Abstract
We previously reported that methylmercury (MeHg) exposure is associated with DNA hypomethylation in the brain stem of male polar bears. Here, we conveniently use archived tissues obtained from controlled laboratory exposure studies to look for evidence that MeHg can disrupt DNA methylation across taxa. Brain (cerebrum) tissues from MeHg-exposed mink (Neovison vison), chicken (Gallus gallus) and yellow perch (Perca flavescens) were analyzed for total Hg levels and global DNA methylation. Tissues from chicken and mink, but not perch, were also analyzed for DNA methyltransferase (DNMT) activity. In mink we observed significant reductions in global DNA methylation in an environmentally-relevant dietary exposure group (1ppm MeHg), but not in a higher group (2ppm MeHg). DNMT activity was significantly reduced in all treatment groups. In chicken or yellow perch, no statistically significant effects of MeHg were observed. Dose-dependent trends were observed in the chicken data but the direction of the change was not consistent between the two endpoints. Our results suggest that MeHg can be epigenetically active in that it has the capacity to affect DNA methylation in mammals. The variability in results across species may suggest inter-taxa differences in epigenetic responses to MeHg, or may be related to differences among the exposure scenarios used as animals were exposed to MeHg through different routes (dietary, egg injection), for different periods of time (19 – 89 days) and at different life stages (embryonic, juvenile, adult).
Keywords: mercury, Hg, epigenetics, ecotoxicology, wildlife, environmental epigenetics, DNA methylation
1.0 INTRODUCTION
Methylmercury (MeHg) is a ubiquitous toxicant that biomagnifies through aquatic food webs (Scheuhammer et al., 2007). Once inside the body, MeHg is effectively absorbed from the bloodstream and can readily enter sensitive tissues such as the brain (Clarkson and Magos, 2006). Numerous studies have documented that under real-world exposure scenarios, MeHg causes adverse effects to the health of individuals and populations of various taxa, including fish (Depew et al., 2012a), birds (Depew et al., 2012b), and mammals (Basu and Head, 2010).
The toxic actions of MeHg are diverse owing to its high affinity for thiol groups (Clarkson and Magos, 2006). A myriad of effects have been described at a molecular and cellular level including changes in neurotransmission, oxidative stress, and gene expression. There is now preliminary evidence that MeHg, like other toxic metals, may cause epigenetic changes (Pilsner et al., 2010). Epigenetics refers to heritable factors affecting gene expression that occur outside of modifications to the DNA sequence itself (Head et al., 2012; Robertson et al., 2000). These include DNA methylation and histone modification. Epigenetic marks are susceptible to environmental influences such as exposure to chemicals, and these changes may be inherited as cells divide mitotically, or even between generations. Epigenetic mechanisms may even help explain the observation of temporal disconnects between exposure and effect, as early-life exposure may leave epigenetic marks that result in adverse health outcomes later in life. This is noteworthy as MeHg is known to have a long latency of effect in many organisms (Basu and Head, 2010; Clarkson and Magos, 2006).
In the biomedical sciences, MeHg-associated changes in epigenetic markers are starting to be found in several experimental model systems. Developmental exposure of mice to MeHg resulted in a range of epigenetic changes in brain-derived neurotrophic factor (BDNF) in male offspring, including increased DNA methylation and altered methylation and acetylation of histones (Onishchenko et al., 2008). These epigenetic changes were associated with depressive symptoms. In female offspring of rats exposed to MeHg throughout pregnancy, reduced hepatic expression of DNA methyltransferases (DNMT1, DNMT3a, DNMT3b) was reported (Desaulniers et al., 2009). DNA methyltransferases are a family of enzymes that establish and maintain patterns of DNA methylation in the genome. An in vitro study using mouse embryonic cells found that acute exposure to Hg2+ impairs histone production and reduces H3-K27 methylation (Gadhia et al., 2012). Such epigenetic findings have been extended to humans as increased methylation of GSTM1/5 promoter in blood was associated with MeHg exposure in women undergoing in vitro fertilization (Hanna et al., 2012), and hypomethylation of SEPP1 was associated with hair Hg levels among a cohort of male dentists (Goodrich et al., 2013). Collectively these aforementioned studies provide founding evidence that mercury compounds can cause epigenetic changes.
Of all organisms, fish-eating wildlife are among those with the greatest exposures to MeHg (Basu and Head, 2010; Scheuhammer et al., 2007). However, little is known about MeHg-associated epigenetic effects in these organisms, (Head et al., 2012; Vandegehuchte and Janssen, 2011). We previously observed reduced global DNA methylation in association with MeHg exposure in brain stem tissues of male polar bears (Pilsner et al., 2010). While the results of this polar bear study were informative, the DNA methylation results were highly variable and likely influenced by a range of known (e.g., other toxicants, health status, age, gender, tissue quality) and unknown factors that could not be well-controlled when studying tissues from wild-caught animals. Given the need to generate data from well-controlled laboratory experiments, the current study was performed to increase understanding of possible MeHg-associated epigenetic changes using a mammalian, avian, and fish model species. All three classes contain species with well-documented sensitivities to MeHg (Scheuhammer et al., 2007). For each class we chose to study a model ecotoxicological test organism, namely mink (Neovison vison), chicken (Gallus gallus domesticus), and yellow perch (Perca flavescens). As part of other research projects, animals of each species were previously exposed in the laboratory to levels of MeHg ranging from ecologically relevant to high. Here we opportunistically used tissues preserved from those studies. The brain tissue (focus: cerebrum) was investigated in each model since this organ is principal target of MeHg (Basu and Head, 2010; Clarkson and Magos, 2006). We examined global genomic DNA methylation using the LUMA assay and the enzymatic activity of DNMT via a commercially available ELISA kit. The guiding hypothesis of this study was that brain global genomic methylation and DNMT enzyme activity would be reduced in association with methylmercury exposure in each of the three model species
2.0 MATERIALS AND METHODS
2.1 Animal Exposure Studies
Mink have been exposed to a range of contaminants in the laboratory largely for the purposes of ecological risk assessment, and in the field they have been shown to accumulate potentially toxic concentrations of MeHg (Basu et al., 2007a). Here, captive juvenile male mink (n = 12 per treatment) were exposed daily to MeHgCl (0, 0.1, 0.5, 1, and 2 mg/kg nominal concentrations in diet) as previously described (Basu et al., 2006, 2007b). Following three months of exposure, animals were euthanized, occipital cortex dissected, immediately frozen on dry ice and stored at −80 °C until analysis. All aspects of this study were approved institutionally by the animal ethics committees of McGill University and the Nova Scotia Agricultural College.
Avian toxicologists have routinely injected chicken eggs with contaminants to address a range of toxicological questions relating to risk assessment in wild birds (Heinz et al., 2009). Here, fertilized white leghorn chicken eggs were obtained from the Michigan State University Poultry Unit. Once received at the University of Michigan School of Public Health, they were air-cell injected with MeHgCl (non-inject, vehicle inject, 0.62, 2.0, 3.2, and 6.4 μg/g egg injected, nominal concentrations; n=9 per treatment) on day 0 of incubation as described elsewhere (Rutkiewicz and Basu, 2013). Eggs were incubated at 37.7°C under 55–60% humidity. On day 19 of embryonic development, embryos were euthanized by decapitation, and the cerebrum regions were dissected from brain and immediately frozen on dry ice and stored at −80 °C until analysis. All aspects of this study were approved by the University of Michigan Committee on Use and Care of Animals.
Perch are known to be sensitive to MeHg in nature (Wiener et al. 2012) and increasing number of studies are utilizing them as a laboratory model (Kwon et al., 2012). Here, female yellow perch were fed pellets containing nominal MeHgCl concentrations of 0, 0.5, 5, and 50 μg/g. Fish were fed twice daily for a period of 4 weeks. Fish were sacrificed and the telencephalon brain region was removed, immediately frozen on dry ice and stored at −80 °C until analyses. For each treatment there were three replicate tanks with five fish per tank, and for the purposes of this study we pooled together all five telencephalons from a given tank. For perch, we measured global genomic DNA methylation but not DNMT activity. All aspects of this study were approved by the University of Wisconsin-Milwaukee Animal Care and Use Committee.
2.2 Mercury analysis
Concentrations of total Hg were measured in the brain tissues of mink, chicken, and perch using methods described previously (Basu et al., 2006; Rutkiewicz and Basu, 2013). For mink and chicken, individual brains were analyzed. For perch, pools of brain from five fish (all from a single tank) were analyzed. Total Hg content in each sample was measured using a Direct Mercury Analyzer 80 (Milestone Inc., Shelton, CT). Analytical accuracy and precision were determined through the use of DOLT-3 standard reference material (National Research Council of Canada) and intermittent analysis of duplicate samples. Across the studies, average recovery (accuracy) was within 10% of expected and precision was less than 9%. Mercury concentrations are reported as μg/g (ppm) wet weight (ww), unless indicated.
2.3 DNA Processing
Genomic DNA was isolated from all tissues using DNeasy kits (Qiagen) according to manufacturer’s instructions. An optional RNAse treatment step was found to be essential for accurate fluorescence-based quantification of the isolated DNA. DNA concentration was measured using a nanodrop spectrophotometer (Thermo Fisher Scientific).
2.4 DNMT Assay
Brain DNMT activity was assessed using a commercially available ELISA kit (EpiQuik™ DNMT Activity, Epigentek Group Inc., Farmingdale, New York, USA) in mink and chicken, but not in perch. Briefly, nuclear extracts were incubated for 1 hr in wells coated with cytosine-rich DNA to enable DNMT-mediated methylation of substrate DNA. The methylated DNA was labeled with an anti-5-methylcytosine antibody, which was then detected colorimetrically following incubation with a secondary antibody. The absorbance values were blank-adjusted and the final DNMT activity was reported as absorbance (OD)/h/mg protein. Assays were performed in duplicate or triplicate and intra-plate variation (as RSD%) was less than 13.3%.
2.5 LUMA Assay
The LUminometric Methylation Assay (LUMA) first described by Karimi et al. (2006) was used to quantify global DNA methylation. LUMA uses a pair of isoschizomers, HpaII (methyl sensitive) and MspI (methyl insensitive), to cleave DNA between the two cytosines of the recognition sequence 5′-CCGG-3′. The isoschizomers are run in parallel reactions with EcoRI as a normalizer for DNA input. Resulting overhangs are quantified by pyrosequencing. This method calculates the percentage of methylation for all CpG sites within the recognition sequence 5′-CCGG-3′ in the genome.
LUMA was carried out according to previously outlined methods (Pilsner et al., 2010). Briefly, each 20 μl reaction contained; DNA, 2μl 10X Tango Buffer (Fermentas), 2.5 U EcoRI, and 5U of either HpaII or MspI (all enzymes supplied by New England Biolabs). The amount of input DNA was 200 ng for mink, and 600 ng for chicken and yellow perch. Input DNA was calibrated for each species in pilot studies. Samples were digested at 37°C for 4 hours followed by a 20 minute heat inactivation at 80°C. Annealing buffer (Qiagen) was added to each sample at a volume of 15 μl, and 30μl of the resulting mixture was aliquoted into a pyrosequencing plate. Overhangs generated by the restriction digest were quantified via pyrosequencing on a Pyromark Q96 instrument (Qiagen). The dispensation order for nucleotides was; GTGTCACATGTGTG. Methylation values were calculated according to the formula: , where G and T are the peak heights for HpaII or MspI (methylation) and EcoRI (input DNA), respectively.
Samples were run in duplicate on each plate. Samples were rejected and run a second time if the coefficient of variation between the two replicates was greater than 5%. Samples producing pyrograms that had non-specific peaks (evidence of degraded DNA) were also rejected.
2.6 Statistics
Analysis of variance (ANOVA) was used to assess the effects of MeHg exposure on epigenetic markers (% DNA methylation, DNMT activity), and pair-wise differences were determined with Tukey’s test. Pearson correlations were used to explore associations between brain Hg levels and epigenetic markers. Statistical analyses were performed using PASW Statistics (V.17.0, Chicago, IL, USA) or SigmaStat (Version 2.03, SPSS Inc., San Rafael, CA, USA). A P-value of <0.05 was considered statistically significant in all tests. All data are presented as mean ± standard deviation unless otherwise indicated.
3.0 RESULTS
3.1 Mink Studies
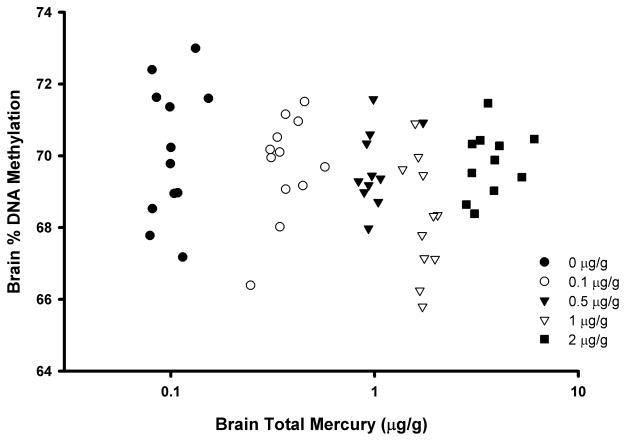
In the occipital cortex of control (unexposed) captive mink, the mean percent DNA methylation for individual samples as determined via the LUMA assay was 70.1 ± 1.9% (Table 1), and ranged between 67.2% and 73.0% for individual control animals. The mean percent DNA methylation was highest in the control group and lowest in the 1ppm MeHg dietary group (68.2 ± 1.6%). This difference between the control group and 1ppm MeHg dietary group was the only pairwise comparison that was of statistical significance (p<0.05). Brain total Hg residue values were available for each of the mink, and the mean values ranged from 0.10 ± 0.02 μg/g in the control group to 3.84 ± 1.0 μg/g in the 2 ppm dietary group. When brain total Hg values were correlated against percent DNA methylation for all mink, there were no significant associations found (Figure 1; r = −0.04, p=0.78). However, there was a significant negative correlation between these two measures (rp= −0.38, p<0.01) following removal of the highest exposed group (2 ppm dietary MeHg).
Table 1.
Mean (± standard deviation) percent DNA methylation and DNMT activity (OD/h/mg) in the occipital cortex brain region of captive mink (Neovison vison) following a 3-month dietary exposure to methylmercury (MeHg). In a given column, superscript letters denote significant (p < 0.05) differences among the treatments via one-way ANOVAs. Sample size is n=9 for % DNA methylation and n=7 for DNMT activity.
| Dietary MeHg (μg/g) | DNA Methylation (%) | DNMT Activity (OD/h/mg) |
|---|---|---|
| 0 | 70.1 ± 1.9a | 5.6 ± 0.7 a |
| 0.1 | 69.7 ± 1.4ab | not measured |
| 0.5 | 69.7 ± 1.0ab | 4.0 ± 0.5b |
| 1 | 68.2 ± 1.6b | 2.5 ± 0.2c |
| 2 | 69.9 ± 1.9ab | 2.6 ± 0.3c |
Figure 1.
Scatterplot (r = −0.04; p=0.78) of total Hg levels and percent methylation of global DNA (as determined via the LUMA assay) in the occipital cortex brain region of captive mink (Neovison vison) following a 3-month exposure to dietary methylmercury (MeHg). Points represent individual mink with different symbols referring to the treatments indicated in the legend.
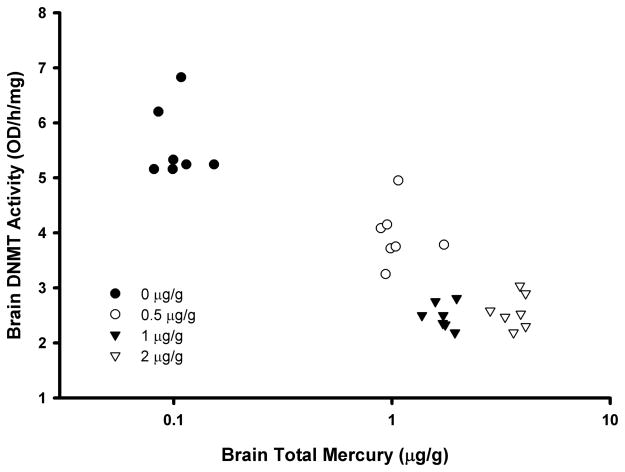
In terms of brain DNMT activity in mink, a subset of 7 randomly selected individuals from four of the five treatment groups was studied. A limited number of wells were available in the ELISA kit, and thus a subset of individuals were chosen from four treatments to enable a range of MeHg exposures to be studied. In the control samples, the mean DNMT activity was 5.60 ± 0.66 OD/h/mg (Table 1). There were MeHg-treatment associated differences in DNMT activity with mean values in all three studied treatment groups being significantly (p<0.01) lower than controls. Mean enzyme levels in the two highest exposure groups were more than 50% lower than the controls. There was a significant negative correlation between brain total Hg and DNMT activity (Figure 2; r = −0.76, p<0.001).
Figure 2.
Scatterplot (r = −0.76; p<0.001) of total Hg levels and DNA methyltransferase activity (as determined via an ELISA assay) in the occipital cortex brain region of captive mink (Neovison vison) following a 3-month exposure to dietary methylmercury (MeHg). Points represent individual mink with different symbols referring to the treatments indicated in the legend.
3.2 Chicken Studies
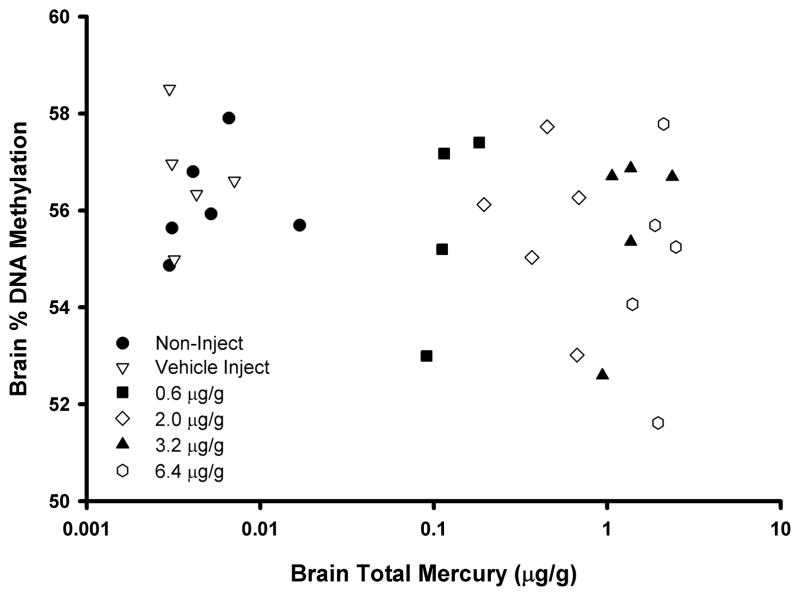
In the cerebrum brain region of control (vehicle-injected) embryonic day 19 chicks, the mean percent DNA methylation as determined via the LUMA assay was 56.2 ± 1.6% (Table 2), and ranged between 53.3% and 58.5% for individual control chicks. There was no difference in mean percent DNA methylation between the vehicle-injected controls and non-injected controls (56.0 ± 0.9%). The mean percent DNA methylation was highest in the control group and lowest in the highest dose group tested (54.7 ± 2.1%), though this difference was not to a level of statistical significance. Brain total Hg residue values were available for some (n=5/dose) of the chicken embryos, and the mean values ranged from 0.005±0.004 μg/g in the control group to 1.97±0.39μg/g in the 6.4 μg/g injected group. Though a negative trend was apparent between brain total Hg values and percent DNA methylation, this was not to a level of statistical significance (Figure 3; r = −0.20; p=0.3).
Table 2.
Mean (± standard deviation) percent DNA methylation and DNMT activity (OD/h/mg) in tissues of embryonic day 19 chickens (Gallus gallus domesticus) that were injected with MeHgCl in ovo on embryonic day 0. The doses refer to μg of MeHg injected per g of egg. Sample sizes vary as indicated. No significant differences were detected.
| DNA Methylation (%) | DNMT Activity (OD/h/mg) | |||
|---|---|---|---|---|
| MeHg Dose | Brain (n=7–9/dose) | Liver (n=7–9/dose) | Heart (n=3–6/dose) | Brain (n=3/dose) |
| No Inject | 56.0 ± 0.9 | 52.1 ± 2.8 | 52.8 ± 1.6 | 2.4 ± 0.5 |
| Vehicle Inject | 56.2 ± 1.6 | 51.2 ± 4.0 | 52.5 ± 1.8 | 2.3 ± 0.4 |
| 0.62 μg/g | 55.8 ± 1.5 | 55.0 ± 2.1 | 54.1 ± 0.7 | 3.0 ± 1.5 |
| 2 μg/g | 55.7 ± 1.9 | 54.9 ± 2.7 | 52.7 ± 0.4 | 3.6 ± 1.1 |
| 3.2 μg/g | 55.5 ± 1.7 | 54.4 ± 3.6 | 53.7 ± 1.6 | 4.2 ± 0.9 |
| 6.4 μg/g | 54.7 ± 2.1 | 52.7 ± 2.5 | 52.9 ± 1.3 | 3.2 ± 1.1 |
Figure 3.
Scatterplot (r = −0.20; p=0.3) of total Hg levels and percent methylation of global DNA (as determined via the LUMA assay) in the cerebrum brain region of embryonic day 19 chickens (Gallus gallus domesticus) that were injected with MeHgCl in ovo on embryonic day 0. Points represent individual birds with different symbols referring to the treatments indicated in the legend.
For chickens, liver and heart were also collected and analyzed via the LUMA assay (Table 2). There was also no MeHg-related difference in % DNA methylation in the liver and heart.
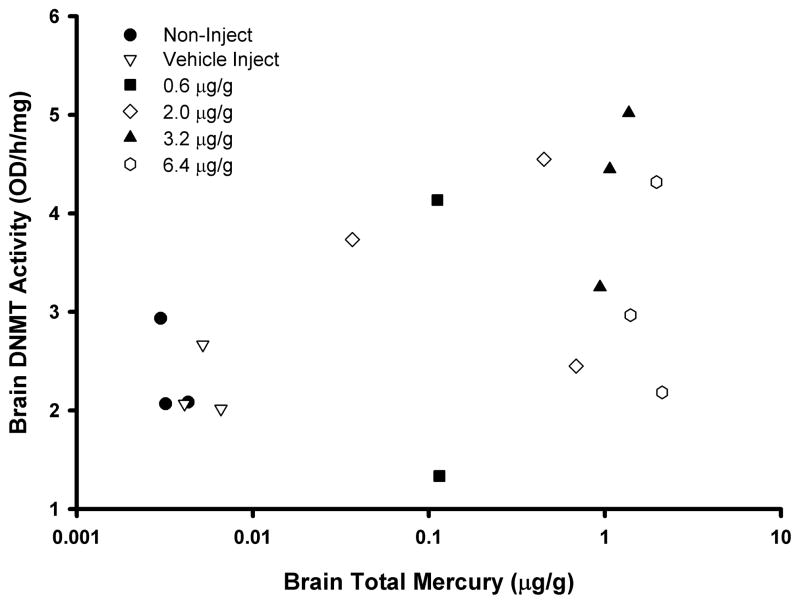
In terms of brain DNMT activity in chicken embryos, a subset of 3 individuals from five groups (both controls, 0.62, 2, 3.2, and 6.4 μg/g) was available for study. There was no difference in activity between the two control groups, and when pooled (n=6) their mean DNMT activity was 2.31 ± 0.39 OD/h/mg (Table 2). Unlike the mink, the mean values in all four studied treatment groups were 31–84% higher than the controls but this was not to a level of statistical significance. Attenuation of DNMT activity was observed in the highest dose group when compared to other treatments. There was no significant correlation between brain total Hg and DNMT activity (Figure 4; r=0.38; p=0.13) when all data were included, though when data from the highest dose was removed a significant correlation was found (r=0.64; p<0.01).
Figure 4.
Scatterplot (r=0.38; p=0.13) of total Hg levels and DNA methyltransferase activity (as determined via an ELISA assay) in the cerebrum brain region of embryonic day 19 chickens (Gallus gallus domesticus) that were injected with MeHgCl in ovo on embryonic day 0. Points represent individual birds with different symbols referring to the treatments indicated in the legend.
3.3 Perch Studies
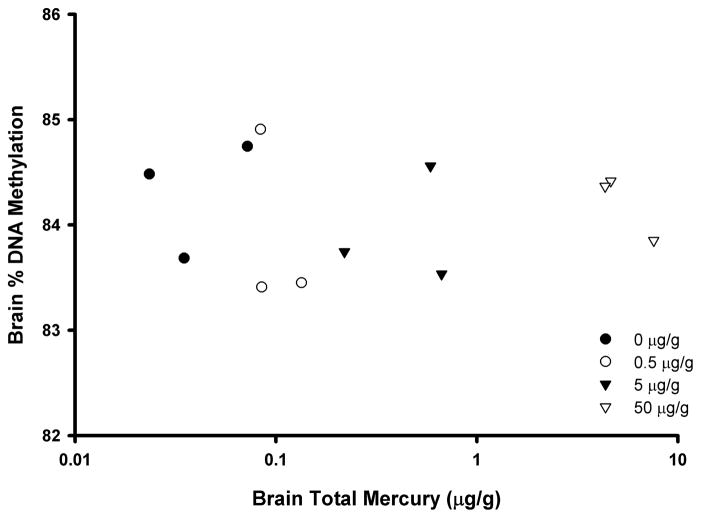
For perch, three replicate pools of five fish each were studied. The mean brain Hg levels across the treatments ranged from 0.04 to 5.52 μg/g. In the telencephalon brain region of control fish, the mean percent DNA methylation as determined via the LUMA assay was 84.3 ± 0.6% in the pooled sample, and ranged between 83.7% and 84.8% (Table 3). There was no MeHg-associated difference in brain percent DNA methylation (Figure 5).
Table 3.
Percent DNA methylation (mean ± standard deviation) in the telencephalon brain region of yellow perch (Perca flavescens) that were fed MeHg for 4 weeks.
| MeHg Diet | % DNA Methylation |
|---|---|
| Control | 84.3 ± 0.6 |
| 0.5 ppm | 83.9 ± 0.9 |
| 5 ppm | 83.9 ± 0.5 |
| 50 ppm | 84.2 ± 0.3 |
Figure 5.
Scatterplot (r=0.04; p=0.90 ) of total Hg levels and percent methylation of global DNA (as determined via the LUMA assay) in the telencephalon brain region of yellow perch (Perca flavescens) that were fed MeHg for 4 weeks. Points represent pools of five fish from a single treatment tank, with different symbols referring to the treatments indicated in the legend.
4.0 DISCUSSION
We looked for evidence that MeHg disrupts global DNA methylation in brain tissue of representative species from three different classes of animals; mammals, birds and fish. This was undertaken by studying a convenience sample of brain tissues from previous MeHg laboratory exposures studies. The data presented here provide evidence that MeHg has the potential to cause epigenetic changes in a mammalian species; DNA hypomethylation and reduced DNMT activity were observed in mink exposed to dietary MeHg. Effects were not observed in chicken or yellow perch, the other two species tested. In the chicken, there were non-significant MeHg-associated trends in both endpoints that warrant further research, whereas in yellow perch, MeHg had no effect on DNA methylation (DNMT activity was not analyzed).
Dietary exposure to 1 ppm MeHg in male mink caused a significant decrease in global DNA methylation as measured by the LUMA assay. These data agree with our previous finding in a mammal that MeHg was associated with DNA hypomethylation in the brain stem of male polar bears (Pilsner et al., 2010). Interestingly, the brain concentrations of Hg observed in this previous study were extremely low, ranging from 0.03 to 0.18 ppm wet weight concentration. Polar bears are top predators and therefore have the potential to accumulate high concentrations of Hg (e.g., liver concentrations can exceed 100 ppm, with corresponding brain levels expected to be >20 ppm), though recent findings suggest that the brain may have mechanisms to minimize Hg exposures (Basu et al., 2009). In spite of this low exposure, an association between brain Hg and DNA methylation was observed in male polar bears. In the current study, DNA hypomethylation was observed in the 1ppm MeHg dose group in mink (corresponds to 1.74 ppm mean brain total mercury, an environmentally-relevant dose), but not at a higher dose of 2 ppm (3.84 ppm mean brain total mercury). Taken together, these findings suggest the possibility that effects of MeHg on DNA hypomethylation occur at relatively low levels of exposure. A larger study with more dietary doses would be necessary to fully characterize the dose-response relationship between MeHg and DNA methylation in mink and to investigate the lack of response at 2ppm MeHg.
We also analyzed DNMT, an enzyme that establishes and maintains patterns of DNA methylation in the cell. Mink DNMT activity was significantly reduced compared to controls in all of the MeHg dose groups tested, including the 2ppm group. The discrepancy between DNA methylation and DNMT activity at 2 ppm MeHg remains unexplained. Nevertheless, the negative association between MeHg and DNMT activity supports our initial hypothesis that MeHg has the potential to disrupt epigenetic processes in mammals, and suggests a partial mechanism for DNA hypomethylation. Mercury has long been known to interact with protein thiols and affect many molecular and cellular targets (Clarkson and Magos, 2006). Mechanisms of action of MeHg are likely to be multi-faceted, but one component may be that it interferes with DNA methylation by interacting with DNMT enzymes. DNA methyltransferases catalyze the transfer of a methyl group from S-adenosyl methionine (SAM) to DNA. Desaulniers et al. (2009) documented reduced hepatic expression of DNMT1 and DNMT3b mRNA in female offspring of rats that were developmentally exposed to MeHg. Reduced methylation of the Cdkn2a gene was found in certain alleles, but other alleles showed no change and there was no MeHg-associated change in global DNA methylation as determined via 5-methyl-deoxycytidine (5mdC) analysis. In another study, MeHg caused downregulation of DNMT3b mRNA and reduced global DNA methylation in primary cultures of rat embryonic cortical neural stem cells (Bose et al., 2013). Given that DNMT serves as critical machinery to create methylation marks, much more research is needed across taxa to understand how stressors such as exposure to environmental contaminants disrupt this process.
The MeHg-associated effects on DNA methylation observed in mink and polar bear were not apparent in the two non-mammalian species, chicken and yellow perch. This could suggest that there are class differences in sensitivity to the epigenetic effects of MeHg, but that there may also be methodological details that could influence this finding. The tissues used for our analyses were conveniently obtained from previous studies, and while the general experimental outlines were similar (environmentally relevant and species-appropriate exposure to MeHg) several methodological details were not consistent among studies. For example, the length of exposure was 3 times shorter for chicken or perch than for mink, and this may have played a role in the lack of a MeHg-associated response in the two non-mammalian species. Gender may be important to consider in future work as the mink were all male (like the polar bears), the perch were all female, while the sex of the chicken embryo was not determined. Additionally, the developmental stage was inconsistent between the three studies. Chickens were exposed as developing embryos, mink were juveniles, and yellow perch were adults (Table 4). Epigenetic marks are established during embryonic development and exposure to chemicals during this period would seem likely to have a different outcome from later–life exposures. The level and route of exposure differed between the three species, but it is interesting to note that the resulting concentrations of brain mercury were similar and overlapping (Table 4).
Table 4.
Summary of findings from the current study.
| Model Species | Brain Total Hg Levels (μg/g, w.w.) | Days of Exposure | Age | Gender | % DNA Methylation | DNMT Activity |
|---|---|---|---|---|---|---|
| Mink | 0.1 – 3.8 | 90 | Juvenile | Male | Significant decrease | Significant decrease |
| Chicken | 0.0 – 2.5 | 18 | Embryos | Mixed | Non-significant decrease | Non-significant increase |
| Perch | 0.0 – 7.6 | 28 | Adult | Female | No apparent change | Not studied |
Aside from MeHg-related effects, there were clear differences in DNA methylation values in the control groups across the three species studied. Significant differences in global DNA methylation were observed with the highest values in yellow perch (84.3 % in controls), followed by mink (70.1 %) and then chicken (56.2 %). This corresponds well with previous work that suggests that DNA methylation is higher in fish than in mammals or birds (Jabbari et al., 1997). Interestingly, DNA methylation data were much less variable from these lab animals (e.g., relative standard deviation in brains from control animals were 2.7% for mink, 1.6% for chicken, 0.7% for perch) whereas wild polar bears varied by approximately 30% (Pilsner et al. 2010). One reason for this inconsistency could be that polar bear tissues were collected under difficult conditions in the field and may not have been of optimal quality. LUMA is very sensitive to sample quality and degraded samples can produce artificially depressed DNA methylation values. Another consideration is that the polar bears experienced a lifetime of exposure to MeHg and other environmental stressors whereas the other species were lab animals raised under more uniform conditions.
This study is one of the first in ecotoxicology to document contaminant-induced changes in DNA methylation and it is important to note that little is known about the significance of global hypomethylation in terms of toxicological outcomes in ecological species. Nevertheless, assessing methylation of DNA across the entire genome has emerged as an important first step in the evaluation of epigenetic impacts caused by stressors. Global DNA hypomethylation is a common outcome measure in cancer biology and in studies of other human diseases (Esteller, 2008; Robertson and Wolffe, 2000). Proper methylation of DNA is critical to ensure structural organization of the genome and modulating gene expression, and thus hypomethylation may result in genomic instability and trigger adverse outcomes (Wilson et al., 2007). It has also been suggested that global DNA hypomethylation may be indicative of a generalized stress response, and that reduced DNA methylation is an early, adaptive and organized response to maintain genomic homeostasis (Szyf, 2011). Beyond global DNA methylation, MeHg exposure has been associated with both increased (Onishchenko et al., 2008; Hanna et al., 2012) and decreased (Gadhia et al., 2012; Goodrich et al., 2013) methylation of specific genes. A closer look at methylation status of individual genes may be necessary to uncover the toxicological consequences of MeHg induced DNA hypomethylation, and this may help increase understanding of the underlying mechanisms by which MeHg causes harm.
The research presented here suggests that MeHg should be considered an epigenetically active compound in that it has the capacity to disrupt DNA methylation in animals exposed under environmentally relevant conditions. This finding suggests new hypotheses and supports emerging findings from biomedical toxicology and epidemiology. However, our results are also limited in some ways that warrant discussion. An effect of MeHg on DNA methylation and DNMT activity was detected in mink, but the effect was not consistent across all doses. No significant effects were seen in chicken or yellow perch, but differences in methodological details in the three studies from which the samples were drawn preclude a true comparison across species. Given that there is relatively little known about contaminant-induced epigenetic effects in any fish and wildlife, our data provide a foundation for future investigation. Important questions in the field of ecotoxicology relating to, for example, remediation efficacy and latency, adaptation, and transgenerational impacts may benefit from a greater understanding of the epigenetic effects of MeHg and other environmental contaminants (Head et al., 2012).
Acknowledgments
This study was funded by a Collaborative Mercury Research Network (COMERN) grant to AS and HMC, Natural Science and Engineering Research Council of Canada (NSERC) grants to HMC and NB, a National Oceanic and Atmospheric Administration grant to JH, and University of Michigan School of Public Health funding to NB. We are thankful to the Canadian Centre for Fur Animal Research (especially Merridy Rankin, Rena Currie, Sarah Gatti-Yorke, Tanya Morse, Cindy Crossman, Jody Muise, and Margot White) for the mink studies, to Krittika Mittal, Jennifer Rutkiewicz, and Mark Bradley for the chicken studies, and to Matthew Pickens for the perch studies. We thank the various anonymous reviewers of this manuscript.
Footnotes
No conflict of interest is declared, and the authors assume sole responsibility for the contents of this manuscript.
References
- Basu N, Scheuhammer AM, Rouvinen-Watt K, Grochowina N, Klenavic K, Evans RD, Chan HM. Methylmercury impairs components of the cholinergic system in captive mink (Mustela vison) Toxicol Sci. 2006;91:202–209. doi: 10.1093/toxsci/kfj121. [DOI] [PubMed] [Google Scholar]
- Basu N, Scheuhammer AM, Bursian S, Rouvinen-Watt K, Elliott J, Chan HM. Mink as a sentinel in environmental health. Environ Res. 2007a;103:130–144. doi: 10.1016/j.envres.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Basu N, Scheuhammer AM, Rouvinen-Watt K, Grochowina N, Evans RD, O’Brien M, Chan HM. Decreased N-methyl-D-aspartic acid (NMDA) receptor levels are associated with mercury exposure in wild and captive mink. Neurotoxicol. 2007b;28 (3):587–593. doi: 10.1016/j.neuro.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Basu N, Scheuhammer AM, Sonne C, Letcher RJ, Born EW, Dietz R. Is dietary mercury of neurotoxicological concern to polar bears (Ursus maritimus)? Environ Toxicol Chem. 2009;28 (1):133–140. doi: 10.1897/08-251.1. [DOI] [PubMed] [Google Scholar]
- Basu N, Head J. Mammalian wildlife as complementary models in environmental neurotoxicology. Neurotox Teratol. 2010;32:114–119. doi: 10.1016/j.ntt.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Bose R, Onishchenko N, Edoff K, Janson Lang AM, Ceccatelli S. Inherited effects of low-dose exposure to methylmercury in neural stem cells. Toxicol Sci. 2012;130:383–390. doi: 10.1093/toxsci/kfs257. [DOI] [PubMed] [Google Scholar]
- Clarkson TW, Magos L. The toxicology of mercury and its chemical compounds. Crit Rev Toxicol. 2006;36 (8):609–662. doi: 10.1080/10408440600845619. [DOI] [PubMed] [Google Scholar]
- Depew D, Basu N, Burgess NM, Campbell LM, Devlin EW, Drevnick P, Hammerschmidt C, Murphy CA, Sandheinrich MB, Wiener JG. Toxicity of dietary methylmercury to fish: Derivation of ecologically meaningful threshold concentrations. Environ Toxiocol Chem. 2012a;31 (7):1536–1547. doi: 10.1002/etc.1859. [DOI] [PubMed] [Google Scholar]
- Depew D, Basu N, Burgess NM, Campbell LM, Evers D, Grasman K, Scheuhammer AM. Derivation of screening benchmarks for dietary methylmercury (MeHg) exposure for the common loon (Gavia immer): Rationale for use in ecological risk assessment. Environ Toxiocol Chem. 2012b;31(10):2399–2407. doi: 10.1002/etc.1971. [DOI] [PubMed] [Google Scholar]
- Desaulniers D, Xiao GH, Lian H, Feng YL, Zhu J, Nakai J, Bowers WJ. Effects of mixtures of polychlorinated biphenyls, methylmercury, and organochlorine pesticides on hepatic DNA methylation in prepubertal female Sprague-Dawley rats. Int J Toxicol. 2009;28 (4):294–307. doi: 10.1177/1091581809337918. [DOI] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Gadhia SR, Calabro AR, Barile FA. Trace metals alter DNA repair and histone modification pathways concurrently in mouse embryonic stem cells. Toxicol Lett. 2012;212 (2):169–179. doi: 10.1016/j.toxlet.2012.05.013. [DOI] [PubMed] [Google Scholar]
- Goodrich JG, Basu N, Franzblau A, Dolinoy D. Mercury biomarkers and DNA methylation among Michigan Dental Professionals. Environ Mol Mutag. 2013 doi: 10.1002/em.21763. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna CW, Bloom MS, Robinson WP, Kim D, Parsons PJ, vom Saal FS, Taylor JA, Steuerwald AJ, Fujimoto VY. DNA methylation changes in whole blood is associated with exposure to the environmental contaminants, mercury, lead, cadmium and bisphenol A, in women undergoing ovarian stimulation for IVF. Hum Reprod. 2012;27 (5):1401–1410. doi: 10.1093/humrep/des038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head JA, Dolinoy DC, Basu N. An introduction to epigenetics for ecotoxicologists. Environ Toxiocol Chem. 2012;31 (2):221–227. doi: 10.1002/etc.1707. [DOI] [PubMed] [Google Scholar]
- Heinz GH, Hoffman DJ, Klimstra JD, Stebbins KR, Kondrad SL, Erwin CA. Species differences in the sensitivity of avian embryos to methylmercury. Arch Environ Contam Toxicol. 2009;56:129–138. doi: 10.1007/s00244-008-9160-3. [DOI] [PubMed] [Google Scholar]
- Jabbari K, Caccio S, Pais de Barros JP, Desgres J, Bernardi G. Evolutionary changes in CpG and methylation levels in the genome of vertebrates. Gene. 1997;205:109–118. doi: 10.1016/s0378-1119(97)00475-7. [DOI] [PubMed] [Google Scholar]
- Karimi M, Johansson S, Ekstrom TJ. Using LUMA: a Luminometric-based assay for global DNA-methylation. Epigenetics. 2006;1:45–48. doi: 10.4161/epi.1.1.2587. [DOI] [PubMed] [Google Scholar]
- Kwon S, Blum JD, Carvan M, Basu N, Head J, Madenjian C, David S. Absence of fractionation of mercury isotopes during trophic transfer of methylmercury to freshwater fish in captivity. Environ Sci Technol. 2012;46 (14):7527–7534. doi: 10.1021/es300794q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishchenko N, Karpova N, Sabri F, Castrén El, Ceccatelli S. Long-lasting depression-like behavior and epigenetic changes of BDNF gene expression induced by perinatal exposure to methylmercury. J Neurochem. 2008;106 (3):1378–1387. doi: 10.1111/j.1471-4159.2008.05484.x. [DOI] [PubMed] [Google Scholar]
- Pilsner JR, Lazarus AL, Nam D-H, Letcher RJ, Sonne C, Dietz R, Basu N. Mercury-associated DNA hypomethylation in polar bear brains via the LUminometric Methylation Assay (LUMA): A sensitive method to study epigenetics in wildlife. Mol Ecol. 2010;19 (2):307–314. doi: 10.1111/j.1365-294X.2009.04452.x. [DOI] [PubMed] [Google Scholar]
- Robertson KD, Wolffe AP. DNA methylation in health and disease. Nat Rev Genet. 2000;1:11–19. doi: 10.1038/35049533. [DOI] [PubMed] [Google Scholar]
- Rutkiewicz J, Basu N. Methylmercury egg injections: Part 1-Tissue distribution of mercury in the white leghorn chicken embryo and hatchling. Ecotox Environ Safe. 2013 doi: 10.1016/j.ecoenv.2013.04.008. Under review. [DOI] [PubMed] [Google Scholar]
- Scheuhammer AM, Meyer MW, Sandheinrich MB, Murray MW. Effects of environmental methylmercury on the health of wild birds, mammals, and fish. Ambio. 2007;36(1):12–18. doi: 10.1579/0044-7447(2007)36[12:eoemot]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Szyf M. The implications of DNA methylation for toxicology: toward toxicomethylomics, the toxicology of DNA methylation. Toxicol Sci. 2011;120(20):235–255. doi: 10.1093/toxsci/kfr024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandegehuchte MB, Janssen CR. Epigenetics and Its Implications for Ecotoxicology. Ecotoxicol. 2011;20:607–624. doi: 10.1007/s10646-011-0634-0. [DOI] [PubMed] [Google Scholar]
- Wiener JG, Sandheinrich MB, Bhavsar SP, Bohr JR, Evers DC, Monson BA, Schrank CS. Toxicological significance of mercury in yellow perch in the Laurentian Great Lakes region. Environ Pollut. 2012;161:350–357. doi: 10.1016/j.envpol.2011.09.025. [DOI] [PubMed] [Google Scholar]
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta. 2007;1775:138–162. doi: 10.1016/j.bbcan.2006.08.007. [DOI] [PubMed] [Google Scholar]