Abstract
Receiver domains control intracellular responses triggered by signal transduction in bacterial two-component systems. Here, we report the solution NMR structure and dynamics of Sma0114 from the bacterium Sinorhizobium meliloti, the first such characterization of a receiver domain from the HWE-kinase family of two-component systems. The structure of Sma0114 adopts a prototypical α5/β5 Rossman-fold but has features that set it apart from other receiver domains. The fourth β-strand of Sma0114 houses a PFxFATGY sequence motif, common to many HWE-kinase-associated receiver domains. This sequence motif in Sma0114 may substitute for the conserved Y-T coupling mechanism, which propagates conformational transitions in the 455 (α4-β5-α5) faces of receiver domains, to prime them for binding downstream effectors once they become activated by phosphorylation. In addition, Sma0114 lacks the fourth α-helix of the consensus 455 face and 15N relaxation data show that it is replaced by a segment that is flexible on the ps-ns timescale. Secondary structure prediction of Sma0114 and other HWE-kinase-associated receiver domains suggests that the absence of helix α4 may be a conserved property of this family. In spite of these differences, Sma0114 has a conserved active site, binds divalent metal ions such as Mg2+ and Ca2+ that are required for phosphorylation, and exhibits μs-ms active site dynamics similar to other receiver domains. Taken together, our results suggest that Sma0114 has a conserved active site but differs from typical receiver domains in the structure of the 455 face that is used to effect signal transduction following activation.
Keywords: response regulator, enzyme dynamics, TCS, binding-site flexibility, Rhizobiales
Phosphorylation is a commonly used mechanism to achieve signal transduction. In bacteria, phosphorylation-mediated signal transduction is carried out by two-component systems. These signaling cascades utilize a sensor histidine kinase that autophosphorylates when triggered by a stimulus, and a response regulator which mediates the downstream output response upon receiving a phosphate from its cognate histidine kinase (1, 2). The output response is maintained while the receiver domain component of the response regulator remains phosphorylated. The lifetime of the phosphorylated state is controlled primarily by the autophosphatase enzymatic activity of the receiver domain.
The two-component system encoded by the genes sma0113 and sma0114 from the bacterium Sinorhizobium meliloti, was identified in a genetic screen for altered succinate-mediated catabolite repression (3). Based on previous work it is expected that Sma0113 and Sma0114 play a role in catabolite repression and polyhydroxy butyrate (PHB) synthesis (3). The tandem genes encode a histidine-tryptophan-glutamate (HWE) histidine kinase (Sma0113), and a single-domain response regulator (Sma0114). HWE-kinases were first described in 2004 (4) and constitute a subclass of the larger histidine kinase superfamily. Compared to the well-characterized histidine kinase family, HWE-kinases have an altered ATP binding site, which lacks the F-box that is normally an integral component of the ATP lid.
The majority of response regulators have receiver and effector domains. The receiver domain acts as a “switch” that undergoes a conformational change in response to phosphorylation at a conserved aspartate (1, 5). The effector domain usually has a DNA binding function that regulates the transcription of its target genes (1). In some cases, including the response regulators CheY, Spo0F, and Sma0114 only the receiver domain is present (6, 7). The roles of single domain response regulators are not as well understood as their two-domain counterparts but there are three known functions. The receiver domain can act directly on a protein effector (e.g. CheY), it can participate in a phospho-relay cascade (e.g. Spo0F), or it can act as a histidine kinases inhibitor (e.g. DivK) (8-10). Structural studies of single-domain response regulators have shown that these are structurally similar in their inactive apo forms but show greater variability in their phosphorylated activated states (11). This is probably because the function of the inactive receiver domain is to receive a phosphate from its cognate histidine kinase, whereas the active phosphorylated enzyme can bind to a wide range of downstream effector proteins (12, 13).
Receiver domains have a conserved α5/β5 Rossman fold, where 5 α-helices surround 5 parallel β-sheets. A conserved pair of acidic residues is located in the loop between strand β1 and helix α1, that forms part of the binding site for a divalent metal cation which is required for stabilizing the incoming phosphate group and forming the acyl-phosphate linkage (5, 14). The site of phosphorylation is a conserved Asp at the C-terminal end of strand β3 (1, 5). The C-terminal end of strand β5 houses a conserved Lys that stabilizes the incoming phosphate group (14). Phosphorylation induces a conformational switch, in which a conserved Thr at the C-terminal end of strand β4 hydrogen bonds with the phosphate group, and causes a rotameric change of an aromatic residue (Tyr or Phe) in strand β5 in a mechanism called ‘Y-T coupling’ (14). Y-T coupling mediates the more global rearrangement of the ‘455 face’ of the enzyme, comprised of the secondary structure elements α4-β5-α5. In Sma0114 the aromatic residue in the Y-T coupling mechanism is replaced by a leucine, and the conserved Thr is part of a PFxFATGY sequence motif that is common in the HWE-kinase-associated family of receiver domains.
The NMR investigations described herein show that the unusual sequence features of Sma0114 lead to unique structural features, primarily affecting the 455 face of the enzyme. 15N relaxation data show that there are also changes in the dynamics of the 455 face of Sma0114 compared to the corresponding regions of other receiver domains (15, 16). By contrast the active site of the enzyme and the ability to bind metals is retained, suggesting that the unusual features of Sma0114 do not alter the activation mechanism but rather the conformational changes of the 455 face accompanying phosphorylation.
Experimental Procedures
NMR Sample Preparation
Recombinant Sma0114 was ligated into a pET28(+) vector and transformed into Escherichia coli BL21 DE3 pLysS cells. Expression and purification of Sma0114 were performed as previously described (17). 15N-Sma0114 and 15N,13C-Sma0114 samples were dissolved in 50 mM Na2HPO4 buffered to pH 6.0. All samples for NMR had 500 μM concentrations of Sma0114, with 0.02% NaN3 to prevent bacterial growth and 1 mM DTT to prevent disulfide formation due to the sole cysteine at position 29 in the protein. For the metal titration with CaCl2 we used a pH 6.0 MES buffer rather than phosphate, to prevent precipitation of Ca2+.
NMR Structure Determination
Chemical shift assignments for Sma0114 have been published previously (17). 3D 15N- and 13C-editted NOESY experiments (18) were used to obtain NOE-based distance restraints. Long-range HNCO (19) and deuterium isotope exchange experiments were used to identify hydrogen bonds. Restraints for the backbone dihedral angles φ and ψ were calculated from the assigned HN, Hα, N, Cα, Cβ and C’ chemical shifts using the program TALOS (20). Additional 3D HNHA data were collected to check φ dihedral angles. Side-chain χ1 dihedral angles and stereospecific assignments for prochiral methylene protons were determined from 3D HNHB data (18) and short-mixing time 2D NOESY spectra (21). Stereospecific assignments for the prochiral methyl groups of Leu and Val residues were obtained from a sample fractionally labeled with 10% 13C-glucose (22). The NMR structure of Sma0114 was calculated using the program X-PLOR (v. 3.851) (23) based on 1627 experimental restraints (Table I). The 20 lowest energy structures have been deposited in the PDB under accession code 2LPM.
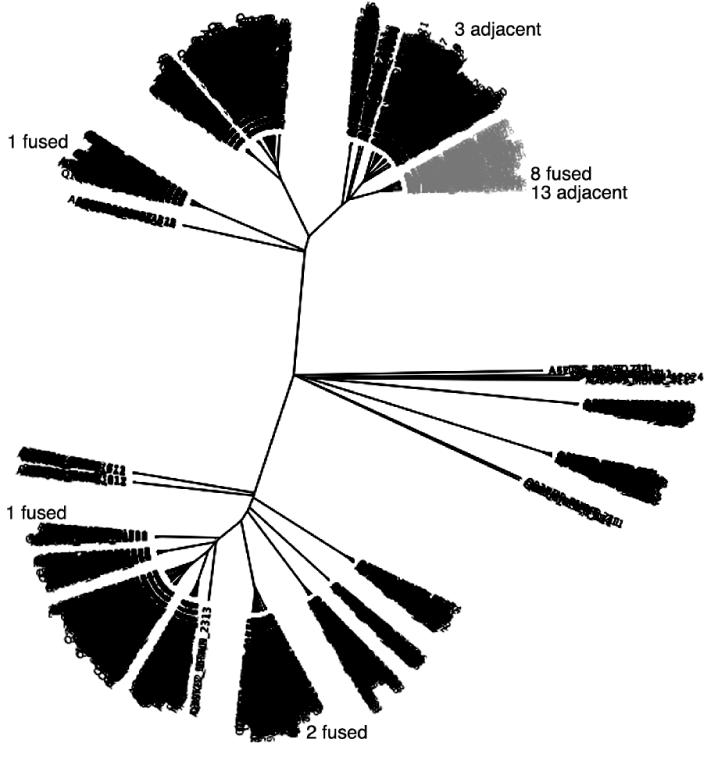
Figure 1.
Phylogentic analysis of HWE-associated response regulators. Maximum likelihood tree of 1792 receiver domains from the order Rhizobiales. The 28 receiver domains from the SMART database that are fused or genetically adjacent to HWE kinases are labeled in the figure. Sma0114 is in the grey cluster.
NMR Relaxation Measurements
Backbone dynamics of Sma0114 were investigated using 15N R1, R2 and 1H-15N NOE experiments. Longitudinal relaxation rates (R1) were characterized using relaxation delays of 0.02, 0.05,0.13,0.21,0.31,0.5,0.71 and 1.0 s. Transverse relaxation rates (R2) were measured using relaxation delays of 0.01,0.03,0.05,0.07,0.09,0.15,0.25 and 0.35 s. 1H-15N NOE values were determined from experiments in which the proton signals were saturated (s) for 4 s and control (c) experiments in which the saturation period was replaced by an equivalent preacquisition delay. Spectra were acquired in an interleaved manner for all three relaxation data sets. Relaxation rates were calculated from least-squares fits of the data to an exponential decay model (Eq. 1) where I is the intensity for the relaxation period τ, I0 is the initial amplitude and R1,2 corresponds to the relaxation rate R1 or R2.
| Equation 1 |
Experimental uncertainties in relaxation parameters were taken as the standard errors of the fits. 1H-15N NOE values were calculated according to Equation 2, where I(s) is the crosspeak intensity in the experiment with saturation (s) and I(c) is the crosspeak intensity without saturation (c).
| Equation 2 |
The errors for the 1H-15N NOE experiment were determined as described previously (24, 25). R1, R2 and 1H-15N NOE values were used as input for Model-Free calculations (26) using the program Tensor2.
Metal Titration Studies
NMR titrations were performed using 15N-labeled samples of Sma0114, over a range of concentrations between 0 and 500 mM MgCl2. Due to the large 150 mM concentration of MgCl2 needed to saturate the enzyme, we also looked at the binding of CaCl2 over a concentration range between 0 and 10 mM. With CaCl2, saturation of Sma0114 was achieved at a metal ion concentration of 1.5 mM.
Bioinformatics Analysis of Sma0114
A sequence alignment of Sma0114 was initially performed against 2036 receiver domains from the order Rhizobiales using the SMART database (v. 6) (27). Sequences with greater than 97% homology to Sma0114 were considered redundant and removed from further analysis. The remaining 1792 sequences were further aligned using MUSCLE (v. 3.8.21). (28). From this alignment, 100 bootstrap replicate maximum-likelihood phylogenies were constructed using RAxML (v. 7.3.0) (29) incorporating the WAG model of amino acid substitutions (30) and estimating the gamma model of rate heterogeneity with 4 discrete rate categories. The bootstrapped trees were mapped onto the best-scoring maximum-likelihood tree.
Analysis of 273 HWE-kinases collected from the MiST 1 database (31) showed that 84 of the kinases had associated receiver domains. Receiver domains were considered to be associated with an HWE-kinase if the genes were fused or immediately upstream or downstream of the kinase. The cognate receiver domains of the 88 HWE-kinases identified in this way, were analyzed using the secondary structure prediction program PsiPred (v 2.6) (32). The final alignment excluded 5 of the 88 sequences because they were less than 100 amino acids in length and did not appear to encode full-length receiver domains. As a control, 8 receiver domain sequences of known structure were employed in the alignment to verify that the α5/β5 motif could be detected accurately by PsiPred (Fig.6C) (32). The sequences used for analysis are provided in Supplementary Table S1.
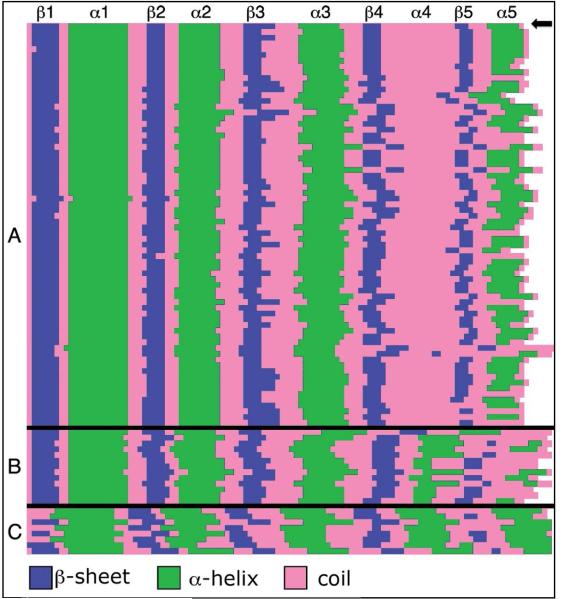
Figure 6.
Secondary structure prediction of HWE-associated receiver domains. Each row corresponds to one receiver domain. (A) Subset of 84 receiver domains with cognate HWE-kinases, all of which are predicted to be missing helix α4. Sma0114 is at the top of the figure labeled with an arrow. (B) Subset of 13 HWE-associated receiver domains predicted to have helix α4. Note that most of the members of this subset are predicted to be missing the alternate helix α5 of the 455 face. (C) Control group of 8 receiver domains of known α5/β5 structures used to verify the secondary structure prediction algorithm.
Results
Phylogenetic Distribution of Receiver Domains Associated with HWE Kinases
In order to place the Sma0114 receiver domain in the context of a family of homologous proteins we characterized the distributions of HWE two-component systems in prokaryotes. The SMART database (v.6) contains 1326 proteins with HWE histidine kinase domains, and the majority of these (70.8%) are found in the α-proteobacteria, with just over half of the total number being found in the order Rhizobiales (65.3%). The SMART database has some 2000 receiver domains from the Rhizobiales order of α-proteobacteria, of which ~1800 are non-redundant. Figure 1 shows a neighbor-joining tree of these receiver domains. There are 28 non-redundant HWE-kinases that are either fused to receiver domains, or that have genes adjacent to ones encoding a HWE histidine kinase domain (Fig. 1). This mapping shows that a majority of 21 HWE-associated receiver domains, including Sma0114, are in a single branch (shown in grey) indicating that they have high sequence similarity. The other receiver domains in the grey branch of the neighbor-joining tree are likely to be cognate response regulators that are physically separated (i.e. not fused or adjacent) from their cognate HWE-kinase.
The NMR Structure of Sma0114 shows Differences from the Receiver Domain Superfamily
Figure 2 shows the NMR structure of Sma0114. The overall fold of the protein is similar to the canonical α5/β5 Rossman-fold of other receiver domains, which is schematically illustrated in Figure 2A. Backbone (Cα, N, C’) traces of the 20 lowest energy NMR structures of Sma0114 are shown in Figure 2B. Structure statistics for the NMR models are given in Table I.
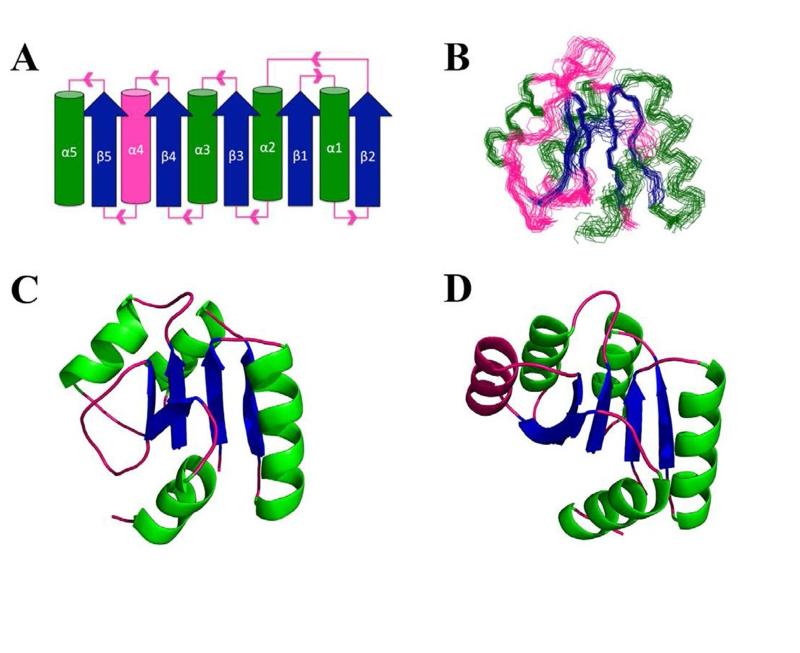
Figure 2.
Fold and NMR structure of Sma0114. (A) Topology diagram for the canonical α5/β5 Rossman fold of receiver domains. Parallel β-sheets (blue) are surrounded by α-helices (green) with pink segments indicating loops. The fourth α-helix which is disordered in Sma0114 is also indicated in pink. (B) Backbone representation of the 20 lowest energy NMR structures. Regular secondary structure elements 9-15 (β1), 19-31 (α1), 34-39 (β2), 42-51 (α2), 55-60 (β3), 70-78 (α3), 81-86 (β4), 102-105 (β5), 110-117 (α5) were used to superpose the structures. (C) NMR structure of Sma0114 closest to the ensemble average. The fourth α-helix is replaced by a disordered segment. (D) NMR structure of Spo0F (PDB accession code 1NAT) (47). The fourth α-helix is shown in pink.
Table I. Statistics for the 20 Lowest Energy NMR Structures of Sma0114.
| NMR Restraints (total) | 1627 | |
| Distance (total) | 1431 | |
| Intraresidue NOEs | 422 | |
| Sequential NOEs | 486 | |
| Short range NOEs (1< |i-j| < 5) | 109 | |
| Long range NOEs (5 < |i-j|) | 414 | |
| Hydrogen bonds (32×2) | 64 | |
| Dihedral (φ 61, ψ 54, χ1 17) | 132 | |
| Residual restraint violations | ||
| NOE (Å)1 | 0.039 ± 0.004 | |
| Dihedral (°)2 | 0.539 ± 0.018 | |
| RMS deviations from ideal geometry | ||
| Bonds (Å) | 0.0040 ± 0.0003 | |
| Angles (°) | 0.73 ± 0.02 | |
| Improper torsions (°) | 0.54 ± 0.02 | |
| van der Waals energy (kcal/mol)3 | 86.28 ± 2.37 | |
| Lennard-Jones energy (kcal/mol) 4 | −83.18 ± 7.40 | |
| Coordinate RMS deviations (Å) | ||
| NMR ensemble to average | Cα, C, N | All Heavy |
| Entire domain (110 residues)5 | 0.94 | 1.46 |
| Excluding loops (72 residues)6 | 0.86 | 1.38 |
Structure contains no NOE violations greater than 0.3Å
Structure contains no dihedral violations greater than 5°
Evdw was calculated using the X-PLOR Frepel function (23) with van derWaals interactions and atomic radii set to 0.8 times their CHARMM (46) values.
EL-J was calculated using the CHARMM empirical energy function (46)
Excluding N-terminus (residues 1-9) and C-terminus (residues 118-123)
Only residues in secondary structure (10-15, 19-31, 34-39, 42-51, 56-60, 70-78, 81-86, 102-105, 108-117)
The NMR structure of Sma0114 closest to the ensemble average is compared with that of a typical receiver domain Spo0F in Figs. 2C and 2D, respectively. The deviations from the canonical receiver domain fold occur primarily along the 455 face of Sma0114. The most conspicuous difference in Sma0114 (Fig. 2C), is that helix α4 is replaced by a disordered segment (residues 88-93). Another difference, is that strand β5 which is shorter than the other β-strands due to two flanking proline residues (P101, P106), turns inward toward the core of the Sma0114 structure (Fig. 2C).
To look more closely for similarities and differences to other receiver domains, we used the DALI server (33) to find the 10 best structural matches to Sma0114. The 10 best matches were all structures of inactive states of receiver domains, and had an average backbone RMSD of 3.17 Å to our Sma0114 structure over an average alignment length of 99 residues. The most prominent difference in Sma0114 is the absence of helix α4. Another important change is that the loop following strand β3 appears to cover and possibly restrict access to the phosphorylation site residue, Asp60, whereas it extends away from the hydrophobic core in the homologous structures.
The alignment to structurally homologous proteins using the DALI server (33) also indicates that the C-terminal portion of the PFxFATGY motif in Sma0114 differs from the corresponding region in the canonical receiver domain fold. The PFxFATGY motif is highlighted in cyan on the NMR structure of Sma0114 in Figure 3A. This motif encompasses all of strand β4 and forms the beginning of the loop between strands β4 and β5. The conserved Thr of the Y-T coupling pathway is part of this motif and is the last residue in strand β4 (shown in purple in Fig. 3A). The partner aromatic residue in the Y-T coupling mechanism is replaced by a Leu103 in strand β5 (shown in black in Fig. 3A). The absence of the conserved aromatic residue has also been seen in the single domain response regulator CheY2 from S. meliloti, an enzyme that has been reported to lack Y-T coupling (34). These changes provide compelling evidence that the prototypical Y-T coupling mechanism is altered in Sma0114.
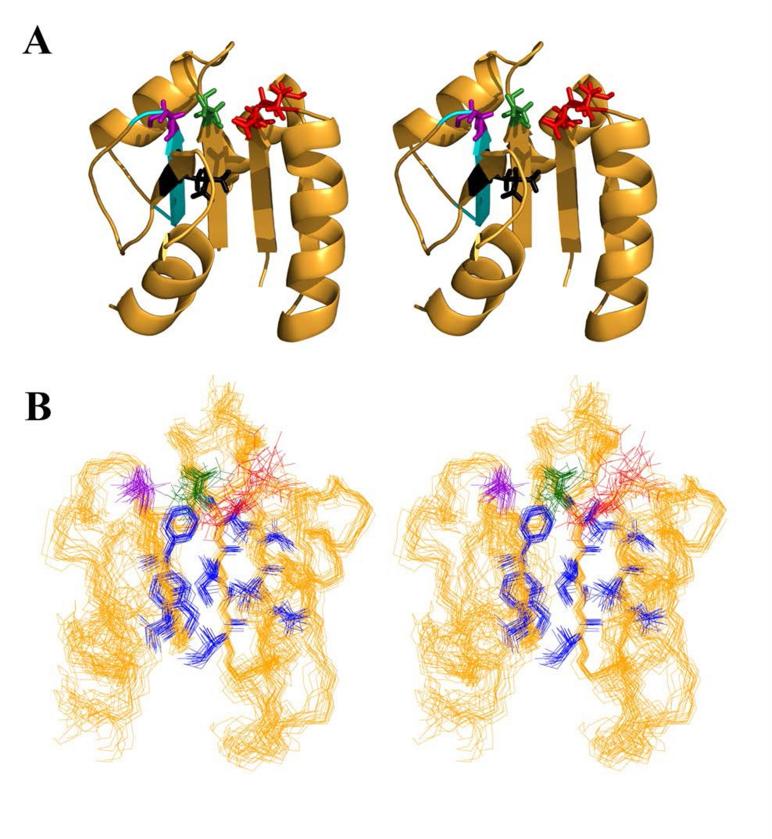
Figure 3.
Active site of Sma0114. (A) Stereo-diagram of the NMR structure of Sma0114 closest to the ensemble average, showing selected side-chains of active site residues: metal-ligands Glu15 and Asp16 in red, the phosphorylation site Asp60 in green, and Thr86 and Leu103 of the Y-T coupling pair in purple and black, respectively. The PFxFATGY motif is shown in cyan. (B) Stereo-view of the 20 lowest energy NMR structures of Sma0114 illustrating the precision of side-chains from different parts of the molecule. Side-chains comprising the hydrophobic core (blue) are well defined while those of active-site residues (color scheme as above) show poorer precision due to R2ex line-broadening.
The two residues of the PFxFATGY motif that form part of the loop immediately following strand β4 (G87 and Y88) show large structural differences in Sma0114 compared to the top 10 structural similarity hits. These two residues are located immediately before the start of the disordered segment that replaces helix α4 in Sma0114. The C-terminal portion of the PFxFATGY motif in Sma0114 superimposes with the N-terminal portion of helix α4 in the 10 closest structural matches. Together with the placement of the Y-T coupling threonine, this suggests that at least the C-terminal part of the PFxFATGY motif belongs to the 455 face of Sma0114.
While the 455 face of the enzyme shows deviations from the receiver domain superfamily, the active site of Sma0114 has features consistent with other receiver domains (Fig. 3A). The metal binding residues, Glu15 and Asp16, are located at a conserved position in the loop between β1 and α1 (7, 11, 13). The phosphorylation site (Asp60) is at the C-terminal end of β3 like in other receiver domains. The hydroxylic residue (Thr86) that hydrogen bonds with the incoming phosphate in the Y-T coupling mechanism is located at the C-terminal end of strand β4. The basic residue Lys105, which typically stabilizes the activated state by forming a salt bridge with the incoming phosphate occurs in a conserved location in strand β5.
Strands β1-β4 and helices α1-α3 have the highest precision in the NMR structure (Fig. 3B). Regions with lower precision occur in the 455 face and include the segments corresponding to helix α4, strand β5, and helix α5. The active site of Sma0114 also has lower precision in the NMR structure, as illustrated in Figure 3B which compares side-chains of residues in the active site and in the hydrophobic core of the protein. The equilibrium shift theory of activation hypothesizes that receiver domains exist in their inactive and active substates simultaneously in solution. Activation via phosphorylation shifts the population from the inactive to the active form (14, 16). It follows that that the active site, which is experiencing exchange between inactive and active conformational substates on a μs-ms timescale, would have a lower precision due to R2ex line broadening. To verify that the lower precision of the active site and 455 face in the NMR structure of Sma0114 is due to genuine flexibility, as it is in other receiver domains (7, 35), we characterized the backbone dynamics of the protein using 15N relaxation data.
NMR Relaxation Data Show Increased Flexibility for the 455 Face and Active Site
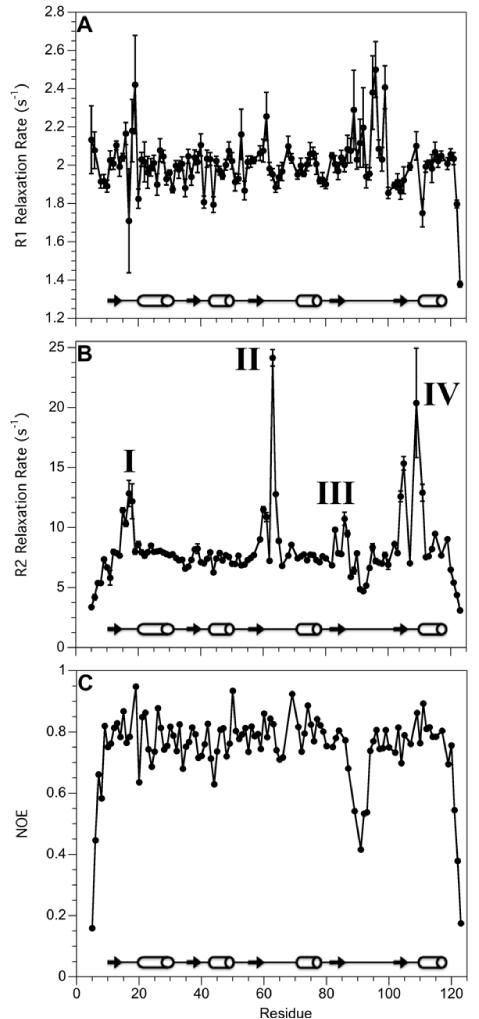
Relaxation data for Sma0114 are shown in Figure 4. The R1 values are roughly constant over the length of the protein with a mean value of 2.00 ± 0.01 s−1, except for raised rates in the region between residues 89 and 99, which also shows lowered 1H-15N NOEs (Fig. 4A). The R2 data reveal four distinct regions that have relaxation contributions greater than the mean value of 8.0 ± 0.3 s−1, as indicated in Fig. 4B. The mean 1H-15N NOE value is 0.75 ± 0.01, close to the theoretical maximum of 0.80 (36). Lowered 1H-15N NOEs are seen for the chain termini and the region between residues 88 and 95 that corresponds to the missing helix α4 (Fig. 4C).
Figure 4.
15N NMR relaxation data for Sma0114. (A) R1 rates. (B) R2 rates. (C) 1H-15N NOE values. Error bars are shown for all data points but in some cases are smaller than the symbols used to depict the data. The secondary structure of Sma0114 is indicated with arrows (β-sheets) and cylinders (α-helices) in each panel.
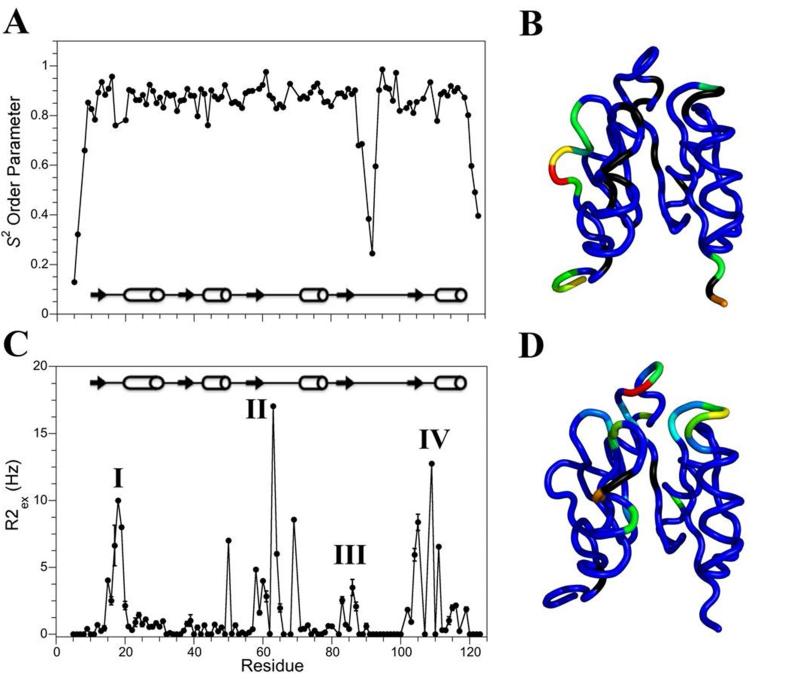
To interpret the relaxation data in terms of backbone dynamics we performed a Model Free analysis (26) using the program Tensor2 (Fig. 5). We obtained a correlation time of 5.1 ns for Sma0114, consistent with a monomeric protein of 13.5 KDa. The S2 values which describe the amplitudes of internal motions on the ps-ns timescale in the Model Free (26) analysis, are shown in Fig. 5A and are mapped on the NMR structure in Fig. 5B. Except for the chain termini, the only region in the protein with low S2 order parameters is the flexible segment that replaces helix α4 (Fig. 5A). The R2ex terms which describe exchange contributions to R2 relaxation from dynamics on the μs-ms timescale are shown in Fig. 5C and are mapped on the NMR structure in Fig. 5D. Residues that experience significant R2ex contributions cluster to four region of the protein: (I) the metal binding site, (II) the phosphorylation site, (III) Thr86 which is predicted to hydrogen bond with an incoming phosphate in the ‘Y-T coupling mechanism’, and (IV) Lys105 which stabilizes the incoming phosphate through a salt-bridge. Thus all four regions with R2ex contributions are in the active site of the enzyme, which has been shown for other receiver domains to be subject to μs-ms timescale interconversion between inactive and active substates as described in the equilibrium shift theory of activation (15, 16).
Figure 5.
Model-free analysis of Sma0114. (A) S2 order parameters. (B) S2 order parameters mapped onto the NMR structure of Sma0114. The color map goes from blue (rigid) to red (flexible). (C) R2ex values. (D) R2ex values mapped onto the NMR structure of Sma0114 with colors ranging from blue (negligible R2ex contributions) to red (large R2ex contributions). Residues for which no data are available are shown in black.
Despite their strong structural homology, receiver domains from various two-component systems exhibit different dynamics on the μs-ms timescale. R2ex terms for Spo0F, an intermediate in the sporulation relay of B. subtilus, are larger for the metal binding site compared to the 455 face of the enzyme (15). Conversely, in NtrC, the nitrogen regulatory response regulator from S. typhimurium, R2ex contributions are large for the 455 face of the enzyme whereas the metal binding site shows no exchange broadening (16, 37). The R2ex profile of Sma0114 differs from Spo0F and NtrC in that it shows exchange broadening in the metal-binding and phosphorylation sites but only in the β5 and α5 regions of the conserved 455 face.
The Family of Receiver Domains Associated with HWE-Kinases is Predicted to Lack Helix α4
The NMR structure of Sma0114 is the first of an HWE-kinase associated response regulator. Due to the unusual features of this enzyme we performed a bioinformatics analysis to determine if the absence of helix α4 is a conserved property of this subfamily. The structure prediction program PsiPred (32) was used to evaluate the secondary structure of 84 receiver domains with cognate HWE-kinase partners. Of the 84 proteins considered, 71 were predicted to lack helix α4 (Fig. 6A). A subset of 13 HWE-associated receiver domains are predicted to have helix α4 but to be missing the last helix α5 (Fig. 6B). This suggests that most of the HWE-associated receiver domains in which helix α4 is present, have compensatory perturbations that disrupt helix α5 of the 455 face. By contrast, all of the α-helices and β-strands of the consensus Rossman fold are accurately predicted by the PsiPred program for a control group of 8 receiver domains of known structure that are not associated with HWE-kinases (Fig. 6C).
Sma0114 binds Mg2+ and Ca2+
We next wanted to see if divalent cations, which are required for the stabilization of the acyl-phosphate linkage (14, 38), are able to bind to Sma0114 in spite of the differences seen for the 455 face. Several groups have used Mg2+ to study the metal-bound states of receiver domains (11, 13, 39). In the case of Sma0114 we found that Mg2+ binds very weakly, with a Kd near 75 mM and saturation at 150 mM Mg2+. The MgCl2 concentration required to stabilize the fully metal bound state leads to a decrease in sensitivity for NMR experiments due to high solution ionic strength, especially for data collection with a cryogenic probe. An additional problem is that the large Mg2+ concentration needed to saturate Sma0114 causes non-specific binding to clusters of acidic residues in the protein. Finally, Sma0114 is more susceptible to aggregation on a timescale of days to weeks needed for a detailed NMR characterization, in the presence of large (> 100 mM) concentrations of MgCl2.
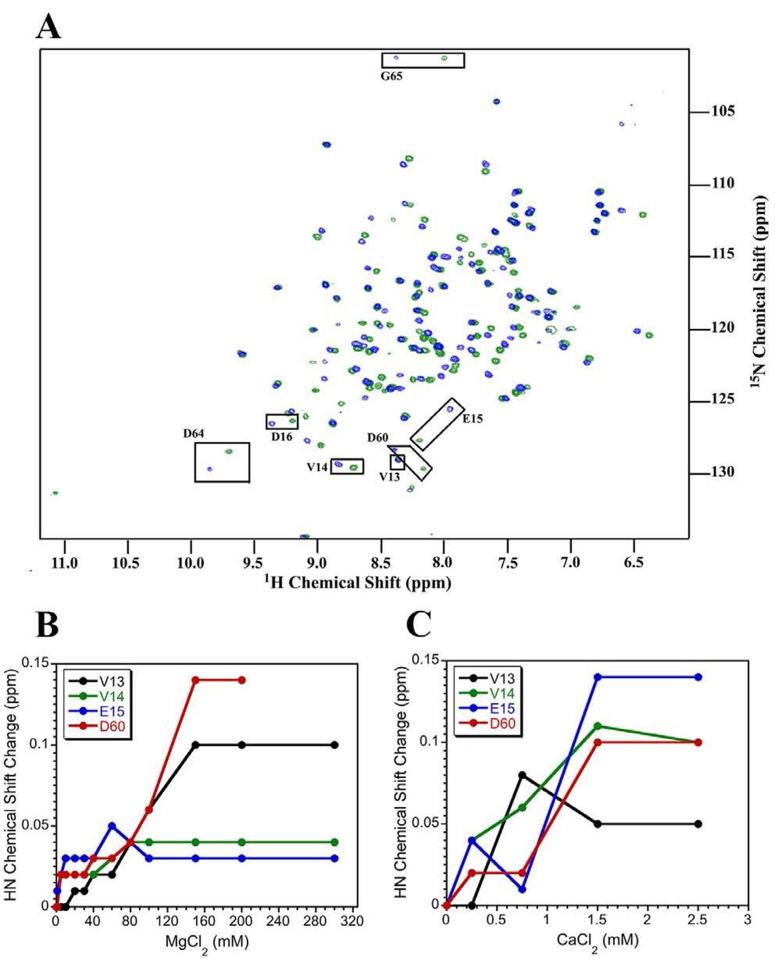
Due to the problems associated with Mg2+, we carried out an NMR titration using CaCl2 as an alternative metal. Superimposed 1H-15N spectra of apo and Ca2+-bound Sma0114 are shown in Figure 7A. In contrast to Mg2+ which has a Kd of 75 mM (Fig. 7B), Sma0114 binds Ca2+ with a ~1 mM Kd, and saturation is achieved at 1.5 mM concentrations of CaCl2 (Fig. 7C). Residues that show the greatest chemical shift changes upon addition of Ca2+ are indicated with boxes in Figure 7A, and for the most part cluster to the active site.
Figure 7.
Metal-binding of Sma0114 followed by NMR spectroscopy. (A) Superposition of 1H-15N-HSQC spectra for the apo (blue) and Ca2+-bound forms of Sma0114 (green). Active site residues that show the largest chemical shift perturbations and are indicated with black boxes. Titration curves for residues as a function of increasing MgCl2 (B) or CaCl2 (C) concentrations.
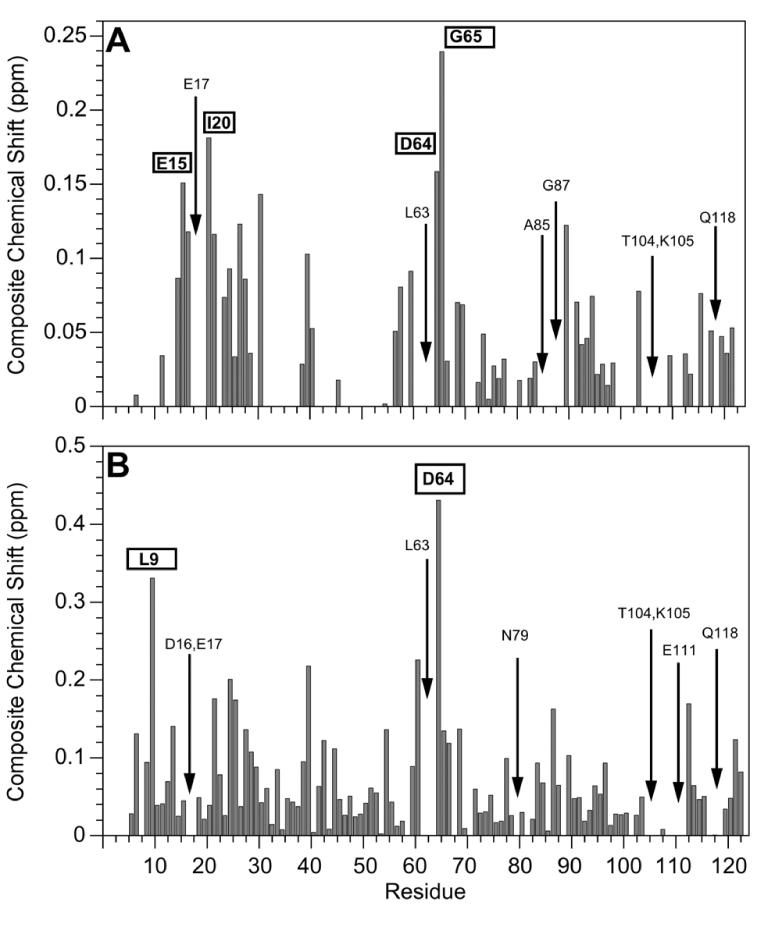
Figure 8 shows the chemical shift perturbations observed in 1H-15N HSQC spectra of Sma0114 due to addition of CaCl2 (Fig. 8A) and MgCl2 (Fig. 8B). The unusually large concentration of MgCl2 necessary to achieve saturation induces non-specific binding to acidic Asp and Glu residues as illustrated by the large number of the chemical shift perturbations and line broadening effects observed (Fig. 8B). By contrast, with CaCl2 perturbations are mostly restricted to the active site of the enzyme (Fig. 8A). Some residues become broadened rather than experiencing chemical shift changes upon addition of CaCl2 or MgCl2, and these are indicated with arrows in Figure 8. They include Glu17, which is one of the ligands for Ca2+, and Lys105, which is part of the active site. Although most studies of metal-bound receiver domains have used Mg2+, Ca2+ was used for structural studies of Spo0A and PhoP (40, 41).
Figure 8.
Perturbations of Sma0114 1H-15N resonances due to metal-binding. Bar graphs show composite HN and N chemical shift changes (ΔHN + 0.1(ΔN)) that occur in the presence of 1.5 mM CaCl2 (A) or 150 mM MgCl2 (B). Residues in boxes indicate amino acids that experience large chemical shift perturbations upon metal binding. Arrows represent residues that broaden beyond detection in the presence of the metals.
Discussion
Several groups have characterized the inactive and active conformations of receiver domains (7, 11, 39). Understanding the structural features of these enzymes has provided a wealth of insight into their functions. Here, we extended the study of the receiver domain family to include an HWE-kinase-associated response regulator. The Sma0114 receiver domain, which may be prototypical of this class, has a conserved active site but shows substantial differences in the 455 face, which is used to propagate signal transduction intracellularly.
The structural and dynamic differences in the 455 face of Sma0114 suggest that this enzyme has an altered recognition interface for binding to downstream effectors. The PFxFATGY motif, which is unique to receiver domains associated with HWE-kinases, forms strand β4 in the Sma0114 structure (Pro81-Thr86) and the rigid part of the following loop (Gly87 and Tyr88). The last residue in the PFxFATGY motif demarcates the start of the Tyr88-Leu93 segment, which replaces helix α4 in Sma0114, and shows low S2 order parameters consistent with a dynamically disordered region. The only other receiver domain for which S2 order parameters have been described is the inactive form of Spo0F (15). In the case of Spo0F, it was shown that except for the chain termini, the protein is rigid on the ps-ns timescale. In typical receiver domains, helix α4 is a key element of the 455 face that undergoes concerted structural rearrangements following phosphorylation to form a new binding surface for downstream effector proteins (11, 35). Not only is helix α4 missing in Sma0114 but our secondary structure prediction analysis suggests that the absence of helix α4, or more rarely helix α5, is a conserved feature of HWE-associated receiver domains. A possible role for flexibility in the segment corresponding to helix α4 is to allow binding of downstream effectors through an induced fit mechanism, thereby controlling the specificity of molecular association.
Residues Pro81 through Gly87 of the PFxFATGY motif, which form strand β4 and the beginning of the following loop, while rigid on the ps-ns timescale have raised R2ex contributions consistent with dynamics on the μs-ms timescale. The exchange contributions to R2 relaxation for these sites, suggest that they experience the dynamic conformational equilibrium between the inactive and active substates of the enzyme, that is a hallmark of receiver domains(7, 14). The PFxFATGY sequence houses the conserved Thr86 of the Y-T coupling mechanism. The partnering aromatic residue for Y-T coupling in Sma0114 is replaced by Leu103. In a previous study of the receiver domain CheY, it was shown that the β4/α4 loop and the N-terminal portion of helix α4 constitute the binding site for downstream effectors (42). Because part of the PFxFATGY motif in Sma0114 occupies the same position in the structure as the β4/α4 loop region in CheY, it is likely that Sma0114 has a similarly reduced binding site for downstream effectors. The PFxFATGY sequence motif in Sma0114, may thus substitute or circumvent the Y-T coupling mechanism typically used in receiver domains. This change could be necessary in Sma0114 because of an altered 455 face, which is missing helix α4 and has additional structural differences in strand β5.
In spite of the differences in the 455 face, Sma0114 retains the ability to bind divalent metal ions needed for activation. The residues in Sma0114 that experience the largest chemical shift perturbations with increasing metal concentrations are consistent with the binding site predicted based on sequence homology to other receiver domains (Fig. 7). Like Spo0F, Sma0114 has a greater affinity for Ca2+ than Mg2+ (43). The 1.5 mM concentration of Ca2+ required to saturate the metal binding site of Sma0114 is larger than the physiological concentration in bacteria (44). A weak metal-binding affinity could function to kinetically stabilize the activated state, by lowering the efficiency of the phosphatase enzymatic activity of the receiver domain, which in turn would effectively increase the lifetime of the phosphorylated activate state (45).
The conservation of active site residues and the ability to bind metals suggest that the Sma0114 receiver domain can become activated by phosphorylation. The increased pliability of the 455 face may help Sma0114 achieve specificity in binding downstream effectors through an induced fit mechanism. We are now in a position to characterize the structure and dynamics of the metal-bound and activated states of Sma0114, which will allow us to obtain a more complete understanding of the structure-function relationships in the HWE-kinase associated class of receiver domains.
Supplementary Material
Acknowledgments
This work was supported by a National Science Foundation Graduate Research Fellowship (GRFP) to S.R.S, a grant from the UConn Research Foundation (UCRF) to A.T.A. and D.J.G. and by DOE grants (DE-FG02-01ER15175 and DE-FG02-06ER15805) to D.J.G.
Abbreviations
- NMR
nuclear magnetic resonance
- HWE
histidine tryptophan glutamate
- NaN3
sodium azide
- DTT
dithiothreitol
- NOE
nuclear overhauser effect
- RMSD
root mean square deviation
- R2ex
relaxation exchange
- Kd
dissociation constant
Footnotes
One table, listing the set of receiver domain sequences used for secondary structure prediction. Supplementary data associated with this article can be found, in the online version, at doi: XXX.
References
- 1.Robinson VL, Buckler DR, Stock AM. A tale of two components: a novel kinase and a regulatory switch. Nat Struct Biol. 2000;7:626–633. doi: 10.1038/77915. [DOI] [PubMed] [Google Scholar]
- 2.Stock JB, Surette MG, McCleary WR, Stock AM. Signal transduction in bacterial chemotaxis. J Biol Chem. 1992;267:19753–19756. [PubMed] [Google Scholar]
- 3.Garcia PP, Bringhurst RM, Arango Pinedo C, Gage DJ. Characterization of a two-component regulatory system that regulates succinate-mediated catabolite repression in Sinorhizobium meliloti. J Bacteriol. 2009;192:5725–5735. doi: 10.1128/JB.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karniol B, Vierstra RD. The HWE histidine kinases, a new family of bacterial two-component sensor kinases with potentially diverse roles in environmental signaling. J Bacteriol. 2004;186:445–453. doi: 10.1128/JB.186.2.445-453.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stock AM, Robinson VL, Goudreau PN. Two-component signal transduction. Annu Rev Biochem. 2000;69:183–215. doi: 10.1146/annurev.biochem.69.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Stock AM, Mottonen JM, Stock JB, Schutt CE. Three-dimensional structure of CheY, the response regulator of bacterial chemotaxis. Nature. 1989;337:745–749. doi: 10.1038/337745a0. [DOI] [PubMed] [Google Scholar]
- 7.Gardino AK, Volkman BF, Cho HS, Lee SY, Wemmer DE, Kern D. The NMR solution structure of BeF(3)(−)-activated Spo0F reveals the conformational switch in a phosphorelay system. J Mol Biol. 2003;331:245–254. doi: 10.1016/s0022-2836(03)00733-2. [DOI] [PubMed] [Google Scholar]
- 8.Foreman R, Fiebig A, Crosson S. The LovK-LovR Two-Component System Is a Regulator of the General Stress Pathway in Caulobacter crescentus. J Bacteriol. 2012;194:3038–3049. doi: 10.1128/JB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenal U, Galperin MY. Single domain response regulators: molecular switches with emerging roles in cell organization and dynamics. Curr Opin Microbiol. 2009;12:152–160. doi: 10.1016/j.mib.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul R, Jaeger T, Abel S, Wiederkehr I, Folcher M, Biondi EG, Laub MT, Jenal U. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell. 2008;133:452–461. doi: 10.1016/j.cell.2008.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hastings CA, Lee SY, Cho HS, Yan D, Kustu S, Wemmer DE. High-resolution solution structure of the beryllofluoride-activated NtrC receiver domain. Biochemistry. 2003;42:9081–9090. doi: 10.1021/bi0273866. [DOI] [PubMed] [Google Scholar]
- 12.Perez E, Samper S, Bordas Y, Guilhot C, Gicquel B, Martin C. An essential role for phoP in Mycobacterium tuberculosis virulence. Mol Microbiol. 2001;41:179–187. doi: 10.1046/j.1365-2958.2001.02500.x. [DOI] [PubMed] [Google Scholar]
- 13.Cho HS, Lee SY, Yan D, Pan X, Parkinson JS, Kustu S, Wemmer DE, Pelton JG. NMR structure of activated CheY. J Mol Biol. 2000;297:543–551. doi: 10.1006/jmbi.2000.3595. [DOI] [PubMed] [Google Scholar]
- 14.Bourret RB. Receiver domain structure and function in response regulator proteins. Curr Opin Microbiol. 2010;13:142–149. doi: 10.1016/j.mib.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feher VA, Cavanagh J. Millisecond-timescale motions contribute to the function of the bacterial response regulator protein Spo0F. Nature. 1999;400:289–293. doi: 10.1038/22357. [DOI] [PubMed] [Google Scholar]
- 16.Gardino AK, Kern D. Functional dynamics of response regulators using NMR relaxation techniques. Methods Enzymol. 2007;423:149–165. doi: 10.1016/S0076-6879(07)23006-X. [DOI] [PubMed] [Google Scholar]
- 17.Sheftic SR, Garcia PP, Robinson VL, Gage DJ, Alexandrescu AT. NMR assignments for the Sinorhizobium meliloti response regulator Sma0114. Biomol NMR Assign. 2010;5:55–58. doi: 10.1007/s12104-010-9266-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cavanagh J, Fairbrother Wayne, J., Palmer Arthur, G., III, Rance Mark., Skelton Nicholas, J. Protein NMR Spectroscopy Principles and Practice (Second Edition) Elsevier Inc.; Amsterdam: 2006. [Google Scholar]
- 19.Jaravine VA, Alexandrescu AT, Grzesiek S. Observation of the closing of individual hydrogen bonds during TFE-induced helix formation in a peptide. Protein Sci. 2001;10:943–950. doi: 10.1110/ps.48501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 21.Case DA, Dyson HJ, Wright PE. Use of chemical shifts and coupling constants in nuclear magnetic resonance structural studies on peptides and proteins. Methods Enzymol. 1994;239:392–416. doi: 10.1016/s0076-6879(94)39015-0. [DOI] [PubMed] [Google Scholar]
- 22.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 23.Brunger AT. Xplor Version 3.8 A system for Crystallography and NMR. Yale University Press; New Haven: 1996. [Google Scholar]
- 24.Nicholson LK, Kay LE, Torchia DA. Protein Dynamics as studied by solution NMR techniques. In: Sarkar S, editor. NMR Spectroscopy and its Application to Biomedical Research. Elsevier Science Publishers; 1994. [Google Scholar]
- 25.Alexandrescu AT, Shortle D. Backbone dynamics of a highly disordered 131 residue fragment of staphylococcal nuclease. J Mol Biol. 1994;242:527–546. doi: 10.1006/jmbi.1994.1598. [DOI] [PubMed] [Google Scholar]
- 26.Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 1. Theory and Validity. J. Am. Chem. Soc. 1982;104:4546–4559. [Google Scholar]
- 27.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 30.Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001;18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 31.Ulrich LE, Zhulin IB. MiST: a microbial signal transduction database. Nucleic Acids Res. 2007;35:D386–390. doi: 10.1093/nar/gkl932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchan DW, Ward SM, Lobley AE, Nugent TC, Bryson K, Jones DT. Protein annotation and modelling servers at University College London. Nucleic Acids Res. 2010;38:W563–568. doi: 10.1093/nar/gkq427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riepl H, Scharf B, Schmitt R, Kalbitzer HR, Maurer T. Solution structures of the inactive and BeF3-activated response regulator CheY2. J Mol Biol. 2004;338:287–297. doi: 10.1016/j.jmb.2004.02.054. [DOI] [PubMed] [Google Scholar]
- 35.Peters G. The effect of Asp54 phosphorylation on the energetics and dynamics in the response regulator protein Spo0F studied by molecular dynamics. Proteins. 2009;75:648–658. doi: 10.1002/prot.22276. [DOI] [PubMed] [Google Scholar]
- 36.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 37.Volkman BF, Lipson D, Wemmer DE, Kern D. Two-state allosteric behavior in a single-domain signaling protein. Science. 2001;291:2429–2433. doi: 10.1126/science.291.5512.2429. [DOI] [PubMed] [Google Scholar]
- 38.Kojetin DJ, Thompson RJ, Benson LM, Naylor S, Waterman J, Davies KG, Opperman CH, Stephenson K, Hoch JA, Cavanagh J. Structural analysis of divalent metals binding to the Bacillus subtilis response regulator Spo0F: the possibility for in vitro metalloregulation in the initiation of sporulation. Biometals. 2005;18:449–466. doi: 10.1007/s10534-005-4303-8. [DOI] [PubMed] [Google Scholar]
- 39.Stock AM, Martinez-Hackert E, Rasmussen BF, West AH, Stock JB, Ringe D, Petsko GA. Structure of the Mg(2+)-bound form of CheY and mechanism of phosphoryl transfer in bacterial chemotaxis. Biochemistry. 1993;32:13375–13380. doi: 10.1021/bi00212a001. [DOI] [PubMed] [Google Scholar]
- 40.Lewis RJ, Muchova K, Brannigan JA, Barak I, Leonard G, Wilkinson AJ. Domain swapping in the sporulation response regulator Spo0A. J Mol Biol. 2000;297:757–770. doi: 10.1006/jmbi.2000.3598. [DOI] [PubMed] [Google Scholar]
- 41.Menon S, Wang S. Structure of the response regulator PhoP from Mycobacterium tuberculosis reveals a dimer through the receiver domain. Biochemistry. 2011;50:5948–5957. doi: 10.1021/bi2005575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guhaniyogi J, Robinson VL, Stock AM. Crystal structures of beryllium fluoride-free and beryllium fluoride-bound CheY in complex with the conserved C-terminal peptide of CheZ reveal dual binding modes specific to CheY conformation. J Mol Biol. 2006;359:624–645. doi: 10.1016/j.jmb.2006.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feher VA, Tzeng YL, Hoch JA, Cavanagh J. Identification of communication networks in Spo0F: a model for phosphorylation-induced conformational change and implications for activation of multiple domain bacterial response regulators. FEBS Lett. 1998;425:1–6. doi: 10.1016/s0014-5793(98)00182-3. [DOI] [PubMed] [Google Scholar]
- 44.Dominguez DC. Calcium signalling in bacteria. Mol Microbiol. 2004;54:291–297. doi: 10.1111/j.1365-2958.2004.04276.x. [DOI] [PubMed] [Google Scholar]
- 45.Feher VA, Zapf JW, Hoch JA, Whiteley JM, McIntosh LP, Rance M, Skelton NJ, Dahlquist FW, Cavanagh J. High-resolution NMR structure and backbone dynamics of the Bacillus subtilis response regulator, Spo0F: implications for phosphorylation and molecular recognition. Biochemistry. 1997;36:10015–10025. doi: 10.1021/bi970816l. [DOI] [PubMed] [Google Scholar]
- 46.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 1983;4:187, 217. [Google Scholar]
- 47.Madhusudan M, Zapf J, Hoch JA, Whiteley JM, Xuong NH, Varughese KI. A response regulatory protein with the site of phosphorylation blocked by an arginine interaction: crystal structure of Spo0F from Bacillus subtilis. Biochemistry. 1997;36:12739–12745. doi: 10.1021/bi971276v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.