patterson N, et al. Clinical characteristics of African Americans vs Caucasian Americans with multiple sclerosis. Neurology. 2004;63:2039–2045. doi: 10.1212/01.wnl.0000145762.60562.5d. [DOI] [PubMed] [Google Scholar]
- 19.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, et al. Methods for high-density admixture mapping of disease genes. Am. J. Hum. Genet. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, Marchini J, Ke X, Monsuur AJ, Whittaker P, Delgado M, et al. A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat. Genet. 2006;38:1166–1172. doi: 10.1038/ng1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baranzini SE, Bernard CC, Oksenberg JR. Modular transcriptional activity characterizes the initiation and progression of autoimmune encephalomyelitis. J. Immunol. 2005;174:7412–7422. doi: 10.4049/jimmunol.174.11.7412. [DOI] [PubMed] [Google Scholar]
- 22.Otaegui D, Mostafavi S, Bernard CC, de Munain AL, Mousavi P, Oksenberg JR, Baranzini SE. Increased transcriptional activity of milk-related genes following the active phase of experimental autoimmune encephalomyelitis and multiple sclerosis. J. Immunol. 2007;179:4074–4082. doi: 10.4049/jimmunol.179.6.4074. [DOI] [PubMed] [Google Scholar]
- 23.Lancaster AK, Single RM, Solberg OD, Nelson MP, Thomson G. PyPop update: a software pipeline for large-scale multilocus population genomics. Tissue Antigens. 2007;69(Suppl. 1):192–197. doi: 10.1111/j.1399-0039.2006.00769.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 25.Thomson G. Mapping disease genes: family-based association studies. Am. J. Hum. Genet. 1995;57:487–498. [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Daly MJ, Sham PC. WHAP: haplotype-based association analysis. Bioinformatics. 2007;23:255–256. doi: 10.1093/bioinformatics/btl580. [DOI] [PubMed] [Google Scholar]
- 27.Valdes AM, McWeeney S, Thomson G. HLA class II DR-DQ amino acids and insulin-dependent diabetes mellitus: application of the haplotype method. Am. J. Hum. Genet. 1997;60:717–728. [PMC free article] [PubMed] [Google Scholar]
- 28.Valdes AM, Thomson G. Detecting disease-predisposing variants: the haplotype method. Am. J. Hum. Genet. 1997;60:703–716. [PMC free article] [PubMed] [Google Scholar]
- 29.Dudbridge F. Pedigree disequilibrium tests for multilocus haplotypes. Genet. Epidemiol. 2003;25:115–121. doi: 10.1002/gepi.10252. [DOI] [PubMed] [Google Scholar]
- 30.Dudbridge F. UNPHASED user guide: technical report 2006/5. Cambridge, UK: MRC Biostatistics Unit; 2006. [Google Scholar]
- 31.Sone T, Tsukamoto K, Hirayama K, Nishimura Y, Takenouchi T, Aizawa M, Sasazuki T. Two distinct class II molecules encoded by the genes within HLA-DR subregion of HLA-Dw2 and Dw12 can act as stimulating and restriction molecules. J. Immunol. 1985;135:1288–1298. [PubMed] [Google Scholar]
- 32.Robbins F, Hurley CK, Tang T, Yao H, Lin YS, Wade J, Goeken N, Hartzman RJ. Diversity associated with the second expressed HLA-DRB locus in the human population. Immunogenetics. 1997;46:104–110. doi: 10.1007/s002510050248. [DOI] [PubMed] [Google Scholar]
- 33.Gregersen JW, Kranc KR, Ke X, Svendsen P, Madsen LS, Thomsen AR, Cardon LR, Bell JI, Fugger L. Functional epistasis on a common MHC haplotype associated with multiple sclerosis. Nature. 2006;443:574–577. doi: 10.1038/nature05133. [DOI] [PubMed] [Google Scholar]
- 34.Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G. Signatures of demographic history and natural selection in the human major histocompatibility complex loci. Genetics. 2006;173:2121–2142. doi: 10.1534/genetics.105.052837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khare M, Mangalam A, Rodriguez M, David CS. HLA DR and DQ interaction in myelin oligodendrocyte glycoprotein-induced experimental autoimmune encephalomyelitis in HLA class II transgenic mice. J. Neuroimmumol. 2005;169:1–12. doi: 10.1016/j.jneuroim.2005.07.023. [DOI] [PubMed] [Google Scholar]
- 36.Marrosu MG, Murru MR, Costa G, Murru R, Muntoni F, Cucca F. DRB1-DQA1-DQB1 loci and MS predisposition in the Sardinian population. Hum. Mol. Genet. 1998;7:1235–1237. doi: 10.1093/hmg/7.8.1235. [DOI] [PubMed] [Google Scholar]
- 37.Mangalam AK, Khare M, Krco C, Rodriguez M, David CS. Identification of T cell epitopes in human proteoplipid protein and induction of experimental autoimmune encephalomyelitis in HLA class II-transgenic mice. Eur. J. Immunol. 2004;34:280–290. doi: 10.1002/eji.200324597. [DOI] [PubMed] [Google Scholar]
- 38.Lang HL, Jacobsen H, Ikemizu S, Andersson C, Harlos K, Madsen L, Hjorth P, Sondergaard L, Svejgaard A, Wucherpfennig K, et al. A functional and structural basis for TCR cross-reactivity in multiple sclerosis. Nat. Immunol. 2002;3:940–943. doi: 10.1038/ni835. [DOI] [PubMed] [Google Scholar]
- 39.Quelvennec E, Bera O, Cabre P, Alizadeh M, Smadja D, Jugde F, Edan G, Semana G. Genetic and functional studies in multiple sclerosis patients from Martinique attest for a specific and direct role of the HLA-DR locus in the syndrome. Tissue Antigens. 2003;61:166–171. doi: 10.1046/j.0001-2815.2002.00008.x. [DOI] [PubMed] [Google Scholar]
- 40.Bierhaus A, Humpert PM, Morcos M, Wendt T, Chavakis T, Arnold B, Stern DM, Nawroth PP. Understanding RAGE, the receptor for advanced glycation end products. J. Mol. Med. 2005;83:876–886. doi: 10.1007/s00109-005-0688-7. [DOI] [PubMed] [Google Scholar]
- 41.Basta G, Lazzerini G, Massaro M, Simoncini T, Tanganelli P, Fu C, Kislinger T, Stern DM, Schmidt AM, De Caterina R. Advanced glycation end products activate endothelium through signal-transduction receptor RAGE: a mechanism for amplification of inflammatory responses. Circulation. 2002;105:816–822. doi: 10.1161/hc0702.104183. [DOI] [PubMed] [Google Scholar]
- 42.Rong LL, Gooch C, Szabolcs M, Herold KC, Lalla E, Hays AP, Yan SF, Yan SS, Schmidt AM. RAGE: a journey from the complications of diabetes to disorders of the nervous system: striking a fine balance between injury and repair. Restor. Neurol. Neurosci. 2005;23:355–365. [PubMed] [Google Scholar]
- 43.Bucciarelli LG, Wendt T, Rong L, Lalla E, Hofmann MA, Goova MT, Taguchi A, Yan SF, Yan SD, Stern DM, Schmidt AM. RAGE is a multiligand receptor of the immunoglobulin superfamily: implications for homeostasis and chronic disease. Cell Mol. Life Sci. 2002;59:1117–1128. doi: 10.1007/s00018-002-8491-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lander HM, Tauras JM, Ogiste JS, Hori O, Moss RA, Schmidt AM. Activation of the receptor for advanced glycation end products triggers a p21(ras)-dependent mitogen-activated protein kinase pathway regulated by oxidant stress. J. Biol. Chem. 1997;272:17810–17814. doi: 10.1074/jbc.272.28.17810. [DOI] [PubMed] [Google Scholar]
- 45.Hofmann MA, Drury S, Fu C, Qu W, Taguchi A, Lu Y, Avila C, Kambham N, Bierhaus A, Nawroth P, et al. RAGE mediates a novel proinflammatory axis: a central cell surface receptor for S100/calgranulin polypeptides. Cell. 1999;97:889–901. doi: 10.1016/s0092-8674(00)80801-6. [DOI] [PubMed] [Google Scholar]
- 46.Foell D, Wittkowski H, Vogl T, Roth J. S100 proteins expressed in phagocytes: a novel group of damage-associated molecular pattern molecules. J. Leukocyte Biol. 2007;81:28–37. doi: 10.1189/jlb.0306170. [DOI] [PubMed] [Google Scholar]
- 47.Yan SD, Chen X, Fu J, Chen M, Zhu H, Roher A, Slattery T, Zhao L, Nagashima M, Morser J, et al. RAGE and amyloid-β peptide neurotoxicity in Alzheimer’s disease. Nature. 1996;382:685–691. doi: 10.1038/382685a0. [DOI] [PubMed] [Google Scholar]
- 48.Hori O, Brett J, Slattery T, Cao R, Zhang J, Chen JX, Nagashima M, Lundh ER, Vijay S, Nitecki D. The receptor for advanced glycation end products (RAGE) is a cellular binding site for amphoterin: mediation of neurite outgrowth and co-expression of rage and amphoterin in the developing nervous system. J. Biol. Chem. 1995;270:25752–25761. doi: 10.1074/jbc.270.43.25752. [DOI] [PubMed] [Google Scholar]
- 49.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sasaki N, Takeuchi M, Chowei H, Kikuchi S, Hayashi Y, Nakano N, Ikeda H, Yamagishi S, Kitamoto T, Saito T, Makita Z. Advanced glycation end products (AGE) and their receptor (RAGE) in the brain of patients with Creutzfeldt-Jakob disease with prion plaques. Neurosci. Lett. 2002;326:117–120. doi: 10.1016/s0304-3940(02)00310-5. [DOI] [PubMed] [Google Scholar]
- 51.Yan SS, Wu ZY, Zhang HP, Furtado G, Chen X, Yan SF, Schmidt AM, Brown C, Stern A, LaFaille J, et al. Suppression of experimental autoimmune encephalomyelitis by selective blockade of encephalitogenic T-cell infiltration of the central nervous system. Nat. Med. 2003;9:287–293. doi: 10.1038/nm831. [DOI] [PubMed] [Google Scholar]
- 52.Sternberg Z, Weinstock-Guttman B, Hojnacki D, Zamboni P, Zivadinov R, Chadha K, Lieberman A, Kazim L, Drake A, Rocco P, et al. Soluble receptor for advanced glycation end products in multiple sclerosis: a potential marker of disease severity. Mult. Scler. 2008;14:759–763. doi: 10.1177/1352458507088105. [DOI] [PubMed] [Google Scholar]
- 53.Hofmann MA, Drury S, Hudson BI, Gleason MR, Qu W, Lu Y, Lalla E, Chitnis S, Monteiro J, Stickland MH, et al. RAGE and arthritis: the G82S polymorphism amplifies the inflammatory response. Genes Immun. 2002;3:123–135. doi: 10.1038/sj.gene.6363861. [DOI] [PubMed] [Google Scholar]
- 54.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]